Abstract
Alginates are commercially valuable and complex polysaccharides composed of varying amounts and distribution patterns of 1–4-linked β-d-mannuronic acid (M) and α-l-guluronic acid (G). This structural variability strongly affects polymer physicochemical properties and thereby both commercial applications and biological functions. One promising approach to alginate fine structure elucidation involves the use of alginate lyases, which degrade the polysaccharide by cleaving the glycosidic linkages through a β-elimination reaction. For such studies one would ideally like to have different lyases, each of which cleaves only one of the four possible linkages in alginates: G-G, G-M, M-G, and M-M. So far no lyase specific for only G-G linkages has been described, and here we report the construction of such an enzyme by mutating the gene encoding Klebsiella pneumoniae lyase AlyA (a polysaccharide lyase family 7 lyase), which cleaves both G-G and G-M linkages. After error-prone PCR mutagenesis and high throughput screening of ∼7000 lyase mutants, enzyme variants with a strongly improved G-G specificity were identified. Furthermore, in the absence of Ca2+, one of these lyases (AlyA5) was found to display no detectable activity against G-M linkages. G-G linkages were cleaved with ∼10% of the optimal activity under the same conditions. The substitutions conferring altered specificity to the mutant enzymes are located in conserved regions in the polysaccharide lyase family 7 alginate lyases. Structure-function analyses by comparison with the known three-dimensional structure of Sphingomonas sp. A1 lyase A1-II′ suggests that the improved G-G specificity might be caused by increased affinity for nonproductive binding of the alternating G-M structure.
Keywords: Carbohydrate, Carbohydrate Metabolism, Enzyme Mutation, Metabolism, Polysaccharide
Introduction
Alginate is a linear polysaccharide widely used in industry and is comprised of 1–4-linked β-d-mannuronic acid (M)2 and its C-5 epimer α-l-guluronic acid (G). The monomers are arranged in blocks of continuous M residues (M-blocks), G residues (G-blocks), or alternating residues (MG-blocks). Alginate is produced by marine brown algae and by bacteria belonging to the genera Azotobacter and Pseudomonas (1–4). Biosynthesis of alginate involves the initial production of polymannuronate followed by the introduction of G residues catalyzed by mannuronan C-5 epimerases (5–7). Pseudomonas alginates differ from those obtained by other sources in lacking G-blocks, and bacterial alginates, as opposed to the seaweed polymers, can be acetylated to various degrees at positions O-2 and/or O-3 on the M residues (8). The material properties of alginates are determined by intrinsic properties like the polymer chain length, acetylation level, and the monomer composition and distribution pattern.
Commercially alginate is harvested from seaweed and has been utilized extensively for a variety of industrial and biotechnological purposes (9–11). In recent years, much attention has been drawn to new and promising applications of alginates in pharmacy and medicine, e.g. in drug or protein delivery, cell encapsulation, tissue regeneration, surgery, and wound management (12–18). With the entry of alginate-based biomaterials into the field of human medicine, the term “tailor-made alginate” has been introduced. This means that one ideally wants to produce and use alginate molecules with defined properties optimally suited for a given application. Hence there is an emerging need for more detailed information concerning the alginate fine structure. Although the total monomer composition and the dyad and triad frequencies can be elucidated by, for example, high resolution NMR spectroscopy (8), there is limited knowledge on the distribution and the absolute length of the various block types.
A promising approach to the challenge of alginate sequence elucidation is the use of lyases to degrade complex alginate molecules followed by analyses on the resulting population of alginate oligomers (19, 20). This concept has become increasingly attractive with the continuous development of chromatographic methods. Ideal lyases in this case are enzymes that cleave only one of the four possible types of linkages (M-M, M-G, G-M, and G-G) in the alginate. For instance, after degradation with an M-M specific lyase, one can obtain information about the amount and length of G-blocks and MG-blocks in the substrate.
Alginate lyases catalyze the degradation of alginate targeting the glycosidic 1 → 4 O-linkages between monomers by a β-elimination mechanism leaving a 4-deoxy-l-erythro-hex-4-enopyranosyluronic acid (often denoted as Δ) as the nonreducing terminal residue (21). Lyases can be isolated from a variety of organisms including algae, marine invertebrates (e.g. mollusks), bacteriophages, and several marine and terrestrial bacterial species (19). The enzymes are generally classified as M- or G-lyases (EC 4.2.2.3 and EC 4.2.2.11) according to their dominating cleaving reaction on M- or G-rich alginates. The majority of lyases characterized at present display activity toward more than one of the four types of linkages.
Klebsiella pneumoniae produces an extracellular alginate lyase, AlyA, which according to the primary structure is classified in the polysaccharide lyase family 7 (PL-7) (22, 23). AlyA is endolytic, acting on G-blocks and MG-blocks where G-M linkages are cleaved in the latter substrate (24). The three-dimensional structure for three of the PL-7 lyases have been determined: A1-II′ from Sphingomonas sp. A1, ALY-1 from Corynebacterium sp. ALY-1, and PA1167 from Pseudomonas aeruginosa PAO1 (25–27). These structures indicate a common basic framework for PL-7 lyases, which is a β-sandwich fold consisting of two β-sheets (SA and SB) creating a deep active cleft that is covered by two flexible loops (L1 and L2). Despite structural similarities, their substrate preferences differ. A1-II′ is reported to have a broad substrate range, acting equally well on G-blocks, M-blocks, and MG-blocks (28). PA1167 has limited activity on G-blocks, acts preferentially on MG-blocks, but also has activity toward M-blocks (27). ALY-1 is reported to be much more active toward a G-rich substrate (81% G) than toward an M-rich (82% M) substrate; however, the activity of ALY-1 toward a specific substrate like MG-blocks has not been reported (29). A1-II, which is another lyase from Sphingomonas sp. A1, is reported to have 2.5–5 times higher activity on polyG than on polyMG (27, 28). Furthermore, the lyase from ATCC43367 is reported to cleave only M-M linkages (30).
In the present work we aimed at modifying the specificity of the K. pneumoniae AlyA to obtain a lyase with preference for cleaving only G-G linkages, a type of specificity that to our knowledge has not been described. AlyA was chosen as a basis because of its inherent high activity on G-G linkages and our previous experience with expression and purification of this lyase (24). To identify the mutants we used in vitro-made alginates with highly defined composition as substrates in a high throughput screening protocol. Mutants with the desired properties were identified, and their enhanced specificity appears to mainly be caused by increased affinity toward nonproductive M-G binding.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Growth Conditions, and DNA Manipulations
Escherichia coli strains DH5α (BRL) and XL10-Gold® (Stratagene) were used as general cloning hosts and for establishing the mutant library, respectively. K. pneumoniae (formerly K. aerogenes, type 25) was used for cloning of the alyA gene. Bacteria were routinely grown at 37 °C in L broth (5 g of yeast extract, 10 g of tryptone, and 10 g of NaCl/liter) or on L agar (L broth supplemented with 20 g of agar/liter). For protein expression, the strains were grown in triple strength L broth (15 g of yeast extract, 30 g of tryptone, and 10 g of NaCl/liter). Growth media were supplemented with ampicillin (100 μg/ml) when appropriate. For growth in 96-well plates, a reduced Hi-Ye medium with the following composition was used: Na2HPO4·2H2O, 12.3 g/liter; KH2PO4, 4.29 g/liter; NH4Cl, 0.43 g/liter; NaCl, 0.71 g/liter; glucose, 2.86 g/liter; yeast extract, 2.86 g/liter; citric acid, 1.43 g/liter; MgSO4, 1.86 mm; Fe(III)-citrate, 118 μm; H3BO3, 21.0 μm; MnCl2, 37.6 μm; EDTA, 9.86 μm; CuCl2, 3.86 μm; Na2MoO4, 4.29 μm; CoCl2, 4.71 μm; and zinc acetate, 17.3 μm. Cultures were induced for protein expression using an induction solution containing: glycerol (99%), 25.8 g/liter; yeast extract, 24 g/liter; and m-toluic acid to a final concentration of 0.5 mm. Standard recombinant DNA protocols were performed as described elsewhere (31). Plasmids were isolated by the Wizard® Plus SV minipreps DNA purification system (Promega). Transformation of XL10-Gold® ultracompetent cells was performed according to the instructions from the manufacturer and for DH5α according to the RbCl transformation protocol (New England BioLabs). DNA sequencing was performed using the BigDye® Terminator version 1.1 cycle sequencing kit (Applied Biosystems).
Vector Constructions and Random and Site-directed Mutagenesis
Primers used for cloning and mutagenesis are given in supplemental Table S1. The alyA gene including the signal sequence was PCR-amplified from the genome of K. pneumoniae and ligated into pGEM-5Zf (Promega) as an NdeI-NotI fragment yielding pAT74. pAT74 was used as template for random- and site-directed mutagenesis using the GeneMorph® II random mutagenesis kit and the QuikChange® II site-directed mutagenesis kit from Stratagene, respectively, and introduction of mutations was verified by sequencing. For protein expression, the wild type or mutant lyase genes were ligated into an expression vector containing the regulatory Pm/xylS promoter system. The vector utilized was identical to pJBphOx-271d (32) except for the ML1–14 mutation in Pm causing increased expression levels (33). As described under “Results,” variants of the eight mutants isolated in the screen were made to decide the contribution of the individual substitutions on the observed phenotype. When possible these constructs were made by subcloning fragments containing the specific mutations into the vector containing wild type alyA; otherwise mutations were reconstructed by site-directed mutagenesis.
Production and Purification of polyMG and polyG
An alginate with a regular poly-alternating structure, polyMG, with FG = 0.47 and FGG = 0 was produced by epimerization of mannuronan with the recombinantly produced mannuronan C-5 epimerase AlgE4 and characterized by NMR as described previously (34). polyG with FG = 0.94 and a degree of polymerization of 18.5 was prepared from Laminaria hyperborea stipes as described elsewhere (21, 35).
Measurement of Alginate Lyase Activity
Alginate lyase activity was determined by monitoring the increase in A230 caused by production of unsaturated uronic acids as the lyase cleaves glycosidic bonds in the polymer chain. Unless otherwise stated, the activity was measured at room temperature in 96-well microtiter plates using a mixture of 240 μl of buffer (50 mm Tris-HCl, 0.2 m NaCl, 1 mm CaCl2, pH 7.5), 60 μl of alginate substrate (4 mg/ml), and 10–20 μl of protein extract. One unit of enzyme activity was defined as the amount of enzyme that increased A230 by 1 unit/min. The lyase activity was determined as the initial activity when the increase in A230 was linear. The lyase activity measurements were performed at least in triplicate.
Robotic Screening of the alyA Mutant Library
The E. coli library was plated on LB-agar in 25 × 25-cm Petri dishes (Corning CLS431301) and incubated overnight at 37 °C. The colonies were picked using a Genetix Q-Pix2 robotic colony picker and transferred to 96-well microplates (Greiner M3186) containing 80 μl of reduced Hi-Ye medium. The microplates were incubated at 30 °C, 900 rpm (3-mm amplitude) and 80% relative humidity. After 24 h, 40 μl of induction solution was added to each well using an Asys Hi-Tech Flexispence microplate dispenser. The microplates were incubated at 37 °C, 900 rpm and 80% relative humidity for 4 h after induction and were frozen at −40 °C prior to analysis. After thawing, the microplates were added 30 μl of B-PER II solution (Pierce) per well, shaken for 30 s (900 rpm, 3 mm amplitude), and incubated at room temperature for 1 h. After incubation, the microplates were shaken (850 rpm, 3-mm amplitude) for 10 min and then centrifuged for 30 min at 3500 × g. For measurement of alginate lyase activity, 384-well microplates (Corning CLS3675) were filled with 50 μl of assay buffer (40 mm MOPS, 20 mm NaCl, 2 mm CaCl2, pH 6.8) containing either polyG or polyMG alginate (0.2 mg/ml). Each 384-well plate contained 192 wells filled with polyG and the rest with polyMG. 4 μl of cell free extract was then added to two wells in the 384-well assay plate, one with each of the two types of alginate. The wells in the 384-well plate were mixed by pipetting (3 × 25 μl), and the absorbance at 230 nm (A230) was read in a Molecular Devices SpectraMax 384+ microplate reader shortly after mixing and after 40 and 120 min of incubation at room temperature. The increase in absorption during incubation was calculated for each type of alginate, and ΔA230 was used for evaluation of the G-G and M-G activity of the enzyme extracts. The tests were performed with parallel cultures in microwell plates (96 parallels) to determine the accuracy of the screening protocols, and the standard deviation was generally below 5% in the enzyme assay. All liquid and microplate handling was performed by a Tecan Genesis RSP 200 robotic liquid handling work station.
Protein Expression and Purification
The cultures were grown in 50 ml of 3× LB medium in 500-ml baffled flasks at 30 °C for 3 h before induction of the Pm promoter with 0.5 mm m-toluic acid. Cultivation was continued for an additional 4 h before harvesting the cells by centrifugation. The cells were disrupted by sonication in 20 mm Na2HPO4, 1 mm CaCl2 (pH 6.8), and centrifuged for 30 min at 20 000 × g. The supernatant was filtered (0.2 μm) and applied on a 5-ml HiTrap HP SP column equilibrated with the same buffer as above. The proteins were eluted by a stepwise NaCl gradient (total gradient from 0 to 1 m) in the same buffer and collected in 5-ml fractions. The fractions were tested for alginate lyase activity, and protein content of active fractions was determined by the Bio-Rad microassay procedure. Bovine serum albumin was used as standard. The purity of lyase-containing fractions was analyzed by SDS-PAGE.
Analysis of Alginate Degradation by High Performance Anion Exchange Chromatography with Pulsed Amperometric Detection (HPAEC-PAD)
Lyase degradation of polyG and polyMG was performed in 50 mm Tris-HCl, 0.2 m NaCl, 1 mm CaCl2, pH 7.5, at room temperature, and the substrate concentration used was 1 mg/ml. To inactivate the lyases, the samples were boiled for 10 min after the addition of 1% SDS. Analysis of lyase-degraded alginate samples was performed using a Dionex BioLC system (Dionex Corp, Sunnyvale, CA) consisting of an AS50 autosampler, an ED40 Electrochemical Detector with a nondisposable gold working electrode, and a GP50 Gradient Pump. All of the samples (1 mg/ml, 25 μl) were injected via a 100-μl loading loop. The oligosaccharides were separated at room temperature by gradient elution with 0–700 mm sodium acetate in 100 mm sodium hydroxide over 80 min on a Dionex IonPac AS4A (4 × 250 mm) anion exchange column connected to an IonPac AG4A (4 × 50 mm) guard column. The flow rate was set to 1 ml/min using waveform A for detection. Data acquisition and analysis were performed using Chromeleon 6.7 software (20). Oligomer standards up to hexamer were produced by fractionation of partially lyase-degraded polyG and polyMG on SEC columns as described earlier (36). The fractions were analyzed with 1H NMR to determine chain length and chemical composition.
Bioinformatic Analysis of Mutations
The experimental three-dimensional structures of Sphingomonas sp. A1 lyase A1-II′ (37) with GGG (Protein Data Bank code 2ZAB) and MMG (Protein Data Bank code 2ZAC) in the substrate binding pocket were downloaded from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (38). The structures were analyzed with ligand-protein contacts (39) and LigPlot (40) and visualized with SwissProtein Data BankViewer (41). Identification and alignment of homologous protein sequences was done with PSI-Blast (42) and ClustalX (43).
RESULTS
Construction and Screening of an alyA Mutant Library for Identification of Enzymes Exhibiting Increased Specificity toward G-G Linkages
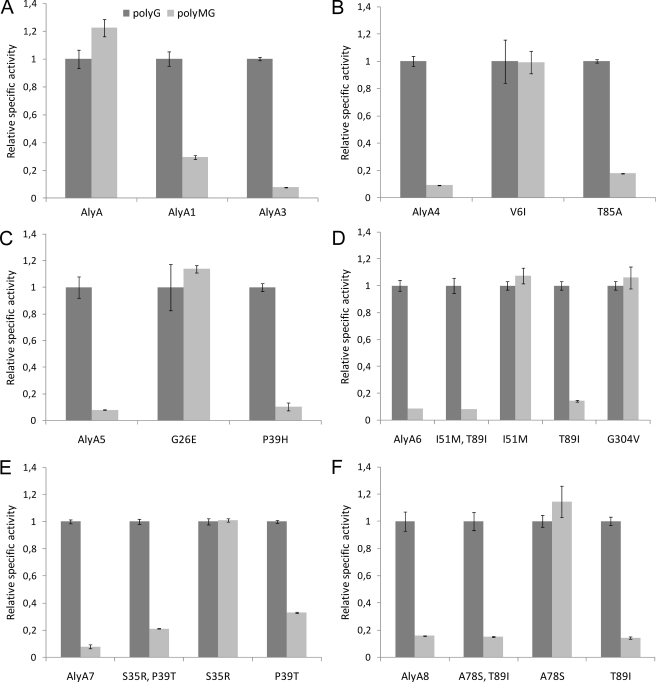
Initial studies showed that the specific activities of AlyA on polyMG and polyG are 937 and 765 units/mg, respectively (Table 1). Because of the defined nature of the substrates, this is considered to be the activity on G-M and G-G linkages, respectively. The undesired activity against G-M bonds is therefore somewhat higher than against the G-G bonds in the wild type enzyme. A library of mutated alyA genes was then constructed by error-prone PCR, and the experimental conditions were adjusted to achieve an average frequency of two to seven nucleotide changes/gene, also verified by sequencing of the lyase gene in 40 randomly selected clones (data not shown). The library (∼108,000 primary clones) was established in E. coli XL10-Gold® cells. 6720 colonies were randomly picked, transferred to 96-well microtiter plates with growth medium, incubated, and induced by m-toluic acid for lyase expression in 96-well plates. Cell-free extract from each culture was prepared and evaluated for enzymatic activity, using polyMG and polyG as substrates. The ratio between the activity against polyMG and polyG for each extract was chosen as the selection criterion in the screen, this value being 1.2 for wild type AlyA. In the primary screen ∼100 clones were identified that displayed a maximum polyMG/polyG ratio of 0.3, and after rescreenings of these clones, eight strains displaying the lowest polyMG/polyG activity ratios were chosen for further characterization. The corresponding variant lyases were denoted AlyA1–8, and DNA sequence analyses showed that none of them were identical (Table 1). The number of deduced amino acid substitutions was either one (AlyA1–3), two (AlyA4–5), or three (AlyA6–8). AlyA2 differed from AlyA1 only by carrying an additional silent mutation at a site corresponding to Asp237. AlyA2 was therefore not included in further studies. Silent mutations were also identified in AlyA3 (Val151), AlyA6 (Thr220), and AlyA7 (Ile80 and Tyr175). Note also that the T89I substitution was found both in AlyA6 and AlyA8 and that Pro39 was substituted to both His (AlyA5) and Thr (AlyA7). This observation correlates well with the finding that these two residues are particularly important for substrate specificity (see below).
TABLE 1.
Activity of wild type AlyA, mutants selected in the screen (AlyA1 and AlyA3–8), and constructed mutants on polyG and polyMG
| Lyase | Amino acid substitution(s) | Activity |
Activity polyMG/activity polyG | |
|---|---|---|---|---|
| polyGa | polyMGa | |||
| units/mg | units/mg | |||
| AlyA (wild type) | 765 ± 50 | 937 ± 47 | 1.2 | |
| AlyA1 | S86L | 31 ± 2 | 9.1 ± 0.4 | 0.3 |
| AlyA3 | S37I | 53.8 ± 0.7 | 4.3 ± 0.1 | 0.1 |
| AlyA4 | V6I, T85A | 23.6 ± 0.9 | 2.16 ± 0.02 | 0.1 |
| V6I | 858 ± 136 | 851 ± 71 | 1.0 | |
| T85A | 24.8 ± 0.3 | 4.41 ± 0.06 | 0.2 | |
| AlyA5 | G26E, P39H | 246 ± 22 | 19.5 ± 0.5 | 0.1 |
| G26E | 692 ± 119 | 787 ± 19 | 1.1 | |
| P39H | 356 ± 10 | 37 ± 11 | 0.1 | |
| AlyA6 | I51M, T89I, G304V | 153 ± 9 | 13.26 ± 0.07 | 0.1 |
| I51M, T89I | 227 ± 13 | 18.8 ± 0.1 | 0.1 | |
| I51M | 731 ± 24 | 786 ± 43 | 1.1 | |
| T89I | 218 ± 7 | 31 ± 2 | 0.1 | |
| G304V | 828 ± 26 | 878 ± 67 | 1.1 | |
| AlyA7 | S35R, P39T, A224V | 43.2 ± 0.7 | 3.4 ± 0.6 | 0.1 |
| S35R, P39T | 32.6 ± 0.6 | 6.87 ± 0.07 | 0.2 | |
| S35R | 443 ± 10 | 448 ± 5 | 1.0 | |
| P39T | 216 ± 2 | 71 ± 1 | 0.3 | |
| AlyA8 | A78S, T89I, A217E | 187 ± 13 | 29.8 ± 0.4 | 0.2 |
| A78S, T89I | 228 ± 15 | 34.3 ± 0.9 | 0.2 | |
| A78S | 820 ± 35 | 938 ± 94 | 1.1 | |
| T89I | 218 ± 7 | 31 ± 2 | 0.1 | |
a A unit is the amount of enzyme required to increase A230 by 1 unit/min. The activity measurements were performed at least in triplicate, and the standard deviations of the means are given. AlyA1 and AlyA3–8 are mutants isolated in the screen. The mutants constructed to analyze the contribution of the separate amino acid substitutions in AlyA1 and AlyA3–8 are listed under each original mutant lyase.
AlyA1 and AlyA3–8 were partially purified by ion exchange chromatography, and the activity against polyG and polyMG was determined. For some enzymes, the level of activity on both substrates was considerably reduced compared with that of the wild type enzyme, but a strongly increased specificity toward G-G linkages was confirmed (Table 1 and Fig. 1). The activity against polyG was highest for AlyA5 (∼32% compared with wild type) and lowest for AlyA4 (∼3%). AlyA4 also displayed the lowest activity against polyMG (∼0.23%), and AlyA8 displayed the highest (∼3%). Based on all of these activity measurements, the enzymes could also be arranged with respect to their specificities against polyG, and these calculations showed that for five of the seven enzymes (AlyA3–7), the ratios between the activities on polyMG and polyG is 0.1, which is ∼12-fold less than for the wild type enzyme. For AlyA8 and AlyA1, the ratios were not equally low (0.2 and 0.3, respectively). Because AlyA5 displayed the highest activity against polyG, it appeared to be the best candidate for further applications in analyses of alginate fine structures, and the biochemical properties of this enzyme was therefore studied in further detail.
FIGURE 1.
Relative specific activity (units/mg) of AlyA wild type (A) and variant enzymes (A–F) against polyG and polyMG. For each lyase, the specific activity against polyG is set to 1. The reaction conditions were 50 mm Tris, pH 7.5, with 0.2 m NaCl and 1 mm CaCl2. The absolute values for the specific activities against each substrate are given in Table 1.
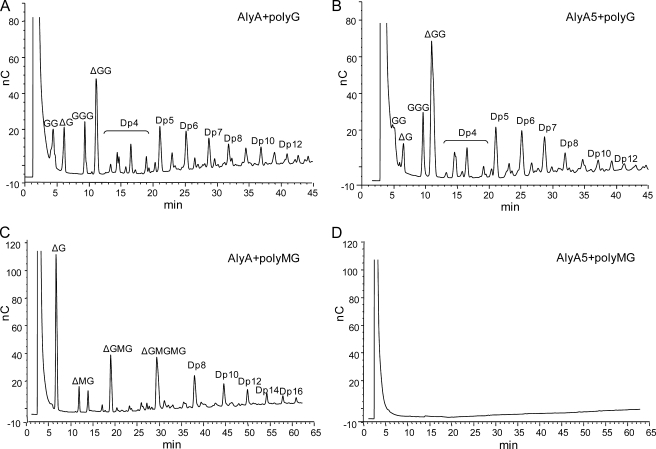
HPAEC-PAD Analysis of Oligosaccharide Pools Generated by Degradation of polyG and polyMG Confirmed That AlyA5 Can Be Used to Selectively Degrade G-blocks
The composition of the oligosaccharide pools after partial degradation of polyG and polyMG with AlyA wild type and AlyA5 was analyzed using HPAEC-PAD. This technique allows for separation of oligomers with a degree of polymerization up to 40. Furthermore, oligomers with an identical degree of polymerization can be separated on the basis of G and M content because elution time increases with increasing M. Likewise, elution time increases with the introduction of unsaturated uronic acids (denoted by Δ in Fig. 2), i.e. GGG elutes from the column before ΔGG.
FIGURE 2.
HPAEC-PAD analysis of oligosaccharides from degraded samples of polyG and polyMG by AlyA wild type (A and C) and AlyA5 (B and D). Degradation (1 mg/ml substrate) was performed in 50 mm Tris with 0.2 m NaCl and 1 mm CaCl2 and was allowed to proceed until A230 = 2 in the polyG reaction mixtures; i.e., polyG was degraded to the same extent by the two enzymes. The corresponding polyMG degradation reactions were stopped at the same time points. Lyases were inactivated by boiling with 1% SDS for 10 min before chromatographic analysis. Δ denotes the unsaturated uronic acid on the nonreducing end of the alginate molecules resulting from lyase degradation. Dpn denotes an oligomer with n residues. The early signal present in all chromatograms is the salt peak. The saturated dimer and trimer (GG and GGG) originate from the reducing end of the polyG molecules.
By controlling enzyme concentrations and incubation times, polyG was degraded with AlyA and AlyA5 to reach the same level of degradation as measured by A230 in each reaction (A230 = 2). The chromatograms (Fig. 2, A and B) show that for both enzymes G-oligomers down to dimers can be detected, and the degradation patterns are similar. The peaks marked Dp4 represent tetramers of different compositions, but the exact composition in each peak could not be identified. The analysis was also repeated under the same reaction conditions, using polyMG as substrate (Fig. 2, C and D). Degradation with AlyA resulted in a pool of oligomers with a degree of polymerization in the entire detection range, whereas for AlyA5 oligomers could not be detected at all. These results therefore confirmed that by keeping the enzyme concentrations at the minimum required for degradation of polyG, AlyA5 can be used to selectively degrade G-blocks into oligomers.
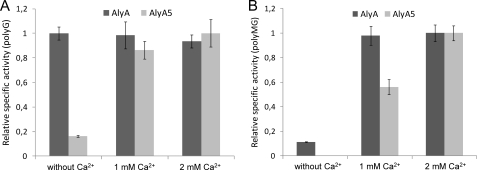
Activity of AlyA5 against polyMG Can Be Further Reduced by Omitting Ca2+ from the Incubation Mixture
For optimal activity, alginate lyases display different requirements for conditions like, for example, ionic strength and the presence of cations (19). Initial studies showed that AlyA activity is optimal on a complex substrate like LF10/60 in a 50 mm Tris buffer at pH 7.5 when supplemented with 0.2 m NaCl and 1 mm Ca2+. However, at the enzyme active site level such a substrate can be seen as a composite of many different substrates (G-blocks, M-blocks, and MG-blocks). It therefore seemed possible that the activity requirements are not the same for the different substructures in the substrate. Because of the defined nature of the substrates used here, this hypothesis could be experimentally tested. AlyA activity against polyG was found not to be affected by the concentration of Ca2+ (0–2 mm). In contrast, AlyA5 activity was stimulated by increasing levels of this cation (Fig. 3A).
FIGURE 3.
Relative specific activity of AlyA (wild type) and AlyA5 against polyG (A) and polyMG (B) with varying concentrations of CaCl2 in the reaction buffer. The basis buffer used was 50 mm Tris, 0.2 m NaCl, pH 7.5. For each lyase, the condition giving the highest activity is set to 1. In the absence of Ca2+, no activity (<1 unit/mg) of AlyA5 on polyMG could be detected.
Interestingly, the activities of both AlyA and AlyA5 on polyMG are strongly stimulated by Ca2+, and in the case of AlyA5 there was no detectable activity in the absence of this cation (Fig. 3B). It could therefore be concluded that by avoiding Ca2+ in the incubation mixture, AlyA5 becomes completely specific for polyG under the given conditions. This Ca2+ dependence of AlyA5 does not appear to be a general feature of the variant enzymes, because AlyA1 was found to behave similarly to AlyA in this respect (data not shown).
Amino Acid Substitutions Responsible for Increased G-G Specificity Are Most Often, but Not Always, Associated with Severe Loss of Total Enzyme Activity
Because AlyA4–8 all contained more than one deduced amino acid substitution, it appeared possible that some substitutions did not contribute to the observed specificity and possibly also led to reduced total activity against polyG. We therefore constructed a series of new mutants aiming at analyzing the contribution of each separate amino acid substitution. The results showed that none of the single substitution enzymes displayed more preferable properties than the originally isolated variants, relative to the criteria used to identify Aly5 as the best candidate (Table 1 and Fig. 1). Many of the variants displayed an activity against both polyG and polyMG that was closer to that of the wild type enzyme (V6I, G26E, I51M, G304V, S35R, and A78S). These substitutions alone therefore did not contribute much to improve the specificity of the enzyme against polyG. In contrast, for AlyA4 and AlyA5, which contained only two substitutions, one of the two substitutions alone (T85A and P39H, respectively) was sufficient to generate most but not all of the improved specificity. AlyA6–8 carry three substitutions each, and T89I and P39T were the best single substitution candidates with respect to the desired specificity. Among these, T89I is the most useful, displaying the lowest activity ratio between polyMG and polyG. Three variants with two substitutions were also made based on the mutations in alyA6–8, and among these I51M/T89I is the most interesting in that it retains quite high activity against polyG while it also displays low activity against polyMG.
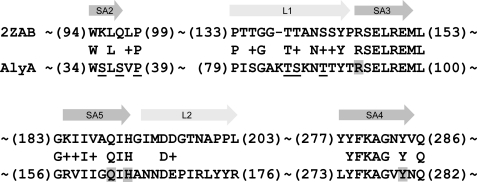
One major conclusion from all of these data is therefore that the P39H and I51M/T89I variants are the most promising candidates among those constructed, and I51M/T89I is very similar to AlyA5 both with respect to specificity and activity against polyG. Another interesting observation is that if one considers enhanced specificity only, substitutions S37I, T85A, and S86L also came out as promising, but with the disadvantage that the activity against polyG is quite low. This leads to the conclusion that the substitutions conferring increased G-G specificity are mainly localized to residues Ser37, Pro39, Thr85, and Thr89 in strand SA2 and flexible loop L1 that are predicted to be present in AlyA (Fig. 4).
FIGURE 4.
Amino acid sequences of strands 2–5 in β-sheet A (SA2–SA5) and flexible loop 1 and 2 (L1 and L2) of AlyA, aligned against lyase A1-II′ (Protein Data Bank code 2ZAB) according to PSI-Blast. SA3–5 constitute the most conserved regions in PL7 family lyases. The numbers in parentheses indicate the positions of the amino acid residues in A1-II′ and AlyA. The underlined residues were found to be substituted in AlyA mutants isolated in the screen, leading to modified specificity of the resulting lyase. Putative catalytic residues are shaded. Please observe that 2ZAB is the Y284F mutant of A1-II′ but that it has been changed back to Tyr284 in the alignment to highlight the conserved active site.
It appeared possible that substitutions from different parent variant enzymes might be combined to improve specificity toward G-G linkages further without excessive loss of activity, and the most promising candidate for such an experiment appeared to involve combination of the P39H, I51M, and T89I substitutions. This combination was constructed, but the resulting enzyme turned out to almost completely lack activity against both polyG and polyMG, indicating a subtle relation between activity and specificity in AlyA.
The Properties of AlyA Variants with Substitutions in Conserved Residues Are Consistent with Structural Predictions
To deduce the molecular mechanisms underlying the properties of the variant lyases described here, we wanted to generate additional support for certain critical assumptions regarding AlyA structure. Iterative PSI-Blast searching with AlyA against the NCBI protein sequence library showed significant similarity to several proteins with known three-dimensional structure; for example, the A1-II′ lyase (Protein Data Bank code 2ZAB) was identified by PSI-Blast as having 22% identity and 35% similarity to AlyA over 284 positions, with an E value of 1.0 × 10−42. Sequence alignment with PA1167, ALY-1, and A1-II′ indicated that Arg93, Gln162, His164, and Tyr280 constitute the catalytic residues of AlyA (26). In agreement with this assumption, we found that alanine substitutions in these residues resulted in enzymes with detectable but very low activity against both polyG and polyMG (0.01–1.3% of wt activity; data not shown). We also found that the low activities are not related to reduced expression levels, because SDS-PAGE analyses showed that the enzymes are produced in amounts similar to the wild type enzyme. Thus, the active site is probably organized as predicted. Furthermore, several residues around and in SA3, SA4, and SA5 are conserved among the PL-7 lyases (Fig. 4), and the effect of introducing alanine substitutions by site-specific mutagenesis in some of these residues was also investigated. Most of these variant enzymes (E95A, I160A, Y274A, F275A, and K276A) displayed reduced activity on both polyG and polyMG (0.2–14% of wild type activity; data not shown), whereas the substrate specificity was not significantly changed. In contrast, the Y91A and R97A substitutions resulted in enzymes with increased G-G and M-G specificity, respectively. The total activities were in both cases low: 64 (polyG) and 19 units/mg (polyMG) for Y91A and 24 (polyG) and 104 units/mg (polyMG) for R97A. These data are therefore consistent with the assumption that the main features of our structural predictions are correct, and possible explanations for the observed changes in substrate specificities have therefore been further elaborated under “Discussion.”
DISCUSSION
To better understand the underlying reasons for the substrate specificities of the variant lyases reported here, we have used sequence alignment between AlyA and A1-II′ (Fig. 4) as a basis for an analysis of protein-substrate interactions. The structures of A1-II′ complexed with GGG and MMG trimers were analyzed for ligand-protein contacts, and the different interaction types were summarized for selected residues (Table 2). The analysis focused on mutations in AlyA with significant effect on specificity or activity. Mutations affecting specificity are according to this analysis mainly associated with subsite +2 and to some degree subsite +3 (Arg97). Mutations affecting subsite +1, on the other hand, seem to mainly interfere with active site residues, generally leading to reduced activity without significant changes in specificity. However, interactions in Table 2 have to be evaluated with some care. Most mutations will lead to subtle structural changes across the protein, and the effect will be difficult to predict. This is illustrated for example by residues Pro39 and Thr85, where mutations lead to increased specificity even though these residues are not predicted to be in contact with substrate.
TABLE 2.
Correlations between mutation effects on substrate binding
The table shows specific interactions between residues and substrates (GGG and MMG) in experimental three-dimensional structures (Protein Data Bank codes 2ZAB and 2ZAC, respectively). Interactions are shown for selected residues in these structures corresponding to residues in AlyA where mutations lead to significant changes in specificity and/or activity. The interactions are shown for subsites +1, +2, and +3 in both structures. The position of each residue in the structure is indicated (strands SA2–SA5, loop L1). Interactions were estimated with ligand-protein contacts and include stabilizing interactions (hydrogen bonds (Hb) and hydrophobic contacts (Ph)), destabilizing interactions (hydrophilic-hydrophobic contacts (HH), acceptor-acceptor contacts (AA)), and other nonclassified interactions (O). The contact surface area is shown for each contact type, and the dominating interactions are shown in bold type. Destabilizing interactions are indicated with negative contact surface areas. The comments in the table indicate possible explanations of the observed effects from the mutations.
| Position | Mutation | Structure | +1 G | +2 G | +3 G | +1 M | +2 M | +3 G | Comments |
|---|---|---|---|---|---|---|---|---|---|
| Reduced activity | |||||||||
| Arg93 | Ala | Arg146 SA3 | Hb (17.3) | Hb (1.7) | Hb (13.3) | Hb (1.7) | Active site | ||
| O (15.0) | O (13.4) | O (5.2) | O (7.6) | ||||||
| Glu95 | Ala | Glu148 SA3 | H-bonds to Arg146 and Gln189 | ||||||
| Ile160 | Ala | Val187 SA5 | May affect loop with Gln189 and His191 | ||||||
| Gln162 | Ala | Gln189 SA5 | Hb (12.6) | Hb (21.0) | Active site | ||||
| AA (−17.8) | AA (−18.2) | ||||||||
| O (3.1) | O (4.1) | ||||||||
| His164 | Ala | His191 SA5 | Hb (16.1) | Hb (0.5) | Hb (13.5) | Hb (1.4) | Active site | ||
| HH (−1.1) | O (2.7) | O (10.1) | O (7.0) | ||||||
| O (17.5) | |||||||||
| Tyr274 | Ala | Tyr278 SA4 | Hb (9.9) | Hb (2.8) | O (21.2) | Hb (2.4) | Stacking interaction with Arg150 and His191 | ||
| O (17.5) | O (0.2) | ||||||||
| Phe275 | Ala | Phe279 SA4 | May affect strand with Tyr278 and Lys280 | ||||||
| Lys276 | Ala | Lys280 SA4 | Hb (3.3) | Hb (16.4) | Hb (17.0) | H-bond to Arg146 | |||
| O (14.5) | O (7.8) | ||||||||
| Tyr280 | Ala | Phe284 SA4 | Ph (8.6) | O (36.2) | Active site (mutated Y284F), stacking interaction with Arg146 | ||||
| O (17.8) | |||||||||
| PolyG preference (with reduced activity) | |||||||||
| Ser37 | Ile | Gln97 SA2 | Hb (19.6) | Hb (5.4) | Hb (18.9) | Hb (6.7) | Increased preference for M in +2 (nonproductive on polyMG) | ||
| O (2.5) | O (3.2) | O (1.6) | |||||||
| Pro39 | His/Thr | Pro99 | May affect loop with e.g. Gln97 | ||||||
| Thr85 | Ala | Thr138 L1 | May affect loop with e.g. Thr139 | ||||||
| Ser86 | Leu | Thr139 L1 | Hb (15.8) | Ph (8.5) | Hb (19.8) | Ph (7.2) | Increased preference for M in +2 (nonproductive on polyMG) | ||
| HH (−5.2) | HH (−7.6) | HH (−6.2) | HH (−4.9) | ||||||
| O (13.4) | O (2.7) | O (8.7) | O (1.7) | ||||||
| Thr89 | Ile/Ala | Ser142 L1 | O (4.7) | Hb (0.2) | May affect loop with Thr139, for example | ||||
| O (6.7) | |||||||||
| Tyr91 | Ala | Tyr144 L1 | O (0.3) | May affect loop with Thr139, for example | |||||
| PolyMG preference (with reduced activity) | |||||||||
| Arg97 | Ala | Arg150 SA3 | HH (−1.2) | Hb (39.0) | Ph (0.2) | Hb (39.9) | Increased preference for M in +3 (productive on polyMG) | ||
| HH (−0.2) | O (6.2) | ||||||||
| O (5.8) | |||||||||
During the lyase catalytic reaction, cleavage of the glycosidic linkage occurs between subsites −1 and +1, with the nonreducing end positioned at −1. AlyA seems to cleave G-M linkages in polyMG by accommodating the substrate with G in −1 and +2 and M in +1 and +3. The altered specificity of the isolated mutants can be explained by considering two main types of possible changes introduced by the substitutions. First, the increased G-G specificity could be a result of increased preference for G or reduced preference for M in subsites +1 and +3. However, this should not reduce the activity toward polyG as much as is observed here. A more likely explanation may therefore be an increased preference for M in +2 and possibly in +4. This would lead to nonproductive binding of polyMG and presumably also lower activity toward polyG because of generally weaker interactions in +2. The effects introduced by the identified mutations support the latter hypothesis. In general, reduced polarity will increase the preference for M, which has a greater potential for hydrophobic interactions than G (44). This can be seen in Ser37 (AlyA3) and Ser86 (AlyA1), both of which show hydrogen bonding toward +2 (Table 2). Substitution to Ile and Leu, respectively, therefore favors M at +2, i.e. nonproductive binding of polyMG. The P39T mutation (AlyA7) seems to exert indirect effects on substrate binding because Pro39 is not in direct contact with the substrate. Pro39 terminates β-strand SA2 (Fig. 2), and P39T may lead to elongation of SA2, which in turn will distort the β-strand and affect hydrogen bonding by Ser37. Similar indirect effects can be seen for residues Thr85 and possibly Thr89 residing in loop L1. Although these residues show only limited direct interaction with substrates, they can exert an indirect effect on residue Ser86 by modifying the properties of the loop. Residue Ser86 resides in L1 and hydrogen bonding to +2 may be lost if the loop conformation is changed. It should be noted that L1 in AlyA contains two serine and four threonine residues, pointing to the importance of potential for polar interactions in this structural element.
In summary, the introduced mutations apparently lead to lyases with reduced ability for interaction with polyMG in a productive manner, i.e. the enzymes are rendered less effective toward this substrate. The activity toward polyG is also reduced, however to a lower extent, and the mutant lyases therefore appear as having increased specificity toward G-G linkages.
We found it intriguing that AlyA5 lyase activity is much more Ca2+-dependent than its parent enzyme, and none of the close homologues of AlyA with known three-dimensional structure include bound Ca2+. In contrast, the more distantly related enzymes β-1,4-glucuronan lyase from the fungus Trichoderma reesei (45) and alginate lyase from Alteromonas sp. (the structure is published as Protein Data Bank entry 1J1T) both include Ca2+. The ions are bound at different positions in these structures and apparently play mainly structural and stabilizing roles. Thus, it is possible that Ca2+ serves a similar function in AlyA5. A substitution in Pro39 may cause significant structural changes in strand SA2 (see above), and Ca2+ might be essential for stabilizing the affected region after mutation.
The aim of the present work was to isolate mutants of AlyA with increased specificity for G-G linkages. To our knowledge the mutants reported here represent the most G-G-specific alginate lyases known at present. It is also an advantage that AlyA5 or its derivatives can be efficiently produced in E. coli, and we are currently aiming at using the mutant enzymes together with other lyases displaying relevant specificities as tools to more efficiently deduce the fine structure of alginates.
Supplementary Material
The work was supported by The Research Council of Norway Project 182695-I40 and by FMC Biopolymer and Algipharma AS.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- M
- β-d-mannuronic acid
- G
- α-l-guluronic acid
- PL-7
- polysaccharide lyase family 7
- MOPS
- 4-morpholinepropanesulfonic acid
- HPAEC-PAD
- high performance anion exchange chromatography with pulsed amperometric detection.
REFERENCES
- 1.Linker A., Jones R. S. (1964) Nature 204, 187–188 [DOI] [PubMed] [Google Scholar]
- 2.Govan J. R., Fyfe J. A., Jarman T. R. (1981) J. Gen. Microbiol. 125, 217–220 [DOI] [PubMed] [Google Scholar]
- 3.Gorin P., Spencer J. (1966) Can. J. Microbiol. 44, 993–998 [DOI] [PubMed] [Google Scholar]
- 4.Cote G., Krull L. (1988) Carbohydr. Res. 181, 143–152 [Google Scholar]
- 5.Haug A., Larsen B. (1969) Biochim. Biophys. Acta 192, 557–559 [DOI] [PubMed] [Google Scholar]
- 6.Haug A., Larsen B. (1971) Carbohydr. Res. 17, 297–308 [DOI] [PubMed] [Google Scholar]
- 7.Ertesvåg H., Høidal H. K., Hals I. K., Rian A., Doseth B., Valla S. (1995) Mol. Microbiol. 16, 719–731 [DOI] [PubMed] [Google Scholar]
- 8.Skjåk-Braek G., Grasdalen H., Larsen B. (1986) Carbohydr. Res. 154, 239–250 [DOI] [PubMed] [Google Scholar]
- 9.Remminghorst U., Rehm B. H. (2006) Biotechnol. Lett. 28, 1701–1712 [DOI] [PubMed] [Google Scholar]
- 10.Onsøyen E. (1996) Carbohydr. Eur. 14, 26–31 [Google Scholar]
- 11.Skjåk-Bræk G., Espevik T. (1996) Carbohydr. Eur. 14, 19–23 [Google Scholar]
- 12.Coviello T., Matricardi P., Marianecci C., Alhaique F. (2007) J. Control. Release 119, 5–24 [DOI] [PubMed] [Google Scholar]
- 13.Zimmermann H., Shirley S. G., Zimmermann U. (2007) Curr. Diab. Rep. 7, 314–320 [DOI] [PubMed] [Google Scholar]
- 14.Hashimoto T., Suzuki Y., Suzuki K., Nakashima T., Tanihara M., Ide C. (2005) J. Mater. Sci. Mater. Med. 16, 503–509 [DOI] [PubMed] [Google Scholar]
- 15.Smelcerovic A., Knezevic-Jugovic Z., Petronijevic Z. (2008) Curr. Pharm. Des. 14, 3168–3195 [DOI] [PubMed] [Google Scholar]
- 16.Petrulyte S. (2008) Dan. Med. Bull. 55, 72–77 [PubMed] [Google Scholar]
- 17.Boateng J. S., Matthews K. H., Stevens H. N., Eccleston G. M. (2008) J. Pharm. Sci. 97, 2892–2923 [DOI] [PubMed] [Google Scholar]
- 18.Thomas S. (2000) J. Wound Care 9, 56–60 [DOI] [PubMed] [Google Scholar]
- 19.Wong T. Y., Preston L. A., Schiller N. L. (2000) Annu. Rev. Microbiol. 54, 289–340 [DOI] [PubMed] [Google Scholar]
- 20.Campa C., Oust A., Skjåk-Braek G., Paulsen B. S., Paoletti S., Christensen B. E., Ballance S. (2004) J. Chromatogr. A 1026, 271–281 [DOI] [PubMed] [Google Scholar]
- 21.Haug A., Larsen B., Smidsrød O. (1967) Acta Chem. Scand. 21, 691–704 [DOI] [PubMed] [Google Scholar]
- 22.Boyd J., Turvey J. R. (1977) Carbohydr. Res. 57, 163–171 [DOI] [PubMed] [Google Scholar]
- 23.Baron A. J., Wong T. Y., Hicks S. J., Gacesa P., Willcock D., McPherson M. J. (1994) Gene 143, 61–66 [DOI] [PubMed] [Google Scholar]
- 24.Haugen F., Kortner F., Larsen B. (1990) Carbohydr. Res. 198, 101–109 [DOI] [PubMed] [Google Scholar]
- 25.Yamasaki M., Ogura K., Hashimoto W., Mikami B., Murata K. (2005) J. Mol. Biol. 352, 11–21 [DOI] [PubMed] [Google Scholar]
- 26.Osawa T., Matsubara Y., Muramatsu T., Kimura M., Kakuta Y. (2005) J. Mol. Biol. 345, 1111–1118 [DOI] [PubMed] [Google Scholar]
- 27.Yamasaki M., Moriwaki S., Miyake O., Hashimoto W., Murata K., Mikami B. (2004) J. Biol. Chem. 279, 31863–31872 [DOI] [PubMed] [Google Scholar]
- 28.Miyake O., Ochiai A., Hashimoto W., Murata K. (2004) J. Bacteriol. 186, 2891–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsubara Y., Kawada R., Iwasaki K., Oda T., Muramatsu T. (1998) J. Protein Chem. 17, 29–36 [DOI] [PubMed] [Google Scholar]
- 30.Chavagnat F., Heyraud A., Colin-Morel P., Guinand M., Wallach J. (1998) Carbohydr. Res. 308, 409–415 [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J., Russel D. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 32.Sletta H., Tøndervik A., Hakvåg S., Aune T. E., Nedal A., Aune R., Evensen G., Valla S., Ellingsen T. E., Brautaset T. (2007) Appl. Environ. Microbiol. 73, 906–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bakke I., Berg L., Aune T. E., Brautaset T., Sletta H., Tøndervik A., Valla S. (2009) Appl. Environ. Microbiol. 75, 2002–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holtan S., Zhang Q., Strand W. I., Skjåk-Braek G. (2006) Biomacromolecules 7, 2108–2121 [DOI] [PubMed] [Google Scholar]
- 35.Haug A., Larsen B., Smidsrød O. (1974) Carbohydr. Res. 32, 217–225 [Google Scholar]
- 36.Ballance S., Holtan S., Aarstad O. A., Sikorski P., Skjåk-Bræk G., Christensen B. E. (2005) J. Chromatogr. 1093, 59–68 [DOI] [PubMed] [Google Scholar]
- 37.Ogura K., Yamasaki M., Mikami B., Hashimoto W., Murata K. (2008) J. Mol. Biol. 380, 373–385 [DOI] [PubMed] [Google Scholar]
- 38.Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. (2000) Nucleic Acids Res. 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sobolev V., Sorokine A., Prilusky J., Abola E. E., Edelman M. (1999) Bioinformatics 15, 327–332 [DOI] [PubMed] [Google Scholar]
- 40.Wallace A. C., Laskowski R. A., Thornton J. M. (1995) Protein Eng. 8, 127–134 [DOI] [PubMed] [Google Scholar]
- 41.Guex N., Peitsch M. C. (1997) Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 42.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson J. D., Gibson T. J., Higgins D. G. (2003) Curr. Prot. Bioinform. 2.3.1–2.3.22 [Google Scholar]
- 44.Chan C., Burrows L. L., Deber C. M. (2004) J. Biol. Chem. 279, 38749–38754 [DOI] [PubMed] [Google Scholar]
- 45.Konno N., Ishida T., Igarashi K., Fushinobu S., Habu N., Samejima M., Isogai A. (2009) FEBS Lett. 583, 1323–1326 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.