Abstract
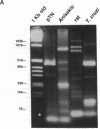
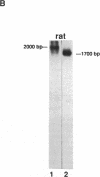
Serine proteases are one of the biologically most important and widely distributed families of enzymes. Isolation of serine protease genes from organisms of widely diverged phylogenetic groups would provide a basis for studying their biological function, the relationship between structure and function, and the molecular evolution of these enzymes. Serine proteases for which little structural information is known are those that are important in the pathogenesis of parasitic nematode and protozoan diseases. Identification and isolation of protease genes from these organisms is a critical first step in understanding their function for the parasite and possibly suggesting innovative approaches to arresting parasitic diseases. Serine protease gene fragments were isolated from genomic DNA of the parasitic nematode Anisakis simplex by using degenerate oligonucleotide primers and the polymerase chain reaction. Primers were designed based upon the consensus sequence of amino acids flanking the active site serine and histidine residues of eukaryotic serine proteases. Four serine protease gene fragments from this parasite were sequenced and one is 67% identical to the rat trypsin II gene. Alignment of these two genes revealed that the intron-exon junctions are conserved between nematode and rat suggesting that this Anisakis serine protease is structurally and functionally similar to rat trypsin. The generality of this approach to identify serine protease genes from genomic DNA of two very divergent species, a parasitic protozoan and a mammal, was also confirmed. Genes for other enzymes or any protein with conserved structural motifs can be identified and isolated using this technology. Using a similar strategy, a cathepsin B-like cysteine (thiol) protease gene fragment was isolated from Caenorhabditis elegans DNA.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bell G. I., Quinto C., Quiroga M., Valenzuela P., Craik C. S., Rutter W. J. Isolation and sequence of a rat chymotrypsin B gene. J Biol Chem. 1984 Nov 25;259(22):14265–14270. [PubMed] [Google Scholar]
- Birktoft J. J., Blow D. M. Structure of crystalline -chymotrypsin. V. The atomic structure of tosyl- -chymotrypsin at 2 A resolution. J Mol Biol. 1972 Jul 21;68(2):187–240. doi: 10.1016/0022-2836(72)90210-0. [DOI] [PubMed] [Google Scholar]
- Björck L., Akesson P., Bohus M., Trojnar J., Abrahamson M., Olafsson I., Grubb A. Bacterial growth blocked by a synthetic peptide based on the structure of a human proteinase inhibitor. Nature. 1989 Jan 26;337(6205):385–386. doi: 10.1038/337385a0. [DOI] [PubMed] [Google Scholar]
- Blow D. M., Birktoft J. J., Hartley B. S. Role of a buried acid group in the mechanism of action of chymotrypsin. Nature. 1969 Jan 25;221(5178):337–340. doi: 10.1038/221337a0. [DOI] [PubMed] [Google Scholar]
- Bode W., Chen Z., Bartels K., Kutzbach C., Schmidt-Kastner G., Bartunik H. Refined 2 A X-ray crystal structure of porcine pancreatic kallikrein A, a specific trypsin-like serine proteinase. Crystallization, structure determination, crystallographic refinement, structure and its comparison with bovine trypsin. J Mol Biol. 1983 Feb 25;164(2):237–282. doi: 10.1016/0022-2836(83)90077-3. [DOI] [PubMed] [Google Scholar]
- Brayer G. D., Delbaere L. T., James M. N. Molecular structure of crystalline Streptomyces griseus protease A at 2.8 A resolution. II. Molecular conformation, comparison with alpha-chymotrypsin and active-site geometry. J Mol Biol. 1978 Sep 5;124(1):261–283. doi: 10.1016/0022-2836(78)90159-6. [DOI] [PubMed] [Google Scholar]
- Brayer G. D., Delbaere L. T., James M. N. Molecular structure of the alpha-lytic protease from Myxobacter 495 at 2.8 Angstroms resolution. J Mol Biol. 1979 Jul 15;131(4):743–775. doi: 10.1016/0022-2836(79)90200-6. [DOI] [PubMed] [Google Scholar]
- Brenner S. The molecular evolution of genes and proteins: a tale of two serines. Nature. 1988 Aug 11;334(6182):528–530. doi: 10.1038/334528a0. [DOI] [PubMed] [Google Scholar]
- Castro C., Craig S. P., Castañeda M. Genome organization and ploidy number in Trypanosoma cruzi. Mol Biochem Parasitol. 1981 Dec 31;4(5-6):273–282. doi: 10.1016/0166-6851(81)90060-8. [DOI] [PubMed] [Google Scholar]
- Codding P. W., Delbaere L. T., Hayakawa K., Hutcheon W. L., James M. N., Jurásek L. The 4.5 Angstrom resolution structure of a bacterial serine protease from Streptomyces griseus. Can J Biochem. 1974 Mar;52(3):208–220. doi: 10.1139/o74-034. [DOI] [PubMed] [Google Scholar]
- Craik C. S., Choo Q. L., Swift G. H., Quinto C., MacDonald R. J., Rutter W. J. Structure of two related rat pancreatic trypsin genes. J Biol Chem. 1984 Nov 25;259(22):14255–14264. [PubMed] [Google Scholar]
- Craik C. S., Roczniak S., Largman C., Rutter W. J. The catalytic role of the active site aspartic acid in serine proteases. Science. 1987 Aug 21;237(4817):909–913. doi: 10.1126/science.3303334. [DOI] [PubMed] [Google Scholar]
- Craik C. S., Rutter W. J., Fletterick R. Splice junctions: association with variation in protein structure. Science. 1983 Jun 10;220(4602):1125–1129. doi: 10.1126/science.6344214. [DOI] [PubMed] [Google Scholar]
- DIXON G. H., GO S., NEURATH H. Peptides combined with 14C-diisopropyl phosphoryl following degradation of 14C-DIP-trypsin with alpha-chymotrypsin. Biochim Biophys Acta. 1956 Jan;19(1):193–195. doi: 10.1016/0006-3002(56)90414-0. [DOI] [PubMed] [Google Scholar]
- Doolittle R. F., Feng D. F. Reconstructing the evolution of vertebrate blood coagulation from a consideration of the amino acid sequences of clotting proteins. Cold Spring Harb Symp Quant Biol. 1987;52:869–874. doi: 10.1101/sqb.1987.052.01.095. [DOI] [PubMed] [Google Scholar]
- Gershenfeld H. K., Weissman I. L. Cloning of a cDNA for a T cell-specific serine protease from a cytotoxic T lymphocyte. Science. 1986 May 16;232(4752):854–858. doi: 10.1126/science.2422755. [DOI] [PubMed] [Google Scholar]
- HARTLEY B. S. AMINO-ACID SEQUENCE OF BOVINE CHYMOTRYPSINOGEN-A. Nature. 1964 Mar 28;201:1284–1287. doi: 10.1038/2011284a0. [DOI] [PubMed] [Google Scholar]
- Hill R. E., Hastie N. D. Accelerated evolution in the reactive centre regions of serine protease inhibitors. Nature. 1987 Mar 5;326(6108):96–99. doi: 10.1038/326096a0. [DOI] [PubMed] [Google Scholar]
- Huber R., Kukla D., Bode W., Schwager P., Bartels K., Deisenhofer J., Steigemann W. Structure of the complex formed by bovine trypsin and bovine pancreatic trypsin inhibitor. II. Crystallographic refinement at 1.9 A resolution. J Mol Biol. 1974 Oct 15;89(1):73–101. doi: 10.1016/0022-2836(74)90163-6. [DOI] [PubMed] [Google Scholar]
- Kraut J. Serine proteases: structure and mechanism of catalysis. Annu Rev Biochem. 1977;46:331–358. doi: 10.1146/annurev.bi.46.070177.001555. [DOI] [PubMed] [Google Scholar]
- LIGHT A., FRATER R., KIMMEL J. R., SMITH E. L. CURRENT STATUS OF THE STRUCTURE OF PAPAIN: THE LINEAR SEQUENCE, ACTIVE SULFHYDRYL GROUP, AND THE DISULFIDE BRIDGES. Proc Natl Acad Sci U S A. 1964 Nov;52:1276–1283. doi: 10.1073/pnas.52.5.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKerrow J. H., Doenhoff M. J. Schistosome proteases. Parasitol Today. 1988 Dec;4(12):334–340. doi: 10.1016/0169-4758(88)90002-6. [DOI] [PubMed] [Google Scholar]
- McKerrow J. H. Parasite proteases. Exp Parasitol. 1989 Jan;68(1):111–115. doi: 10.1016/0014-4894(89)90016-7. [DOI] [PubMed] [Google Scholar]
- Neurath H. Evolution of proteolytic enzymes. Science. 1984 Apr 27;224(4647):350–357. doi: 10.1126/science.6369538. [DOI] [PubMed] [Google Scholar]
- Neurath H. The versatility of proteolytic enzymes. J Cell Biochem. 1986;32(1):35–49. doi: 10.1002/jcb.240320105. [DOI] [PubMed] [Google Scholar]
- Newport G. R., McKerrow J. H., Hedstrom R., Petitt M., McGarrigle L., Barr P. J., Agabian N. Cloning of the proteinase that facilitates infection by schistosome parasites. J Biol Chem. 1988 Sep 15;263(26):13179–13184. [PubMed] [Google Scholar]
- Portnoy D. A., Erickson A. H., Kochan J., Ravetch J. V., Unkeless J. C. Cloning and characterization of a mouse cysteine proteinase. J Biol Chem. 1986 Nov 5;261(31):14697–14703. [PubMed] [Google Scholar]
- Rappolee D. A., Mark D., Banda M. J., Werb Z. Wound macrophages express TGF-alpha and other growth factors in vivo: analysis by mRNA phenotyping. Science. 1988 Aug 5;241(4866):708–712. doi: 10.1126/science.3041594. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Sarkis G. J., Kurpiewski M. R., Ashcom J. D., Jen-Jacobson L., Jacobson L. A. Proteases of the nematode Caenorhabditis elegans. Arch Biochem Biophys. 1988 Feb 15;261(1):80–90. doi: 10.1016/0003-9861(88)90106-3. [DOI] [PubMed] [Google Scholar]
- Sawyer L., Shotton D. M., Campbell J. W., Wendell P. L., Muirhead H., Watson H. C. The atomic structure of crystalline porcine pancreatic elastase at 2.5 A resolution: comparisons with the structure of alpha-chymotrypsin. J Mol Biol. 1978 Jan 15;118(2):137–208. doi: 10.1016/0022-2836(78)90412-6. [DOI] [PubMed] [Google Scholar]
- Sloane B. F., Rozhin J., Hatfield J. S., Crissman J. D., Honn K. V. Plasma membrane-associated cysteine proteinases in human and animal tumors. Exp Cell Biol. 1987;55(4):209–224. doi: 10.1159/000163420. [DOI] [PubMed] [Google Scholar]
- Stroud R. M. A family of protein-cutting proteins. Sci Am. 1974 Jul;231(1):74–88. doi: 10.1038/scientificamerican0774-74. [DOI] [PubMed] [Google Scholar]
- Stroud R. M., Kay L. M., Dickerson R. E. The structure of bovine trypsin: electron density maps of the inhibited enzyme at 5 Angstrom and at 2-7 Angstron resolution. J Mol Biol. 1974 Feb 25;83(2):185–208. doi: 10.1016/0022-2836(74)90387-8. [DOI] [PubMed] [Google Scholar]
- Tulinsky A., Vandlen R. L., Morimoto C. N., Mani N. V., Wright L. H. Variability in the tertiary structure of alpha-chymotrypsin at 2.8-A resolution. Biochemistry. 1973 Oct 9;12(21):4185–4192. doi: 10.1021/bi00745a023. [DOI] [PubMed] [Google Scholar]
- Vasquez J. R., Evnin L. B., Higaki J. N., Craik C. S. An expression system for trypsin. J Cell Biochem. 1989 Mar;39(3):265–276. doi: 10.1002/jcb.240390306. [DOI] [PubMed] [Google Scholar]
- Wright C. S., Alden R. A., Kraut J. Structure of subtilisin BPN' at 2.5 angström resolution. Nature. 1969 Jan 18;221(5177):235–242. doi: 10.1038/221235a0. [DOI] [PubMed] [Google Scholar]