Abstract
The mammalian SPRED (Sprouty-related protein with an EVH1 domain) proteins include a family of three members, SPRED1–3. Currently, little is known about their biochemistry. The best described, SPRED1, has been shown to inhibit the Ras/ERK pathway downstream of Ras. All three SPREDs have a cysteine-rich domain (CRD) that has high homology to the CRD of the Sprouty family of proteins, several of which are also Ras/ERK inhibitors. In the belief that binding partners would clarify SPRED function, we assayed for their associated proteins. Here, we describe the direct and endogenous interaction of SPRED1 and SPRED2 with the novel kinase, DYRK1A. DYRK1A has become the subject of recent research focus as it plays a central role in Caenorhabditis elegans oocyte maturation and egg activation, and there is strong evidence that it could be involved in Down syndrome in humans. Both SPRED1 and SPRED2 inhibit the ability of DYRK1A to phosphorylate its substrates, Tau and STAT3. This inhibition occurs via an interaction of the CRD of the SPREDs with the kinase domain of DYRK1A. DYRK1A substrates must bind to the kinase to enable phosphorylation, and SPRED proteins compete for the same binding site to modify this process. Our accumulated evidence indicates that the SPRED proteins are likely physiological modifiers of DYRK1A.
Keywords: Dual Specificity Kinase, p53, Receptor Tyrosine Kinase, STAT Transcription Factor, Tau, DYRK1A, FGFR, SPRED, STAT3, Sprouty
Introduction
The discovery of the major components in the canonical receptor tyrosine kinase (RTK)2/Ras/ERK pathway was a defining sequence of events in the field of signal transduction. Later, other pathways were discovered, and a substantial degree of cross-talk between these pathways was characterized. Subsequent research also established an array of proteins that collaborated with the core proteins in the RTK/Ras/ERK pathway to up-regulate, down-regulate, or strategically position pathway components to various cellular locations (1, 2). Aberrant activation of the RTK/Ras/ERK pathway is known to be involved in the progression of various human cancers (3–6).
One feedback down-regulator of the RTK/Ras/ERK pathway was discovered in a screen for genes involved in tracheal branching in Drosophila (7, 8). The derived protein, named Sprouty (Spry) because of its effect on tracheal branching when absent, was the founding member of a family of mammalian Sprouty proteins, Sprouty1–4 (9, 10). All Sprouty proteins have a unique, highly conserved Cys-rich C-terminal region, which was later identified in another protein family known as SPRED (Sprouty-related proteins with EVH1 domain), consisting of three mammalian members, of which SPRED1 and -2 are the most commonly studied (7, 11). In addition to its cysteine-rich domain (CRD), SPRED proteins contain an N-terminal Ena/vasodilator-stimulated phosphoprotein homology-1 (EVH1) domain and a small c-Kit-binding domain (12, 13). Although both SPRED1 and -2 share more than 60% sequence identity and are both membrane-associated proteins, their expression patterns are markedly different in various tissues and cell types (11, 13). Furthermore, SPRED2 has been implicated in the regulation of secretion pathways, although there is no evidence that SPRED1 has a similar role (14).
SPREDs, like Sprouty proteins, also down-regulate the RTK/Ras/ERK pathway by a mechanism that involves Ras and Raf but is seemingly distinct from the postulated Sprouty down-regulatory mechanisms (11, 15–20). There is a probable misconception that both families inhibit the RTK/Ras/ERK pathway via the cysteine-rich domain they both possess. Rather, accumulated binding evidence centered on SPRED and Sprouty proteins indicates that the CRD domain is a common site for protein interaction, including the kinases Raf1 and Tesk1 (21, 22). In a preliminary screen for other proteins associated with SPRED1, SPRED2, and Sprouty2, we detected DYRK1A. Aranda et al. (23) also described the association of DYRK1A with Sprouty2, and this interaction appears to mildly impact on the ability of Sprouty2 to inhibit the FGFR/Ras/ERK pathway.
The dual specificity tyrosine-regulated kinases (DYRKs) are subfamilies of protein kinases that phosphorylate target proteins on serine/threonine residues within the RPX(S/T)P motif (24). Their kinase activity depends on the autophosphorylation of a Tyr-Xaa-Tyr (YXY) motif in the activation loop of the catalytic domain, and hence DYRK activity is independent of an upstream tyrosine kinase (25–29). The DYRK family is classified into three subfamilies, DYRK1, YAK1, and DYRK2, based on phylogenetic analysis (30–32). The DYRK1 subfamily is animal-specific and represented by the mammalian DYRK1, minibrain (mnb) in Drosophila, and mbk-1 in Caenorhabditis elegans (28, 33–35). There are at least seven mammalian DYRK family members as follows: DYRK1A, DYRK1B (also known as Mirk), DYRK1C, DYRK2, DYRK3, DYRK4 (31), with DYRK1A being the most extensively characterized. It is located on chromosome 21 in the Down syndrome critical region, and various collated data indicate that the kinase may play a role in Down syndrome (33, 36, 37). In addition to the conserved catalytic domain, DYRK1A also contains two nuclear localization signals (NLSs) as follows: a classical bipartite NLS at the N terminus and a complex NLS within the catalytic domain (38). The kinase domain is followed by a PEST domain and then a series of histidine repeats that target DYRK1A to the splicing factor compartment (38). Substrates of DYRK1A (some only characterized in vitro) include the following: STAT3, EF2B, Tau, Gli1, CREB, dynamin, glycogen synthase, members of the Forkhead family (FKHR), as well as several splicing factors, including cyclin L2 and SF3b/SAP155 (29, 39–46). The binding parameters involved in substrate interaction with DYRK family members have not been explored.
In this study, we investigate the binding of SPRED proteins to DYRK1A and the downstream effects of this interaction. Both SPRED proteins affect the ability of DYRK1A to phosphorylate its substrates. The kinase activity of DYRK1A was not inhibited by the interaction with the SPRED proteins. However, both SPRED1 and -2 appeared to bind to a common binding site on DYRK1A in competition with recognized substrate proteins. Knockdown experiments indicated that SPRED proteins could modify the interaction of DYRK1A with its substrates and concomitantly affect downstream signaling events.
EXPERIMENTAL PROCEDURES
Plasmids
Full-length DYRK1A and Tau amplified from mouse brain cDNA were cloned into pXJ40-HA and pXJ40-myc mammalian expression vectors, respectively. FGFR1, FLAG-tagged human Spry2, and mouse SPRED1 and -2 full-length constructs have been described earlier (47, 48). The DYRK1A kinase-dead K179R mutant was generated by site-directed mutagenesis using the proofreading Pfu DNA polymerase (Promega, Madison, WI). The pXJ40-myc-hSTAT3 was kindly provided by Dr. X. Cao (Institute of Molecular and Cell Biology, Singapore).
Antibodies and Reagents
Mouse and rabbit anti-FLAG, agarose-conjugated anti-FLAG M2 beads, rabbit anti-HA and anti-SPRED2 used in knockdown experiments, and harmine were purchased from Sigma. Sheep anti-SPRED1 was from R & D Systems (Minneapolis, MN). Rabbit anti-SPRED2 was purchased from Abnova Corp. (Taipei, Taiwan). Mouse anti-β-actin and rabbit anti-acetyl-p53 (Lys-382) were from Abcam (Cambridge, UK). Rat anti-HA was from Roche Applied Science. Mouse monoclonal antibodies against Myc and Tau, rabbit antibodies against FGFR1, Myc, and DYRK1A, and a goat antibody against DYRK1A were from Santa Cruz Biotechnology (Santa Cruz, CA). Affinity-purified rabbit polyclonal antibodies against SPRED1 and -2 were generated against Limulus polyphemus hemocyanin-conjugated peptide CMWKNDLERDDTD-amide and CATDSSSNSSQKREPT-amide of SPRED1 and -2 proteins, respectively (BioGenes GmbH, Berlin, Germany). A mouse monoclonal antibody against STAT3 was from BD Transduction Laboratories, and a mouse antibody against phospho-STAT3 (Ser-727) was from Cell Signaling Technology (Beverly, MA). Rabbit anti-phospho-Tau (Thr-212) was purchased from Invitrogen. Mouse monoclonal anti-p53 (DO-1) was a kind gift from Prof. Sir David Lane (Agency of Science Technology and Research, Singapore).
Yeast Two-hybrid Screen
To identify the interacting partners of SPREDs and Spry2, yeast two-hybrid screens were performed using the MatchMaker III system from Clontech, according to the manufacturer's instructions. The full-length human Spry2 and mouse SPREDs were subcloned into the pGBK-T7 vector and used as baits on pretransformed mouse brain and mouse embryo libraries, respectively, cloned into pACT2 vector. Plasmids of positive clones were isolated as described previously (48), and positive in-frame clones were further subcloned into pXJ40 vector for expression in mammalian cells.
Cell Lines and Transfection
All cell lines used were purchased from ATCC (Manassas, VA). HEK293 and PC-3 cells were maintained in RPMI 1640 medium, supplemented with 10% fetal bovine serum and 2 mm l-glutamine. All transfections were carried out using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions.
Immunoprecipitation and Immunoblotting
Immunoprecipitation and immunoblotting were carried out essentially as described previously (49). Cells were harvested 24 h post-transfection in HEPES lysis buffer (20 mm HEPES (pH 7.4) 137 mm NaCl, 1 mm EGTA, 1.5 mm MgCl2, 10% (v/v) glycerol, 1% Triton X-100, a mixture of protease inhibitors (Complete protease inhibitor, Roche Applied Science), and 1 mm Na3VO4). Cell lysates were then used for immunoprecipitation and subsequent immunoblotting. Quantification of immunoblots was performed with GS-800-calibrated densitometer (Bio-Rad). All Western blot data shown are representatives of at least three separate individual experiments, unless otherwise stated. For endogenous interaction, PC-3 cells were harvested using HEPES lysis buffer, and the cell lysates were subjected to immunoprecipitation using either rabbit DYRK1A or rabbit SPRED1 and -2 antibodies. The resulting immunoprecipitates were separated by SDS-PAGE and blotted with sheep anti-SPRED1 and rabbit anti-SPRED2 or goat anti-DYRK1A to detect the proteins.
Cell Viability Assay
HEK293 cells, transfected with the indicated plasmids, were treated with 5 μm etoposide (Sigma) for 24 h. Cell viability was subsequently measured using the Cell Proliferation Reagent WST-1 system (Roche Applied Science) according to the manufacturer's protocol.
Small Interfering RNA Knockdown
A pool of small interfering RNAs (siRNA) against human SPRED1, SPRED2, and control siRNA (ON-TARGETplus Nontargeting Pool) were purchased from Dharmacon RNAi Technologies (ON-TARGETplus SMARTpool L-016638-00-0005, SMARTpool L-018590-00-0005, and D-001810-10-05, respectively). The siRNA constructs were transfected into 293 cells at about 40% confluency using the Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. Cells were transfected with various cDNA plasmids 48 h post-transfection of the siRNA as stated in the figure legend of the specific experiment. The cells were harvested 24 h after the second transfection and subjected to immunoblotting as described above.
Alkaline Phosphatase Treatment
Cell lysates of 293 cells transfected with FLAG-SPRED2, myc-Raf1, and HA-WT DYRK1A or HA-KD DYRK1A were harvested in lysis buffer without sodium orthovanadate and incubated with calf intestinal phosphatase from New England Biolabs (Beverly, MA) for 2 h at 37 °C, using BSA as a control. Immunoprecipitation using agarose-conjugated anti-FLAG M2 beads was then performed on these lysates, as described earlier.
Statistical Analysis
Two-tailed, unpaired Student's t tests were carried out to determine whether the results differed significantly between samples within an experiment. Differences were considered significant at a significance level of p < 0.05, and the data are presented as the mean ± S.E. of n number of independent experiments.
RESULTS
SPRED1 and -2 Interact with DYRK1A through Their CRD
Previous reports have demonstrated that several kinases interact with Spry and SPRED proteins (21, 22, 48, 50, 51). We used yeast two-hybrid screens to identify proteins that bind to Spry2, SPRED1, and SPRED2 (supplemental Fig. 1), and we obtained preliminary evidence that DYRK1A binds to all three proteins. The binding of Spry2 to DYRK1A was also recently described by Aranda et al. (23). We decided to characterize the binding parameters of SPRED proteins and their consequences on cell function and to use Spry2 binding for comparison, where appropriate.
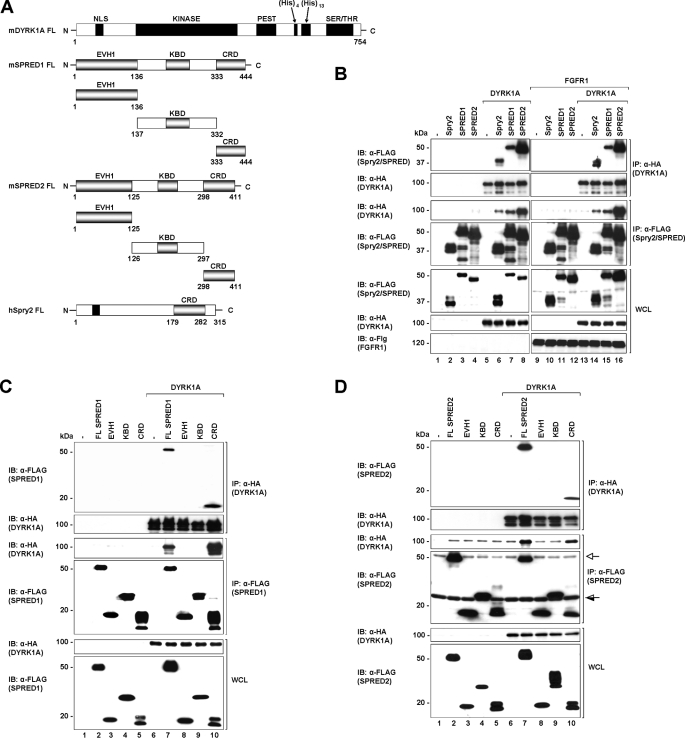
A schematic diagram of the domains of the full-length proteins as well as the SPRED truncation constructs used in subsequent experiments is shown in Fig. 1A. To verify the yeast two-hybrid interactions, HEK293 cells were co-transfected with plasmids encoding full-length HA-tagged DYRK1A and FLAG-tagged SPRED1, SPRED2, or Spry2 (as control). Co-immunoprecipitation assays confirmed that DYRK1A interacts with SPRED1 and -2 (Fig. 1B, top and 3rd panels, lanes 7 and 8), demonstrating that DYRK1A is a common interacting partner of both Spry and SPRED families. Interestingly, the interaction between DYRK1A and SPRED2 (Fig. 1B, top and 3rd panels, lane 8) is comparatively stronger than the interaction between DYRK1A with SPRED1 (Fig. 1B, top and 3rd panels, lane 7) or Spry2 (Fig. 1B, top and 3rd panels, lane 6). SPRED and Spry proteins have been reported to be negative regulators of cell signaling mediated by growth factors, such as FGF, through interactions with different proteins (13, 52) As such, we wanted to determine whether FGF receptor activation altered the interaction between SPREDs and DYRK1A. The results shown in Fig. 1B (1st and 3rd panels, right and left) indicate that the interaction between SPREDs and DYRK1A was independent of FGFR1 activation.
FIGURE 1.
SPRED1 and -2 interact with DYRK1A through the Cys-rich domain. A, schematic diagram showing the domain structures of human Spry2, mouse SPRED1, mouse SPRED2, and mouse DYRK1A. CRD, NLS, KINASE (kinase catalytic domain), PEST (domain rich in proline, glutamate, serine, and threonine), His (histidine repeat sequence), SER/THR (serine/threonine-rich region), EVH1 (Ena-VASP homology-1 domain), and KBD (c-Kit binding domain). B, HEK293 cells were transfected with the indicated plasmids (FLAG-Spry2, FLAG-SPRED1, FLAG-SPRED2, and HA-DYRK1A) or a control pXJ40 vector. 24 h post-transfection cell lysates were subjected to immunoprecipitation (IP) using anti-FLAG or rat anti-HA. The immunoprecipitates were separated on SDS-PAGE and immunoblotted with the antibodies indicated on the left. Whole cell lysates (WCL) were immunoblotted (IB) to verify equal protein expression levels in all the samples tested. C and D, different fragments of SPRED1 (C) and SPRED2 (D) as indicated in A were co-expressed with DYRK1A in 293 cells. Lysates were subjected to immunoprecipitation followed by immunoblotting of the immunoprecipitates and WCL samples with the antibodies indicated to the left. FL, full-length protein; open arrow indicates the immunoglobulin heavy chain (SPRED2 band is the darker band that runs marginally lower than the immunoglobulin heavy chain). Half-open arrow indicates the immunoglobulin light chain (the c-Kit-binding domain band is above the immunoglobulin light chain). There is a faint nonspecific band that runs across the immunoprecipitated DYRK1A in the anti-FLAG immunoprecipitated complex (3rd panel, lanes 7 and 10; the DYRK1A band in these samples show as higher intensity bands).
To map the region of SPREDs that interact with DYRK1A, a series of FLAG-tagged truncation constructs of SPRED1 and -2 (Fig. 1A) were prepared and used in co-immunoprecipitation assays with DYRK1A. We observed that both SPRED1 and -2 specifically interacted with DYRK1A through their CRD, independent of FGFR1 activation (Fig. 1, C and D).
Kinase Domain of DYRK1A Is Central to Its Interactions with SPRED1 and -2
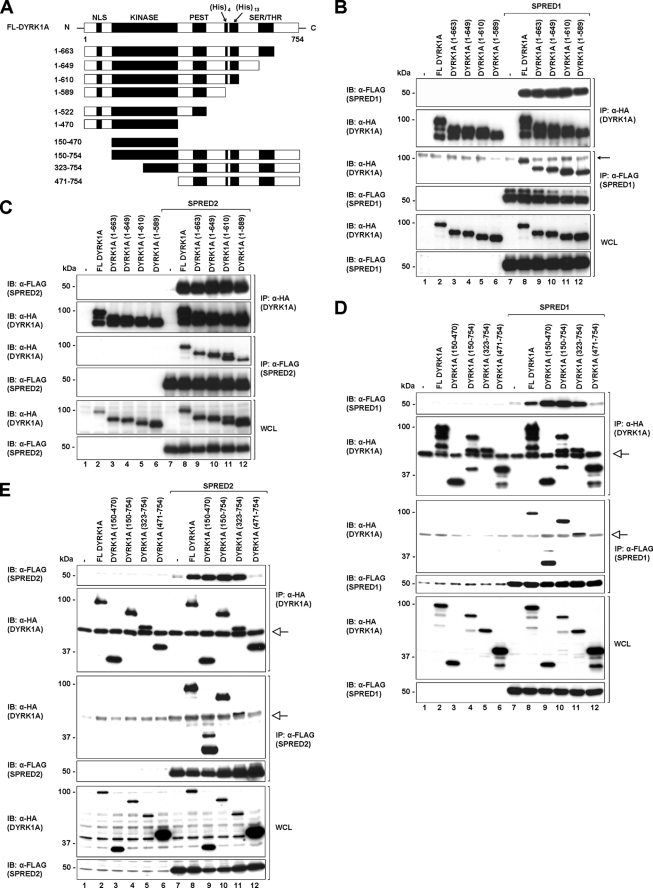
A previous report identified that Spry2 interacts with the histidine repeat sequence in DYRK1A (23). To ascertain which domains on DYRK1A were required for its interaction with SPRED proteins, we first confirmed the binding site for Spry2 on DYRK1A. Three C-terminal truncated constructs of DYRK1A (1–649, 1–610, and 1–589) were prepared, and their interactions with full-length FLAG-tagged Spry2 were assessed (Fig. 2A). Consistent with previous findings, Spry2 interacted with the truncated mutants of DYRK1A that retain the histidine repeat sequence (1–649 and 1–610) as well as full-length DYRK1A but not with the mutant lacking the histidine repeat sequence (1–589) (supplemental Fig. 2).
FIGURE 2.
SPRED1 and -2 interact with the kinase domain of DYRK1A. A, schematic diagram showing full-length and different truncation constructs of DYRK1A that were used in the subsequent studies. B and C, full-length, C-terminal truncated mutants (1–663, 1–649, 1–610, and 1–589) of DYRK1A and full-length SPRED1 (B) and SPRED2 (C) were transfected in 293 cells. Cell lysates were immunoprecipitated (IP) with FLAG or rat HA antibodies. Immunoprecipitates and WCL were resolved by SDS-PAGE and immunoblotted (IB) with the antibodies indicated on the left. Arrow indicates a nonspecific band. The band of the FL DYRK1A protein in the anti-FLAG complex seen in lane 8 (B, 3rd panel) is higher in intensity compared with the nonspecific band. D and E, 293 cells were co-transfected with full-length DYRK1A, the kinase domain of DYRK1A(150–470), and three N-terminal truncates of DYRK1A (150–754, 323–754, and 471–754) as indicated. Cell lysates were immunoprecipitated with anti-FLAG and rat anti-HA, and the precipitated proteins were separated on SDS-PAGE and analyzed by Western blotting techniques with the indicated antibodies. Open arrow indicates the immunoglobulin heavy chain.
Using the same three constructs and an additional construct that retains the Ser/Thr domain in the C-terminal region of DYRK1A(1–663) (Fig. 2A), we then employed co-immunoprecipitation experiments to determine the region on DYRK1A where SPRED1 and -2 bind. Surprisingly, SPRED1 and -2 interacted with all four of the truncated proteins, including DYRK1A(1–589), which does not interact with Spry2, suggesting that the histidine repeat sequence of DYRK1A is not necessary for its interaction with SPRED1 or -2 (Fig. 2, B and C). To further delineate the SPRED-binding site on DYRK1A, two more C-terminal truncated mutants were prepared, as shown in Fig. 2A (1–522 and 1–470). Again, SPRED1 and -2 still bound to these protein fragments (supplemental Fig. 3, A and B). This indicated that the binding site for the SPRED proteins was likely to be within the N terminus of DYRK1A. We then created three N-terminal truncated constructs containing varying proportions of the kinase domain (150–754, 323–754, and 471–754) and one containing only the kinase domain of DYRK1A(150–470) (Fig. 2A) to test their association with SPRED1 and -2. Based on the co-immunoprecipitation assays, both SPRED1 and -2 showed comparatively strong binding to the kinase domain of DYRK1A(150–470) (Fig. 2, D and E, lane 9), as well as to the other two truncated proteins containing at least a partial kinase domain (150–754 and 323–754) (Fig. 2, D and E, lanes 10 and 11). However, SPREDs do not bind to the DYRK1A truncated mutant lacking the kinase domain (471–754) (Fig. 2, D and E, lane 12). On the other hand, Spry2 interacted with all the truncated DYRK1A proteins, apart from the one containing only the kinase domain (supplemental Fig. 4, lane 9). These findings indicate that the SPRED proteins have a different DYRK1A binding location compared with Spry2.
SPRED1 and -2 Interact Endogenously and Directly with DYRK1A
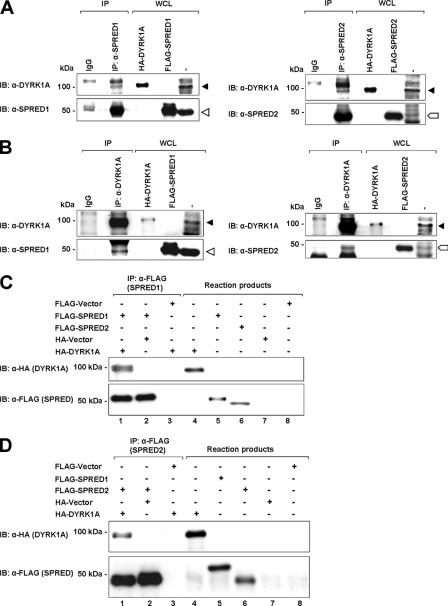
For the interaction between DYRK1A and SPRED1 or -2 to be physiologically relevant, binding at endogenous levels should occur. The human prostate cancer cell line PC3 expresses relatively high levels of DYRK1A (53), and it was therefore selected for endogenous interaction assays. Endogenous SPRED1 and -2 were immunoprecipitated from PC3 cell lysates and assayed for the presence of DYRK1A. DYRK1A co-immunoprecipitated with both SPRED1 and -2 (Fig. 3A, closed triangle, DYRK1A; open triangle, SPRED1; block arrow, SPRED2), indicating the formation of an endogenous complex. To further verify this, the reciprocal immunoprecipitation of endogenous DYRK1A was performed, and the immunoprecipitates were assayed for the presence of SPRED1 and -2. As expected, both endogenous SPRED1 and -2 showed binding to endogenous DYRK1A (Fig. 3B).
FIGURE 3.
SPRED1 and -2 interact endogenously and directly with DYRK1A. A, PC-3 cell lysates were immunoprecipitated with anti-SPRED1 and -2 antibodies, and the immunoprecipitated (IP) proteins and the WCL were subjected to SDS-PAGE and immunoblotted (IB) with anti-DYRK1A antibody. B, PC-3 cell lysates were immunoprecipitated with anti-DYRK1A antibody. The immunoprecipitates and the WCL were separated on SDS-PAGE and analyzed by immunoblotting with anti-SPRED1 and -SPRED2, respectively. Closed triangle indicates DYRK1A band; open triangle indicates SPRED1; block arrow indicates SPRED2. The HA-tagged DYRK1A and FLAG-tagged SPRED1 and -2 in the WCL are used as positive controls to indicate that the correct protein bands are present in the immunocomplex. C and D, direct association of DYRK1A and SPRED1 and -2 in vitro. Binding of in vitro translated HA-tagged DYRK1A to FLAG-tagged SPRED1 (C) and SPRED2 (D) proteins in the transcription and translation (TnT) assay. TnT reaction products and anti-FLAG immunoprecipitates were subjected to SDS-PAGE and immunoblotted with anti-HA and anti-FLAG.
We then sought to determine whether the interaction between DYRK1A and the SPRED proteins was direct. To investigate this, we performed a rabbit reticulocyte in vitro transcription and translation (TnT) assay. FLAG-tagged SPRED proteins were immunoprecipitated from the reaction product using FLAG antibody. HA-tagged DYRK1A was found to co-immunoprecipitate with both FLAG-tagged SPRED1 (Fig. 3C) and FLAG-SPRED2 (Fig. 3D). However, neither HA vector nor FLAG vector co-immunoprecipitated with FLAG-SPRED1/2 or HA-DYRK1A, respectively. These results indicate that DYRK1A interacts directly with SPRED proteins.
SPRED1 and -2 Inhibit DYRK1A-mediated Cell Proliferation by Promoting Acetylation of p53
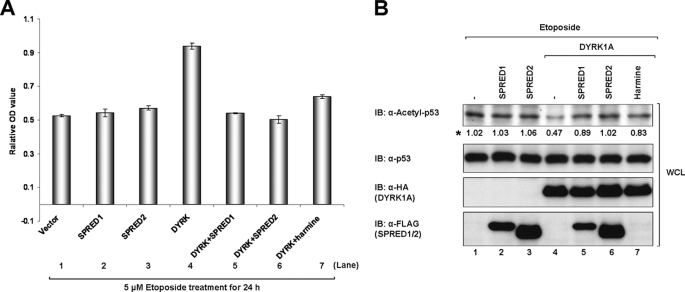
We next wanted to ascertain whether SPRED proteins were able to modulate the reported function of DYRK1A in cell proliferation through their interaction. As a tumor suppressor, p53 limits cell proliferation by inducing cell cycle arrest and apoptosis in response to DNA damage (54, 55). The acetylation of p53 at the C-terminal Lys-382 enhances its transcription associated with cell cycle arrest and apoptosis (55, 56). However, the deacetylation of p53, which represses p53 activity, often leads to inappropriate cell growth, increased cell survival, and genetic instability (57, 58). It has been reported that DYRK1A can induce cell proliferation by promoting the deacetylation of p53 (59). To address whether SPRED1 and -2 would in turn affect this function of DYRK1A, we transfected 293 cells with the indicated expression constructs and then treated the cells with etoposide, a DNA-damaging reagent that induces p53 acetylation (59). Consistent with the previous report (59), DYRK1A promoted cell proliferation following etoposide treatment (Fig. 4A, bar 4) by inhibiting the acetylation of p53 (Fig. 4B, lane 4). However, when SPRED1 or -2 was co-expressed with DYRK1A, both proteins suppressed the DYRK1A-induced proliferation of 293 cells (Fig. 4A, bars 5 and 6) and reversed the acetylation levels of p53 (Fig. 4B, lanes 5 and 6).
FIGURE 4.
SPRED1 and -2 inhibit DYRK1A-mediated cell proliferation by suppressing p53 deacetylation. A, 293 cells were transfected with combination of FLAG-tagged SPRED1, SPRED2 and HA-tagged DYRK1A. 24 h post-transfection, the cells were treated with 5 μm etoposide, or both 5 μm etoposide and 100 nm harmine for 24 h. Cell viability of the treated cells was analyzed as described under “Experimental Procedures” (n = 3, p < 0.05). B, levels of acetylated p53, total p53, HA-DYRK1A, and FLAG-SPRED1 and -2 in the WCL mentioned in A were determined by Western analysis with the indicated antibodies. Asterisk indicates the quantification results of acetyl-p53. IB, immunoblot.
The low molecular weight alkaloid harmine has a history as a psychoactive drug, and it was recently discovered in an extensive kinase assay system to be a highly specific inhibitor for DYRK family members, especially DYRK1A (60, 61), but the mode of action of harmine on DYRK1A is currently unknown. Here, we employed harmine as a known inhibitor of DYRK1A activity, to determine the degree to which the SPRED proteins were inducing p53 acetylation. As expected, 100 nmol/liter harmine inhibited DYRK1A-induced proliferation (Fig. 4A, bar 7) and antagonized DYRK1A-induced deacetylation of p53 (Fig. 4B, lane 7), verifying that DYRK1A is at least partly responsible for inhibiting p53 acetylation and the subsequent cell cycle arrest.
SPRED1, -2, and Spry2 Disrupt the DYRK1A-directed Substrate Phosphorylation
To further investigate the effect of SPRED proteins on the modification of DYRK1A function, we sought to examine whether SPRED proteins could disrupt the phosphorylation of two previously reported substrates of DYRK1A, Tau and STAT3.
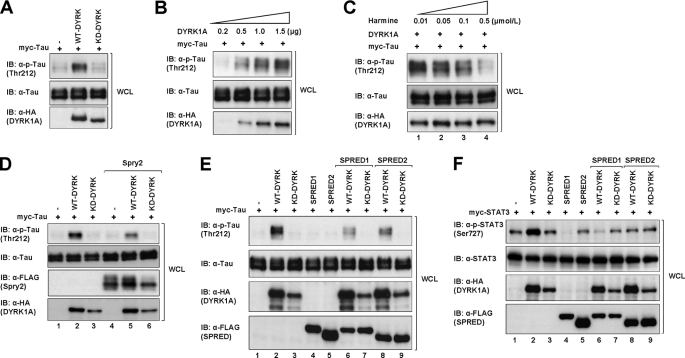
Others have reported that DYRK1A phosphorylates Tau in cells on Thr-212 (42, 43). We first sought to confirm this observation by transfecting HEK293 cells with Myc-tagged Tau and either the wild type DYRK1A (WT-DYRK1A) or a DYRK1A mutant harboring a K179R mutation of the ATP-binding site, which renders the protein kinase dead (KD-DYRK1A). It was observed that Tau was significantly phosphorylated by WT-DYRK1A compared with the vector control and KD-DYRK1A (Fig. 5A). To further confirm this, DYRK1A was co-expressed in increasing levels in 293 cells with a constant level of Tau. We observed that Tau phosphorylation was increased in parallel with the increasing amounts of DYRK1A (Fig. 5B and supplemental Fig. 5A), suggesting that DYRK1A phosphorylates Tau in a dose-dependent manner. Next, DYRK1A and Tau-transfected cells were treated with increasing concentrations of harmine for 30 min (Fig. 5C and supplemental Fig. 5B). Similar to the effects seen with the KD-DYRK1A, Tau phosphorylation was inhibited by harmine in a dose-dependent manner and was almost completely suppressed by 0.5 μm harmine (Fig. 5C, lane 4), indicating that harmine is able to inhibit the kinase-directed phosphorylation of Tau on Thr-212. We then assessed the effect of SPRED1, -2, and Spry2 on Tau phosphorylation by DYRK1A (Fig. 5, D and E, and supplemental Fig. 5, C and D). Interestingly, Tau phosphorylation was significantly reduced in the presence of both Spry2 and WT-DYRK1A (Fig. 5D, lane 5), as compared with WT-DYRK1A alone (Fig. 5D, lane 2). Similarly, SPRED1 and -2 also strongly inhibited Tau phosphorylation (Fig. 5E, lanes 6 and 8, respectively, and supplemental Fig. 5D).
FIGURE 5.
SPRED1, SPRED2, and Spry2 disrupt the DYRK1A-directed substrate phosphorylation. A, cell lysates from 293 cells transfected with the indicated plasmids (WT-DYRK1A, KD-DYRK1A, and vector control) were subjected to SDS-PAGE and immunoblotted with anti-phospho-Tau Thr-212 (p-Tau Thr-212), anti-Tau, and anti-HA. B, cell lysates from 293 cells transfected with increasing amounts of DYRK1A and a constant amount of Tau were subjected to SDS-PAGE and immunoblotted (IB) with anti-phospho-Tau (p-Tau Thr-212), anti-Tau, and anti-HA. C, 293 cells were transfected with constant amount of WT-DYRK1A and Tau. The cells were treated with increasing concentrations of harmine for 30 min after 24 h post-transfection. The cell lysates were subjected to SDS-PAGE and immunoblotted with the antibodies indicated on the left. D, cell lysates from 293 cells co-transfected with WT-, KD-DYRK1A, Spry2, and Tau were subjected to SDS-PAGE and immunoblotted with the indicated antibodies. E, cell lysates from 293 cells transfected with combinations of WT-, KD-DYRK1A, SPRED1, SPRED2, and Tau were subjected to SDS-PAGE and immunoblotted with the antibodies shown on the left. The relative quantities of phospho-Tau for each lane in B–E (top panel) were indicated in the bar charts (supplemental Fig. 5, A–D, respectively). F, cell lysates from 293 cells co-transfected with WT-, KD-DYRK1A, SPRED1, SPRED2, and STAT3 were subjected to SDS-PAGE and immunoblotted with the indicated antibodies. The relative levels of phospho-STAT3 are indicated in the bar chart (supplemental Fig. 5E).
To further verify the modifying effect of SPRED, another substrate of DYRK1A, STAT3, was employed in a parallel set of experiments. Similar to the results obtained using Tau, STAT3 was significantly phosphorylated by WT-DYRK1A (Fig. 5F, lane 2) relative to the vector control and KD-DYRK1A (Fig. 5F, lanes 1 and 3, respectively). Furthermore, the WT-DYRK1A-induced phosphorylation of STAT3 was decreased in the presence of SPRED1 and -2 (Fig. 5F, lanes 6 and 8, and supplemental Fig. 5E).
There are two possible explanations regarding the decreased phosphorylation of DYRK1A substrates in the presence of SPRED proteins: 1) binding inhibits the catalytic activity of the kinase domain or 2) binding precedes phosphorylation and SPREDs compete for this binding. We next examined the first of these two possibilities.
SPRED Proteins Do Not Inhibit the Kinase Activity of DYRK1A
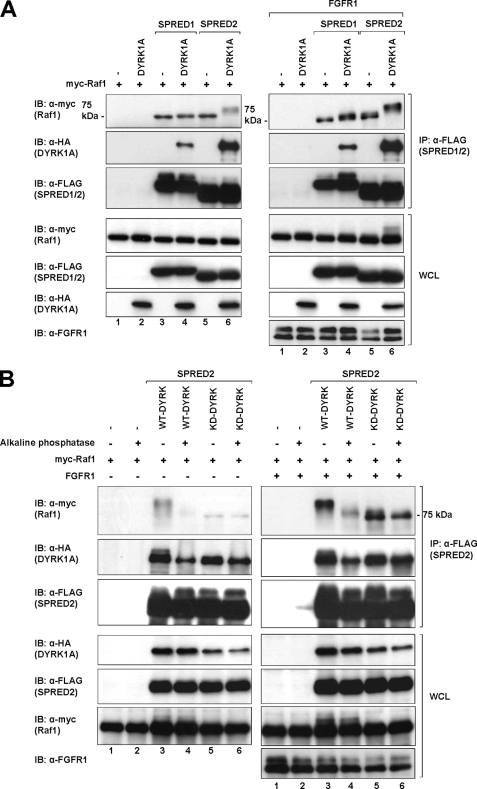
While investigating a related project, we noticed that upon FGFR1 activation there was a distinct mobility shift in Raf1 levels on SDS-polyacrylamide gels separating lysates of SPRED2 immunoprecipitations. Interestingly, this mobility shift of Raf1 only occurred in SPRED2 precipitates (Fig. 6A, top right and top left panels, lane 6), but not in SPRED1 precipitates (Fig. 6A, top right and top left panels, lane 4), and additionally it required the presence of DYRK1A (Fig. 6A, top right and top left panels, lane 6). There is considerable precedence to show that certain residues on particular proteins become atypically retarded when separated on SDS-polyacrylamide gels due to phosphorylation events, Spry2 being a case in point (62, 63). If, in this case, Raf1 is indeed retarded in the gel due to phosphorylation, this experiment would indicate that SPRED2, the strongest binder in our experiments to DYRK1A, does not inhibit the kinase activity of DYRK1A. This is also assuming that Raf1 is a substrate of DYRK1A.
FIGURE 6.
SPRED proteins do not inhibit the kinase activity of DYRK1A. A, HEK293 cells were transfected with the indicated plasmids (FLAG-SPRED1/2, HA-DYRK1A, and myc-Raf1) or a control pXJ40 vector. 24 h post-transfection cell lysates were subjected to immunoprecipitation (IP) using anti-FLAG. The immunoprecipitates were separated on SDS-PAGE and immunoblotted with the antibodies indicated on the left. Whole cell lysates (WCL) were immunoblotted (IB) to verify equal protein expression levels in all the samples tested. B, HEK293 cells were transfected with the indicated plasmids (FLAG-SPRED2, HA-WT-DYRK1A or HA-KD-DYRK1A, and myc-Raf1). Lysates obtained using the above setup were treated with alkaline phosphatase or BSA as a control and resolved by SDS-PAGE as mentioned in A.
Lysates from cells transfected with Raf1 and DYRK1A in combination with SPRED2 were treated with alkaline phosphatase and subjected to immunoblot analysis, using untreated samples as a control. Following alkaline phosphatase treatment, the mobility shift of Raf1 is no longer apparent (Fig. 6B, top left and top right panels, lane 4), indicating that the mobility shift is due to a phosphorylation event. In making this observation, there is a possibility that SPRED2, DYRK1A, or Raf1 may associate with another kinase that causes this mobility shift of Raf1. To demonstrate that the Raf1 band shifting results from an active DYRK1A, we incorporated KD-DYRK1A into the experiment as described above (Fig. 6B, top left and top right panels, lane 5). The result clearly demonstrates that there is no mobility shift of Raf1 with KD-DYRK1A. This demonstrates the gel retardation of Raf1 in the presence of DYRK1A and SPRED2 is a phosphorylation event specific to DYRK1A activation but requiring the presence of SPRED2. We currently do not know the specific site of phosphorylation on Raf1, but this is the subject of further investigation. Pending the identification of the phosphorylation site on Raf1, there is strong evidence that the association with SPRED2, at least, does not inhibit the kinase activity of DYRK1A.
SPRED1 and -2 Compete with Tau for Binding to DYRK1A
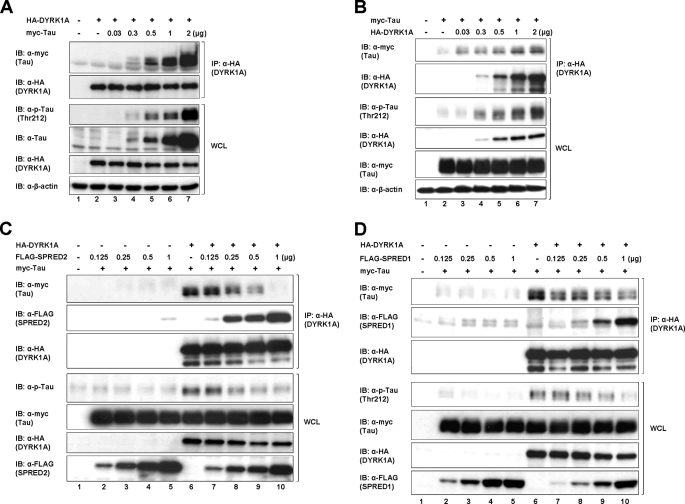
The second possible explanation for the inhibition of DYRK1A substrate phosphorylation is that SPRED1 and -2 compete for obligatory binding of substrates preceding phosphorylation. We first set up an experiment to demonstrate that the binding of Tau to DYRK1A was “dose-responsive” (Fig. 7A). With a constant level of DYRK1A and an increasing amount of Tau, we observed that the amount of Tau precipitating with DYRK1A paralleled the levels of Tau that were transfected. Furthermore, the degree of Tau phosphorylation mirrored the binding (Fig. 7A, 3rd panel), suggesting that phosphorylation is dependent on prior binding. The reciprocal experiment using increasing amounts of DYRK1A and a constant level of Tau showed the same trend (Fig. 7B). Based on these results, we concluded that any protein that competes for binding to DYRK1A, whether it is a substrate or not, will down-regulate the phosphorylation of bona fide DYRK1A substrates. To demonstrate the possible competition by SPRED2 with Tau, an experiment was set up where constant amounts of DYRK1A and Tau were transfected into cells with increasing amounts of SPRED2 (Fig. 7C). Proteins were precipitated using anti-HA (DYRK1A), and the resultant blots were probed with antibodies as shown in Fig. 7C. Two features from the data are noteworthy: 1) as the amount of SPRED2 bound to DYRK1A increases (Fig. 7C, 2nd panel), the amount of Tau binding diminishes (Fig. 7C, 1st panel), and 2) the degree of Tau phosphorylation again mirrors the amount of Tau bound to DYRK1A (Fig. 7C, 4th panel). The same experiment was repeated with SPRED1 in place of SPRED2, and a similar outcome occurred (Fig. 7D). From the data we conclude that SPRED proteins bind to a similar domain on DYRK1A as Tau, and possibly other substrates, and the hierarchy of binding will depend on the concentration of the competing proteins and the relative affinity for the DYRK1A-binding site. Substrates that are successfully competed off do not get phosphorylated, resulting in a disruption of downstream signaling. Such competition could be physiologically relevant.
FIGURE 7.
SPRED1 and -2 compete with Tau for binding to DYRK1A. A, HEK293 cells were transfected with constant amount of the HA-DYRK1A plasmids and an increasing amount of myc-Tau. Lysates were immunoprecipitated (IP) using anti-HA and subjected to SDS-PAGE immunoblot (IB) analysis as indicated in Fig. 1. B, HEK293 cells were transfected with constant amounts of myc-Tau and an increasing amount of HA-DYRK1A. Lysates were immunoprecipitated using anti-HA, separated on SDS-PAGE, and immunoblotted using the antibodies indicated on the left. C, HEK293 cells were transfected with constant amounts of HA-DYRK1A and myc-Tau together with an increasing amount of FLAG-SPRED2 plasmids. Lysates were immunoprecipitated using anti-HA and subjected to SDS-PAGE and immunoblotted analysis as indicated. D, HEK293 cells were treated as described in C using FLAG-SPRED1 instead of FLAG-SPRED2. Cell lysates were likewise treated and analyzed as in C.
Knockdown of SPRED1 and -2 Increases the DYRK1A-dependent Phosphorylation of Tau and STAT3
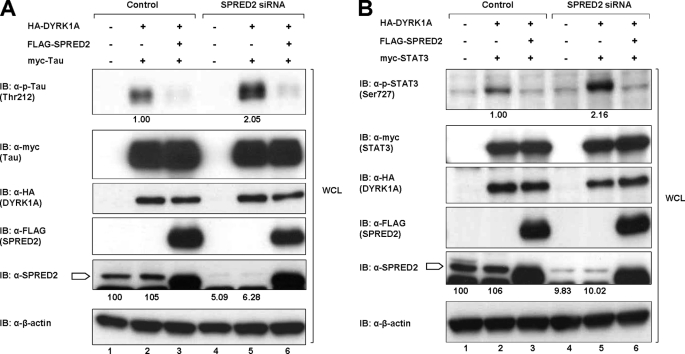
To demonstrate a likely physiological plausibility, we set up experiments to knock down either SPRED1 or SPRED2 in 293 cells and to assess the effect on Tau phosphorylation. A pool of siRNA, as described under “Experimental Procedures,” was employed in each case. Endogenous SPRED2 was decreased to less than 10% of original levels (Fig. 8A, 5th panel, lanes 4 and 5) with a concomitant 100% increase of Tau phosphorylation (Fig. 8A, top panel, lane 5). Similarly, there was a 1.5-fold increase in Tau phosphorylation when SPRED1 was knocked down (supplemental Fig. 6A, top panel, lane 5). In each case, when SPRED was again overexpressed, Tau phosphorylation levels were substantially diminished (Fig. 8A, top panel, lanes 3 and 6, and supplemental Fig. 6A, lanes 3 and 6). Parallel experiments with STAT3 in place of Tau (Fig. 8B and supplemental Fig. 6B) yielded similar results. These data collectively demonstrate that knocking down endogenous SPRED proteins significantly impacts on the degree of DYRK1A-directed phosphorylation of both Tau and STAT3 even when these known substrates were present in a greater concentration.
FIGURE 8.
Knockdown of SPRED2 increases the DYRK1A-dependent phosphorylation of Tau and STAT3. A, 293 cells were transfected with vector control, nonspecific siRNA, or a pool of four SPRED2 siRNAs using Lipofectamine 2000 as described under “Experimental Procedures.” 48 h post-transfection of the siRNA, the cells were transfected with the other cDNA plasmids as indicated. Cells were harvested 24 h after the second transfection. Cell lysates were separated on SDS-PAGE and immunoblotted (IB) with antibodies indicated on the left. B, 293 cells were treated in the same way as described in A, but with the replacement of myc-Tau with myc-STAT3. Cell lysates were subsequently treated similarly as in A. Block arrow indicates SPRED2.
DISCUSSION
In our long term quest to determine a function for the SPRED (and Sprouty) proteins, we have characterized the interaction between the novel kinase DYRK1A and SPRED1 or SPRED2. During the course of our studies, the interaction between Sprouty2 and DYRK1A was described, and where appropriate, we have used this as a comparative control (23).
The SPRED proteins interact directly with DYRK1A at the kinase domain via their conserved CRD. The SPRED-DYRK1A interaction prevents DYRK1A from phosphorylating its substrate proteins, as demonstrated with Tau and STAT3. Presumably, the associated physiological effects that are governed by the activation of these substrates would likewise be affected. We demonstrated that the inhibition of Tau phosphorylation was due to competitive binding between SPRED proteins and Tau or STAT3 for DYRK1A, rather than inhibition of the kinase activity, per se. There is a paucity of data on the binding of substrates to the DYRK family proteins, when compared with other kinases. Certain kinases, such as ERK1/2, have a specialist-binding domain, where all interacting upstream and downstream kinases and phosphatases must first dock before interacting with the catalytic site (64). DYRK kinases are evolutionarily related to ERKs, so they may have evolved a similar binding strategy (24, 65). It is also likely that there may be specialized inhibitory proteins that bind to ERK or DYRK but are not necessarily substrates. Currently, our mass spectrometry analysis results show that SPRED2 is a likely substrate of DYRK1A (supplemental Fig. 7). Work is ongoing in this respect but such information does not significantly impact on the evidence presented above, which centers on the binding parameters between these two proteins.
It is notable that the majority of proteins that interact with SPREDs, and indeed Sprys, do so via the CRD. In particular, several kinases that have been demonstrated to interact with SPREDs and Sprys to date do so at least in part through the CRD (48, 51). Raf1 strongly interacts with SPRED proteins and less so with Spry proteins via their respective CRDs (22).3 Both SPRED and Spry families interact with Tesk1, again via their CRDs, resulting in the inhibition of the kinase (21, 48). Furthermore, Spry2 interacts with and inhibits PKCδ via a complex mechanism and multipoint binding that involves the CRD of that protein (66). It is notable that each interaction results in modifying the downstream effectors of these kinases. Although there is no universal mechanism, there is apparently a degree of specificity and, thus far, a parallel outcome.
The physiological functions of DYRK analogs, and in particular DYRK1A, have not been fully characterized. Several years ago, there was a lack of data on this novel kinase and its apparent mainly nuclear localization (38, 67). It is believed to translocate out of the nucleus because a number of its substrates have a cytosolic localization. Both SPRED proteins and DYRK1A are strongly expressed in the brain and notably in similar regions (11, 14, 67), adding credibility to the hypothesis that SPRED proteins are modifiers of DYRK1A function. Within individual cells, DYRK1A is mainly located in the nucleus, whereas SPRED proteins are located in the cytosol or on the plasma membrane (38, 67). A number of the substrates for DRYK1A are located in the cytosol, and this is where SPRED proteins likely exert their effect on the kinase.
The essence of a kinase is in the substrates it phosphorylates. Phosphorylated proteins will often initiate or propagate a signal by activating enzymes or initiating the formation of a binding complex that controls specific cellular events. Kinases are subject to various controls, and here we describe a possible control mechanism for DYRK1A by SPRED family proteins.
Supplementary Material
Acknowledgments
We thank Dr. Chow Soah Yee for experimental advice and critical review of the manuscript, Ee Hunck THO and Audrey Qin IYU for technical assistance, and Daniel Yim and Drs. Martina Quintanar-Audelo and Cherlyn Ng for proofreading. We thank Dr. Sze Siu Kwan, Newman from School of Biological Sciences, Nanyang Technological University, Singapore, for helping us with the mass spectrometry analysis.
This work was supported by Agency of Science and Technology Research, A*Star, Singapore.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–7.
D. Li, R. A. Jackson, P. Yusoff, and G. R. Guy, unpublished data.
- RTK
- receptor tyrosine kinase
- WCL
- whole cell lysates
- CRD
- cysteine-rich domain
- NLS
- nuclear localization signal.
REFERENCES
- 1.Schlessinger J. (2000) Cell 103, 211–225 [DOI] [PubMed] [Google Scholar]
- 2.Schlessinger J. (2002) Cell 110, 669–672 [DOI] [PubMed] [Google Scholar]
- 3.Zwick E., Bange J., Ullrich A. (2001) Endocr.-Relat. Cancer 8, 161–173 [DOI] [PubMed] [Google Scholar]
- 4.Shaw R. J., Cantley L. C. (2006) Nature 441, 424–430 [DOI] [PubMed] [Google Scholar]
- 5.Midgley R. S., Kerr D. J. (2002) Crit. Rev. Oncol. Hematol. 44, 109–120 [DOI] [PubMed] [Google Scholar]
- 6.Malumbres M., Barbacid M. (2003) Nat. Rev. Cancer 3, 459–465 [DOI] [PubMed] [Google Scholar]
- 7.Hacohen N., Kramer S., Sutherland D., Hiromi Y., Krasnow M. A. (1998) Cell 92, 253–263 [DOI] [PubMed] [Google Scholar]
- 8.Casci T., Vinós J., Freeman M. (1999) Cell 96, 655–665 [DOI] [PubMed] [Google Scholar]
- 9.Tefft J. D., Lee M., Smith S., Leinwand M., Zhao J., Bringas P., Jr., Crowe D. L., Warburton D. (1999) Curr. Biol. 9, 219–222 [DOI] [PubMed] [Google Scholar]
- 10.de Maximy A. A., Nakatake Y., Moncada S., Itoh N., Thiery J. P., Bellusci S. (1999) Mech. Dev. 81, 213–216 [DOI] [PubMed] [Google Scholar]
- 11.Bundschu K., Walter U., Schuh K. (2007) BioEssays 29, 897–907 [DOI] [PubMed] [Google Scholar]
- 12.Kato R., Nonami A., Taketomi T., Wakioka T., Kuroiwa A., Matsuda Y., Yoshimura A. (2003) Biochem. Biophys. Res. Commun. 302, 767–772 [DOI] [PubMed] [Google Scholar]
- 13.Wakioka T., Sasaki A., Kato R., Shouda T., Matsumoto A., Miyoshi K., Tsuneoka M., Komiya S., Baron R., Yoshimura A. (2001) Nature 412, 647–651 [DOI] [PubMed] [Google Scholar]
- 14.Engelhardt C. M., Bundschu K., Messerschmitt M., Renné T., Walter U., Reinhard M., Schuh K. (2004) Histochem. Cell Biol. 122, 527–538 [DOI] [PubMed] [Google Scholar]
- 15.Edwin F., Anderson K., Ying C., Patel T. B. (2009) Mol. Pharmacol. 76, 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guy G. R., Jackson R. A., Yusoff P., Chow S. Y. (2009) J. Endocrinol. 203, 191–202 [DOI] [PubMed] [Google Scholar]
- 17.Cabrita M. A., Christofori G. (2008) Angiogenesis 11, 53–62 [DOI] [PubMed] [Google Scholar]
- 18.Mason J. M., Morrison D. J., Basson M. A., Licht J. D. (2006) Trends Cell Biol. 16, 45–54 [DOI] [PubMed] [Google Scholar]
- 19.Kim H. J., Bar-Sagi D. (2004) Nat. Rev. Mol. Cell Biol. 5, 441–450 [DOI] [PubMed] [Google Scholar]
- 20.Guy G. R., Wong E. S., Yusoff P., Chandramouli S., Lo T. L., Lim J., Fong C. W. (2003) J. Cell Sci. 116, 3061–3068 [DOI] [PubMed] [Google Scholar]
- 21.Tsumura Y., Toshima J., Leeksma O. C., Ohashi K., Mizuno K. (2005) Biochem. J. 387, 627–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasaki A., Taketomi T., Kato R., Saeki K., Nonami A., Sasaki M., Kuriyama M., Saito N., Shibuya M., Yoshimura A. (2003) Nat. Cell Biol. 5, 427–432 [DOI] [PubMed] [Google Scholar]
- 23.Aranda S., Alvarez M., Turró S., Laguna A., de la Luna S. (2008) Mol. Cell. Biol. 28, 5899–5911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Himpel S., Tegge W., Frank R., Leder S., Joost H. G., Becker W. (2000) J. Biol. Chem. 275, 2431–2438 [DOI] [PubMed] [Google Scholar]
- 25.Kentrup H., Becker W., Heukelbach J., Wilmes A., Schürmann A., Huppertz C., Kainulainen H., Joost H. G. (1996) J. Biol. Chem. 271, 3488–3495 [DOI] [PubMed] [Google Scholar]
- 26.Himpel S., Panzer P., Eirmbter K., Czajkowska H., Sayed M., Packman L. C., Blundell T., Kentrup H., Grötzinger J., Joost H. G., Becker W. (2001) Biochem. J. 359, 497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lochhead P. A., Sibbet G., Morrice N., Cleghon V. (2005) Cell 121, 925–936 [DOI] [PubMed] [Google Scholar]
- 28.Lochhead P. A., Sibbet G., Kinstrie R., Cleghon T., Rylatt M., Morrison D. K., Cleghon V. (2003) Biochem. J. 374, 381–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiechmann S., Czajkowska H., de Graaf K., Grötzinger J., Joost H. G., Becker W. (2003) Biochem. Biophys. Res. Commun. 302, 403–408 [DOI] [PubMed] [Google Scholar]
- 30.Becker W., Joost H. G. (1999) Prog. Nucleic Acids Res. Mol. Biol. 62, 1–17 [DOI] [PubMed] [Google Scholar]
- 31.Becker W., Weber Y., Wetzel K., Eirmbter K., Tejedor F. J., Joost H. G. (1998) J. Biol. Chem. 273, 25893–25902 [DOI] [PubMed] [Google Scholar]
- 32.Tatebe H., Nakano K., Maximo R., Shiozaki K. (2008) Curr. Biol. 18, 322–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guimerá J., Casas C., Pucharcòs C., Solans A., Domènech A., Planas A. M., Ashley J., Lovett M., Estivill X., Pritchard M. A. (1996) Hum. Mol. Genet. 5, 1305–1310 [DOI] [PubMed] [Google Scholar]
- 34.Raich W. B., Moorman C., Lacefield C. O., Lehrer J., Bartsch D., Plasterk R. H., Kandel E. R., Hobert O. (2003) Genetics 163, 571–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pang K. M., Ishidate T., Nakamura K., Shirayama M., Trzepacz C., Schubert C. M., Priess J. R., Mello C. C. (2004) Dev. Biol. 265, 127–139 [DOI] [PubMed] [Google Scholar]
- 36.Guimera J., Casas C., Estivill X., Pritchard M. (1999) Genomics 57, 407–418 [DOI] [PubMed] [Google Scholar]
- 37.Altafaj X., Dierssen M., Baamonde C., Martí E., Visa J., Guimerà J., Oset M., González J. R., Flórez J., Fillat C., Estivill X. (2001) Hum. Mol. Genet. 10, 1915–1923 [DOI] [PubMed] [Google Scholar]
- 38.Alvarez M., Estivill X., de la Luna S. (2003) J. Cell Sci. 116, 3099–3107 [DOI] [PubMed] [Google Scholar]
- 39.Matsuo R., Ochiai W., Nakashima K., Taga T. (2001) J. Immunol. Methods 247, 141–151 [DOI] [PubMed] [Google Scholar]
- 40.Woods Y. L., Rena G., Morrice N., Barthel A., Becker W., Guo S., Unterman T. G., Cohen P. (2001) Biochem. J. 355, 597–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mao J., Maye P., Kogerman P., Tejedor F. J., Toftgard R., Xie W., Wu G., Wu D. (2002) J. Biol. Chem. 277, 35156–35161 [DOI] [PubMed] [Google Scholar]
- 42.Woods Y. L., Cohen P., Becker W., Jakes R., Goedert M., Wang X., Proud C. G. (2001) Biochem. J. 355, 609–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryoo S. R., Jeong H. K., Radnaabazar C., Yoo J. J., Cho H. J., Lee H. W., Kim I. S., Cheon Y. H., Ahn Y. S., Chung S. H., Song W. J. (2007) J. Biol. Chem. 282, 34850–34857 [DOI] [PubMed] [Google Scholar]
- 44.Yang E. J., Ahn Y. S., Chung K. C. (2001) J. Biol. Chem. 276, 39819–39824 [DOI] [PubMed] [Google Scholar]
- 45.Park J., Yang E. J., Yoon J. H., Chung K. C. (2007) Mol. Cell. Neurosci. 36, 270–279 [DOI] [PubMed] [Google Scholar]
- 46.Huang Y., Chen-Hwang M. C., Dolios G., Murakami N., Padovan J. C., Wang R., Hwang Y. W. (2004) Biochemistry 43, 10173–10185 [DOI] [PubMed] [Google Scholar]
- 47.Lao D. H., Chandramouli S., Yusoff P., Fong C. W., Saw T. Y., Tai L. P., Yu C. Y., Leong H. F., Guy G. R. (2006) J. Biol. Chem. 281, 29993–30000 [DOI] [PubMed] [Google Scholar]
- 48.Chandramouli S., Yu C. Y., Yusoff P., Lao D. H., Leong H. F., Mizuno K., Guy G. R. (2008) J. Biol. Chem. 283, 1679–1691 [DOI] [PubMed] [Google Scholar]
- 49.Yusoff P., Lao D. H., Ong S. H., Wong E. S., Lim J., Lo T. L., Leong H. F., Fong C. W., Guy G. R. (2002) J. Biol. Chem. 277, 3195–3201 [DOI] [PubMed] [Google Scholar]
- 50.Nonami A., Taketomi T., Kimura A., Saeki K., Takaki H., Sanada T., Taniguchi K., Harada M., Kato R., Yoshimura A. (2005) Genes Cells 10, 887–895 [DOI] [PubMed] [Google Scholar]
- 51.Johne C., Matenia D., Li X. Y., Timm T., Balusamy K., Mandelkow E. M. (2008) Mol. Biol. Cell 19, 1391–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mardakheh F. K., Yekezare M., Machesky L. M., Heath J. K. (2009) J. Cell Biol. 187, 265–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maenz B., Hekerman P., Vela E. M., Galceran J., Becker W. (2008) BMC Mol. Biol. 9, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lakin N. D., Jackson S. P. (1999) Oncogene 18, 7644–7655 [DOI] [PubMed] [Google Scholar]
- 55.Shen Y., White E. (2001) Adv. Cancer Res. 82, 55–84 [DOI] [PubMed] [Google Scholar]
- 56.Yamaguchi H., Woods N. T., Piluso L. G., Lee H. H., Chen J., Bhalla K. N., Monteiro A., Liu X., Hung M. C., Wang H. G. (2009) J. Biol. Chem. 284, 11171–11183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Juan L. J., Shia W. J., Chen M. H., Yang W. M., Seto E., Lin Y. S., Wu C. W. (2000) J. Biol. Chem. 275, 20436–20443 [DOI] [PubMed] [Google Scholar]
- 58.Kirsch D. G., Kastan M. B. (1998) J. Clin. Oncol. 16, 3158–3168 [DOI] [PubMed] [Google Scholar]
- 59.Guo X., Williams J. G., Schug T. T., Li X. (2010) J. Biol. Chem. 285, 13223–13232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seifert A., Allan L. A., Clarke P. R. (2008) FEBS J. 275, 6268–6280 [DOI] [PubMed] [Google Scholar]
- 61.Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S., Alessi D. R., Cohen P. (2007) Biochem. J. 408, 297–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lao D. H., Yusoff P., Chandramouli S., Philp R. J., Fong C. W., Jackson R. A., Saw T. Y., Yu C. Y., Guy G. R. (2007) J. Biol. Chem. 282, 9117–9126 [DOI] [PubMed] [Google Scholar]
- 63.DaSilva J., Xu L., Kim H. J., Miller W. T., Bar-Sagi D. (2006) Mol. Cell. Biol. 26, 1898–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Biondi R. M., Nebreda A. R. (2003) Biochem. J. 372, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miyata Y., Nishida E. (1999) Biochem. Biophys. Res. Commun. 266, 291–295 [DOI] [PubMed] [Google Scholar]
- 66.Chow S. Y., Yu C. Y., Guy G. R. (2009) J. Biol. Chem. 284, 19623–19636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martí E., Altafaj X., Dierssen M., de la Luna S., Fotaki V., Alvarez M., Pérez-Riba M., Ferrer I., Estivill X. (2003) Brain Res. 964, 250–263 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.