Abstract
The molecular basis of resistance to β-lactams and β-lactam-β-lactamase inhibitor combinations in the KPC family of class A enzymes is of extreme importance to the future design of effective β-lactam therapy. Recent crystal structures of KPC-2 and other class A β-lactamases suggest that Ambler position Trp105 may be of importance in binding β-lactam compounds. Based on this notion, we explored the role of residue Trp105 in KPC-2 by conducting site-saturation mutagenesis at this position. Escherichia coli DH10B cells expressing the Trp105Phe, -Tyr, -Asn, and -His KPC-2 variants possessed minimal inhibitory concentrations (MICs) similar to E. coli cells expressing wild type (WT) KPC-2. Interestingly, most of the variants showed increased MICs to ampicillin-clavulanic acid but not to ampicillin-sulbactam or piperacillin-tazobactam. To explain the biochemical basis of this behavior, four variants (Trp105Phe, -Asn, -Leu, and -Val) were studied in detail. Consistent with the MIC data, the Trp105Phe β-lactamase displayed improved catalytic efficiencies, kcat/Km, toward piperacillin, cephalothin, and nitrocefin, but slightly decreased kcat/Km toward cefotaxime and imipenem when compared to WT β-lactamase. The Trp105Asn variant exhibited increased Kms for all substrates. In contrast, the Trp105Leu and -Val substituted enzymes demonstrated notably decreased catalytic efficiencies (kcat/Km) for all substrates. With respect to clavulanic acid, the Kis and partition ratios were increased for the Trp105Phe, -Asn, and -Val variants. We conclude that interactions between Trp105 of KPC-2 and the β-lactam are essential for hydrolysis of substrates. Taken together, kinetic and molecular modeling studies define the role of Trp105 in β-lactam and β-lactamase inhibitor discrimination.
Keywords: β-lactamase, KPC-2, carbapenemase, carbapenem discrimination, clavulanic acid
Introduction
Carbapenems are traditionally used as the “last-line” of therapy for complex nosocomial infections.1,2 However, the recent emergence of class A β-lactamases capable of hydrolyzing carbapenems threatens this class of β-lactam antibiotics.3–6 One of these β-lactamases, KPC-2, is becoming a major healthcare threat in the United States and throughout the world.5,7,8 KPC-2 manifests a very broad substrate profile including penicillins, extended-spectrum cephalosporins, cephamycins, and carbapenems.9,10 Moreover, Papp-Wallace et al. showed that this enzyme is resistant to β-lactamase inhibitors (clavulanic acid, sulbactam, and tazobactam), which are typically given in combination with a β-lactam for the treatment of infections caused by β-lactamase producing pathogens.11 Understanding the structural requirements that allow for this broad spectrum is critical for future β-lactam and β-lactamase inhibitor design.
When the sequence and crystal structure of KPC-2 is contrasted to homologous β-lactamases (CTX-M-1, SHV-1, and TEM-1), several active site residues that are unique and/or in distinctive positions in KPC-2 are evident.2,12 On the basis of this comparative structural analysis, we were compelled to (i) explain the carbapenem specificity of this class A enzyme, (ii) evaluate the role of individual contact residues in β-lactam specificity, and (iii) determine the amino acid sequence requirements for inhibitor resistance. We previously assessed the role of Ambler position13 Thr237 in KPC-2 and found that this residue is necessary for selected cephalosporinase and carbapenemase activity.14 In addition, we discovered that Thr237 discriminates between clavulanic acid and the penicillin sulfone inhibitors (sulbactam and tazobactam).14 In this study, we chose to examine the role of another important active site residue, Ambler position Trp105 in KPC-2 β-lactamase.
Trp105 is positioned at the entrance to the β-lactamase active site in class A enzymes (Fig. 1). In contrast, SHV-1, TEM-1, and CTX-M-1 possess a Tyr at position 105 (Table I). Several studies examined the role of position 105 in the binding and hydrolysis of β-lactams in TEM-1. In TEM-1, Tyr105 is believed to stabilize the β-lactam through intermolecular interactions (i.e., hydrophobic and van der Waals) with the β-lactam ring or side chain of substrates.15–17 In addition, Tyr105 in TEM-1 demonstrates a role in substrate discrimination. Specifically, several Tyr105 variants in TEM-1 maintain elevated penicillin minimal inhibitory concentrations (MICs) when expressed in Escherichia coli. Remarkably, these same variants do not show increased cephalosporin MICs when expressed in the same background.15 For penicillin resistance, aromatic residues (e.g., Phe) and small amino acids (e.g., Gly) can substitute for Tyr at 105. In contrast, only aromatic residues permit cephalosporin resistance.
Figure 1.
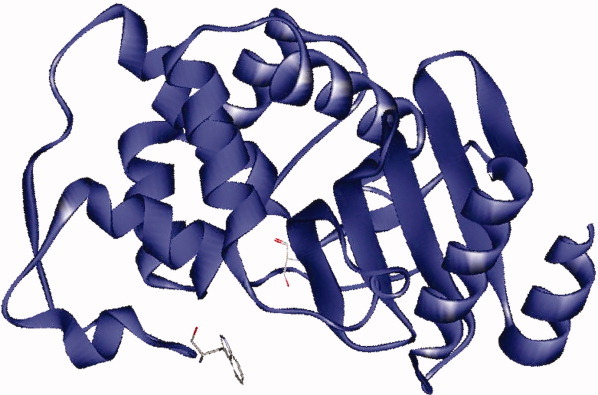
Structure of KPC-2 (PDB# 20V5) with catalytic residue Ser70 and active site residue Trp105 represented.
Table I.
Sequence Alignment of Ambler Positions 103–107 in Various Class A β-Lactamases
| β-lactamase | 103 | 104 | 105 | 106 | 107 |
|---|---|---|---|---|---|
| KPC-2 | Val | Pro | Trp | Ser | Pro |
| SME-1 | Glu | Tyr | His | Ser | Pro |
| NmcA | Glu | Phe | His | Ser | Pro |
| SFC-1 | Glu | Pro | His | Ser | Pro |
| IMI-1 | Glu | Phe | His | Ser | Pro |
| BIC-1 | Glu | Pro | His | Ser | Pro |
| GES-1 | Val | Glu | Trp | Ser | Pro |
| TEM-1 | Val | Glu | Tyr | Ser | Pro |
| SHV-1 | Val | Asp | Tyr | Ser | Pro |
| CTX-M-1 | Val | Asn | Tyr | Asn | Pro |
Other carbapenemases, such as SME-1, NmcA, SFC-1, IMI-1, BIC-1, and GES-2,4-6 possess a histidine at position 105 (Table I). In the case of SME-1 β-lactamase, ampicillin and imipenem resistance are maintained by substitutions of aromatic residues at His105.16 Surprisingly, the His105Arg variant in SME-1 displays increased resistance to cefotaxime. Thus, substrate specificity is also influenced by residue 105 in SME-1.
Based on careful examination of the structure of KPC-2 complexed with a highly charged molecule, N,N-Bis(2-hydroxyethyl)glycine (bicine),2 we rationalized that Trp105 plays a key role in substrate discrimination. The experiments performed herein demonstrate that Ambler position Trp105 plays a role in defining carbapenem discrimination in KPC-2. In addition, we found that amino acids with the ability to form similar intermolecular interactions (i.e., hydrophobic and van der Waals) maintain MICs against all tested β-lactams and β-lactam-β-lactamase inhibitor combinations and that Trp105 also discriminates among each of the inhibitors. For example, several variants display a “clavulanic acid-resistant, sulfone-susceptible” phenotype. Overall, our results highlight the important role of Trp105 in substrate and inhibitor interactions in KPC-2 β-lactamase.
Results
Mutagenesis, DNA sequencing, and β-lactamase expression
The nucleotides encoding Trp105 in blaKPC-2 were mutagenized in the pBC SK (+) blaKPC-2 vector to obtain the other 19 amino acids. One hundred colonies were screened for mutations encoding position 105 of blaKPC-2. All 19 mutants were identified and expressed in the same genetic background, E. coli DH10B cells.
A polyclonal anti-KPC-2 antibody was next used to assess levels of steady-state protein expression of KPC-2 β-lactamase in mid-log phase. Based on previous analyses, Trp105 does not lie within one of the three major epitopes detected by our polyclonal anti-KPC-2 antibody.14 Steady-state expression levels revealed that all 19 variants are expressed in E. coli DH10B cells (Fig. 2).
Figure 2.

Immunoblot using a polyclonal anti-KPC-2 antibody to measure KPC-2 protein expression from the pBC SK(+)blaKPC-2 plasmid in whole E. coli DH10B cell extracts of WT (KPC-2) and the 19 variants at position 105.
Susceptibility testing
After obtaining all KPC variant β-lactamases expressed in the same genetic background, we conducted susceptibility testing by the agar dilution method using the following β-lactams: penicillins (ampicillin and piperacillin), cephalosporins (cephalothin and cefotaxime), and carbapenems (imipenem, ertapenem, meropenem, and doripenem) (Table II). This approach allowed us to determine the effect of substituting Trp105 with the 19 other amino acids.
Table II.
Minimal Inhibitory Concentrations of β-Lactams and β-Lactam-β-Lactamase Inhibitor Combinations (mg/L)
| AMPa | PIPa | CEFa | CTXa | IMPa | ERTa | MEMa | DORa | AMP-CLAVab | AMP-SULab | acPIP-TZB | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K. pneumoniae 1534 | 8192 | 1024 | 1024 | 16 | 8 | 8 | 4 | 4 | 50/32 | 50/512 | 512/64 | |
| E. coli DH10B pBC SK(+) blaKPC-2 (WT)d | 256 | 128 | 256 | 2 | 0.5 | 0.25 | 0.25 | 0.25 | 50/4 | 50/128 | 64/8 | |
| aae | aa side chain volf | AMPa | PIPa | CEFa | CTXa | IMPa | ERTa | MEMa | DORa | AMP-CLAVab | AMP-SULab | PIP-TZBab |
| Trp105Ala | 28.5 | 256 | 64 | 128 | 1 | 1 | 0.125 | 0.06 | 0.125 | 50/16 | 50/64 | 32/4 |
| Trp105Gly | 128 | 16 | 128 | 0.25 | 0.5 | 0.06 | 0.06 | 0.06 | 50/16 | 50/8 | 16/2 | |
| Trp105Ile | 106.6 | 64 | 16 | 64 | 0.5 | 0.25 | 0.06 | 0.06 | 0.125 | 50/4 | 50/1 | 16/2 |
| Trp105Leu | 106.6 | 32 | 4 | 64 | 0.5 | 0.5 | 0.06 | 0.06 | 0.125 | 50/1 | 50/1 | 8/1 |
| Trp105Met | 102.8 | 128 | 32 | 128 | 1 | 0.5 | 0.125 | 0.06 | 0.125 | 50/16 | 50/8 | 32/4 |
| Trp105Val | 79.9 | 128 | 16 | 64 | 0.25 | 0.5 | 0.06 | 0.06 | 0.125 | 50/16 | 50/4 | 16/2 |
| Trp105Cys | 48.4 | 64 | 16 | 64 | 0.25 | 0.25 | 0.06 | 0.06 | 0.06 | 50/1 | 50/8 | 16/2 |
| Trp105Asn | 54.0 | 512 | 128 | 256 | 4 | 1 | 0.25 | 0.125 | 0.25 | 50/16 | 50/256 | 64/8 |
| Trp105Pro | 52.6 | 128 | 16 | 64 | 0.5 | 0.5 | 0.125 | 0.125 | 0.125 | 50/8 | 50/8 | 16/2 |
| Trp105Gln | 83.7 | 64 | 8 | 128 | 0.5 | 0.5 | 0.06 | 0.06 | 0.125 | 50/16 | 50/8 | 8/1 |
| Trp105Ser | 28.9 | 128 | 16 | 128 | 1 | 0.5 | 0.06 | 0.06 | 0.06 | 50/16 | 50/32 | 16/2 |
| Trp105Thr | 56.0 | 128 | 16 | 64 | 1 | 0.25 | 0.06 | 0.06 | 0.125 | 50/16 | 50/8 | 16/2 |
| Trp105Phe | 129.8 | 256 | 64 | 256 | 2 | 1 | 0.125 | 0.125 | 0.25 | 50/16 | 50/64 | 64/8 |
| Trp105Tyr | 133.5 | 512 | 64 | 256 | 2 | 1 | 0.25 | 0.125 | 0.5 | 50/8 | 50/128 | 64/8 |
| Trp105Asp | 51.0 | 64 | 2 | 64 | 0.25 | 0.5 | 0.125 | 0.06 | 0.25 | 50/16 | 50/64 | 4/0.5 |
| Trp105Glu | 78.3 | 32 | 4 | 64 | 0.25 | 0.5 | 0.06 | 0.06 | 0.125 | 50/2 | 50/8 | 4/0.5 |
| Trp105His | 93.1 | 256 | 32 | 128 | 4 | 0.5 | 0.125 | 0.125 | 0.125 | 50/16 | 50/256 | 64/8 |
| Trp105Lys | 108.5 | 8 | 4 | 64 | 0.25 | 0.25 | 0.06 | 0.06 | 0.06 | 50/1 | 50/1 | 4/0.5 |
| Trp105Arg | 113.3 | 16 | 8 | 64 | 1 | 0.25 | 0.06 | 0.06 | 0.06 | 50/1 | 50/1 | 8/1 |
| E. coli DH10B | 1 | 2 | 4 | 0.06 | 0.25 | 0.06 | 0.06 | 0.06 | 50/1 | 50/1 | 4/0.5 | |
| E. coli DH10B pBC SK(+) | 1 | 2 | 4 | 0.06 | 0.25 | 0.06 | 0.06 | 0.06 | 50/1 | 50/1 | 4/0.5 | |
β-Lactam and β-Lactam-β-lactamase inhibitor combination abbreviations: AMP, ampicillin; PIP, piperacillin; CEF, cephalothin; CTX, cefotaxime; IMP, imipenem; ERT, ertapenem; MEM, meropenem; DOR, doripenem; AMP-CLAV, ampicillin-clavulanic acid; AMP-SUL, ampicillin-sulbactam; PIP-TZB, piperacillin-tazobactam.
Ampicillin was maintained at a constant concentration of 50 mg/L and clavulanic acid and sulbactam concentrations were varied.
Piperacillin-tazobactam, both were varied at a ratio 8:1.
All 19 variants at 105 are expressed from pBC SK (+) blaKPC-2 in E. coli DH10B.
Amino acid substitution.
Volume of amino acid side chain substitution relative to the volume of the side chain of glycine. The side chain of tryptophan (WT) is 167.7.14
The MIC results for the 19 variant strains and comparison strains are summarized in Table II. The parent strain K. pneumoniae 1534 and E. coli DH10B control strains containing wild type (WT) blaKPC-2 displayed elevated MICs against all the β-lactams tested, whereas E. coli DH10B control strains lacking blaKPC-2 exhibited lower MICs.
With respect to the penicillin and cephalosporin substrates, the Trp105Phe, -His, -Asn, and -Tyr variants expressed from pBC SK(+)blaKPC-2 in E. coli DH10B rendered MICs within two doubling dilutions of WT for all substrates (ampicillin 256–512 mg/L, piperacillin 32–128 mg/L, cephalothin 128–256 mg/L, and cefotaxime 2–4 mg/L). We note some flexibility for retaining cephalothin MICs, because all 19 variants when produced by E. coli DH10B pBC SK(+)blaKPC-2 are within two doubling dilutions (64–256 mg/L) of WT.
Next, we found that the carbapenem MICs for WT and the 19 variants expressed in E. coli DH10B from pBC SK(+)blaKPC-2 varied slightly (within two doubling dilutions of the E. coli DH10B control strains). Unexpectedly, 14 of 19 variants maintained WT MICs to imipenem: the Trp105Ala, -Asp, -Glu, -Phe, -Gly, -His, -Leu, -Met, -Asn, -Pro, -Gln, -Ser, -Val, and -Tyr variants displayed MICs ≥ to WT (0.5 mg/L).
To expand the sensitivity of the carbapenem susceptibility testing, we engineered the variants at Ambler position Trp105 in a different construct, the pBR322-catI-blaKPC-2 plasmid; this vector was previously characterized by our group (data not shown).
We obtained 16 of the 19 variants at Ambler position 105 and proceeded to test carbapenem MICs in this alternate expression system. Based on susceptibility testing, it is clear that the basal expression level of blaKPC-2 is higher in the pBR322-catI-blaKPC-2 plasmid than in the pBC SK(+) blaKPC-2 plasmid. The MICs for all 16 variants were correspondingly higher when expressed from pBR322-catI-blaKPC-2 plasmid in E. coli DH10B cells (Table III). The Trp105Phe and -His variants produced from pBR322-catI-blaKPC-2 plasmid in E. coli DH10B maintained WT level imipenem MICs (4 mg/L), whereas 12 of 16 variants displayed MICs within one dilution of WT (2 mg/L). In contrast, no variants exhibited MICs similar to WT for meropenem, ertapenem, or doripenem; the Trp105Phe was within one dilution of WT for meropenem and ertapenem at 1 mg/L. We interpret these results to mean that carbapenems MIC differences may be due to the relative permeability/stability of the carbapenems compounds as the control strain E. coli DH10B without blaKPC has a higher MIC for imipenem at 0.25 mg/L versus 0.06 mg/L for the other carbapenems.
Table III.
Minimal Inhibitory Concentrations of Carbapenems (mg/L) for Variants Expressed from pBR322-catI-blaKPC-2 in E. coli DH10B
| Strain | IMPa | MEMa | ERTa | DORa |
|---|---|---|---|---|
| E. coli DH10B pBR322-catI-blaKPC-2a | 4 | 2 | 2 | 4 |
| Trp105Ala | 2 | 0.25 | 0.5 | 0.5 |
| Trp105Ile | 2 | 0.25 | 0.5 | 0.5 |
| Trp105Leu | 2 | 0.25 | 0.25 | 0.5 |
| Trp105Met | 2 | 0.5 | 0.5 | 1 |
| Trp105Val | 2 | 0.25 | 0.5 | 0.5 |
| Trp105Cys | 2 | 0.125 | 0.5 | 0.5 |
| Trp105Asn | 2 | 0.5 | 0.5 | 1 |
| Trp105Pro | 2 | 0.25 | 0.5 | 0.5 |
| Trp105Gln | 2 | 0.25 | 0.25 | 0.5 |
| Trp105Ser | 2 | 0.25 | 0.25 | 0.5 |
| Trp105Thr | 2 | 0.25 | 0.5 | 0.5 |
| Trp105Phe | 4 | 1 | 1 | 1 |
| Trp105Glu | 2 | 0.5 | 0.25 | 1 |
| Trp105His | 4 | 0.5 | 0.5 | 0.5 |
| Trp105Lys | 1 | 0.125 | 0.125 | 0.25 |
| Trp105Arg | 1 | 0.125 | 0.25 | 0.25 |
| E. coli DH10B | 0.25 | 0.06 | 0.06 | 0.06 |
β-lactam abbreviations: IMP, imipenem; MEM, meropenem; ERT, ertapenem; and DOR, doripenem.
In an effort to further assess the impact of the substitutions at position Trp105 in KPC-2, MICs were also determined for β-lactam-β-lactam inhibitor (ampicillin-clavulanic acid, ampicillin-sulbactam, and piperacillin-tazobactam) combinations (Table II). We found that many variants demonstrated increased ampicillin-clavulanic acid MICs. The Trp105Ala, -Asp, -Phe, -Gly, -His, -Met, -Asn, -Pro, -Glu, -Ser, -Thr, -Val, and -Tyr displayed ampicillin-clavulanic acid MICs of ampicillin 50 mg/L and clavulanic acid 8 mg/L to ampicillin 50 mg/L and clavulanic acid 16 mg/L while the WT level was ampicillin 50 mg/L and clavulanic acid 4 mg/L. For ampicillin-sulbactam, the Trp105His, -Asn, and -Tyr variants exhibited MICs similar to WT (ampicillin 50 mg/L and sulbactam 128 mg/L to ampicillin 50 mg/L and sulbactam 256 mg/L). For piperacillin-tazobactam, Trp105Phe, -His, -Asn, and -Tyr maintain WT MICs (piperacillin 64 mg/L and tazobactam 8 mg/L).
Kinetics of KPC-2 with substrates and inhibitors
To understand the biochemical correlates of the phenotypic changes by single amino acid substitutions at position 105, we determined the steady-state kinetic parameters of various substrates and inhibitors with purified WT KPC-2 and the Trp105Phe, -Asn, -Leu, and -Val variants. We studied the Trp105Phe and Trp105Asn variants because when these mutations were expressed in E. coli DH10B cells, the strains maintained high MICs to most substrates and substrate–inhibitor combinations. We studied the Trp105Leu variant, as this substitution rendered the E. coli host strain more susceptible to most β-lactams than WT. The Trp105Val variant was selected for further analysis to explain the differences in MICs between ampicillin-clavulanic acid versus ampicillin-sulbactam and piperacillin-tazobactam.
The Trp105Phe variant displayed maintained or slightly improved catalytic efficiencies compared with WT for piperacillin, cephalothin, and nitrocefin (Table IV). The catalytic efficiency for cephalothin was enhanced 2.6-fold, reflected by a nearly 5-fold increase in kcat. For this variant, cefotaxime and imipenem catalytic efficiencies were decreased by 1.4- and 1.6-fold, respectively because of decreased kcats.
Table IV.
KPC-2 W105 Variant Substrate Kinetics
| Km (μM) | kcat (s−1) | kcat/Km (μM−1 s−1) | Fold change kcat/Km | ΔΔGcat (kcal/mol) | |
|---|---|---|---|---|---|
| Piperacillin | |||||
| WT | 15 ± 2 | 51 ± 6 | 3.3 ± 0.1 | 1 | |
| Trp105Phe | 24 ± 4 | 112 ± 6 | 4.7 ± 0.2 | 1.4 | −0.2 |
| Trp105Asn | 34 ± 5 | 89 ± 2 | 2.6 ± 0.2 | −1.3 | 0.1 |
| Trp105Leu | >100 | N/Da | <0.0002b | N/Da | N/Da |
| Trp105Val | 37 ± 4 | 50 ± 2 | 1.4 ± 0.1 | −2.4 | 0.5 |
| Cephalothin | |||||
| WT | 14 ± 2 | 44 ± 2 | 3.1 ± 0.1 | 1 | |
| Trp105Phe | 27 ± 5 | 219 ± 5 | 8.1 ± 0.2 | 2.6 | −0.6 |
| Trp105Asn | 47 ± 4 | 219 ± 2 | 4.7 ± 0.1 | 1.3 | −0.3 |
| Trp105Leu | 177 ± 24 | 175 ± 9 | 1.0 ± 0.1 | −3.1 | 0.7 |
| Trp105Val | 127 ± 9 | 194 ± 9 | 1.5 ± 0.1 | −2.1 | 0.4 |
| Nitrocefin | |||||
| WT | 6 ± 1 | 52 ± 2 | 8.5 ± 0.1 | 1.0 | |
| Trp105Phe | 11 ± 2 | 96 ± 4 | 8.7 ± 0.2 | 1.0 | 0 |
| Trp105Asn | 24 ± 3 | 104 ± 5 | 4.3 ± 0.1 | −2.0 | 0.4 |
| Trp105Leu | 61 ± 5 | 97 ± 16 | 1.6 ± 0.2 | −5.3 | 1.0 |
| Trp105Val | 62 ± 11 | 103 ± 6 | 1.7 ± 0.2 | −5.0 | 1.0 |
| Cefotaxime | |||||
| WT | 207 ± 30 | 142 ± 15 | 0.7 ± 0.1 | 1.0 | |
| Trp105Phe | 200 ± 17 | 102 ± 6 | 0.5 ± 0.1 | −1.4 | 0.2 |
| Trp105Asn | >200 | N/Da | <0.0004b | N/Da | N/Da |
| Trp105Leu | >200 | N/Da | <0.0001b | N/Da | N/Da |
| Trp105Val | >200 | N/Da | <0.00006b | N/Da | N/Da |
| Imipenem | |||||
| WT | 19 ± 4 | 19 ± 1 | 1.0 ± 0.1 | 1.0 | |
| Trp105Phe | 15 ± 3 | 9 ± 1 | 0.6 ± 0.2 | −1.7 | 0.3 |
| Trp105Asn | 33 ± 2 | 3.1 ± 0.1 | 0.09 ± 0.01 | −11.1 | 1.4 |
| Trp105Leu | 24 ± 5 | 1.5 ± 0.1 | 0.06 ± 0.01 | −16.7 | 1.7 |
| Trp105Val | 83 ± 3 | 5.5 ± 0.1 | 0.07 ± 0.01 | −14.3 | 1.6 |
| Doripenem | |||||
| WT | 5.5 ±1 | 0.73 ± 0.07 | 0.13 ± 0.01 | 1 | |
| Trp105Leu | 4.8 ± 1 | 0.75 ± 0.07 | 0.21 ± 0.02 | 1.6 | −0.3 |
Km was very high, thus values could not be determined.
kcat/Km estimated from the equation v = (kcat/Km) × [E] × [S].
The Trp105Asn variant demonstrated slightly higher Kms (≤4-fold, for all substrates). Notably, the Km for cefotaxime (>200 μM) could not be determined. In contrast, this variant displayed increased kcats for piperacillin, cephalothin, and nitrocefin with a corresponding increase in catalytic efficiency for cephalothin of 1.3-fold. Moreover, the catalytic efficiency and kcat for imipenem were decreased.
When compared with WT, the Trp105Leu and -Val variants exhibited considerably decreased catalytic efficiencies attributed mostly to increased Kms for all substrates, varying by up to 12.6-fold for Trp105Leu and cephalothin. In addition to increased Kms for imipenem in the case of the Trp105Val variant, both variants demonstrated decreased kcats.
The contribution of each substitution at Ambler position 105 was determined by comparing change in activation energy, ΔΔGcat, of the variant enzymes to that of WT KPC-2. These calculations allow us to determine the activation energy required to proceed from the free enzyme and β-lactam to the transition state. In this context, we see that imipenem kcat is affected the most by substitutions at Trp105. Using this energetic consideration, more energy is required to achieve the transition state for the variant enzymes when hydrolyzing imipenem; thus, the variants are not “catalytically favorable” (Table IV). Interestingly, the Trp105Asn and -Phe variants demonstrated activation energy gains for the reaction with cephalothin and, therefore, would be “catalytically favorable.”
We noted that the susceptibility determinations with Trp105Phe and -Asn variants expressed in E. coli DH10B cells exhibited increased ampicillin-clavulanic acid MICs by two doubling dilutions. Accordingly, the Trp105Phe and -Asn variants demonstrate notably increased Kis for clavulanic acid by 6.0- and 20.0-fold with slightly increased partition ratios by 2.0- and 2.5-fold, respectively (Table V). Compared with WT, the Trp105Phe and -Asn variants interact similarly with sulbactam and tazobactam varying only slightly by ≤1.9-fold in Ki and ≤2.0-fold for their partition ratios.
Table V.
KPC-2 W105 Variant Inhibitor Kineticsa
| Clavulanic acid | Sulbactam | Tazobactam | |
|---|---|---|---|
| Ki (μM) | |||
| WT | 11 ± 1 | 167 ± 16 | 74 ± 7 |
| Trp105Phe | 61 ± 6 | 177 ± 17 | 113 ± 10 |
| Trp105Asn | 234 ± 20 | 86 ± 8 | 85 ± 8 |
| Trp105Val | 107 ± 10 | 63 ± 6 | 160 ± 16 |
| Partition ratio (15 min) | |||
| WT | 2000 | 1500 | 250 |
| Trp105Phe | 4000 | 2000 | 250 |
| Trp105Asn | 5000 | 1500 | 125 |
| Trp105Val | 7500 | 125 | 125 |
Nitrocefin is the substrate used for inhibitor kinetics.
When produced by E. coli DH10B cells, the Trp105Val variant's rank order of potency for inhibitor MICs is ampicillin-clavulanic acid ≪ piperacillin-tazobactam < ampicillin-sulbactam. The Trp105Val variant exhibits a 10-fold increase in Ki and 3.8-fold increase in the partition ratio of clavulanic acid. In addition, the Trp105Val variant is more susceptible to sulbactam than tazobactam as evidenced by its decreased partition ratio from 1500 molecules of sulbactam by WT KPC-2 to 125 by the Trp105Val variant. Because the Trp105Leu variant's ampicillin and piperacillin MICs were below 50 mg/L, inhibitor kinetics were not determined for this variant.
Molecular modeling
Using our biochemical analysis as a foundation, we performed molecular modeling and molecular dynamics simulation (MDS). Our aim here was to understand the differences between imipenem and doripenem as substrates for WT and the Trp105Leu variant. We conducted this analysis on the apoenzyme, KPC-2, as well as KPC-2 and the Trp105Leu variant complexed with imipenem and doripenem.
The apoenzyme
During a 150 ps apoenzyme simulation, we find that the side chain of residue 105 is not flexible, rotating at most 15–20° and shifting <1.0 Å from Ser70. This rigidity may play a role in restricting the movement of substrates in the active site and restricting the overall flexibility of the active site cavity.
KPC-2 and imipenem
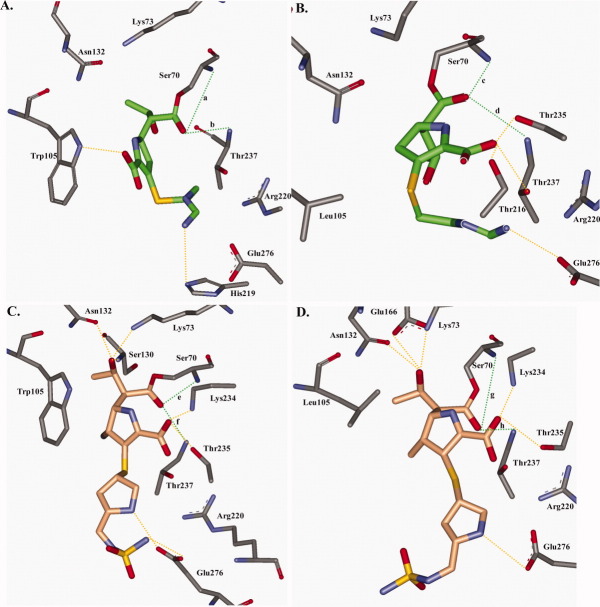
In the initial docking complex of KPC-2 with imipenem after the formation of the bond between Ser70 and imipenem and before MDS, the carboxylate of imipenem seems to interact via hydrogen bonds with Thr237 and Arg220.14 However after MDS, hydrogen bonding interactions can occur between the indole of Trp105 and the carboxylate of imipenem. In addition, at times hydrolyzed β-lactam ring of imipenem is positioned within van der Waals interaction distance of the bicyclic ring of Trp105 [Fig. 4(A)]. MDS further reveals that the R2 side chain of imipenem is flexible and can interact through hydrogen bonds with His219, and the R1 side chain is oriented toward Thr237.
Figure 4.
Molecular representations with the lowest potential energy of a total of 50 frames after MDS for each β-lactamase-β-lactam complex are presented. All potential hydrogen bonds that occurred during MDS are denoted in yellow, except the hydrogen bonds between the carbapenem's carbonyl oxygen and backbone nitrogens of Ser70 and Thr237 which are denoted in green and are labeled a-h corresponding to designations presented in Figure 6. A. KPC-2 with imipenem (green); B. The Trp105Leu variant with imipenem (green); C. KPC-2 with doripenem (orange); D. The Trp105Leu variant with doripenem (orange).
To assess the stability of the imipenem-KPC-2 complex, we generated heat maps that represent the changes in hydrogen bonding interactions between imipenem and all residues found in KPC-2 during the MDS time course [Fig. 5(A)]. From this analysis, we learn that imipenem in KPC-2 makes few hydrogen bonding interactions compared with (a) doripenem in KPC-2 and (b) imipenem and doripenem in the Trp105Leu variant.
Figure 5.
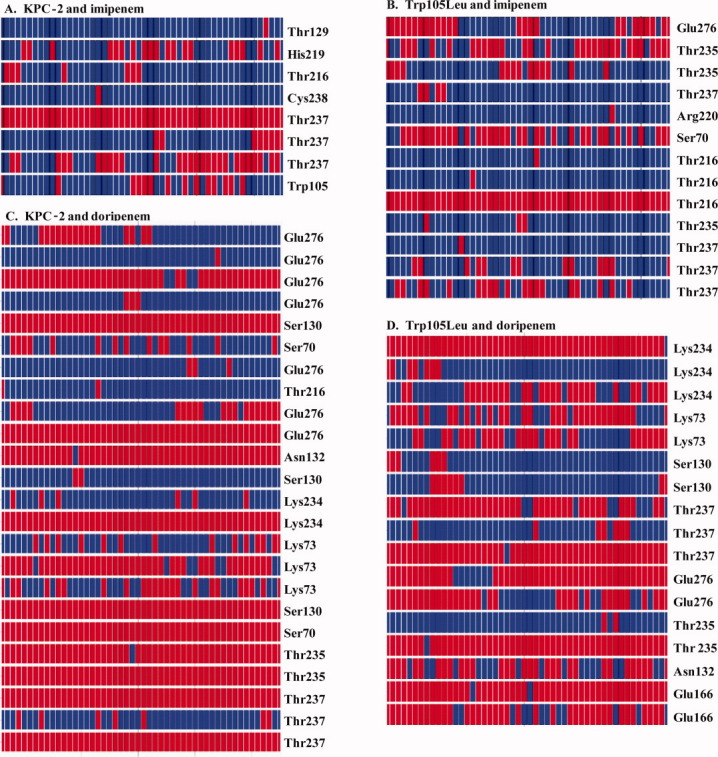
Heat plots revealing the movement of all possible hydrogen bonding interactions during the trajectory between the β-lactam and β-lactamase (red denotes a hydrogen bond and blue denotes no hydrogen bond) A. KPC-2 and imipenem; B. Trp105Leu variant and imipenem; C. KPC-2 and doripenem; and D. Trp105Leu and doripenem.
Trp105Leu and imipenem
In TEM-1, a Tyr105Asp variant alters the movement or flexibility of active site residues far from Tyr105 compared with WT TEM-1; this interpretation correlates with the decreased catalytic efficiency of this variant. Based on nuclear magnetic resonance analysis, Doucet et al. showed that substitutions at Tyr105 in TEM-1 also alter the overall internal protein motion.15
In the Trp105Leu variant model of KPC-2 with imipenem, Leu shifts into the active site. This results in narrowing of the catalytic center and widening the opening or entrance of the enzyme [Figure 4(B)]. As a result, the R1 side chain of imipenem would shift 3 Å to compensate while a loss of a hydrogen bond between imipenem's carboxylate and position 105 appears to occur. Here, the carboxylate interacts among Thr216, Thr235, or Thr237. Because the R2 side chain of imipenem is flexible, hydrogen bonding interactions can occur between Glu276 and the R2 side chain [Figure 4(B)]. During MDS, the carbonyl of imipenem is positioned in the oxyanion hole exhibiting nominal flexibility unlike with WT KPC-2 and imipenem during MDS [Fig. 6(B)]. Interestingly, because of the alterations in the overall active site, the number of hydrogen bonding interactions between residues present in the Trp105Leu variant and imipenem are actually increased [Fig. 5(B)]. Kinetic observations reveal that the kcat/Km for imipenem is ∼10 fold decreased in the Trp105 variant compared with WT KPC-2 (Table IV). Also, an additional ∼1.7 kcal/mol compared with WT is required for imipenem turnover by the Trp105Leu variant.
Figure 6.
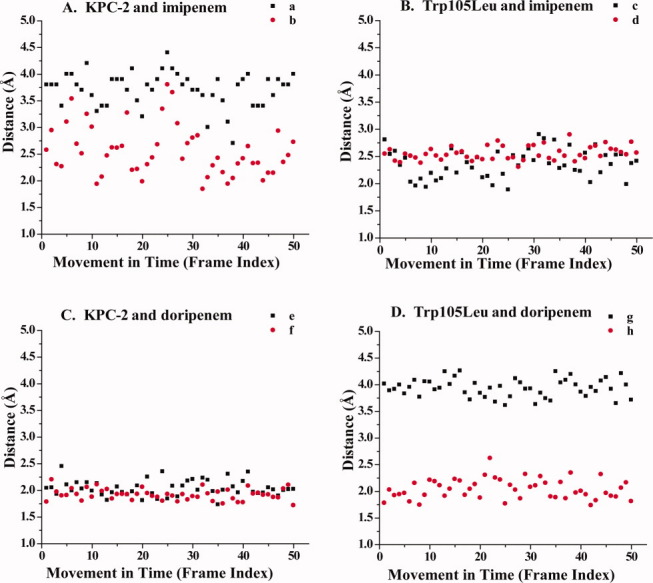
Hydrogen bond distances in Å reveal the movement of the carbapenem's carbonyl oxygen between the backbone nitrogens of Ser70 (black) and Thr237 (red) during time. Letter designations (a–h) correspond to the hydrogen bonds present in Figure 4. A. KPC-2 and imipenem; B. Trp105Leu variant and imipenem; C. KPC-2 and doripenem; and D. Trp105Leu and doripenem.
KPC-2 and doripenem
During the simulation of KPC-2 with doripenem, we were not able to detect interactions between Trp105 and the β-lactam [Figure 4(C)]. We suspect that this is due to the bulk and rigidity of doripenem's R2 side chain. The carboxylate of doripenem during MDS can form hydrogen bonds with either Lys234 or Thr235, the R1 side chain with either Lys73, Ser130, or Asn132, and the R2 side chain with Glu276 [Figure 4(C)]. In all representations, the carbonyl of doripenem is positioned in the oxyanion hole with minimal movement similar to the Trp105Leu variant with imipenem [Figure 6(C)]. Compared with KPC-2 with imipenem, the number of hydrogen bonding interactions between the KPC-2 and doripenem are increased [Figs. 4(A,C) and 5(A,C)]. Doripenem's kcat/Km is decreased by 10-fold compared with imipenem in the WT enzyme (Table IV).18
Trp105Leu and doripenem
The model of the Trp105Leu variant with doripenem shares similarities to the WT KPC-2 model with doripenem [Figs. 4(C,D) and 5(C,D)]. The carboxylate of doripenem again can form hydrogen bonds with either Lys234 or Thr235, the R1 side chain of doripenem with either Lys73, Asn132, or Glu166. The R2 side chain makes hydrogen bonding interactions with Glu276. Because of movement of the R1 side chain of doripenem, the carbonyl of doripenem shifts away from Ser70, but the hydrogen bonding network of the carbonyl to Ser70 and Thr237 is stable, which is similar to what is observed with Trp105Leu and imipenem and WT KPC-2 and doripenem during MDS [Fig. 6(D)]. Interestingly, the kcat/Km of doripenem for the Trp105Leu variant is similar to the WT enzyme and the transition state is more energetically favorable by ∼0.3 kcal/mol (Table IV).
Discussion
Biochemical properties
Revealing structure–function relationships in KPC is of paramount importance as the design of novel β-lactams and β-lactamase inhibitors will need to be based on understanding the contribution of different active site residues to molecular recognition. Our results illustrate that the biochemical properties of Ambler position Trp105 in the KPC-2 β-lactamase are a major determinant in substrate and inhibitor specificity. This insight is first evident when we examine MICs. The Trp105Phe, -His, -Asn, and -Tyr β-lactamases expressed in E. coli DH10B display maintained or increased MICs for most β-lactams and β-lactam-β-lactamase inhibitor combinations compared with WT. Because -Phe, -His, -Asn, and -Tyr have similar biochemical properties (i.e., hydrophobic or can make van der Waals interactions), maintaining MICs equivalent to WT is expected. We also note that the MICs are significantly lower for the remainder of the variants; other amino acid substitutions lead to either diminished interactions between Ambler position 105 or the β-lactam or result in lack of favorable interactions for binding and catalysis. In addition, we discover a novel role for Ambler position 105 in the discrimination of β-lactamase inhibitors (clavulanic acid vs. the sulfones).
Inhibitor discrimination
We were first intrigued that structural flexibility at position 105 in KPC-2 exists for retaining ampicillin-clavulanic acid MICs because many variants (13 of 19) display increased MICs against this β-lactam β-lactamase inhibitor combination. The β-lactamases when purified also demonstrate elevated Kis. Loss of an aromatic residue at Ambler position 105 is postulated to play a role in clavulanic acid resistance in other class A enzymes,19 thus strengthening the importance of our observation (increased ampicillin-clavulanic acid MICs). In contrast, to maintain ampicillin-sulbactam and piperacillin-tazobactam inhibitor MICs, a residue that is capable of making similar biochemical interactions as Trp105 is necessary. Papp-Wallace et al. previously observed this “clavulanic acid-resistant, sulfone-susceptible” phenotype with the variants of KPC-2 at position 237 expressed in E. coli.14 We attribute this similar phenotype at Ambler position 105 to important differences in the reaction mechanisms of clavulanic acid and the sulfones with KPC-2 and its variants. For instance, clavulanic acid's partition ratio is higher than for the sulfones with the variants and Kis for clavulanic acid varying significantly more than for the sulbactam and tazobactam. We hypothesize that this difference may be attributed to the oxapenam backbone of clavulanic acid versus the sulfonyl group of sulbactam and tazobactam [Fig. 3(C)]. Further investigation of the “clavulanic acid-resistant, sulfone-susceptible” phenotype of KPC-2 variants is ongoing.
Figure 3.
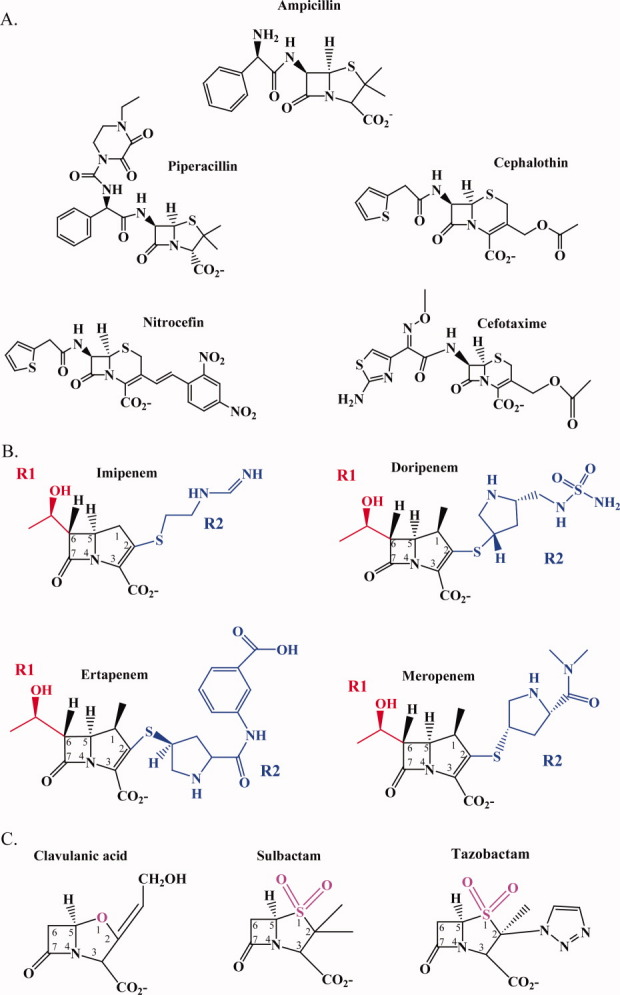
Chemical structures of the β-lactams and β-lactamase inhibitors used in this study: A. Penicillins and cephalosporins; B. Carbapenems (red denotes R1 side chain and blue denotes R2 side chain); C. β-Lactamase inhibitors (oxapenam backbone of clavulanic acid, magenta and sulfonyl backbone of sulbactam and tazobactam, magenta).
Carbapenem discrimination
The differences between the kinetic observations of WT KPC-2 and the Trp105Leu variant with imipenem and doripenem and their molecular representations led us to hypothesize that the deacylation rate, k3, is decreased for the Trp105Leu variant with imipenem and doripenem and for KPC-2 with doripenem as compared with KPC-2 with imipenem [Eq. (1)]. This decrease in k3 would correlate with the increased number of hydrogen bonding interactions between these representations as opposed to KPC-2 with imipenem, and the stability of the hydrogen bonding network within the oxyanion hole. The rate constant k3 has been shown to be the rate-determining step in imipenem hydrolysis by class A β-lactamases (NMC-A and TEM-1).20,21 In GES β-lactamases, k3 is rate-limiting in imipenem hydrolysis for GES-5, whereas compared with GES-1, k2 is rate limiting.22 Investigations are ongoing to determine the differences in hydrolysis of these carbapenems by WT KPC-2.
Conclusions
Our biochemical and molecular analyses brings us to three major conclusions. First, we established that Ambler position 105 is important for the recognition of substrates in KPC-2 and only amino acids that can make similar intermolecular interactions (i.e., hydrophobic and van der Waals) maintain β-lactam and β-lactam-β-lactamase inhibitor MICs. As was suggested by the crystal structure with the trapped N,N-Bis(2-hydroxyethyl)glycine molecule, the side chain of 105 is necessary for binding and maintaining catalytic activity, which is most likely due to stabilization of β-lactam in the active site before and during hydrolysis.2 Thus, the biochemical properties of the amino acid side chain at Ambler position 105 are important for activity; the volume of the amino acid side chains does not seem to play a role (Table II).
Second, we find that ampicillin-clavulanic acid resistance is enhanced with substitutions at position 105. Many variants at 105 had increased ampicillin-clavulanic acid MICs which is primarily due to increased Kis and increased partition ratios. Thus, interactions with 105 may be important for inhibition of KPC-2 and should be considered for future inhibitor design. Interestingly, the sulfone inhibitors behave differently; when combined with a β-lactam a similar amino acid at 105 is required for increased MICs, whereas other amino acids result in lower MICs. Surprisingly, residue 105 seems to be able to discriminate between different inhibitors.
Finally, we discerned that imipenem's R2 side chain might be more flexible in the KPC-2 active site while doripenem may be more rigid. This difference seems to have a major influence on hydrolysis of these substrates because doripenem can form more stable hydrogen bonding network interactions with the KPC-2 active site than imipenem.
Taken together these findings define a unique class A β-lactamase (KPC) that is poised to hydrolyze a broad range of β-lactams (both substrates and inhibitors). Clearly, the future design of carbapenems and mechanism-based inhibitors must take into account the extraordinary complexity of the hydrogen bonding network in the active site. Comparative studies of similar enzymes may yield very important insights into the evolutionary journey of this unique group. Our current study, as a part of a series of investigations, makes us to reflect on selective carbapenem and inhibitor-resistant phenotype of KPC. We wonder whether the clinical use of clavulanic acid for 2 decades or the presence of other inhibitors in nature has driven the evolution of an enzyme able to hydrolyze carbapenems as part of its “extended-spectrum” profile. Such an evolutionary “accident” would confer a clear selective advantage and suggest that β-lactamases will continue to evolve unexpected substrate profiles.
Materials and Methods
Bacterial strains and plasmids
K. pneumoniae possessing blaKPC-2 and E. coli with the pBR322-catI- blaKPC-2 plasmid were a kind gift from Dr. Fred Tenover from the Centers for Disease Control and Prevention, Atlanta, GA.9 The cloning of blaKPC-2 into pBC SK (+) phagemid (Stratagene, La Jolla, CA) and into pET24a(+) plasmid (Novagen, Darmstadt, Germany) is previously described.14 WT and Trp105 variants of pBR322-catI-blaKPC-2 and pBC SK (+) blaKPC-2 were expressed in E. coli DH10B cells (Invitrogen, Carlsbad, CA). E. coli Origami™2 DE3 pLys cells (Novagen) were used as a host strain for pET24a(+)-blaKPC-2A22 with WT and Trp105Phe, -Asn, -Leu, and -Val variants to ensure formation of KPC-2′s disulfide bond.
Site-saturation mutagenesis
The Quikchange® XL site-directed mutagenesis kit (Stratagene) was used to perform site-saturation mutagenesis at Ambler position 105 using the manufacturer's protocol as previously described.14 Degenerate primers were designed to generate all 19 substitutions at 105. In addition, site-directed mutagenesis primers were designed for the Trp105Phe, -Asn, -Leu, and -Val variants. The pBC SK (+) blaKPC-2 and pBR322-catI-blaKPC-2 vectors were used as templates for PCR with the site-saturation degenerate primer sets and the pET24a(+)-blaKPC-2A22 plasmid was used with the site-directed mutagenesis primer sets. The site-saturation mutant plasmids were electroporated into E. coli DH10B. All 19 E. coli DH10B variants at 105 were obtained for the pBC SK (+) blaKPC-2 plasmid. However, only 16/19 E. coli DH10B variants at 105 were generated for pBR322-catI-blaKPC-2 plasmid; Trp105Asp, Trp105Gly, and Trp105Tyr were not obtained. E. coli Origami™2 DE3 pLys cells were transformed with the Trp105Phe,-Asn,-Leu, and -Val variant plasmids for protein expression.
Antibiotic susceptibility
Lysogeny broth (LB) agar dilution MICs, according to Clinical and Laboratory Standards Institute, were used to phenotypically characterize K. pneumoniae expressing blaKPC-2, E. coli DH10B, E. coli DH10B with blaKPC-2 and the blaKPC-2 105 variants, as previously described.11,23 MICs for β-lactams and β-lactam-β-lactamase inhibitor combinations were generated using a “Steers Replicator™.” The structures of compounds used in this study are presented in Figure 3.
β-Lactamase purification
The KPC-2 β-lactamase and Trp105Phe,-Asn,-Leu, and -Val variants were purified from E. coli Origami™2 DE3 pLys cells carrying the pET24a(+)-blaKPC-2A22 plasmid with Trp105Phe,-Asn,-Leu, and -Val mutations as previously described.11,14 Briefly, cells were grown in super optimal broth, pelleted, and frozen for 18 h at −20°C. Pellets were subjected to stringent periplasmic fractionation, and crude extracts were used for preparative isoelectric focusing. Preparative isoelectric focusing fractions were concentrated and further purified using Äkta fast protein liquid chromatography. The purity of the enzymes after sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was estimated at ≥95% and further analyzed according to mass spectrometry.
Immunoblotting
E. coli DH10B cells from all 19 variants and WT were grown in LB for immunoblotting, as described previously.14 Cell pellets were resuspended directly into SDS-PAGE loading dye, run on a SDS-PAGE gel, and transferred to a polyvinylidene fluoride membrane. The membrane was blocked overnight in 5.0% bovine serum albumin (BSA) (Amresco, Solon, OH) in 20 mM Tris-HCl pH 7.4 with 150 mM NaCl (Tris buffered saline [TBS]). The membrane was washed with TBS and incubated with polyclonal anti-KPC-2 antibody from rabbits (Sigma) in 5.0% BSA in TBS. The membrane was washed with TBS with 0.05% Tween-20 (TBST) and incubated with protein G-HRP conjugate (Biorad). The blot was washed again with TBST and then developed using ECL™ developing kit (GE Healthcare Life Sciences) according to the manufacturer's instructions.
Kinetics
Steady-state kinetic parameters were determined using an Agilent™ (Santa Clara, CA) 8453 Diode Array spectrophotometer as previously described.11,14 The extinction coefficient used for doripenem: Δɛ = −11,460 M−1cm−1 at 299 nm. Briefly, each assay was completed in 10 mM PBS at pH 7.4 at room temperature with the enzyme (E) to substrate (S) concentrations set to establish pseudo first-order kinetics, where the concentration of enzyme is negligible [Eq. (1)].
| (1) |
In this model, formation of the Michaelis complex, E:S, is represented by the dissociation constant, K, which is equivalent to k−1/k1. k2 is the first-order rate constant for the acylation step, or formation of E–S. k3 is the rate constant for the hydrolysis of the acyl-enzyme to form E + P, where P is product.
Using Enzfitter™ (Biosoft Corporation, Ferguson, MO), a nonlinear least square fit of the initial reaction rates to the Henri Michaelis-Menten equation determined the kinetic parameters, Vmax and Km [Eq. (2)]:
| (2) |
To determine Ki of the inhibitors (I) for the enzyme, a direct competition assay under steady state conditions was performed. Nitrocefin (NCF) was used as the reporter substrate (S) at a final concentration of 100 μM. Inverse initial steady-state velocities (1/v0) were plotted against inhibitor concentration to obtain a straight line. Measuring the initial steady-state velocity immediately after mixing yielded a Ki which approximates Km if we assume that under these conditions the inhibitors behaved as competitive inhibitors. Ki (observed) was determined by dividing the value for the y-intercept by the slope of line. The data were corrected to account for the affinity of NCF (S) for the β-lactamase [Eq. (3)].
| (3) |
Partition ratios or rate constant for product formation compared to the rate constant for inactivation (kcat/kinact) for KPC-2 were obtained by incubating KPC-2 with increasing concentrations of inhibitor at room temperature for 15 min in 10 mM PBS, pH 7.4. The ratio of inhibitor to enzyme (I:E) necessary to inhibit the hydrolysis of NCF by greater than 90% was determined.
ΔΔGcat is the activation energy that reflects the energy required for binding, and bond breaking and formation. Activation energies were determined as previously described using the following equation.14
| (4) |
Molecular modeling
The crystal coordinates of KPC-2 (PDB accession code 2OV5) were used to construct acyl-enzyme models of imipenem and doripenem with the WT KPC-2 β-lactamase and the Trp105Leu variant, as previously described using Discovery Studio 2.1 (DS 2.1, Accelrys, Inc. San Diego, CA) molecular modeling software.11 The Trp105Leu variant was built by substituting the residue at position 105. The imipenem and doripenem structures were constructed using Fragment Builder tools and were minimized using a Standard Dynamics Cascade protocol of DS 2.1. The hydrolyzed imipenem and doripenem were automatically docked into the active site of KPC-2 β-lactamase using the Flexible Docking module of DS 2.1. The protocol allowed docking of flexible carbapenem in the flexible active site of KPC-2. After docking, a covalent bond was introduced between Ser70 and C7 of the hydrolyzed β-lactam. The most favorable pose of imipenem and doripenem was chosen and the complex was created.
The acyl-enzyme complexes of the Trp105Leu variant with imipenem and doripenem were created in the same way. To check the stability and look for possible conformational changes of the β-lactamase-ligand complex, MDS was conducted. Each stage of MDS was performed for 5000 steps for a total of 15.0 ps. The initial temperature was 50K. To establish a trajectory the complex was heated to a final temperature of 300K. The heating stage was followed by an equilibration stage at 300K and finally the production stage of the trajectory. During the production stage, snapshots of the trajectory were taken every 100 steps for a total of 50 snapshots or frames out of the 5000 total steps. After each stage, the potential and total energy versus time was monitored to make sure that the system was equilibrated and when the plateau was reached the simulation was stopped. The trajectory RMSD relative to the starting structure was in the 1 Å range. In addition to the acyl-enzyme complexes, MDS was also performed on the apo-KPC-2 for a longer time (150 ps) to monitor movements by Trp105.16 After MDS, the 50 frames of the trajectory were evaluated using the analysis module of DS 2.1. The snapshot with the lowest potential energy was chosen for each β-lactamase-β-lactam complex and these models are presented in Figure 4. Heat plots revealing the movement of all possible hydrogen bonding interactions during the trajectory between the β-lactam and β-lactamase of the four acyl-enzyme complexes were created (Fig. 5). In addition, the movement of the carbonyl oxygen of the β-lactam between the backbone nitrogens of Ser70 and Thr237 of the β-lactamase were monitored, and the data are plotted as distance in Å versus the frame index or movement in time for each acyl-enzyme complex (Fig. 6).
During the simulation of KPC-2 with imipenem, the carbonyl of imipenem is positioned in the oxyanion hole. However, carbonyl of imipenem is very flexible within the WT KPC-2 active site, as evidenced when monitoring the hydrogen bonding distances between the backbone nitrogens of Ser70 and Thr237 and the carbonyl oxygen of imipenem during MDS [Fig. 6(A)].
Acknowledgments
The authors thank Dr. Michael E. Harris and Dr. Sarah Drawz for critical review of this manuscript. R.A.B. received research and speaking invites from various pharmaceutical companies.
Glossary
Abbreviations:
- WT
wild type
- MDS
molecular dynamics simulation
- MIC
minimal inhibitory concentration
- LB
lysogeny broth
- BSA
bovine serum albumin
- TBS
Tris Buffered Saline
References
- 1.Kattan JN, Villegas MV, Quinn JP. New developments in carbapenems. Clin Microbiol Infect. 2008;14:1102–1111. doi: 10.1111/j.1469-0691.2008.02101.x. [DOI] [PubMed] [Google Scholar]
- 2.Ke W, Bethel CR, Thomson JM, Bonomo RA, van den Akker F. Crystal structure of KPC-2: insights into carbapenemase activity in class A beta-lactamases. Biochemistry. 2007;46:5732–5740. doi: 10.1021/bi700300u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Girlich D, Poirel L, Nordmann P. Novel ambler class A carbapenem-hydrolyzing {beta}-lactamase, from a Pseudomonas fluorescens in the Seine River, Paris, France. Antimicrob Agents Chemother. 2009;54:328–332. doi: 10.1128/AAC.00961-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henriques I, Moura A, Alves A, Saavedra MJ, Correia A. Molecular characterization of a carbapenem-hydrolyzing class A beta-lactamase, SFC-1, from Serratia fonticola UTAD54. Antimicrob Agents Chemother. 2004;48:2321–2324. doi: 10.1128/AAC.48.6.2321-2324.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poirel L, Weldhagen GF, Naas T, De Champs C, Dove MG, Nordmann P. GES-2, a class A beta-lactamase from Pseudomonas aeruginosa with increased hydrolysis of imipenem. Antimicrob Agents Chemother. 2001;45:2598–2603. doi: 10.1128/AAC.45.9.2598-2603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Endimiani A, Hujer AM, Perez F, Bethel CR, Hujer KM, Kroeger J, Oethinger M, Paterson DL, Adams MD, Jacobs MR, Diekema DJ, Hall GS, Jenkins SG, Rice LB, Tenover FC, Bonomo RA. Characterization of blaKPC-containing Klebsiella pneumoniae isolates detected in different institutions in the Eastern USA. J Antimicrob Chemother. 2009;63:427–437. doi: 10.1093/jac/dkn547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oelschlaeger P, Ai N, Duprez KT, Welsh WJ, Toney JH. Evolving carbapenemases: can medicinal chemists advance one step ahead of the coming storm? J Med Chem. 2010;53:3013–3027. doi: 10.1021/jm9012938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yigit H, Queenan AM, Rasheed JK, Biddle JW, Domenech-Sanchez A, Alberti S, Bush K, Tenover FC. Carbapenem-resistant strain of Klebsiella oxytoca harboring carbapenem-hydrolyzing beta-lactamase KPC-2. Antimicrob Agents Chemother. 2003;47:3881–3889. doi: 10.1128/AAC.47.12.3881-3889.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papp-Wallace KM, Bethel CR, Distler AM, Kasuboski C, Taracila M, Bonomo RA. Inhibitor resistance in the KPC-2 beta-lactamase, a preeminent property of this class A beta-lactamase. Antimicrob Agents Chemother. 2010;54:890–897. doi: 10.1128/AAC.00693-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petrella S, Ziental-Gelus N, Mayer C, Renard M, Jarlier V, Sougakoff W. Genetic and structural insights into the dissemination potential of the extremely broad-spectrum class A beta-lactamase KPC-2 identified in an Escherichia coli strain and an Enterobacter cloacae strain isolated from the same patient in France. Antimicrob Agents Chemother. 2008;52:3725–3736. doi: 10.1128/AAC.00163-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ambler RP. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 14.Papp-Wallace KM, Taracila MS, Endimiani A, Hujer AM, Hujer KM, Distler AM, Hornick JM, Bonomo RA. Substrate selectivity and a novel role in inhibitor discrimination by position 237 in the KPC-2 β-lactamase. Antimicrob Agents Chemother. 2010;54:2867–2877. doi: 10.1128/AAC.00197-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doucet N, De Wals PY, Pelletier JN. Site-saturation mutagenesis of Tyr-105 reveals its importance in substrate stabilization and discrimination in TEM-1 beta-lactamase. J Biol Chem. 2004;279:46295–46303. doi: 10.1074/jbc.M407606200. [DOI] [PubMed] [Google Scholar]
- 16.Doucet N, Pelletier JN. Simulated annealing exploration of an active-site tyrosine in TEM-1 beta-lactamase suggests the existence of alternate conformations. Proteins. 2007;69:340–348. doi: 10.1002/prot.21485. [DOI] [PubMed] [Google Scholar]
- 17.Doucet N, Savard PY, Pelletier JN, Gagne SM. NMR investigation of Tyr105 mutants in TEM-1 beta-lactamase: dynamics are correlated with function. J Biol Chem. 2007;282:21448–21459. doi: 10.1074/jbc.M609777200. [DOI] [PubMed] [Google Scholar]
- 18.Queenan AM, Shang W, Flamm R, Bush K. Hydrolysis and inhibition profiles of beta-lactamases from molecular classes A to D with doripenem, imipenem and meropenem. Antimicrob Agents Chemother. 2009;54:565–569. doi: 10.1128/AAC.01004-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Page MG. b-Lactamase inhibitors. Drug Resist Updat. 2000;3:109–125. doi: 10.1054/drup.2000.0137. [DOI] [PubMed] [Google Scholar]
- 20.Mourey L, Miyashita K, Swaren P, Bulychev A, Samama JP, Mobashery S. Inhibition of the NMC-A β-lactamase by a penicillanic acid derivative and the structural bases for the increase in substrate profile of this antibiotic resistance enzyme. J Am Chem Soc. 1998;120:9382–9383. [Google Scholar]
- 21.Taibi P, Mobashery S. Mechanism of turnover of imipenem by the TEM .beta.-lactamase revisited. J Am Chem Soc. 1995;117:7600–7605. [Google Scholar]
- 22.Frase H, Shi Q, Testero SA, Mobashery S, Vakulenko SB. Mechanistic basis for the emergence of catalytic competence against carbapenem antibiotics by the GES family of beta-lactamases. J Biol Chem. 2009;284:29509–29513. doi: 10.1074/jbc.M109.011262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.CLSI. CLSI Performance Standards for Antimicrobial Susceptibility Testing. Eighteen Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute; 2007. CLSI M100-S17. [Google Scholar]