Abstract
VPS4 proteins are AAA+ ATPases required to form multivesicular bodies, release viral particles, and complete cytokinesis. They act by disassembling ESCRT-III heteropolymers during or after their proposed function in membrane scission. Here we show that purified human VPS4A is essentially inactive but can be stimulated to hydrolyze ATP by ESCRT-III proteins in a reaction that requires both their previously defined MIT interacting motifs and ∼50 amino acids of the adjacent sequence. Importantly, C-terminal fragments of all ESCRT-III proteins tested, including CHMP2A, CHMP1B, CHMP3, CHMP4A, CHMP6, and CHMP5, activated VPS4A suggesting that it disassembles ESCRT-III heteropolymers by affecting each component protein. VPS4A is thought to act as a ring-shaped cylindrical oligomer like other AAA+ ATPases, but this has been difficult to directly demonstrate. We found that concentrating His6-VPS4A on liposomes containing Ni2+-nitrilotriacetic acid-tagged lipid increased ATP hydrolysis, confirming the importance of inter-subunit interactions for activity. We also found that mutating pore loops expected to line the center of a cylindrical oligomer changed the response of VPS4A to ESCRT-III proteins. Based on these data, we propose that ESCRT-III proteins facilitate assembly of functional but transient VPS4A oligomers and interact with sequences inside the pore of the assembled enzyme. Deleting the N-terminal MIT domain and adjacent linker from VPS4A increased both basal and liposome-enhanced ATPase activity, indicating that these elements play a role in autoinhibiting VPS4A until it encounters ESCRT-III proteins. These findings reveal new ways in which VPS4 activity is regulated and specifically directed to ESCRT-III polymers.
Keywords: ATPases, Endosomes, Enzyme Mechanisms, Membrane Trafficking, Protein-Protein Interactions, AAA+ ATPases, ESCRT Machinery, Multivesicular Body
Introduction
Multivesicular bodies (MVBs)2 are endosomes that contain intralumenal vesicles generated by invagination from the endosomal membrane. The MVB is an intermediate compartment en route to the lysosome; surface receptors destined for degradation are sorted into intralumenal vesicles and delivered to the lysosome. Impairment in this pathway leads to many cellular problems, including prolonged signaling of receptors normally silenced by internalization into the MVB (reviewed in Refs. 1–5). Four ESCRT (endosomal sorting complexes required for transport) complexes (ESCRT-0, -I, -II, and -III) are responsible for cargo sorting and vesicle formation in the MVB pathway. The ESCRT complexes also play essential roles in topologically similar membrane remodeling events including enveloped virus budding and cell abscission at the end of cytokinesis (6–8).
ESCRT-III differs from the other ESCRT complexes in that it is a loosely defined polymer that assembles when its subunits are recruited to the appropriate membrane and bind to each other (9). There are seven ESCRT-III-related proteins in yeast and 12 in human cells. Human ESCRT-III proteins are referred to either as orthologues of their yeast counterparts or as CHMPs (charged multivesicular body proteins). To standardize our discussion, we will primarily use the CHMP nomenclature in this article. All of the ESCRT-III proteins are thought to share a common core tertiary structure consisting of six α-helices (α1–α6) arranged around a 7-nm helical hairpin formed by α1 and α2 (10–12). Despite this similarity, the proteins are not functionally redundant and operate in a defined order (13–15).
Current thinking is that ESCRT-III helps to create and ultimately sever the neck that connects vesicles or other structures budding away from the cytosol to their originating membrane. ESCRT-III polymers are filaments with the potential to induce or stabilize membrane curvature needed for processes in which the ESCRT machinery participates (16, 17). Recent reconstitution studies show that ESCRT-III alone can drive formation and release of lumenal vesicles into giant unilamellar liposomes (14, 18). The only protein absolutely required for this reaction is Snf7p (CHMP4 A, B, and C in humans), which in yeast is also the most abundant ESCRT-III protein (13). Various models inspired by electron microscopic images of ESCRT-III assemblies have emerged to explain how ESCRT-III polymers may help generate vesicles but clearly much has yet to be learned (14, 16, 19, 20).
Normal function of ESCRT-III both in vivo and in vitro depends on the continued availability of individual ESCRT-III proteins to incorporate into growing polymers. Enzymes that belong to the AAA+ (ATPases associated with a variety of cellular activities) family of VPS4 proteins maintain a pool of free proteins by disassembling ESCRT-III polymers (21). Removing or inhibiting VPS4 proteins in vivo blocks ESCRT-dependent processes (22, 23) and all of the ESCRTs accumulate in large membrane-associated complexes (9, 21). VPS4 is highly conserved throughout evolution, and is encoded by one gene (vps4) in the yeast Saccharomyces cerevisiae and two genes (VPS4A and VPS4B) in humans. Each of these enzymes consists of an amino-terminal MIT (microtubule interacting and transport) domain connected by a linker to a canonical AAA+ domain with a unique β domain and C-terminal helix, all of which have been visualized by x-ray crystallography (24–28). Cryo-EM studies of yeast Vps4p in its ATP-bound state indicate that the yeast enzyme forms a two-tiered hexameric ring similar to other AAA+ proteins (28–30), although comparable structures have not been observed using mammalian VPS4A or VPS4B. The MIT domains of VPS4 proteins bind to sequence elements known as MIT interacting motifs (MIMs) at or near the C termini of ESCRT-III proteins (31–33). Interactions between these motifs provide a link between VPS4 proteins and the ESCRT-III machinery and are required for normal cargo sorting in yeast (31, 33, 34). However, how these interactions lead to ESCRT-III polymer disassembly is unknown. Furthermore, how VPS4 activity is controlled and coordinated with the presence of ESCRT-III polymers remains unclear.
Taking advantage of the fact that mammalian VPS4 enzymes have extremely low basal ATPase activity and do not readily assemble into detectable oligomeric structures, we find that C-terminal fragments of all ESCRT-III proteins potently stimulate the ATPase activity of the enzyme when MIM motifs and an additional ∼50 amino acids of primarily acidic sequence are present. This stimulation relieves intrinsic autoinhibition mediated by N-terminal regulatory elements in VPS4A, allowing the enzyme to assume its active oligomeric state. These data show that human VPS4A is regulated at least in part by exposure to its ESCRT-III complex substrate.
EXPERIMENTAL PROCEDURES
Materials
Homopolymers of poly-l-lysine (15–30 kDa), poly-l-glutamic acid (20–40 kDa), poly-l-threonine (5–15 kDa), and poly-d-glutamic acid (15–50 kDa) were obtained from Sigma, resuspended in water, and dialyzed against buffer (20 mm Tris, pH 7.6, 100 mm NaCl) prior to use. MIM1 (ADADLEERLKNLRRD) and control (ADADLEEEAKNLRRD) peptides were purchased from GenScript (Piscataway, NJ) and confirmed by mass spectrometry. Peptides were resuspended in reaction buffer before use. Dioleoyl-phosphatidylcholine (DOPC) and nickel-1,2-dioleoyl-sn-glycero-3-([N-(5-amino-1-carboxypentyl)iminodiacetic acid]succinyl)-nickel salt (Ni2+-NTA-DOGS) were purchased from Avanti Polar Lipids (Alabaster, AL). All other chemicals were from Sigma.
Plasmids and Mutagenesis
Plasmids encoding His6-NSF (35), His6-αSNAP (36), His6-p97 (37), full-length and truncated FLAG-CHMP2A (38), and GST-CHMP fragments (39) have been previously described. GFP-VPS4A was a kind gift from Wes Sundquist (University of Utah), pGEX-Vps4p from Markus Babst (University of Utah), and pGEX-spastin and pGEX-α tubulin from Brett Lauring (Columbia University).
To generate His6-VPS4A we amplified DNA encoding VPS4A and cloned it into pET28a (Novagen, Madison WI) between BamHI and XhoI sites. Similarly, His6-VPS4B and His6Vps4p have the encoding DNA between these sites in pET28a. Point mutations were introduced as indicated by QuikChange site-directed mutagenesis (Stratagene, La Jolla CA). Truncated VPS4A proteins were generated by PCR amplification followed again by cloning between BamHI and XhoI sites in pET28a. His6-spastin was created by introducing DNA encoding the short isoform of human spastin as a BamHI-EcoRI fragment into pET28a. ESCRT-III protein fragments and point mutants were similarly generated by PCR-mediated amplification of the indicated coding sequence followed by subcloning into pET28a (for His6-tagged proteins) or pGEX4T-1 (for GST-tagged proteins). All were amplified from plasmids encoding full-length ESCRT-III proteins as described (38, 39).
To swap MIM1 sequences between the C-terminal fragments of CHMP1B and CHMP3, DNA encoding CHMP3(115–210) or CHMP1B(106–186) was blunt end ligated to DNA encoding the MIM from the other protein (CHMP1B(187–199) and CHMP3(211–222)). Chimeras were again cloned into pET28a between BamHI and XhoI sites. Syntaxin-MIM1 and tubulin-MIM1 fusions were prepared by introducing a SpeI restriction site into rat syntaxin1A and murine α2-tubulin to allow excision of fragments encoding syntaxin residues 159–242 and tubulin residues 348–453. These were fused with intervening residues TS to CHMP1B MIM1 residues 187–199. DNA sequence of the coding region of all proteins and mutants was verified by sequencing at the Nucleic Acid Chemistry Laboratory (Washington University).
Protein Expression and Purification
Freshly transformed BL21(DE3) Escherichia coli grown in 0.5–1 liter of Terrific Broth were induced to express the indicated protein by adding 0.4 μm isopropyl 1-thio-β-d-galactopyranoside and shaking for 3 h at room temperature. To purify His6-VPS4 proteins, bacterial pellets were lysed in 20 mm HEPES, pH 7.6, 100 mm NaCl, 10% glycerol, 1 mm DTT, 1 mm ATP, 2 mm MgCl2, 1 mm PMSF by incubation with 2.5 mg of lysozyme followed by sonication. Triton X-100 was added to 0.25%, samples were incubated for 15 min, and then clarified by centrifugation at 12,000 × g for 20 min. Ni2+-NTA-agarose (Qiagen, Valencia, CA) was added together with 5 mm imidazole and incubated for 1 h at 4 °C. Unbound material was removed by washing in lysis buffer supplemented with 1 m NaCl, after which resin was incubated in lysis buffer for 20 min at room temperature to release contaminating bacterial ATPases. Proteins were eluted from washed resin by sequential addition of 50, 200, and 400 mm imidazole in a buffer appropriate for the next purification step. VPS4A and VPS4B were further purified on a MonoS column (GE Healthcare) in 50 mm PIPES, pH 6.9, 10% glycerol, 1 mm DTT, and 0.1–1 m NaCl. Yeast Vps4p protein was further purified on MonoQ (GE Healthcare) in 20 mm Tris, pH 8.0, 1 mm DTT, 10% glycerol, and 0.1–1 m NaCl. Pooled fractions containing purified enzyme were concentrated if necessary to create stocks of ∼1 mg/ml, quantitated using Bradford reagent (Bio-Rad) with BSA as a standard, aliquoted, and snap frozen for storage. Size exclusion chromatography to monitor oligomer size was performed on Superdex 75 or 200 10/30 columns (GE Healthcare) in 50 mm HEPES, pH 7.6, 150 mm KCl, 5% glycerol, 0.5 mm ATP, 3 mm MgCl2, and 1 mm DTT. His6-NSF and His6-p97 were prepared essentially as previously described (37). His6-spastin was grown and purified as above for VPS4A, except that protein was expressed in BL21-CodonPlus(DE3)-RIL E. coli (Stratagene, La Jolla, CA) and fractions containing His6-spastin after elution from the MonoS column were further purified using a Superdex 75 size exclusion column.
ESCRT-III proteins and fragments were prepared using the same general protocol, with variations after elution from Ni2+-NTA-agarose or glutathione-Sepharose (Amersham Biosciences) depending on the protein or protein fragment in question. C-terminal ESCRT-III fragments were further purified on a MonoQ column. Size exclusion chromatography for preparative and analytic purposes used Superdex 200 10/30 or Superdex 75 10/30 columns in 20 mm HEPES, pH 7.4, 150 mm NaCl, 1 mm DTT, and 5% glycerol. All proteins used in ATPase assays were tested for ATPase activity and shown to be free of contaminating ATPases.
ATPase Assay
ATPase assays measured released phosphate based on the formation of colored phosphomolybdate complexes with the cationic dye malachite green (40). Reactions were set up by aliquoting 25 μl containing appropriately diluted ESCRT-III protein, liposome, or amino acid homopolymer in 20 mm Tris, pH 7.6, 100 mm KOAc into wells of a 96-well plate. VPS4A (or other enzyme as indicated) was diluted into 20 mm Tris, pH 7.6, 100 mm KOAc, 10 mm MgCl2, and 2 mm ATP. 25 μl of this solution was added to each well to start the ATPase reactions. Enzyme concentration in standard reactions was 0.2 μm except in the liposome experiments where the final concentration was 0.1 μm. Reactions were incubated for 5 or 10 min at 37 °C, and stopped by adding 100 μl of malachite green color reagent (14 mm ammonium molybdate, 1.3 m HCl, 0.1% Triton X-100, and 1.5 mm malachite green) followed by 50 μl of 21% citric acid. ATP hydrolysis in these reactions was linear with time as long as ATP consumption was limited to 5% or less, a condition that was met in all reported assays. In a few cases (indicated), reactions were performed at 23.5 °C to slow ATP hydrolysis. Within 30 min of adding the malachite green reagent, absorbance at 660 nm was measured using a Synergy HT plate reader (Bio-Tek, Winooski, VT). Sodium phosphate standards were included on each ATPase reaction plate to quantitate the released phosphate. Data were fit to the Hill equation (Y = (Ymax × Xh)/[Km(app) + Xh])) using Prism4 (GraphPad, San Diego CA). Statistical comparisons were done using t tests performed in Prism4.
Liposomes
Liposomes were prepared and used essentially as described (41, 42). Briefly, Ni2+-NTA-DOGS and DOPC in chloroform were mixed at the desired ratio in 1 mg of total lipid and dried under nitrogen. Lipids were then lyophilized and stored at −20 °C under nitrogen. For use, lipid mixtures were resuspended in 1 ml of diethyl ether and then mixed with 1 ml of buffer (20 mm Tris, pH 7.6, 100 mm KOAc). The mixture was emulsified by sonicating (80 kHz and 80 W power in a water bath, Laboratory Supplies Co., Hicksville, NY) and placed in a rotary evaporator to remove ether. The resulting multilamellar liposomes were passed through 200-nm polycarbonate membranes in a mini-extruder (Avanti Polar Lipids, Alabaster, AL) to generate small unilamellar liposomes ∼120 nm in diameter. Phospholipids were quantitated as described (43). Liposome preparations typically contained 0.8–0.9 mg/ml of phospholipids. Extruded liposomes were stored at 4 °C prior to use. Surface area calculations used to estimate the effective protein concentration on the liposome surface were as described (41, 42).
RESULTS
Mammalian VPS4 Enzymes Have Low Basal ATPase Activity
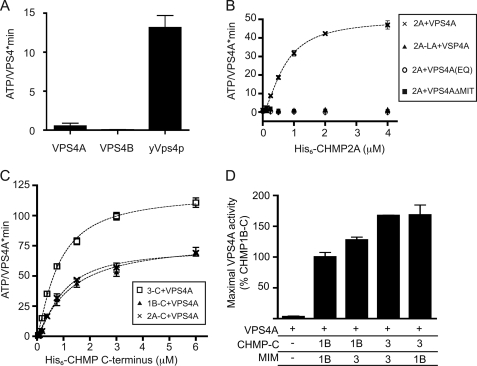
To study the regulation of mammalian VPS4 enzymes, we set out to characterize purified VPS4A and VPS4B/SKD1. His6-VPS4A had low but reproducible basal ATPase activity, whereas His6-VPS4B had no detectable activity (Fig. 1A). Yeast His6-Vps4p assayed under identical conditions hydrolyzed ATP at a rate of 13 ATP/Vps4·min, similar to what has previously been reported (21, 44). The low hydrolytic activity of mammalian VPS4 enzymes relative to yeast Vps4p and other AAA+ ATPases prompted us to explore how they might be activated by interaction with other factors.
FIGURE 1.
Effect of ESCRT-III proteins that contain MIM1 motifs on VPS4A activity. A, ATP hydrolysis by human VPS4A, VPS4B, and yeast Vps4p assessed under standard reaction conditions of 0.2 μm enzyme and 1 mm ATP at 37 °C. B, effect of CHMP2A on ATP hydrolysis by VPS4A. ×, wild-type VPS4A with varying concentration of wild-type CHMP2A (Vmax 50 ATP/VPS4A·min); triangles, wild-type VPS4A and CHMP2A L216A/L219A; circles, VPS4A E228Q mutant with wild-type CHMP2A; boxes, VPS4AΔMIT mutant with CHMP2A. C, effect of C-terminal CHMP fragments on ATP hydrolysis by VPS4A. Boxes, CHMP3-C (amino acids 115–222), Vmax of 118 ATP/VPS4A·min; ×, CHMP2A-C (amino acids 117–222), Vmax of 72 ATP/VPS4A·min; triangles, CHMP1B-C (amino acids 106–199) with Vmax of 78 ATP/VPS4A·min. D, effect of chimeric CHMP-MIM proteins on ATP hydrolysis by VPS4A. Shown is the activity of VPS4A in response to 6 μm of the indicated protein, scaled to the activity reached with CHMP1B-C (100%). The activity promoted by CHMP1B-C differs compared with CHMP1B-3 (p = 0.046), as does the activity promoted by CHMP3-C versus CHMP1B-3 (p = 0.0067); the activity promoted by CHMP3-C is not significantly different from that of CHMP3–1B. Error bars indicate S.D. from one experiment conducted in duplicate or triplicate; all experiments were repeated three times.
MIM1-containing ESCRT-III Proteins Activate VPS4A
A characteristic of many AAA+ ATPases is that their ATPase activity increases when they engage substrate proteins (45). We therefore examined the effects of ESCRT-III proteins on VPS4 activity, focusing our studies on the widely expressed VPS4A isoform. We first tested CHMP2A, one of two human Vps2 homologues, because Vps2 plays an important role in enabling VPS4 function both in vivo (13) and in vitro (14). It also contains a well studied MIM1 motif at its C terminus (32, 33). Strikingly, full-length CHMP2A stimulated ATP hydrolysis by 200 nm VPS4A in a concentration-dependent manner to an apparent Vmax of 50 ATP/VPS4A·min (Fig. 1B, ×). CHMP2A did not stimulate VPS4A when the enzyme contained an E228Q mutation in its Walker B motif (Fig. 1B, circles) or was missing its MIT domain (Fig. 1B, squares). Moreover, VPS4A did not respond to CHMP2A with a binding-defective L216A/L219A mutation in its MIM1 motif (Fig. 1B, triangles). These results show that known interacting elements, MIT and MIM1, bring VPS4A and CHMP2A together to promote ATP hydrolysis.
We next examined the effect of other ESCRT-III proteins that contain recognizable MIM1 motifs, CHMP1B and CHMP3 (32, 33). These proteins stimulated VPS4A ATPase activity to anywhere between 2 and >30 ATP/VPS4A·min, but we found that the results varied from experiment to experiment and were not reproducible (data not shown). We therefore wondered if variations in conformational state, i.e. how “open” or “closed” the proteins are, might account for these differences (10, 38, 46). CHMP3 in particular has been shown to adopt functionally distinct open and closed states that could differ in whether the surface of the MIM1 motif that binds VPS4A is exposed (12, 47).
To exclude problems with conformational variability, we turned to studying fragments of these ESCRT-III proteins lacking their two most N-terminal helices (α1 and α2) and thus unable to adopt the closed conformation that depends on interactions between N- and C-terminal domains (38, 46, 48). We purified His6-tagged C-terminal fragments of CHMP2A, CHMP1B, and CHMP3 and confirmed that they did not assemble into polymers by size exclusion chromatography on a Superdex 75 column (data not shown). When added to VPS4A, each fragment stimulated ATP hydrolysis in a concentration-dependent and reproducible manner (Fig. 1C). Notably, the CHMP3 fragment stimulated VPS4A to a higher apparent Vmax than did the CHMP2A or CHMP1B fragments. We wondered whether the MIM1 or other sequences in CHMP3 were responsible for its effect on VPS4A. Prior studies suggested that the CHMP3 MIM1 has a lower affinity for the VPS4 MIT domain than do those of CHMP2A and CHMP1B (32, 33). Swapping MIM1 motifs between CHMP1B and CHMP3 fragments to create CHMP3-1B and CHMP1B-3 chimeras showed that both the MIM1 motif and sequences elsewhere in the fragments are involved in activating VPS4A (Fig. 1D).
C-terminal Fragments of All Subclasses of ESCRT-III Proteins Activate VPS4A
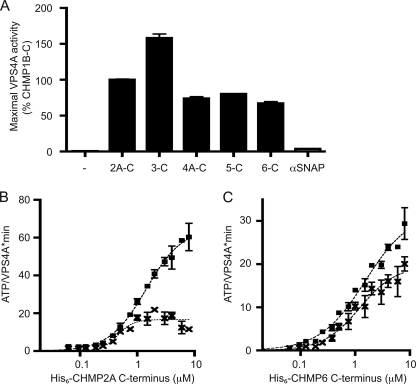
An important question for understanding the dynamics of ESCRT-III polymers is whether VPS4 proteins act directly on each component or instead act only on a subset of the components to disassemble the entire polymer. This is a particularly significant issue with regard to the major Snf7/CHMP4 components of ESCRT-III because of the low affinity of the interaction between the Snf7 MIM2 and the VPS4 MIT domains (13, 31). To compare the ability of different ESCRT-III proteins to activate VPS4A, we prepared C-terminal fragments representing each of the six major groups of ESCRT-III proteins. Surprisingly, all stimulated VPS4A ATPase activity, whereas the unrelated protein α-SNAP did not (Fig. 2A). Given the known role of MIM motifs in binding to VPS4 proteins, we also explored the effects of mutating critical residues in these motifs (31, 32). A MIM1 L216A/L219A mutation in CHMP2A-C decreased VPS4A activation (Fig. 2B). Mutating the MIM2 in CHMP6-C with a L170D,V173D replacement also reduced stimulation of ATPase activity (Fig. 2C). Interestingly, neither MIM mutation completely eliminated VPS4A stimulation, again pointing to a role for elements outside of the MIMs in activating VPS4A. CHMP5 does not contain a canonical MIM motif, but as will be discussed elsewhere has an internal sequence similar to MIM1 that may be responsible for stimulating VPS4A.3 Overall, the fact that C-terminal fragments from all subfamilies of ESCRT-III proteins promote increased ATP hydrolysis suggests that VPS4A may act independently on each subunit of ESCRT-III.
FIGURE 2.
All six subclasses of ESCRT-III proteins stimulate VPS4A ATPase activity. A, maximal ATP hydrolysis by VPS4A in the presence of C-terminal fragments of CHMP1B(106–199), CHMP2A(117–222), CHMP3(115–222), CHMP4A(119–222), CHMP5(121–219), CHMP6(118–201), or α-SNAP. Assays were conducted as described in the legend to Fig. 1C; maximal activity for each protein was scaled to a CHMP1B-C control present on each assay plate. B, effect of mutating the MIM1 motif in CHMP2A-C. Squares, ATP hydrolysis by VPS4A in the presence of wild-type CHMP2A-C. ×, ATP hydrolysis by VPS4A in the presence of MIM1 mutant CHMP2A-L216A/L219A. C, effect of mutating the MIM2 motif in CHMP6-C. Squares, ATP hydrolysis by VPS4A in the presence of wild-type CHMP6-C. ×, ATP hydrolysis by VPS4A in the presence of MIM2 mutant CHMP6-L170D,V173D. Error bars indicate S.D. from duplicate reactions of one experiment; all experiments were repeated three times.
Both MIMs and Adjacent ESCRT-III Sequences Play a Role in Activating VPS4A
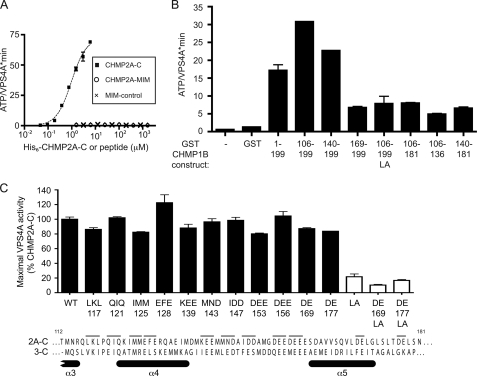
To define the sequence(s) in ESCRT-III proteins that stimulate VPS4A, we set out to find the minimal activating fragment in one of these proteins. Given that MIM motifs bind VPS4 proteins (31–33) and are needed to recruit it to endosomes in vivo (34), we began by asking whether these might also be sufficient to stimulate ATPase activity. We synthesized a 15-amino acid peptide corresponding to a canonical MIM1. Notably, adding even extremely high concentrations of this peptide or a mutated variant to VPS4A had no effect on ATPase activity (Fig. 3A). To define additional required elements, we prepared CHMP1B fragments of increasing length and tested their effect on VPS4A (Fig. 3B). A 30-amino acid fragment containing the C-terminal MIM1 (GST-CHMP1B(169–199)) increased ATPase activity, and this stimulation was further enhanced by progressive inclusion of N-terminal sequences in longer fragments. Fragments lacking a functional C-terminal MIM1 (e.g. GST-CHMP1B(106–181), (106–136), (140–181), and (106–199LA)) also had some effect on the ATPase activity of VPS4A. Taken together, these results again suggest that elements both within and outside of the MIM motif affect VPS4A activity.
FIGURE 3.
Both MIM1 and adjacent sequences are required to activate VPS4A. A, effect of a peptide encompassing the CHMP2A MIM1 (residues 208–222, ADADLEERLKNLRRD, circles), a mutant variant (ADADLEEEAKNLRRD, ×), and the CHMP2A-C fragment (residues 117–222, boxes) on ATP hydrolysis by VPS4A. B, effect of CHMP1B fragments on ATP hydrolysis by VPS4A. Activity in the presence of 6 μm of the indicated GST-CHMP1B fragment is shown. The MIM1 motif is present at the extreme C terminus of CHMP1B, and is missing when residues 182–199 are deleted. The LA mutant also contains a L188A/L192A MIM1 mutation. C, effect of point mutations in CHMP2A-C on stimulation of VPS4A. Clusters of two or three residues (thin bars above CHMP2A sequence) were replaced with alanine and tested for their effects on VPS4A. Mutants are named by the residues changed and the numerical position of the first mutated residue. LA mutants (white bars) also contain a L216A/L219A MIM1 mutation. Alignment of CHMP2A-C with CHMP3-C shows the position of α-helices observed in the crystal structure of CHMP3 and predicted in CHMP2A (10). Error bars indicate S.D. from duplicate reactions of one experiment; all experiments were repeated three times.
To determine whether specific sequences help activate VPS4A we replaced groups of three residues with alanine throughout a CHMP2A C-terminal fragment (Fig. 3C). Although there are a few highly conserved residues, the most obvious shared characteristic of the ESCRT-III C-terminal fragments is an abundance of acidic residues, in many cases clustered into patches (10, 49). We therefore focused on changing acidic residues, especially in the region between α4 and α5 because this region in CHMP1B had provided the largest increase in stimulation of VPS4A (Fig. 3B). Surprisingly, none of these alanine replacements reduced the stimulation of CHMP2A-C by VPS4A more than ∼20% (Fig. 3C). To specifically ask about effects of these alanine replacements on MIM1-independent stimulation of VPS4A, we also mutated two pairs in the context of a CHMP2A-C MIM1 mutant (L216A/L219A). As shown with white bars in Fig. 3B, there appeared to be some decrease associated with the loss of negatively charged residues in this context, but these differences were not found to be statistically significant. This suggests that no single short determinant within CHMP2A, at least among those analyzed here, is responsible for activating VPS4A and may explain how all ESCRT-III proteins engage VPS4A, despite their low overall sequence identity.
Role for Acidic Residues in Activating VPS4A
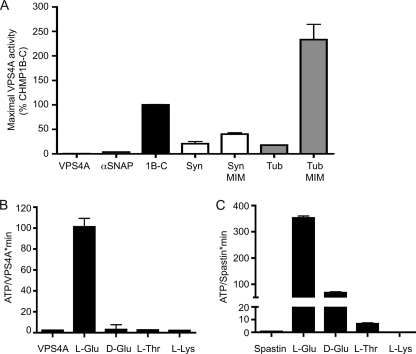
Given the lack of a well defined sequence in the ESCRT-III proteins responsible for stimulating VPS4A, we asked more generally about the specificity with which proteins engage this enzyme. α-SNAP had no effect on VPS4A (Figs. 2A and 4A) despite the fact that it enhanced ATPase activity of NSF, its partner AAA+ protein (data not shown). To further probe requirements for VPS4A activation, we chose two unrelated fragments similar to the stimulatory ESCRT-III fragments in length, a fragment of syntaxin 1A (residues 159–242) and a C-terminal fragment of α-tubulin (residues 348–453). Both are expected to have some α-helical content (50, 51) similar to ESCRT-III proteins (8, 10, 12). On their own, each had a small but statistically significant effect on VPS4A (Fig. 4A). However, with a 15-residue MIM1 motif fused to the C terminus we found that both enhanced ATPase activity (Fig. 4A). In fact, the α-tubulin-MIM1 fusion protein stimulated VPS4A to hydrolyze more ATP than did ESCRT-III proteins. Both α-tubulin and the ESCRT-III C-terminal fragments contain clusters of acidic residues, and a particularly long stretch of acidic residues at the extreme C terminus of α-tubulin is important for its interaction with spastin, a microtubule severing AAA+ ATPase closely related to VPS4 proteins (52). Our results suggest a model in which initial MIT-MIM interactions deliver acidic polypeptides to the AAA+ domain of VPS4A.
FIGURE 4.
Role for acidic residues in VPS4A activation. A, effect of heterologous proteins on ATP hydrolysis by VPS4A. Fragments tested include syntaxin1A (residues 159–242, Syn) and α2 tubulin (residues 348–453, Tub) alone or fused to the MIM1 of CHMP1B (residues 186–199, Syn-MIM and Tub-MIM). Maximal rate of ATP hydrolysis by VPS4A was determined as described in the legend to Fig. 1C and is shown scaled to that of CHMP1B-C included on each plate for standardization. Syntaxin and α2 tubulin fragments on their own stimulate VPS4A activity (p = 0.0032 and 0.007, respectively) to levels similar to that of CHMP1B missing its MIM1 motif (Fig. 3). Adding the MIM1 motif greatly increases the effect of these fragments on VPS4A, again similar to what is seen for CHMP1B. B, effect of amino acid homopolymers including poly-l-glutamic acid (20–40 kDa), poly-d-glutamic acid (20–40 kDa), poly-l-threonine (8 kDa), and poly-l-lysine (15–30 kDa) on ATP hydrolysis by VPS4A. C, effect of amino acid homopolymers on ATP hydrolysis by spastin. Error bars indicate S.D. from duplicate reactions of one experiment; all experiments were repeated three times.
Further support for the idea that acidic residues play a special role in activating VPS4 and related enzymes came from experiments with amino acid homopolymers. Poly-l-glutamic acid greatly increased VPS4A ATPase activity, whereas other homopolymers including poly-l-lysine and poly-l-threonine did not (Fig. 4B). Stimulation by poly-l-glutamic acid was stereospecific because poly-d-glutamic acid had no effect. Poly-l-glutamic acid also specifically stimulated ATP hydrolysis by spastin (Fig. 4C), but had little or no effect on two other AAA+ proteins, NSF and p97 (data not shown). Previous studies of the effects of homopolymeric amino acids on AAA+ proteins have shown that ClpB and Hsp104 are selectively stimulated by poly-l-lysine (53, 54). The activating effect of acidic amino acids is therefore specific to the subset of AAA+ ATPases that includes VPS4 proteins and spastin.
Activation of His6-tagged VPS4A on Ni2+-NTA Containing Liposomes
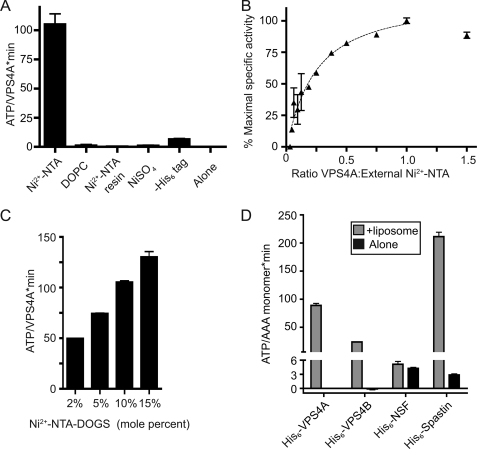
Based on the fact that AAA+ ATPases almost always function as oligomers (55, 56) and on earlier studies showing that yeast Vps4p is a dodecamer when ATP hydrolysis is blocked (26, 28), we wondered whether ESCRT-III proteins activate VPS4A by promoting oligomer assembly. Assembled ESCRT-III could provide a scaffold that concentrates VPS4A and thereby promotes oligomerization. However, the C-terminal ESCRT-III fragments studied above do not polymerize and we were unable to detect VPS4A oligomers by gel filtration in either the absence or presence of the fragments (supplemental Fig. S1). To determine whether VPS4A is nonetheless turned on by enhancing inter-subunit interactions, we turned to a technique used previously to greatly increase enzyme concentration and study inter-subunit activation of the epidermal growth factor receptor kinase in which His6-tagged enzyme is bound to the surface of liposomes containing Ni2+-NTA-DOGS (41). Adding His6-VPS4A to liposomes containing 5% Ni2+-NTA-DOGS increased ATPase activity to levels similar to those seen upon interaction with ESCRT-III proteins (Fig. 5A). This stimulation required the His6 tag on VPS4A and Ni2+-NTA-DOGS in the liposomes, and adding Ni2+-NTA-Sepharose or Ni2+ alone to VPS4A did not enhance ATPase activity (Fig. 5A). Maximum stimulation occurred when the ratio of surface-exposed Ni2+-NTA moieties and added His6-VPS4A molecules was ∼1:1 (Fig. 5B). Importantly, maximal activity was strongly affected by the density of Ni2+-NTA-DOGS in the liposomes over a range of 2–15 mol %, with increased density of tagged lipid yielding increased VPS4A activity (Fig. 5C). Note that the mol % of Ni2+-NTA-DOGS was varied by adding a constant amount of this lipid to increasing amounts of DOPC during preparation of the liposomes. Based on surface area, we estimate that the effective concentration of VPS4A on these liposomes ranged from 2 to 18 mm (see “Experimental Procedures”). Ni2+-NTA-DOGS liposomes also activated the AAA+ ATPase spastin, which is known to transiently oligomerize, but had no effect on the stably oligomerized AAA+ ATPase NSF (Fig. 5D). The fact that VPS4A is activated by binding to these liposomes indicates that intersubunit interactions promote ATP hydrolysis. The fact that maximal activation required a 1:1 ratio of His6-VPS4A to Ni2+-NTA-DOGS suggesting that the two-layered ring structure seen in studies of yeast Vps4p (26, 28) may not be required for VPS4A ATPase activity.
FIGURE 5.
Activation of His6-VPS4A by liposomes containing Ni2+-NTA-DOGS. A, effect of the indicated reagent or manipulation on ATP hydrolysis by 0.2 μm VPS4A: from left to right, liposomes containing 5% Ni2+-NTA-conjugated to DOGS (with molar excess of Ni2+-NTA compared with His6 tag on VPS4A), liposomes containing DOPC only, Ni2+-NTA-Sepharose, 0.5 μm NiSO4, VPS4A with its His6 tag removed together with Ni2+-NTA containing liposomes, and VPS4A alone. B, specific activity of ATP hydrolysis by VPS4A when increasing amounts of enzyme are added to a fixed amount of liposomes containing 5% Ni2+-NTA-conjugated DOGS. C, effect of changing the density of Ni2+-NTA-DOGS present in liposomes (from 2 to 15 mol %, adjusted with varying amounts of DOPC) on VPS4A activity. D, effects of liposomes containing 5% Ni2+-NTA-DOGS on AAA+ ATPases including VPS4A, VPS4B, spastin, and NSF. Basal activity of each enzyme is shown with black bars and activity in the presence of Ni2+-NTA containing liposomes with gray bars. Error bars indicate S.D. from one experiment conducted in duplicate or triplicate; all experiments were repeated three times unless otherwise noted.
Effect of Pore Loop Mutations on VPS4 Activity Supports Function as Ring-shaped Oligomer
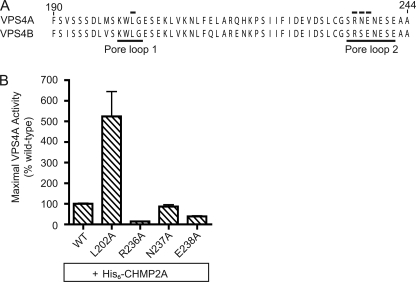
If VPS4A functions as a transient cylindrical oligomer, we would expect loops exposed in the central pore (pore loop 1 and pore loop 2) to interact with substrates to promote unfolding as has been found for other AAA+ proteins (24, 52, 57–62). Both loops have previously been implicated in normal VPS4 function based on the observation that mutations in either one create proteins with inhibitory effects on HIV budding (24, 26). To ask directly about the role of pore loops in engaging ESCRT-III proteins, we mutated residues in them and tested the response of the mutant enzymes to added ESCRT-III proteins. None of the pore loop mutations affected basal activity (data not shown). However, a pore loop 1 mutant in which the central leucine was replaced with alanine (L202A) hydrolyzed more ATP in response to added CHMP2A than did wild-type enzyme, whereas pore loop 2 R254A mutant was less stimulated by CHMP2A than wild-type VPS4A (Fig. 6). Other pore loop 2 mutations had little or no effect on ESCRT-III-stimulated ATPase activity. These data reveal a connection between ESCRT-III proteins and central pore loop residues in VPS4A, and are consistent with a role for these pore loops in ESCRT-III disassembly.
FIGURE 6.
Pore loops regulate response to ESCRT-III proteins in vitro. A, sequence alignment of human VPS4A and VPS4B indicating the position of two pore loops (thin lines (24, 26)). Numbers correspond to VPS4A sequence and residues mutated are indicated by black bars. B, effect of the indicated VPS4A mutations on His6-CHMP2A stimulated ATPase activity, scaled to the maximal response of wild-type VPS4A to His6-CHMP2A. For all experiments, error bars indicate S.D. from duplicate reactions, n = 3.
Regulating VPS4A Activity: Mechanisms Responsible for Controlling Autoinhibition
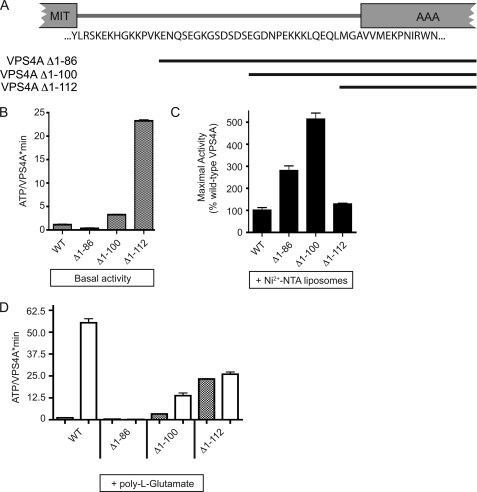
There is a large difference between the concentration of VPS4A activated in solution by ESCRT-III protein fragments (200 nm, Figs. 1–3) and the concentration of VPS4A needed to promote activation on the Ni2+-NTA liposome surface (low mm, Fig. 5). This suggests that ESCRT-III proteins change something in VPS4A to activate it. Because the only clearly defined binding sites for ESCRT-III proteins in VPS4 are in the N-terminal MIT domain (31–33), we asked whether this domain or the linker that connects it to the core AAA+ domain might be involved in regulating VPS4A activity. A previous report of nucleotide-dependent changes in the protease sensitivity of the linker (25) led us to suspect that it in particular might play a role in regulating VPS4A activity. We therefore deleted the MIT domain together with increasing segments of the linker to create three truncated enzymes (Fig. 7A). Basal activity was substantially increased in enzyme lacking the entire MIT and linker region (Fig. 7B) although this was not because of assembly into a stable oligomer (supplemental Fig. S1). This supports the hypothesis that elements within the linker help maintain the low basal activity of VPS4A, likely acting to autoinhibit the VPS4A. Further support for this hypothesis came from examining the effects of Ni2+-NTA containing liposomes on these mutants. All but the shortest deletion mutant had substantially increased maximal activity when concentrated on the liposomes (Fig. 7C). We suspect that the constraints imposed by tethering the shortest mutant to the liposome prevented it from reaching the maximal activity seen with the longer mutants.
FIGURE 7.
The linker connecting MIT and AAA+ domains autoinhibit VPS4A activity. A, schematic diagram of VPS4A showing the residues that connect the MIT and the AAA+ domain (24, 70). Position of deletion mutants are indicated by black bars. B, basal ATPase activity of VPS4A N-terminal mutants assayed under standard conditions using 0.2 μm enzyme. C, effect of Ni2+-NTA liposomes on ATPase activity of indicated mutants. D, effect of 50 μm poly-l-glutamate on ATPase activity of the indicated mutants. For all experiments error bars indicate S.D. from duplicate reactions, n = 3.
To gain insight into how inhibitory effects of the linker might be regulated we asked how the truncated enzymes respond to stimulatory factors as above. (We omitted analysis of CHMP fragments as these had no effect at attainable concentrations when VPS4A is missing its MIT domain (Fig. 1B and not shown).) All of the mutants were not as well stimulated by poly-l-glutamic acid as was full-length VPS4A (Fig. 7D), despite their increased basal and liposome-stimulated activity (above). The complete lack of response of VPS4AΔ1–86 to poly-l-glutamic acid suggests that partial deletion of the linker may eliminate or bury sequences that normally respond to acidic amino acids. These results suggest that interactions between acidic ESCRT-III residues and sequences in VPS4A, in particular in the linker connecting the MIT and AAA+ domains, are involved in regulating the intrinsic autoinhibition of the enzyme. The relatively low conservation in these sequences between human and yeast proteins may account for differences in the degree to which VPS4 enzymes from different species are autoinhibited.
DISCUSSION
VPS4-like proteins control the assembly of the ESCRT machinery and its association with the endosomal membrane, and as the only energy consuming enzymes directly implicated in the ESCRT pathway have for a long time been thought to play a key role in creating vesicles within the MVB (21, 63). Although human VPS4A is essentially inactive in solution, we find that it can be stimulated at physiologically relevant concentrations to hydrolyze ATP following interaction with ESCRT-III proteins. Our data suggests that this activation is the result of two effects of ESCRT-III proteins on VPS4A: enabling functional enzyme assembly and engaging the core catalytic machinery of the enzyme. Full-length CHMP2A was the most potent activator of VPS4A as might have been expected based on the privileged role of Vps2p in recruiting Vps4p in vivo and in vitro (13, 14, 64). Importantly, however, C-terminal fragments of all ESCRT-III proteins stimulated VPS4A suggesting that in the end the enzyme is likely to act on each subunit of an ESCRT-III polymer. Activation of VPS4A by ESCRT-III fragments required more than their previously defined MIM motifs (Fig. 3) pointing to roles for additional contacts between the ESCRT-III proteins and VPS4A. The sequences required for activation include a region that we previously identified as a candidate secondary interaction site for VPS4A in CHMP2A and CHMP1B (39). We propose that MIT-MIM engagement allows VPS4A to associate with and act upon additional sequences in the C-terminal halves of ESCRT-III proteins, ultimately driving ESCRT-III disassembly accompanied by subunit release and recycling.
Given that replacing blocks of three residues with alanines throughout a C-terminal ESCRT-III fragment did not significantly affect VPS4A activation (Fig. 3), it seems likely that once initiated, ESCRT-III processing by VPS4A does not require uniquely defined sequences. This may explain how VPS4A acts on all 12 ESCRT-III proteins in human cells despite their rather divergent sequences. In fact, AAA+ proteins that unfold proteins for proteolysis operate by recognizing short targeting motifs and then processing any attached polypeptide (58). A well characterized example is the 11-residue ssrA tag that is appended to proteins stalled on ribosomes in bacteria and then recognized by AAA+ protein ClpX for unfolding and delivery to the protease ClpP (65). Despite the lack of unique sequence requirements, we found that acidic residues play a special role in stimulating VPS4A activity. In addition to acidic ESCRT-III fragments, both poly-l-glutamic acid and the acidic C terminus of α-tubulin fused to a MIM1 motif were effective activators of VPS4A (Fig. 4). Common to these proteins is the presence of clustered acidic residues, which could activate VPS4A by any of several mechanisms including relieving linker-dependent autoinhibition (Fig. 7) and engaging residue(s) within the central pore of the enzyme (Fig. 6). Interestingly, the AAA+ protein most closely related to VPS4 is spastin, an enzyme that severs microtubules in a reaction dependent on the acidic C termini of tubulin subunits (52, 66). Like VPS4A, it is robustly activated by acidic homopolymers (Fig. 4) extending the substantial structural and functional parallels between these enzymes.
A complicating factor in studying how VPS4A activity is regulated is the superposition of transient oligomer assembly and substrate engagement. Previous studies have shown that yeast enzyme can be trapped as a dodecamer when ATP hydrolysis is blocked by mutation of the essential Walker B glutamic acid residue (21, 26, 44). Although we were able to reproduce these results using yeast proteins (data not shown), we were unable to detect equivalent oligomers using human enzymes (supplemental Fig. S1 and data not shown). Two pieces of evidence suggest that nonetheless human VPS4A functions as an oligomer similar to other AAA+ proteins (45). First, we found that VPS4A could be activated independently of interacting proteins by increasing its concentration to high levels on liposomes using a combination of Ni2+-NTA-conjugated lipids and His6-VPS4A (Fig. 5). The concentration-dependent activation reached a maximal level that correlated with the density of tagged lipid in the liposomes, supporting a role for intersubunit interactions in hydrolysis. Second, mutating key residues in pore loops known to function at the center of hexameric rings formed by other AAA+ ATPases (62, 67) perturbs VPS4 function in cells (24, 26)4 and in the experiments reported here altered the response of the enzyme to ESCRT-III proteins in vitro (Fig. 6). The observed differences in ATP hydrolysis (increased hydrolysis by a pore loop 1 mutant (L202A) and decreased hydrolysis by a pore loop 2 mutant (R254A) after adding CHMP2A) parallel effects of similar mutations in other enzymes including ClpX, and may arise from changes in engagement between the pore loops and substrate (59). Taken together, our data support the idea that VPS4A functions as an oligomer similar in its transient nature to yeast Vps4p (68) and also spastin and katanin (52, 69).
The large difference in the concentration of VPS4A needed to stimulate its ATPase activity on the surface of Ni2+-NTA containing liposomes (> millimolar) compared with solution-based activation by ESCRT-III fragments (< micromolar) suggests that the ESCRT-III fragments must induce a fundamental change in VPS4A to promote assembly of active enzyme. Our deletion studies (Fig. 7) indicated that individual VPS4A monomers are significantly autoinhibited by elements present within the linker between their MIT and AAA+ domain as well as by interactions within the AAA+ domain that involve communication with the pore loops of the enzyme. Relief of these and other possible autoinhibitory interactions is likely to underlie the robust activation of VPS4A upon interaction with its ESCRT-III substrates. The details of the activating changes, which may include not only exposure of the interfaces responsible for subunit-subunit interactions but also changes in the relative positioning of the AAA+ large α/β and small α-helical subdomains (which are known to vary based on comparisons between currently available subunit crystal structures (28)) will require future structural analysis.
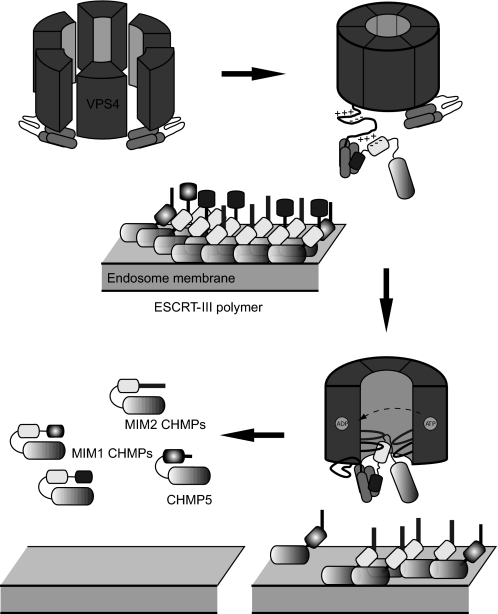
A summary of our working model for VPS4A activation and ESCRT-III disassembly is provided in Fig. 8. Autoinhibited VPS4A subunits interact with ESCRT-III proteins when they are present in their open or assembled conformation on the endosomal membrane. These interactions relieve the autoinhibition of VPS4A and thereby allow assembly of the functional VPS4A oligomer, modeled here as a hexameric ring. Interactions between VPS4A and ESCRT-III involve both the MIM and acidic sequences near the C termini of ESCRT-III proteins and several regions of VPS4A including its N-terminal MIT domains (light gray), a linker (black), and the central pore (gray) of the main AAA+ domain. Once brought into proximity by these interactions, acidic sequences within ESCRT-III proteins enter the central pore of the enzyme where they engage pore loops as shown. By analogy to other AAA+ ATPases, ATP-triggered movement of these loops is likely to provide a physical force that ultimately disengages the ESCRT-III subunit from the polymer, releasing it from the membrane into the cytoplasm. Whether a VPS4A ring persists between processing different ESCRT-III subunits remains an open question. In future studies it will be important to couple our measurements of ATP hydrolysis to a quantitative assay of polymer disassembly, and establish whether ESCRT-III proteins are completely unfolded and translocated through a VPS4A pore or instead are released following a less global conformational change. All together, our studies provide new insight into how this unique AAA+ ATPase responds to its substrates in an appropriately regulated fashion, and will guide future development of reagents to manipulate VPS4A activity and thereby modulate the ESCRT pathway.
FIGURE 8.
Model showing proposed activation of VPS4A followed by ESCRT-III polymer disassembly. VPS4A is either a monomer or a transient oligomer in solution (top left), but likely functions as a hexameric or dodecameric oligomer (21, 44, 55, 71). Interactions with membrane-associated ESCRT-III polymers containing open ESCRT-III proteins activate VPS4 at least in part by relieving intrinsic autoinhibition to shift its equilibrium to favor the oligomeric and active species (top right). Through conformational changes induced by ATP hydrolysis, ESCRT-III proteins then enter the central pore of VPS4A where pore loops mediate a chaperone-like activity, which ultimately returns the ESCRT-III proteins to their closed conformations in the cytoplasm (bottom right) (56, 72). This activity applies to all subgroups of ESCRT-III proteins, represented schematically here based on the identity of their MIM motif (bottom left).
Supplementary Material
Acknowledgments
We thank Teresa Naismith for help with site-directed mutagenesis, Soomin Shim for pilot studies and helpful discussions, John Monsey and Ron Bose for help with liposome preparation and helpful discussions, and all members of the Hanson lab for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM076686.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
S. Shim, S. A. Merrill, and P. I. Hanson, unpublished data.
S. Shim, unpublished data.
- MVB
- multivesicular body
- ESCRT
- endosomal sorting complex required for transport
- CHMP
- charged multivesicular body protein
- AAA+
- ATPase associated with a variety of cellular activities
- MIT
- microtubule interacting and transport
- MIM
- MIT interacting motif
- DOPC
- dioleoyl-phosphatidylcholine
- Ni2+-NTA
- nickel-nitrilotriacetic acid
- DOGS
- nickel-1,2-dioleoyl-sn-glycero-3-([N-(5-amino-1-carboxypentyl)iminodiacetic acid] succinyl)-nickel salt
- PIPES
- 1,4-piperazinediethanesulfonic acid
- NSF
- N-ethyl maleimide-sensitive factor, protein encoded by the NSF gene.
REFERENCES
- 1.Hanson P. I., Shim S., Merrill S. A. (2009) Curr. Opin. Cell Biol. 21, 568–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlton J. G., Martin-Serrano J. (2009) Biochem. Soc. Trans. 37, 195–199 [DOI] [PubMed] [Google Scholar]
- 3.Kaksonen M. (2008) J. Cell Biol. 180, 1059–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saksena S., Sun J., Chu T., Emr S. D. (2007) Trends Biochem. Sci. 32, 561–573 [DOI] [PubMed] [Google Scholar]
- 5.Luzio J. P., Pryor P. R., Bright N. A. (2007) Nat. Rev. Mol. Cell Biol. 8, 622–632 [DOI] [PubMed] [Google Scholar]
- 6.Agromayor M., Carlton J. G., Phelan J. P., Matthews D. R., Carlin L. M., Ameer-Beg S., Bowers K., Martin-Serrano J. (2009) Mol. Biol. Cell 20, 1374–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bajorek M., Morita E., Skalicky J. J., Morham S. G., Babst M., Sundquist W. I. (2009) Mol. Biol. Cell 20, 1360–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lata S., Schoehn G., Solomons J., Pires R., Göttlinger H. G., Weissenhorn W. (2009) Biochem. Soc. Trans. 37, 156–160 [DOI] [PubMed] [Google Scholar]
- 9.Babst M., Katzmann D. J., Estepa-Sabal E. J., Meerloo T., Emr S. D. (2002) Dev. Cell 3, 271–282 [DOI] [PubMed] [Google Scholar]
- 10.Muzioł T., Pineda-Molina E., Ravelli R. B., Zamborlini A., Usami Y., Göttlinger H., Weissenhorn W. (2006) Dev. Cell 10, 821–830 [DOI] [PubMed] [Google Scholar]
- 11.Xiao J., Chen X. W., Davies B. A., Saltiel A. R., Katzmann D. J., Xu Z. (2009) Mol. Biol. Cell 20, 3514–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bajorek M., Schubert H. L., McCullough J., Langelier C., Eckert D. M., Stubblefield W. M., Uter N. T., Myszka D. G., Hill C. P., Sundquist W. I. (2009) Nat. Struct. Mol. Biol. 16, 754–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teis D., Saksena S., Emr S. D. (2008) Dev. Cell 15, 578–589 [DOI] [PubMed] [Google Scholar]
- 14.Wollert T., Wunder C., Lippincott-Schwartz J., Hurley J. H. (2009) Nature 458, 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghazi-Tabatabai S., Obita T., Pobbati A. V., Perisic O., Samson R. Y., Bell S. D., Williams R. L. (2009) Biochem. Soc. Trans. 37, 151–155 [DOI] [PubMed] [Google Scholar]
- 16.Hanson P. I., Roth R., Lin Y., Heuser J. E. (2008) J. Cell Biol. 180, 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lata S., Schoehn G., Jain A., Pires R., Piehler J., Gottlinger H. G., Weissenhorn W. (2008) Science 321, 1354–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Im Y. J., Wollert T., Boura E., Hurley J. H. (2009) Dev. Cell 17, 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saksena S., Wahlman J., Teis D., Johnson A. E., Emr S. D. (2009) Cell 136, 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fabrikant G., Lata S., Riches J. D., Briggs J. A., Weissenhorn W., Kozlov M. M. (2009) PLoS Comput. Biol. 5, e1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Babst M., Wendland B., Estepa E. J., Emr S. D. (1998) EMBO J. 17, 2982–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nickerson D. P., West M., Odorizzi G. (2006) J. Cell Biol. 175, 715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nickerson D. P., West M., Henry R., Odorizzi G. (2010) Mol. Biol. Cell 21, 1023–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott A., Chung H. Y., Gonciarz-Swiatek M., Hill G. C., Whitby F. G., Gaspar J., Holton J. M., Viswanathan R., Ghaffarian S., Hill C. P., Sundquist W. I. (2005) EMBO J. 24, 3658–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao J., Xia H., Yoshino-Koh K., Zhou J., Xu Z. (2007) J. Mol. Biol. 374, 655–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonciarz M. D., Whitby F. G., Eckert D. M., Kieffer C., Heroux A., Sundquist W. I., Hill C. P. (2008) J. Mol. Biol. 384, 878–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inoue M., Kamikubo H., Kataoka M., Kato R., Yoshimori T., Wakatsuki S., Kawasaki M. (2008) Traffic 9, 2180–2189 [DOI] [PubMed] [Google Scholar]
- 28.Landsberg M. J., Vajjhala P. R., Rothnagel R., Munn A. L., Hankamer B. (2009) Structure 17, 427–437 [DOI] [PubMed] [Google Scholar]
- 29.Hartmann C., Chami M., Zachariae U., de Groot B. L., Engel A., Grütter M. G. (2008) J. Mol. Biol. 377, 352–363 [DOI] [PubMed] [Google Scholar]
- 30.Yu Z., Gonciarz M. D., Sundquist W. I., Hill C. P., Jensen G. J. (2008) J. Mol. Biol. 377, 364–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kieffer C., Skalicky J. J., Morita E., De Domenico I., Ward D. M., Kaplan J., Sundquist W. I. (2008) Dev. Cell 15, 62–73 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 32.Stuchell-Brereton M. D., Skalicky J. J., Kieffer C., Karren M. A., Ghaffarian S., Sundquist W. I. (2007) Nature 449, 740–744 [DOI] [PubMed] [Google Scholar]
- 33.Obita T., Saksena S., Ghazi-Tabatabai S., Gill D. J., Perisic O., Emr S. D., Williams R. L. (2007) Nature 449, 735–739 [DOI] [PubMed] [Google Scholar]
- 34.Shestakova A., Hanono A., Drosner S., Curtiss M., Davies B. A., Katzmann D. J., Babst M. (2010) Mol. Biol. Cell 21, 1059–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lauer J. M., Dalal S., Marz K. E., Nonet M. L., Hanson P. I. (2006) J. Biol. Chem. 281, 14823–14832 [DOI] [PubMed] [Google Scholar]
- 36.Marz K. E., Lauer J. M., Hanson P. I. (2003) J. Biol. Chem. 278, 27000–27008 [DOI] [PubMed] [Google Scholar]
- 37.Dalal S., Rosser M. F., Cyr D. M., Hanson P. I. (2004) Mol. Biol. Cell 15, 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shim S., Kimpler L. A., Hanson P. I. (2007) Traffic 8, 1068–1079 [DOI] [PubMed] [Google Scholar]
- 39.Shim S., Merrill S. A., Hanson P. I. (2008) Mol. Biol. Cell 19, 2661–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lill R., Dowhan W., Wickner W. (1990) Cell 60, 271–280 [DOI] [PubMed] [Google Scholar]
- 41.Zhang X., Gureasko J., Shen K., Cole P. A., Kuriyan J. (2006) Cell 125, 1137–1149 [DOI] [PubMed] [Google Scholar]
- 42.Monsey J., Shen W., Schlesinger P., Bose R. (2010) J. Biol. Chem. 285, 7035–7044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stewart J. C. (1980) Anal. Biochem. 104, 10–14 [DOI] [PubMed] [Google Scholar]
- 44.Azmi I., Davies B., Dimaano C., Payne J., Eckert D., Babst M., Katzmann D. J. (2006) J. Cell Biol. 172, 705–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanson P. I., Whiteheart S. W. (2005) Nat. Rev. Mol. Cell Biol. 6, 519–529 [DOI] [PubMed] [Google Scholar]
- 46.Zamborlini A., Usami Y., Radoshitzky S. R., Popova E., Palu G., Göttlinger H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 19140–19145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lata S., Roessle M., Solomons J., Jamin M., Gottlinger H. G., Svergun D. I., Weissenhorn W. (2008) J. Mol. Biol. 378, 818–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin Y., Kimpler L. A., Naismith T. V., Lauer J. M., Hanson P. I. (2005) J. Biol. Chem. 280, 12799–12809 [DOI] [PubMed] [Google Scholar]
- 49.Leung K. F., Dacks J. B., Field M. C. (2008) Traffic 9, 1698–1716 [DOI] [PubMed] [Google Scholar]
- 50.Nogales E., Wolf S. G., Downing K. H. (1998) Nature 391, 199–203 [DOI] [PubMed] [Google Scholar]
- 51.Misura K. M., Gonzalez L. C., Jr., May A. P., Scheller R. H., Weis W. I. (2001) J. Biol. Chem. 276, 41301–41309 [DOI] [PubMed] [Google Scholar]
- 52.White S. R., Evans K. J., Lary J., Cole J. L., Lauring B. (2007) J. Cell Biol. 176, 995–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cashikar A. G., Schirmer E. C., Hattendorf D. A., Glover J. R., Ramakrishnan M. S., Ware D. M., Lindquist S. L. (2002) Mol Cell 9, 751–760 [DOI] [PubMed] [Google Scholar]
- 54.Strub C., Schlieker C., Bukau B., Mogk A. (2003) FEBS Lett. 553, 125–130 [DOI] [PubMed] [Google Scholar]
- 55.Ogura T., Whiteheart S. W., Wilkinson A. J. (2004) J. Struct. Biol. 146, 106–112 [DOI] [PubMed] [Google Scholar]
- 56.Augustin S., Gerdes F., Lee S., Tsai F. T., Langer T., Tatsuta T. (2009) Mol. Cell 35, 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DeLaBarre B., Christianson J. C., Kopito R. R., Brunger A. T. (2006) Mol. Cell 22, 451–462 [DOI] [PubMed] [Google Scholar]
- 58.Barkow S. R., Levchenko I., Baker T. A., Sauer R. T. (2009) Chem. Biol. 16, 605–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin A., Baker T. A., Sauer R. T. (2008) Nat. Struct. Mol. Biol. 15, 1147–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Farbman M. E., Gershenson A., Licht S. (2008) Biochemistry 47, 13497–13505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zolkiewski M. (2006) Mol. Microbiol. 61, 1094–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park E., Rho Y. M., Koh O. J., Ahn S. W., Seong I. S., Song J. J., Bang O., Seol J. H., Wang J., Eom S. H., Chung C. H. (2005) J. Biol. Chem. 280, 22892–22898 [DOI] [PubMed] [Google Scholar]
- 63.Babst M., Sato T. K., Banta L. M., Emr S. D. (1997) EMBO J. 16, 1820–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Azmi I. F., Davies B. A., Xiao J., Babst M., Xu Z., Katzmann D. J. (2008) Dev. Cell 14, 50–61 [DOI] [PubMed] [Google Scholar]
- 65.Kim Y. I., Burton R. E., Burton B. M., Sauer R. T., Baker T. A. (2000) Mol. Cell 5, 639–648 [DOI] [PubMed] [Google Scholar]
- 66.Roll-Mecak A., Vale R. D. (2008) Nature 451, 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hinnerwisch J., Fenton W. A., Furtak K. J., Farr G. W., Horwich A. L. (2005) Cell 121, 1029–1041 [DOI] [PubMed] [Google Scholar]
- 68.Dimaano C., Jones C. B., Hanono A., Curtiss M., Babst M. (2008) Mol. Biol. Cell 19, 465–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hartman J. J., Vale R. D. (1999) Science 286, 782–785 [DOI] [PubMed] [Google Scholar]
- 70.Scott A., Gaspar J., Stuchell-Brereton M. D., Alam S. L., Skalicky J. J., Sundquist W. I. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13813–13818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ogura T., Wilkinson A. J. (2001) Genes Cells 6, 575–597 [DOI] [PubMed] [Google Scholar]
- 72.Zhang X., Wigley D. B. (2008) Nat. Struct. Mol. Biol. 15, 1223–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.