Abstract
Antiretroviral cytidine deaminase APOBEC3G, which is abundantly expressed in peripheral blood lymphocytes and macrophages, strongly protects these cells against HIV-1 infection. The HIV-1 Vif protein overcomes this antiviral effect by enhancing proteasome-mediated APOBEC3G degradation and is key for maintaining viral infectivity. The 579-bp-long vif gene displays high genetic diversity among HIV-1 subtypes. Therefore, it is intriguing to address whether Vif proteins derived from different subtypes differ in their viral defense activity against APOBEC3G. Expression plasmids encoding Vif proteins derived from subtypes A, B, C, CRF01_AE, and CRF02_AG isolates were created, and their anti-APOBEC3G activities were compared. Viruses produced from cells expressing APOBEC3G and Vif proteins from different subtypes showed relatively different viral infectivities. Notably, subtype C-derived Vif proteins tested had the highest activity against APOBEC3G that was ascribed to its increased binding activity, for which the N-terminal domain of the Vif protein sequences was responsible. These results suggest that the biological differences of Vif proteins belonging to different subtypes might affect viral fitness and quasispecies in vivo.
Keywords: Human Immunodeficiency Virus, Innate Immunity, Viral Genetics, Viral Protein, Viral Replication, APOBEC3G, Vif, Subtype
Introduction
Among the seven human APOBEC33 cytidine deaminase proteins (from A to H) that act as intrinsic restriction factors against endogenous and exogenous retroviruses (1–9), APOBEC3G provides the most potent retroviral restriction in vitro and in vivo (10–12). This host protein is abundantly expressed in peripheral blood mononuclear cells (PBMCs) and macrophages. APOBEC3G deaminates deoxycytidine to deoxyuridine in nascent viral minus-strand cDNA, thereby inducing G-to-A hypermutations during reverse transcription (13–16). It also partially restricts viral replication in a deamination-independent fashion, mainly by blocking DNA synthesis (17–20). Human immunodeficiency virus type 1 (HIV-1) is armed with Vif3 protein, which induces proteasome-mediated APOBEC3G degradation (21–24) via a mechanism involving the Cullin5 (Cul5)-containing E3 ubiquitin ligase (25–28). As a result, Vif protein reduces virion incorporation of APOBEC3G in virus-producer cells (29–32), leading to efficient reverse transcription in the target cells. By the same mechanism, Vif protein can inactivate APOBEC3F and APOBEC3DE, which are expressed in PBMCs and suppress Vif-deficient HIV-1 infection to a lesser extent than does APOBEC3G (33–35).
Several in vivo studies have demonstrated that APOBEC3G-induced G-to-A hypermutation is frequently observed in patient-derived proviral DNA (36–42) even in the presence of full-length but polymorphic vif genes (43). The vif genes also have high in vivo genetic variability (11, 44–46) and subtype-dependent amino acid substitutions (47). These findings imply that the sequence diversity of vif genes (possibly in a subtype-dependent manner) might harbor differential levels of anti-APOBEC3G activity. Among the strains tested in the present study, Vif protein derived from subtype C strains harbored the most robust anti-APOBEC3G activity. This activity was determined by the N-terminal region of the protein, which bound APOBEC3G more efficiently than subtype B-derived Vif. Consistent with this, subtype B-based viruses carrying subtype C-derived Vif proteins were rarely deaminated in primary lymphocytes endogenously expressing APOBEC3G. These results indicate that the sequence variability of Vif proteins dependent on HIV-1 subtypes leads to differential anti-APOBEC3G activity, presumably resulting in differential levels of HIV-1 fitness and viral progeny diversity.
EXPERIMENTAL PROCEDURES
Viruses
The HIV-1 isolates utilized in this study were registered in GenBankTM and included: subtype A, UG029-A3 (#AB098332), UG031-A1 (#AB098330), UG031-A2 (#AB098331), 92RW025A (#AB287376), and 92UG037 (#AB253428); subtype B, NL4-3 (48), JRFL (49), SF2 (50), BaL (51), and 01JPDR3884 (#AB289589); subtype C, 02ZMJCC05 (#AB254155), 02ZMJMC18 (#AB254156), 02ZM109C31 (#AB573087), 02ZM112C23 (#AB254145), 02ZMDBC33 (#AB254153), and 02ZMGNC46 (#AB573088); CRF01_AE, 93TH051 (#AB220944), 93TH057AE18 (#AB253424), 93TH060 (#AB220946), 93TH062 (#AB220947), and 93TH065 (#AB220948); CRF02_AG, 03GH178AG1 (#AB572922), 03GH180AG13 (#AB572923), GH184AG25 (#AB286860), GHNJ188 (#AB231896), and 97GH_AG2 (#AB052867). Amino acid alignments of these Vif proteins are shown in supplemental Fig. S1.
DNA Construction
The HIV-1 proviral constructs pNL4-3 (48), Vif-deficient HIV-1 proviral indicator construct pNL-Luc-F(−)E(−) (34), Rev expression plasmid pCA-Rev (52), vesicular stomatitis virus glycoprotein (VSV-G) expression vector pHIT/G (53), and HA-tagged human APOBEC3G expression plasmid pCA-hA3G-HA (8) were previously described. To create a C-terminal FLAG-tagged expression plasmid, a synthetic double-stranded oligonucleotide NotI linker harboring an MscI site upstream of the FLAG epitope (sense, 5′-GGC CTA TGG CCA CGA TTA TAA AGA CGA TGA CGA CAA GTA GAG C-3′; antisense, 5′-GGC CGC TCT ACT TGT CGT CAT CGT CTT TAT AAT CGT GGC CAT A-3′) was inserted into the NotI site of the mammalian expression vector pCAGGS (54), in which the preexisting MscI site was disrupted for further cloning. To confer Rev-dependent expression on the expression plasmids, the pNL4-3-derived Rev-responsive element (RRE; nucleotide 7759–7992) was PCR-amplified, digested with NotI, and inserted into the FLAG-tagged expression plasmid, resulting in pCAGGS-FLAG-RRE. HIV-1 vif genes derived from the different subtypes described above were PCR-amplified, digested with KpnI and MscI, and cloned into pCAGGS-FLAG-RRE.
To create chimeric constructs between the Vif proteins of NL4-3 and 02ZMDBC33 (representative of subtypes B and C, respectively), the KpnI-PflMI fragment (corresponding to Vif residues 1–87) of pC-NLvif-FLAG-RRE (NL-Vif) or pC-DBvif-FLAG-RRE (DB-Vif) was replaced with that of DB-Vif or NL-Vif, respectively, resulting in NL/DB-Vif or DB/NL-Vif, respectively. DB(38–87)-Vif consisting of the N-terminal NL-Vif region (residues 1–37), the middle DB-Vif region (residues 38–87), and the C-terminal NL-Vif region downstream of the PflMI site was created with overlapping PCR-based cloning using NL-Vif and DB/NL-Vif as templates. Similarly, chimeric constructs between the N-terminal DB-Vif region (residues 1–37, 1–34, 1–31, and 1–23) and the C-terminal NL-Vif region, which were designated DB(1–37)-Vif, DB(1–34)-Vif, DB(1–31)-Vif, and DB(1–23)-Vif, were created using DB-Vif and NL-Vif as templates, respectively. DB(9–37)-Vif consisting of the DB-Vif region of residues 9–37 with the NL-Vif backbone was created with the QuikChange site-directed mutagenesis kit (Stratagene) using DB(1–37)-Vif as a template.
To introduce the Lys-17 → Arg or Lys-19 → Arg mutations into DB(1–31)-Vif to create DB(1–31)K17R-Vif or DB(1–31)K19R-Vif, respectively, QuikChange site-directed mutagenesis was performed using DB(1–31)-Vif as a template. The Vif-chimeric HIV-1 proviral constructs pNL-DBvif and pNL-DB(1–31)vif were created with overlapping PCR-based cloning using pNL4-3 and DB-Vif as templates, respectively. A Vif-deficient HIV-1 proviral construct pNL-Δvif was generated by introducing a stop codon linker at the PflMI site of pNL4-3. The T7 epitope-tagged (55) APOBEC3G plasmid pCA-hA3G-T7E was generated by inserting an APOBEC3G fragment amplified from pCA-hA3G-HA (8) into a modified pCAGGS expression vector carrying a C-terminal T7 epitope tag.
To create the Elongin C expression plasmid pC-EloC-HA, total RNA from H9 cells was subjected to reverse transcription followed by amplification with specific oligonucleotides. An amplified Elongin C fragment was cloned into modified pCAGGS carrying a C-terminal HA tag. To generate the Cul5 expression plasmid pC-Cul5, total RNA isolated from MOLT-4 cells was subjected to RT-PCR amplification of the Cul5 gene using specific oligonucleotides, and an amplified Cul5 fragment was cloned into pCAGGS. All constructs were verified using an ABI model 3130 Genetic Analyzer (Applied Biosystems).
Phylogenetic Analysis
Patient-derived vif genes and the vif sequences of reference strains representing the different genetic subtypes were aligned using the ClustalW program (56). A phylogenetic tree, constructed by the neighbor-joining method with branching order reliability determined by the bootstrap approach, was implemented with the ClustalW program. Genetic distances were estimated by the Kimura 2-parameter method (57).
Cell Maintenance, Transfections, and Protein Analyses
To confirm Vif protein expression, the 293T cells, maintained in Dulbecco's modified Eagle's medium supplemented with 10% FCS, were cotransfected with 200 ng of pCA-Rev, 200 ng of FLAG-tagged Vif expression plasmids, and empty vector up to 1 μg of total DNA by using the FuGENE 6 transfection reagent (Roche Applied Science). Cell extracts from the transfected cells were subjected to Western blot analysis using the anti-FLAG mouse monoclonal antibody M2 (Sigma).
Virion Production, APOBEC3G Degradation, and Viral Infectivity Assay
To prepare VSV-G-pseudotyped HIV-1 luciferase reporter viruses, 3.5 × 105 293T cells were cotransfected with 35 ng of pCA-hA3G-HA, 0.1 μg of the VSV-G expression plasmid pHIT/G, 8 ng of the Vif expression plasmids, and 0.87 μg of an empty vector together with 1 μg of pNL-Luc-F(-)E(-) using FuGENE 6. Sixteen hours later cells were washed with phosphate-buffered saline, and 1 ml of fresh complete medium was added. After 24 h, supernatants were treated with 37.5 units/ml DNase I (Roche Applied Science) for 37 °C for 30 min and then harvested.
To analyze the level of Vif-degraded APOBEC3G, cells were lysed and subjected to Western blot analysis using the anti-HA mouse monoclonal antibody HA-7 (Sigma). The p24 antigen levels in viral supernatants were measured by an HIV-1 p24-antigen capture enzyme-linked immunosorbent assay (Advanced BioScience Laboratories). To determine the viral infectivity, 3.5 × 104 293T cells were incubated with 1 ng of p24 antigen of the HIV-1 supernatants. After 48 h, cells were lysed in 100 μl of lysis buffer. The firefly luciferase activity was determined with a Centro LB960 (Berthold) luminometer.
Immunoprecipitation
The 293T cells (7 × 105) were cotransfected using FuGENE 6 with 200 ng of pCA-hA3G-T7E, 64 ng of pC-Vif-FLAG-RRE, 64 ng of pCA-Rev, and empty vector up to 2 μg of total DNA. After 36 h, transfected cells were treated with 20 μm of MG-132 (Calbiochem) for 9 h and suspended in 500 μl of radioimmunoprecipitation assay buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 20 μm MG-132, and complete protease inhibitor mixture (Roche Applied Science)). The resultant lysates were clarified by brief centrifugation, precleared with 30 μl of rProtein A-Sepharose Fast Flow (GE Healthcare) for 1 h at 4 °C, and then incubated with an anti-FLAG M2 Affinity Gel (Sigma). After 1 h at 4 °C, the immune complexes were extensively washed with radioimmunoprecipitation assay buffer. Equal aliquots of the total and bound fractions were subjected to gel electrophoresis and transferred to a nitrocellulose membrane. The membranes were probed with an anti-T7 tag mouse monoclonal antibody (Novagen), an anti-FLAG rabbit polyclonal antibody (Sigma), or an anti-β-actin mouse monoclonal antibody AC-74 (Sigma). The signal intensity of the immunoprecipitated APOBEC3G protein on Western blots was quantified using the LAS-3000 imaging system (Fujifilm).
G-to-A Mutation Assay
Freshly isolated PBMCs (1 × 106) were stimulated with phytohemagglutinin (3 μg/ml) for 72 h. After 293T transfection with the proviral DNA clones pNL4-3, pNL-DBvif, NL-DB(1–31)vif, and pNL-Δvif, PBMCs were infected overnight with 50 ng of the resulting four viruses. Cells were cultured in the presence of interleukin-2 (10 units/ml). After 12 days, the total cellular DNAs from infected cells were isolated using DNeasy (Qiagen). Fragments of the 3′ region of the envelope gene (nucleotides 8127–8756, which has a high G-to-A mutational frequency (58)) were PCR-amplified with High Fidelity DNA Polymerase (Roche Applied Science) and cloned into the TOPO TA-cloning vector pCR4 (Invitrogen). The nucleotide sequence was determined to compare the G-to-A mutational frequency using ABI3130 (ABI).
RESULTS
Phylogenetic Analysis of the HIV-1 Accessory Gene vif Derived from Different Subtypes
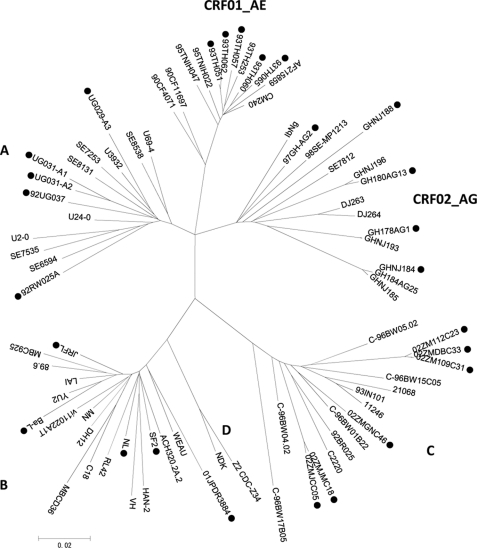
HIV-1 Vif expression plasmids were generated by PCR-amplifying known and recently isolated vif genes derived from the worldwide prevalent subtypes A, B, C, CRF01_AE, and CRF_02AG. Five subtype A (UG029-A3, UG031-A1, UG031-A2, 92RW025A, and 92UG037), five subtype B (NL, SF2, JRFL, BaL, and DR3884), six subtype C (02ZMJCC05, 02ZMJMC18, 02ZM109C31, 02ZM112C23, 02ZMDBC33, and 02ZMGN46), five subtype CRF01_AE (93TH051, 93TH057, 93TH060, 93TH062, and 93TH065), and five subtype CRF_02AG strains (03GH178, 03GH180C13, 03GH184AG25, GHNJ188, and 97GH_AG2) were used. The genes were cloned into FLAG-tagged mammalian expression plasmids, sequenced, and genetically compared. Fig. 1 shows the phylogenetic analysis results for the HIV-1 vif genes utilized in this study. This tree clearly classifies each subtype based on difference between the vif sequences.
FIGURE 1.
Sequence variation of HIV-1 accessory gene vif. An unrooted phylogenetic tree based on HIV-1 vif sequences from group M reference strains was generated by the neighbor-joining method. The vif sequences from the major subtypes (A, B, C, D, CRF01_AE, and CRF02_AG) containing reference strains, and isolates tested in this study are included. Different subtype-derived vif sequences selected for functional testing are indicated by ●.
Anti-APOBEC3G Activities of HIV-1 Vif Proteins Differ in a Subtype-dependent Manner
Because the vif sequences appear to be variable (11, 44–47), we examined whether Vif proteins derived from different subtypes with the sequence diversity shown here would show differential levels of anti-APOBEC3G activity. To do this we first determined the optimal doses of the APOBEC3G and Vif expression plasmids. The mRNA levels of APOBEC3G endogenously expressed in three donor-derived PBMCs were compared with levels expressed in cells transfected with the serially diluted APOBEC3G expression plasmid. Levels of Vif protein physiologically expressed from the Vif-positive NL4-3 proviral construct were compared with levels expressed from the serially diluted NL-Vif expression plasmid. Real-time RT-PCR revealed that introduction of 25 ng of APOBEC3G plasmid into 293T cells reproduced the endogenous expression level of APOBEC3G in PBMCs (supplemental Fig. S2A).
Infectivity enhancement by the Vif protein was evaluated from the proviral construct or from the expression plasmid in the presence of a fixed amount of the APOBEC3G plasmid (as determined above). The optimal dose of the Vif plasmid that reflected its physiological expression was 8 ng (supplemental Fig. S2E). Using the same approach, the endogenous expression level of APOBEC3F in PBMCs was also determined (supplemental Fig. S2B). The corresponding dose (∼0.8 ng) of the APOBEC3F expression plasmid showed only a ∼30% reduction of Δvif virus infectivity, whereas that of APOBEC3G showed an ∼400-fold reduction (supplemental Fig. S2, D and C, respectively). These findings suggest that the endogenous APOBEC3F level is significantly less potent than that of APOBEC3G in inhibiting HIV-1 infection, as previously and very recently described (12, 59, 60).
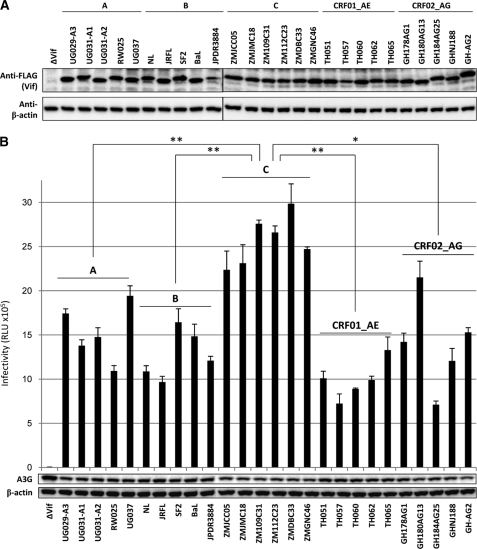
Vif protein expression in the plasmid-transfected cells was confirmed by immunoblotting using anti-FLAG antibodies (Fig. 2A). Vif functional testing was performed using viruses from 293T cells cotransfected with a Vif-Env-double-defective HIV-1 construct and a VSV-G expression plasmid together with optimal doses of APOBEC3G and various subtype-derived Vif expression plasmids. Most Vif proteins derived from subtypes A, B, CRF01_AE, and CRF_02AG showed non-significant but somewhat differential activity levels based on infectivity profiles (which represent the anti-APOBEC3G activity) (Fig. 2B). In contrast, subtype C-derived Vif proteins showed exclusively high anti-APOBEC3G activity levels. The Vif-mediated infectivity enhancements generally correlated with the proteasomal degradation levels of APOBEC3G protein (Fig. 2B). Compared with Vif from other subtypes, subtype C-derived Vif proteins seemed to harbor higher activity against the antiviral APOBEC3G protein.
FIGURE 2.
Functional testing to compare the anti-APOBEC3G activity of Vif proteins from different subtypes. A, Western blot analysis was performed using extracts from 293T cells transfected with combinations of plasmids encoding Rev and RRE-carrying/FLAG-tagged Vif derived from five major subtypes. Antibodies specific for FLAG and β-actin were used. Note that a larger amount (200 ng) of Vif expression plasmids was used to confirm the expression of Vif proteins, whereas the optimal amount (8 ng) was used for virus preparation in B. B, 293T cells were cotransfected with a luciferase-based Vif/Env-deficient HIV-1, VSV-G, and HA-tagged APOBEC3G expression plasmids together with plasmids encoding FLAG-tagged Vif derived from different subtypes. After 40 h, cell lysate and viral supernatant were harvested. To examine the proteasomal degradation of APOBEC3G in the virus-producing cells, the lysate was subjected to Western blot analysis (shown below the bar graph) using monoclonal antibodies against HA or β-actin. Supernatants normalized by the p24 antigen of VSV-G-pseudotyped luciferase viruses were incubated with 293T cells for an additional 48 h. Cells were then lysed and subjected to a luciferase assay. Results are representative of at least five independent triplicate experiments. RLU, relative light units. Data shown are the mean ± S.D.; *, p < 0.005, **, p < 0.001, t test.
The N-terminal Domain of Subtype C-derived Vif Confers a Robust Anti-APOBEC3G Activity
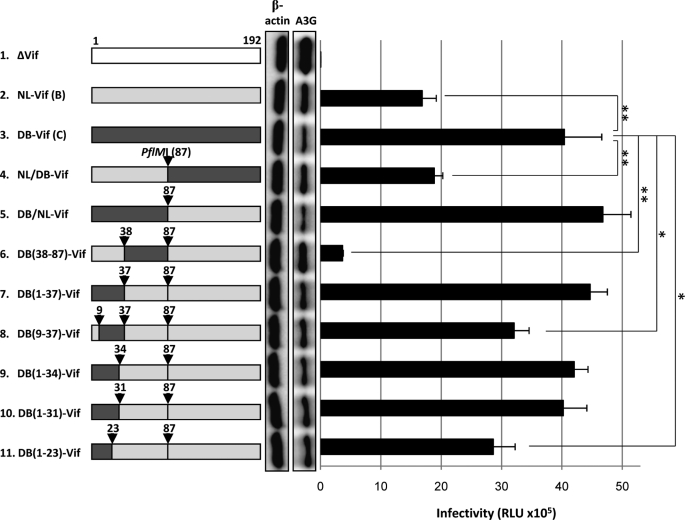
To identify the smallest segment of subtype C-derived Vif protein needed to control anti-APOBEC3G activity, chimeric Vif constructs were created between subtypes B (derived from NL4-3) and C (derived from 02ZMDBC33) utilizing a preexisting restriction enzyme site PflMI commonly located in the middle of Vif. The chimeric Vif construct carrying N-terminal residues 1–87 of subtype C-derived Vif (DB/NL-Vif) displayed an even higher activity than the construct carrying C-terminal residues 88–189 of subtype C-derived Vif (NL/DB-Vif) (Fig. 3, lanes 4 and 5). Therefore, we dissected the N-terminal half of subtype C-derived Vif by substituting the upstream (residues 1–37) or downstream (residues 38–87) half of this fragment for the corresponding region of subtype B-Vif using the overlapping PCR method (resulting in DB(1–37)-Vif and DB(38–87)-Vif). N-terminal residues 1–37 appeared to determine the subtype C-derived Vif activity against APOBEC3G (Fig. 3, lanes 6 and 7). Narrowing the N-terminal region of subtype C-derived Vif protein from residues 1 to 37 to residues 9–37, 1–34, 1–31, or 1–23 revealed that the N-terminal region up to position 31 was crucial for the full anti-APOBEC3G activity seen in subtype C-derived Vif proteins (Fig. 3, lanes 8–11). The viral infectivity levels were consistent with those of APOBEC3G degradation (Fig. 3), as observed in Fig. 2B.
FIGURE 3.
Mapping the determinant of the enhanced anti-APOBEC3G activity of subtype C-derived Vif protein. Vif chimeras between subtypes B and C (depicted on the left) were first tested for the ability of Vif proteins to induce proteasomal degradation of APOBEC3G (as shown on the left side of the bar graph) by performing Western blot analysis as described in Fig. 2B. Functional testing for the anti-APOBEC3G activity of the chimeric Vif proteins in the single-round replication assay is also described in Fig. 2B. Results are representative of at least four independent triplicate experiments. RLU, relative light units. Data shown are the mean ± S.D.; *, p < 0.05; **, p < 0.01, t test.
Among the cluster of amino acids residues 1–31 of subtype C-derived Vif, 8L and 31V were found to be critical based on the chimeric experiments described above (Fig. 3; see also supplemental Fig. S3A). Although 8L is highly specific for the subtype C-derived Vif sequences available from the Los Alamos HIV sequence data base (www.hiv.lanl.gov), 31V is not subtype C-specific but is conserved in ∼40% of subtype C-derived Vif sequences in the data base. To address whether the critical amino acids would include 17K (which is not found in other subtypes but is conserved in ∼50% of subtype C-derived Vif sequences, the other half of which carry 17R; supplemental Fig. S3A) and 19K (which is rather rare in subtype C-derived Vif sequences in the data base, most of which normally carry 19R; supplemental Fig. S3A), the lysine at position 17 or 19 was replaced with arginine. The results indicated that 17K was important, whereas19K was replaceable with arginine (supplemental Fig. S3, B and C). Thus, the critical amino acid cluster not present in NL is the N-terminal cluster of 8L, 17K, 19K/R, 22N (highly conserved in non-B subtypes), 23S (conserved in all subtypes but NL), and 31V.
N-terminal Region of Subtype C-derived Vif Harbors Higher APOBEC3G Binding Activity
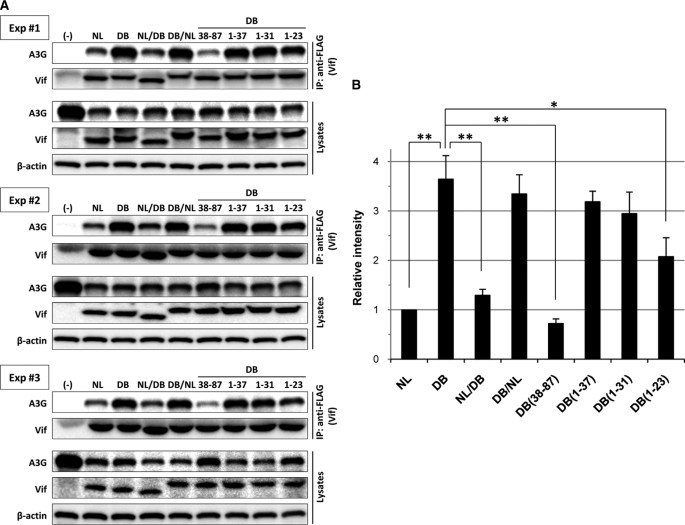
To examine whether subtype C-derived Vif or B-Vif carrying the N-terminal 1–31 fragment of subtype C-derived Vif could bind to APOBEC3G more efficiently than subtype B-Vif, we analyzed the interaction between APOBEC3G and Vif by coimmunoprecipitation. In three independent experiments (Figs. 4, A and B), the chimeric B-Vif proteins carrying the N-terminal half up to residues 1–31 of subtype C-derived Vif (DB/NL, DB(1–37), DB(1–34), and DB(1–31)) and DB-Vif showed higher levels of APOBEC3G binding than did the B-Vif protein. The binding levels of Vif proteins to APOBEC3G were consistent with the infectivity enhancement levels. Thus, the enhanced anti-APOBEC3G activity of subtype C-derived Vif protein is determined by its higher APOBEC3G binding activity at N-terminal amino acids 1–31. As expected, interactions with components of the E3 ubiquitin ligase complexes Cul5 and Elongin C were equivalent among all Vif proteins tested. This is consistent with the conservation of highly conserved SOCS box and the upstream cysteines (Cys-114 and Cys-133) required for E3 ligase complex assembly (supplemental Fig. S4).
FIGURE 4.
Binding activity of chimeric Vif proteins to APOBEC3G. A, 293T cells were cotransfected with the T7 epitope-tagged APOBEC3G expression plasmid, Rev expression plasmid, and RRE-carrying/FLAG-tagged chimeric Vif expression plasmids were immunoprecipitated (IP) with an anti-FLAG monoclonal antibody. The resulting complexes were analyzed by immunoblotting with monoclonal antibodies to the T7 epitope or with polyclonal antibodies to FLAG to detect APOBEC3G and Vif proteins, respectively (upper two panels in each experiment). Cell lysate aliquots were also analyzed by immunoblotting in parallel for T7 epitope and FLAG together with β-actin (lower three panels). Results of three independent experiments are individually shown as Experiments 1, 2, and 3. B, binding activity of chimeric Vif proteins to APOBEC3G was quantified based on the band intensity of the immunoprecipitated APOBEC3G protein. Results are the mean ± S.D. of three experiments. *, p < 0.01; **, p < 0.001, t test.
Subtype C-derived Vif Strongly Protects Viral Genomes from G-to-A Mutations Caused by Endogenous Levels of APOBEC3G Expression
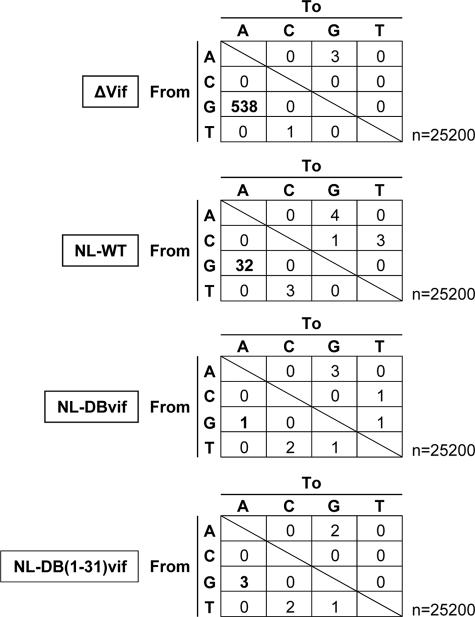
Finally, we addressed whether the anti-APOBEC3G activity of the Vif protein would correlate with the frequency of viral G-to-A mutations caused by endogenous APOBEC3G in PBMCs multiply infected by replication-competent viruses. We created the pNL4-3-based full-length proviral DNA harboring the entire or N-terminal (residues 1–31) domain of the subtype C-derived Vif sequence to compare their viral G-to-A mutation rate of the resulting viruses with those of NL4-3 wild-type (WT) (used as a subtype B reference) and with Vif-deficient viruses. Freshly isolated PBMCs were stimulated for 72 h with phytohemagglutinin and interleukin-2 and were infected with NL, NL-DBvif, NL-DB(1–31)vif, or NL-Δvif viruses produced from 293T cells transfected with their proviral DNA clones. At 12 days post-infection, the total cellular DNAs were extracted from the infected cells. The DNAs were subjected to PCR amplification targeting the envelope region followed by TA-cloning. Forty clones each from four different infected cells were sequenced.
The endogenous level of APOBEC3G expression in the PBMCs efficiently induced the G-to-A mutation of Vif-deficient viruses. As expected, envelope sequences (nucleotides 8137–8766) derived from viruses carrying the entire or N-terminal domain of subtype C-derived Vif displayed lower G-to-A mutation rates than those from NL4-3 WT viruses (Fig. 5). However, viruses with subtype C-derived Vif sequences did not show distinct growth kinetics from NL4-3 WT viruses over the time course of the experiment (supplemental Fig. S5). Thus, in primary lymphocytes expressing endogenous APOBEC3G levels, viruses harboring the subtype C-derived Vif protein can maintain the viral nucleotide sequences with less frequent G-to-A mutations than those harboring the subtype B-derived Vif.
FIGURE 5.
Analysis of G-to-A mutational frequency of proviral DNAs in PBMCs. Viruses from pNL4-3-based full-length proviral DNA clones carrying vif-deficient (Δvif), WT, subtype C-derived (DBvif) or B/C-chimeric (DB(1–31)vif) vif genes were used to infect freshly isolated and phytohemagglutinin-stimulated PBMCs. Infected cells were harvested and lysed for total DNA extraction at 12 days after infection. An amplified HIV-1 env fragment was cloned and analyzed for the G-to-A mutational frequency by sequencing. Numbers of substitutions are depicted for each viral infection of PBMCs.
DISCUSSION
To date most functional analyses of HIV-1 Vif proteins against APOBEC3G have been based on subtype B-derived Vif, and none has focused on the effect of subtypic differences on anti-APOBEC3G activity. The present study demonstrated that Vif proteins derived from subtype C strains had the most enhanced activity against APOBEC3G among the subtypes tested. Based on experiments using chimeric Vif constructs between subtypes B and C, the enhanced anti-APOBEC3G activity observed in subtype C-derived Vif was determined by residues 1–31, which positively regulated the binding activity for APOBEC3G. Consistent with this observation, these residues were also related to resistance to the G-to-A mutation at an endogenous level of APOBEC3G expression.
The worldwide prevalence of subtype C viruses, including recombinant forms, is >50% among HIV-1-infected individuals (see the UNAIDS website). This finding implies that the viruses per se might display characteristics that are distinct from other subtypes. In vitro functional testing confirmed our hypothesis that subtype C-derived Vif proteins display different activity against APOBEC3G than those derived from other subtypes. Several lines of evidence suggest the uniqueness of subtype C viruses, such as their extra copy of the NF-κB enhancer element in the long terminal repeat region (61, 62), their low replication level in macrophages and CD4-postitive lymphocytes (63–65) with reduced pathogenic fitness (66), their predominant CCR5 tropism (63, 67–69), and the fact that autologous neutralizing antibody induction is associated with shorter V1-to-V5 envelope lengths (70, 71).
Our new finding of the robust anti-APOBEC3G activity of subtype C-derived Vif might be restricted to certain regional populations of subtype C strains. Among the identified cluster of residues 1–31 critical for robust anti-APOBEC3G activity, 31V is not subtype C-specific but is conserved in ∼40% of subtype C-derived Vif proteins. This conservation is particularly high in Indian (80%, n = 15) and Zambian (77%, n = 18) subtype C-derived Vif sequences (available as interpatient samples from the data base). Conservation of 17K, which is specific for and conserved in ∼50% of all subtype C-derived Vif sequences, also depends on the sampling country (e.g. Botswana, 62% (n = 45); India, 93% (n = 15); Tanzania, 50% (n = 20); South Africa, 48% (n = 269); Zambia, 61% (n = 18)). Based on the data base and chimeric/mutational analyses shown in Fig. 3, residues 19K/R, 23S (conserved in all subtypes but NL), 22N (conserved in non-B subtypes), and 8L (highly specific for subtype C), completely conserved in subtype C-derived Vif sequences, were not sufficient for the full activity of subtype C-derived Vif proteins. This full activity likely requires the existence of 31V and 17K described above, both of which are conserved in all of our Zambian subtype C samples, and in 44% of Zambian (n = 18) or 73% of Indian (n = 15) subtype C-derived Vif sequences in the data base. It is likely that the observed subtype C viruses carrying fully active Vif are regionally circulating strains.
Still, the central findings of the present study are consistent with the previous results reported by Janini et al. (38), who have provided some evidence in favor of the hypothesis that subtype C HIV-1 is relatively protected from APOBEC3G effects in vivo. In the study they evaluated the presence of hypermutated HIV-1 sequences in proviral DNA from the PBMCs of 53 patients using a screening method that identified AT-rich sequences. They showed that the hypermutation occurred in 57% of subtype A, 67% of subtype D, 44% of subtype B, and 21% of subtype C infections and that subtype C hypermutated sequences (in 3 of 14 patients) also had lower levels of the G-to-A mutation than other subtype sequences, although the differences did not reach statistical significance because of the small sample size. To confirm these in vivo observations, further analysis involving large-scale sequencing of patient derived-subtype C proviral DNA will be necessary (note that the G-to-A hypermutation is rarely detectable in plasma RNA sequences (40–42)).
Several groups have identified distinct APOBEC3G binding domains of Vif (11, 72–81). It should be noted that the N-terminal amino acid cluster identified here that underlies the increased APOBEC3G binding activity of subtype C-derived Vif partially overlaps with previously identified domains, e.g. the N-terminal 21WxSLVK26 (76) or 23SLVx4Yx9Y40 (77) motif. It seems likely that additional changes of the N-terminal amino acids of Vif protein surrounding these reported motifs might result in a previously unknown gain of enhanced binding activity of this protein to APOBEC3G, as observed in the subtype C-derived Vif proteins tested. This needs to be further elucidated by structure-based analyses of Vif proteins.
Consistent with our observation that subtype C-derived Vif proteins showed higher anti-APOBEC3G activity than subtype B-derived Vif proteins, a much lower G-to-A mutational rate was observed in viruses carrying subtype C-derived Vif than in NL4-3 WT viruses at endogenous APOBEC3G expression levels in PBMCs. Sadler et al. (82) recently reported that sublethal G-to-A mutation levels induced by APOBEC3G even in the presence of Vif allowed the viruses to yield sufficiently replication-proficient viral progeny with a highly diverse pool of quasispecies. In this context it would be intriguing to conjecture that subtype C viruses might not be as flexible in modulating HIV-1 fitness or in increasing diversity as subtype B viruses, which have moderate but not robust Vif activity in APOBEC3G neutralization. Further studies are required to test this hypothesis.
Supplementary Material
Acknowledgment
We thank Yuko Sakamoto for technical assistance.
This work was supported by grants from the Ministry of Health, Labor, and Welfare of Japan (Research on HIV/AIDS; H21-009) and from the Ministry of Education, Science, Technology, Sports and Culture of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- APOBEC3
- apolipoprotein B mRNA editing enzyme catalytic polypeptide-like protein 3
- PBMC
- peripheral blood mononuclear cell
- Vif
- virion infectivity factor
- VSV-G
- vesicular stomatitis virus glycoprotein
- RRE
- Rev-responsive element
- Cul5
- Cullin5.
REFERENCES
- 1.Langlois M. A., Neuberger M. S. (2008) J. Virol. 82, 4660–4664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sheehy A. M., Gaddis N. C., Choi J. D., Malim M. H. (2002) Nature 418, 646–650 [DOI] [PubMed] [Google Scholar]
- 3.Yu Q., Chen D., König R., Mariani R., Unutmaz D., Landau N. R. (2004) J. Biol. Chem. 279, 53379–53386 [DOI] [PubMed] [Google Scholar]
- 4.Delebecque F., Suspène R., Calattini S., Casartelli N., Saïb A., Froment A., Wain-Hobson S., Gessain A., Vartanian J. P., Schwartz O. (2006) J. Virol. 80, 605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stenglein M. D., Harris R. S. (2006) J. Biol. Chem. 281, 16837–16841 [DOI] [PubMed] [Google Scholar]
- 6.Bogerd H. P., Wiegand H. L., Hulme A. E., Garcia-Perez J. L., O'Shea K. S., Moran J. V., Cullen B. R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8780–8785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muckenfuss H., Hamdorf M., Held U., Perkovic M., Löwer J., Cichutek K., Flory E., Schumann G. G., Münk C. (2006) J. Biol. Chem. 281, 22161–22172 [DOI] [PubMed] [Google Scholar]
- 8.Kinomoto M., Kanno T., Shimura M., Ishizaka Y., Kojima A., Kurata T., Sata T., Tokunaga K. (2007) Nucleic Acids Res. 35, 2955–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sasada A., Takaori-Kondo A., Shirakawa K., Kobayashi M., Abudu A., Hishizawa M., Imada K., Tanaka Y., Uchiyama T. (2005) Retrovirology 2, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bishop K. N., Holmes R. K., Sheehy A. M., Davidson N. O., Cho S. J., Malim M. H. (2004) Curr. Biol. 14, 1392–1396 [DOI] [PubMed] [Google Scholar]
- 11.Simon V., Zennou V., Murray D., Huang Y., Ho D. D., Bieniasz P. D. (2005) PLoS Pathog. 1, e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zennou V., Bieniasz P. D. (2006) Virology 349, 31–40 [DOI] [PubMed] [Google Scholar]
- 13.Zhang H., Yang B., Pomerantz R. J., Zhang C., Arunachalam S. C., Gao L. (2003) Nature 424, 94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mangeat B., Turelli P., Caron G., Friedli M., Perrin L., Trono D. (2003) Nature 424, 99–103 [DOI] [PubMed] [Google Scholar]
- 15.Harris R. S., Bishop K. N., Sheehy A. M., Craig H. M., Petersen-Mahrt S. K., Watt I. N., Neuberger M. S., Malim M. H. (2003) Cell 113, 803–809 [DOI] [PubMed] [Google Scholar]
- 16.Shindo K., Takaori-Kondo A., Kobayashi M., Abudu A., Fukunaga K., Uchiyama T. (2003) J. Biol. Chem. 278, 44412–44416 [DOI] [PubMed] [Google Scholar]
- 17.Newman E. N., Holmes R. K., Craig H. M., Klein K. C., Lingappa J. R., Malim M. H., Sheehy A. M. (2005) Curr. Biol. 15, 166–170 [DOI] [PubMed] [Google Scholar]
- 18.Guo F., Cen S., Niu M., Saadatmand J., Kleiman L. (2006) J. Virol. 80, 11710–11722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo K., Wang T., Liu B., Tian C., Xiao Z., Kappes J., Yu X. F. (2007) J. Virol. 81, 7238–7248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mbisa J. L., Barr R., Thomas J. A., Vandegraaff N., Dorweiler I. J., Svarovskaia E. S., Brown W. L., Mansky L. M., Gorelick R. J., Harris R. S., Engelman A., Pathak V. K. (2007) J. Virol. 81, 7099–7110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sheehy A. M., Gaddis N. C., Malim M. H. (2003) Nat. Med. 9, 1404–1407 [DOI] [PubMed] [Google Scholar]
- 22.Marin M., Rose K. M., Kozak S. L., Kabat D. (2003) Nat. Med. 9, 1398–1403 [DOI] [PubMed] [Google Scholar]
- 23.Stopak K., de Noronha C., Yonemoto W., Greene W. C. (2003) Mol. Cell 12, 591–601 [DOI] [PubMed] [Google Scholar]
- 24.Mehle A., Strack B., Ancuta P., Zhang C., McPike M., Gabuzda D. (2004) J. Biol. Chem. 279, 7792–7798 [DOI] [PubMed] [Google Scholar]
- 25.Yu X., Yu Y., Liu B., Luo K., Kong W., Mao P., Yu X. F. (2003) Science 302, 1056–1060 [DOI] [PubMed] [Google Scholar]
- 26.Mehle A., Goncalves J., Santa-Marta M., McPike M., Gabuzda D. (2004) Genes Dev. 18, 2861–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu Y., Xiao Z., Ehrlich E. S., Yu X., Yu X. F. (2004) Genes Dev. 18, 2867–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi M., Takaori-Kondo A., Miyauchi Y., Iwai K., Uchiyama T. (2005) J. Biol. Chem. 280, 18573–18578 [DOI] [PubMed] [Google Scholar]
- 29.Alce T. M., Popik W. (2004) J. Biol. Chem. 279, 34083–34086 [DOI] [PubMed] [Google Scholar]
- 30.Svarovskaia E. S., Xu H., Mbisa J. L., Barr R., Gorelick R. J., Ono A., Freed E. O., Hu W. S., Pathak V. K. (2004) J. Biol. Chem. 279, 35822–35828 [DOI] [PubMed] [Google Scholar]
- 31.Zennou V., Perez-Caballero D., Göttlinger H., Bieniasz P. D. (2004) J. Virol. 78, 12058–12061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schäfer A., Bogerd H. P., Cullen B. R. (2004) Virology 328, 163–168 [DOI] [PubMed] [Google Scholar]
- 33.Wiegand H. L., Doehle B. P., Bogerd H. P., Cullen B. R. (2004) EMBO J. 23, 2451–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng Y. H., Irwin D., Kurosu T., Tokunaga K., Sata T., Peterlin B. M. (2004) J. Virol. 78, 6073–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dang Y., Wang X., Esselman W. J., Zheng Y. H. (2006) J. Virol. 80, 10522–10533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fitzgibbon J. E., Mazar S., Dubin D. T. (1993) AIDS Res. Hum. Retroviruses 9, 833–838 [DOI] [PubMed] [Google Scholar]
- 37.Borman A. M., Quillent C., Charneau P., Kean K. M., Clavel F. (1995) Virology 208, 601–609 [DOI] [PubMed] [Google Scholar]
- 38.Janini M., Rogers M., Birx D. R., McCutchan F. E. (2001) J. Virol. 75, 7973–7986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koulinska I. N., Chaplin B., Mwakagile D., Essex M., Renjifo B. (2003) AIDS Res. Hum. Retroviruses 19, 1115–1123 [DOI] [PubMed] [Google Scholar]
- 40.Russell R. A., Moore M. D., Hu W. S., Pathak V. K. (2009) Retrovirology 6, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bailey J. R., Williams T. M., Siliciano R. F., Blankson J. N. (2006) J. Exp. Med. 203, 1357–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kieffer T. L., Kwon P., Nettles R. E., Han Y., Ray S. C., Siliciano R. F. (2005) J. Virol. 79, 1975–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pace C., Keller J., Nolan D., James I., Gaudieri S., Moore C., Mallal S. (2006) J. Virol. 80, 9259–9269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wieland U., Hartmann J., Suhr H., Salzberger B., Eggers H. J., Kühn J. E. (1994) Virology 203, 43–51 [DOI] [PubMed] [Google Scholar]
- 45.Wieland U., Seelhoff A., Hofmann A., Kühn J. E., Eggers H. J., Mugyenyi P., Schwander S. (1997) J. Gen. Virol. 78, 393–400 [DOI] [PubMed] [Google Scholar]
- 46.Jacobs G. B., Nistal M., Laten A., van Rensburg E. J., Rethwilm A., Preiser W., Bodem J., Engelbrecht S. (2008) AIDS Res. Hum. Retroviruses 24, 991–994 [DOI] [PubMed] [Google Scholar]
- 47.Stephens E. B., Singh D. K., Pacyniak E., McCormick C. (2001) AIDS Res. Hum. Retroviruses 17, 169–177 [DOI] [PubMed] [Google Scholar]
- 48.Adachi A., Gendelman H. E., Koenig S., Folks T., Willey R., Rabson A., Martin M. A. (1986) J. Virol. 59, 284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koyanagi Y., O'Brien W. A., Zhao J. Q., Golde D. W., Gasson J. C., Chen I. S. (1988) Science 241, 1673–1675 [DOI] [PubMed] [Google Scholar]
- 50.Shioda T., Levy J. A., Cheng-Mayer C. (1991) Nature 349, 167–169 [DOI] [PubMed] [Google Scholar]
- 51.Gartner S., Markovits P., Markovitz D. M., Kaplan M. H., Gallo R. C., Popovic M. (1986) Science 233, 215–219 [DOI] [PubMed] [Google Scholar]
- 52.Iwabu Y., Fujita H., Kinomoto M., Kaneko K., Ishizaka Y., Tanaka Y., Sata T., Tokunaga K. (2009) J. Biol. Chem. 284, 35060–35072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fouchier R. A., Meyer B. E., Simon J. H., Fischer U., Malim M. H. (1997) EMBO J. 16, 4531–4539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Niwa H., Yamamura K., Miyazaki J. (1991) Gene 108, 193–199 [DOI] [PubMed] [Google Scholar]
- 55.Lutz-Freyermuth C., Query C. C., Keene J. D. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 6393–6397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kimura M. (1980) J. Mol. Evol. 16, 111–120 [DOI] [PubMed] [Google Scholar]
- 58.Yu Q., König R., Pillai S., Chiles K., Kearney M., Palmer S., Richman D., Coffin J. M., Landau N. R. (2004) Nat. Struct. Mol. Biol. 11, 435–442 [DOI] [PubMed] [Google Scholar]
- 59.Mulder L. C., Ooms M., Majdak S., Smedresman J., Linscheid C., Harari A., Kunz A., Simon V. (2010) J. Virol. 84, 9613–9617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miyagi E., Brown C. R., Opi S., Khan M., Goila-Gaur R., Kao S., Walker R. C., Jr., Hirsch V., Strebel K. (2010) J. Virol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Montano M. A., Novitsky V. A., Blackard J. T., Cho N. L., Katzenstein D. A., Essex M. (1997) J. Virol. 71, 8657–8665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salminen M. O., Johansson B., Sönnerborg A., Ayehunie S., Gotte D., Leinikki P., Burke D. S., McCutchan F. E. (1996) AIDS Res. Hum. Retroviruses 12, 1329–1339 [DOI] [PubMed] [Google Scholar]
- 63.Björndal A., Sönnerborg A., Tscherning C., Albert J., Fenyö E. M. (1999) AIDS Res. Hum. Retroviruses 15, 647–653 [DOI] [PubMed] [Google Scholar]
- 64.Pollakis G., Abebe A., Kliphuis A., Chalaby M. I., Bakker M., Mengistu Y., Brouwer M., Goudsmit J., Schuitemaker H., Paxton W. A. (2004) J. Virol. 78, 2841–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ball S. C., Abraha A., Collins K. R., Marozsan A. J., Baird H., Quiñones-Mateu M. E., Penn-Nicholson A., Murray M., Richard N., Lobritz M., Zimmerman P. A., Kawamura T., Blauvelt A., Arts E. J. (2003) J. Virol. 77, 1021–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abraha A., Nankya I. L., Gibson R., Demers K., Tebit D. M., Johnston E., Katzenstein D., Siddiqui A., Herrera C., Fischetti L., Shattock R. J., Arts E. J. (2009) J. Virol. 83, 5592–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Esbjörnsson J., Månsson F., Martínez-Arias W., Vincic E., Biague A. J., da Silva Z. J., Fenyö E. M., Norrgren H., Medstrand P. (2010) Retrovirology 7, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ndung'u T., Sepako E., McLane M. F., Chand F., Bedi K., Gaseitsiwe S., Doualla-Bell F., Peter T., Thior I., Moyo S. M., Gilbert P. B., Novitsky V. A., Essex M. (2006) Virology 347, 247–260 [DOI] [PubMed] [Google Scholar]
- 69.Cilliers T., Nhlapo J., Coetzer M., Orlovic D., Ketas T., Olson W. C., Moore J. P., Trkola A., Morris L. (2003) J. Virol. 77, 4449–4456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gray E. S., Moore P. L., Choge I. A., Decker J. M., Bibollet-Ruche F., Li H., Leseka N., Treurnicht F., Mlisana K., Shaw G. M., Karim S. S., Williamson C., Morris L. (2007) J. Virol. 81, 6187–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li B., Decker J. M., Johnson R. W., Bibollet-Ruche F., Wei X., Mulenga J., Allen S., Hunter E., Hahn B. H., Shaw G. M., Blackwell J. L., Derdeyn C. A. (2006) J. Virol. 80, 5211–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dang Y., Wang X., York I. A., Zheng Y. H. (2010) J. Virol. 84, 8561–8570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dang Y., Davis R. W., York I. A., Zheng Y. H. (2010) J. Virol. 84, 5741–5750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Russell R. A., Smith J., Barr R., Bhattacharyya D., Pathak V. K. (2009) J. Virol. 83, 1992–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pery E., Rajendran K. S., Brazier A. J., Gabuzda D. (2009) J. Virol. 83, 2374–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dang Y., Wang X., Zhou T., York I. A., Zheng Y. H. (2009) J. Virol. 83, 8544–8552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen G., He Z., Wang T., Xu R., Yu X. F. (2009) J. Virol. 83, 8674–8682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yamashita T., Kamada K., Hatcho K., Adachi A., Nomaguchi M. (2008) Microbes Infect. 10, 1142–1149 [DOI] [PubMed] [Google Scholar]
- 79.He Z., Zhang W., Chen G., Xu R., Yu X. F. (2008) J. Mol. Biol. 381, 1000–1011 [DOI] [PubMed] [Google Scholar]
- 80.Donahue J. P., Vetter M. L., Mukhtar N. A., D'Aquila R. T. (2008) Virology 377, 49–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Russell R. A., Pathak V. K. (2007) J. Virol. 81, 8201–8210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sadler H. A., Stenglein M. D., Harris R. S., Mansky L. M. (2010) J. Virol. 84, 7396–7404 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.