Abstract
Cytoskeletal regulation of cell adhesion is vital to the organization of multicellular structures. The focal adhesion protein zyxin emerged as a key regulator of actin assembly because zyxin recruits Enabled/vasodilator-stimulated phospho-proteins (Ena/VASP) to promote actin assembly. Zyxin also localizes to the sites of cell-cell adhesion and is thought to promote actin assembly with Ena/VASP. Using shRNA targeted to zyxin, we analyzed the roles of zyxin at adhesive contacts. In zyxin-deficient cells, the actin assembly at both focal adhesion and cell-cell adhesion was limited, but their migration rate was unchanged. Cell spreading on E-cadherin-coated surfaces and the formation of cell clusters were slower for zyxin-deficient cells than wild type cells. By ablating a single cell within a cell monolayer, we quantified the rate of wound closure driven by a contractile circumferential actin ring. Zyxin-deficient cells failed to recruit VASP to cell-cell junctions at the wound edge and had a slower wound closure rate than wild type cells. Our results suggest that, by recruiting VASP, zyxin regulates actin assembly at the sites of force-bearing cell-cell adhesion.
Keywords: Actin, Cell Adhesion, Cell Migration, Cell-Cell Interaction, Epithelial Cell, E-cadherin, VASP, Zyxin
Introduction
Cell adhesion plays a central role during multicellular organization by interacting with surrounding extracellular matrix and neighboring cells. Adhesion proteins are abundant, but they alone do not sufficiently provide the necessary function and strength of cell adhesion. At cell-extracellular matrix and cell-cell junctions, the actin cytoskeleton promotes the formation and stabilization of adhesive contacts. The regulation of actin assembly is therefore crucial to the regulation of cell adhesion.
Despite the presence of distinct adhesive receptors, some actin-binding proteins are shared by both cell-extracellular and cell-cell junctions. Zyxin is one such protein (1, 2). Interestingly, the localization of zyxin is mechano-sensitive. Zyxin is recruited to actin stress fibers upon uniaxial stretching of cells on a compliant substrate (3) or the application of external force on dorsal cell surface (4). In addition, stretch-induced actin polymerization at focal adhesion sites has been shown to depend on zyxin (5). Because of its unique ability to sense external mechanical cues and regulate actin polymerization, zyxin is an attractive candidate for regulating cell adhesion.
Although zyxin does not interact directly with actin, zyxin contains binding sites for other actin-binding proteins. The N terminus of zyxin contains an ActA proline-rich domain that binds to the Enabled/vasodilator-stimulated phospho-protein (Ena2/VASP) family (6) and an α-actinin binding site (7, 8) whereas the C terminus contains three LIM domains. Internal deletion of the α-actinin binding domain disrupts zyxin localization to focal adhesions (8, 9). However, the expression of the LIM domains alone is sufficient for targeting zyxin to focal adhesions (9). The proline-rich Ena/VASP binding domain is not required for the localization of zyxin to focal adhesions (9), but it is essential for the localization of Ena/VASP to focal adhesions (5, 6, 10). Furthermore, zyxin promotes VASP-dependent actin assembly when zyxin is enriched at the membrane (6) or mitochondria (11). VASP is thought to compete with actin-capping protein for binding to the barbed ends of actin filaments, thus promoting actin filament elongation (12). By recruiting Ena/VASP to focal adhesions, zyxin regulates focal adhesion assembly and actin organization.
Previous studies focused on zyxin at focal adhesion sites, but little is known about the roles of zyxin at cell-cell adhesion. Both α-actinin binding domain and LIM domains of zyxin have the ability to localize to cell-cell contacts (13). The overexpression of zyxin without LIM domains (zyxinΔLIM) enhances cell aggregation, but proline-to-alanine mutations in the proline-rich domain or the LIM domains of zyxin alone do not have such effects (13). Yet, the overexpression of zyxinΔLIM with the proline-to-alanine mutations reduces the enhanced cell cluster formation observed in zyxinΔLIM-overexpressing cells, suggesting that the LIM domains negatively regulate the function of Ena/VASP binding sites (13).
Ena/VASP has been shown to be important for the integrity of cell-cell adhesion and the underlying actin organization. For example, Ena/VASP-null mice suffer vascular defects, likely due to the disruption of endothelial barrier function (14). In primary keratinocytes, the expression of dominant negative VASP results in the disruption of adhesion zipper formation at cell-cell contacts (15). Furthermore, sequestration of Ena/VASP proteins to the mitochondria results in reduced actin level at cell-cell contacts and cadherin-based adhesion plaques (16). Together, all previous results point to the intimate functional relationship between zyxin and Ena/VASP in facilitating actin assembly at the sites of focal adhesion and cell-cell adhesion.
We sought to analyze the role of zyxin as a regulator of VASP localization and actin assembly at sites of cell-cell adhesion using epithelial cells stably expressing zyxin shRNA. In zyxin-deficient cells, actin assembly was reduced at both sites of focal adhesion and cell-cell adhesion. Zyxin-deficient cells had no migration defects, but exhibited slower cell spreading on an E-cadherin-coated surface and cell cluster formation. Furthermore, the monolayer of zyxin-deficient cells was unable to recruit VASP to cell-cell junctions at the wound edge and effectively close a small circular wound. Together, our results suggest that zyxin regulates E-cadherin-mediated cell-cell adhesion by facilitating actin assembly.
EXPERIMENTAL PROCEDURES
Cell Lines and Reagents
Madin-Darby canine kidney cells were cultured in low glucose Dulbecco's modified Eagle's medium with 10% fetal bovine serum. Cells were fixed in PBS containing 3% paraformaldehyde, 0.3% Triton X-100 for 10 min. Antibodies used were zyxin B71 (17), zyxin (Abcam), VASP (BD Biosciences), E-cadherin (clone 36; BD Biosciences), vinculin (hVIN-1; Sigma), paxillin (clone 349; BD Biosciences), α-catenin (15D9; Alexis Biochemicals), and β-catenin (clone 14; BD Biosciences). The GFP intensity in pSuper.gfp-expressing cells was relatively weak, thus enabling secondary label in the same channel. For actin assembly assays, cells were permeabilized prior to fixation with 2 mg/ml saponin and incubated with 2.4 μm Alexa Fluor 568-labeled actin (Invitrogen) in rinsing buffer (1% BSA, 1 mm ATP, 20 mm HEPES, pH 7.3, 138 mm KOAc, 4 mm MgCl2) for 10 min. The GFP-tagged zyxin plasmid (18) was transfected into Madin-Darby canine kidney cells and G418 antibiotic-resistant stable cells were subcloned to homogeneity and analyzed with Western blotting (supplemental Fig. S2). The phenotype of zyxin-GFP expressing cells closely mimics that of wild type cells.
shRNA
To knock down zyxin, a canine-specific sequence (5′-GACAAGAACTTCCACATGA-3′ (shRNA1)) was inserted into the pSuper.gfp/neo expression vector (Oligoengine, Seattle, WA) and transfected using Lipofectamine 2000 (Invitrogen). This sequence provided the highest level of zyxin knockdown (<5% of wild type zyxin level) among six other sequences targeted to different regions of the zyxin mRNA. The zyxin-deficient clone generated from this sequence was used in the majority of this study. A vector containing the scrambled RNA sequence (5′-CACATAAGGTCCATACGAA-3′) was generated for negative controls. Other shRNA sequences shown in this study are: 5′-ACCACAGCCCAGGGAGAAG-3′ (shRNA2) and 5′-CATCACCTGCTTCACCTGC-3′ (shRNA3). The G418-resistant stable cells were subcloned to homogeneity and analyzed with Western blotting (supplemental Fig. S2).
Microscopy and Live-cell Imaging
All samples were imaged on a Zeiss AxioObserver equipped with a Yokogawa spinning disk confocal system, 10×, 40×, and 63× objectives, 488- and 561-nm solid-state lasers, and a CoolSNAP HQ camera. The microscope system was controlled by Slidebook software (Intelligent Imaging Innovations). Live cells were imaged on glass bottom dishes (MatTek, Ashland, MA) in a temperature-controlled chamber at 37 °C.
Cell Migration Assay
For cell sheet migration, cells were plated at high density in a collagen-coated glass bottom dish containing a thin polydimethylsiloxane strip. After 40 min, the polydimethylsiloxane strip was removed to create an open space between the monolayers. Unlike pipette scraping to create a wound, the polydimethylsiloxane strip does not remove collagen coating. The average migration rate was determined from 10 fields of view for each cell type. For single cell migration, cells were plated at low density on a collagen-coated glass bottom dish for 30 min before placing on the microscope for imaging. Data were analyzed for 46 wild type cells and 62 zyxin-knockdown cells. Migrating cells were imaged every 10 min for 12 h.
Cell Spreading and Hanging Drop Assay
Cell spreading on coverslips was imaged every 10 min for at least 10 h. The surface was coated with the purified extracellular domain of E-cadherin (2 μg/ml; R&D Systems). Cell spreading was quantified by measuring the cell area at each time point. For the hanging drop assay, cells were suspended in medium (25 μl at 250,000 cells/ml) and incubated upside down for 0, 1, 2, or 3 h, then triturated 30 times through a pipette tip. Quantification of cell cluster size was performed using a filtering and thresholding algorithm in ImageJ.
Wound Healing Assay
To create a small circular wound, a single cell in the cell monolayer was selectively ablated using a photo-ablation system (Intelligent Imaging Innovation), and imaged every minute for 1 h. The area of the wound opening was quantified using ImageJ and analyzed in Microsoft Excel.
RESULTS
Zyxin-deficient Cells Migrate Efficiently as a Cell Sheet
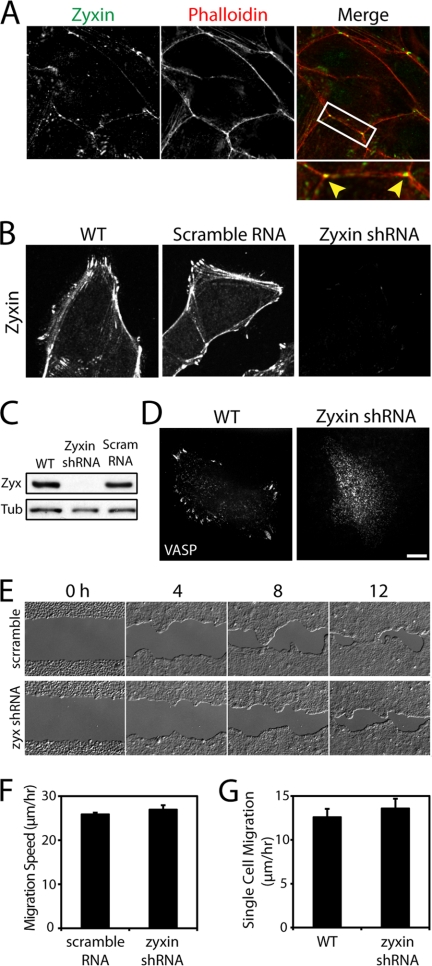
Zyxin is a focal adhesion protein that also localizes to cell-cell junctions of simple epithelial (Madin-Darby canine kidney) cells. Zyxin was immunolocalized along cell-cell contacts and often concentrated at multicellular junctions of wild type cells (Fig. 1A). To evaluate the roles of zyxin in the formation of cell adhesion, we stably silenced zyxin in Madin-Darby canine kidney cells using shRNA and subcloned stable cells to establish a homogeneous population. According to immunofluorescence analysis, no detectable zyxin labeling was observed in the zyxin-knockdown cells (Fig. 1B). Based on Western blot analysis, the lowest zyxin protein level in the knockdown cells was <5% of the wild type level (Fig. 1C; see other clones in supplemental Fig. S2).
FIGURE 1.
Zyxin-knockdown cells migrate efficiently as a cell sheet. A, zyxin localized to cell-cell and multicellular junctions. B, zyxin localized to the sites of focal adhesion and cell-cell adhesion and along actin fibers in wild type (WT) and scramble RNA-transfected cells. Zyxin was not detectable in zyxin-knockdown cells. C, Western blot analysis of wild type, zyxin shRNA, or scramble RNA-transfected cells. D, VASP localized to focal adhesion sites of wild type cells, but remained in the cytoplasm of zyxin shRNA-transfected cells. Scale bar, 10 μm. E, time lapse images show zyxin-knockdown and scramble RNA cells closing a large gap in a monolayer. F, zyxin-knockdown cells close the wounds at a slightly faster average rate than scramble RNA transfectants, but the difference is not statistically significant. Data represent the average migration rate ± S.E. (error bars). n = 10 wounds. G, single cell migration analysis shows zyxin-knockdown cells migrating on collagen-coated surface at a slightly faster speed than normal cells, but the difference is not statistically significant. Data represent the average single cell migration speed ± S.E. n = 46 wild type cells and 62 zyxin-knockdown cells.
Consistent with previous observations in zyxin-null fibroblasts (10), zyxin-knockdown cells failed to recruit VASP to focal adhesion sites (Fig. 1D), but retained the ability to form actin bundles (supplemental Fig. S1A). The tips of actin bundles were often enriched with actin in wild type cells, whereas no actin accumulation was observed in zyxin-knockdown cells (supplemental Fig. S1A, inset), suggesting that actin assembly may be compromised at focal adhesion sites in the absence of zyxin. To directly assess actin polymerization activity, we incubated saponin-permeabilized cells with fluorescently labeled actin (Fig. S1B). Wild type cells incorporated exogenous actin to the tips of actin bundles and along actin fibers, whereas zyxin-knockdown cells did not (supplemental Fig. S1B). Consistent with previous studies (5, 10), these results suggest that zyxin-mediated VASP recruitment is critical for actin assembly at focal adhesion.
Surprisingly, the limited actin assembly of zyxin-deficient cells did not affect migration rates on a collagen-coated surface (Fig. 1, E and F). This is in contrast to the enhanced migration rate observed for zyxin-null fibroblasts (10). The difference could not be attributed to the relative strength of cell-cell adhesion in fibroblasts and epithelial cells because single cell migration of zyxin-knockdown epithelial cells was similar to wild type cells (Fig. 1G). The monolayer of zyxin-deficient cells migrated as a cohesive cell sheet without any defects in cell-cell adhesion between neighboring cells. These data suggest that cell-cell junctions between zyxin-deficient cells are sufficiently strong to maintain cell-cell interactions during collective cell migration.
Cadherin-mediated Cell Adhesion Depends on Zyxin
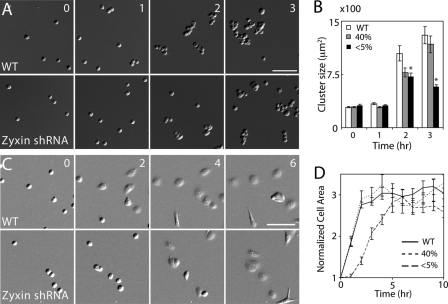
Despite the absence of zyxin, zyxin-knockdown cells formed a cell monolayer without any obvious morphological defects and migrated efficiently as a cell sheet. We sought to analyze cell-cell adhesion in the absence of cell-extracellular matrix adhesion to expose any defects in cell-cell adhesion. To assay for the formation of cell-cell adhesion, we suspended cells in a small volume of medium and quantified the multicellular clustering. In this hanging drop assay, the clustered cells were triturated to disrupt loosely adhered cells. Wild type cells rapidly clustered whereas zyxin-knockdown cells remained relatively dispersed after 2–3 h (Fig. 2, A and B), suggesting that the cell-cell adhesion is weaker in zyxin-deficient cells.
FIGURE 2.
Cadherin-mediated adhesion of zyxin-knockdown cells is weak. A, cell cluster formation of wild type (WT) and zyxin-knockdown cells. Both wild type and zyxin-knockdown cells remained as single cells, doublets, and triplets after 1 h. By 2 and 3 h, wild type cells remained in large clusters despite rigorous pipetting. In contrast, zyxin-knockdown cells could be broken into smaller clusters. B, quantification of cluster size. Data represent the mean ± S.E. (error bars). n > 130. *, p < 0.001 (relative to wild type). C, spreading of wild type and zyxin-knockdown cells after 0, 2, 4, and 6 h on E-cadherin-coated surface. D, quantification of cell spreading. Cell area was normalized to the initial time point. Most wild-type cells were spread out by 2 h. In contrast, zyxin-knockdown cells took 6 h to spread out to the same degree. n = 6 fields of view. Time is in hours. Scale bars, 100 μm.
To isolate the role of zyxin-mediated actin assembly at cadherin junctions, we analyzed cadherin-mediated cell spreading where actin polymerization is essential for membrane extensions. On E-cadherin-coated substrates, zyxin-knockdown cells adhered but spread out slower than wild type cells (Fig. 2, C and D). Although most wild type cells spread out within 2 h, zyxin-knockdown cells took at least 6 h to spread out to the same degree. Because most cells eventually adhere and spread out on E-cadherin substrate, zyxin may play a more significant role in the early stages of cell adhesion by facilitating actin assembly at the leading edge.
The slow formation of cell-cell adhesion of zyxin-knockdown cells may be due to either reduced actin assembly or weak adhesive complex. Because zyxin-knockdown cells have lower E-cadherin levels (supplemental Fig. S2), it is possible that the delay in cell spreading on an E-cadherin-coated substrate could be due to weak adhesion. However, the partial zyxin-knockdown cells (∼40% of the wild type zyxin level with normal E-cadherin level (supplemental Fig. S2)) also had weaker cell adhesion than wild type cells (Fig. 2B). In addition, on E-cadherin-coated substrate, zyxin-knockdown cells can form cadherin plaques (see below), suggesting that there are sufficient levels of E-cadherin for the formation of cadherin junctions.
Actin Assembly at E-cadherin Adhesion Plaques and Puncta Requires Zyxin
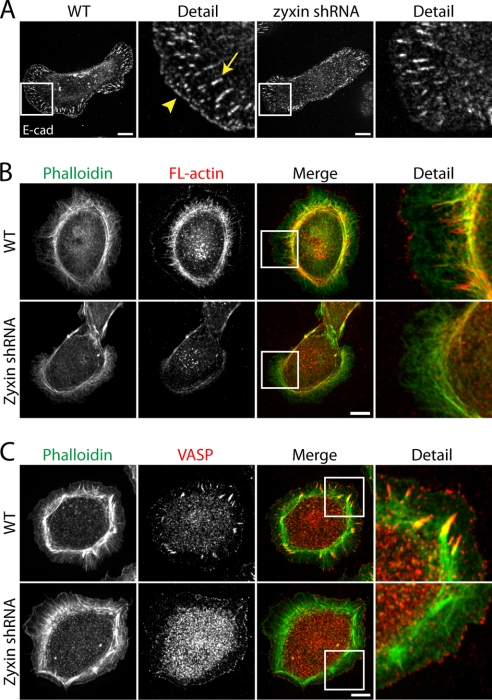
When wild type cells adhere on an E-cadherin-coated surface, endogenous E-cadherin formed focal adhesion-like plaques (Fig. 3A, arrow, and supplemental Fig. S3A) and smaller E-cadherin puncta along the leading edge of lamellipodia (Fig. 3A, arrowhead, and supplemental Fig. S3A) (16, 19). These E-cadherin adhesion plaques did not contain integrin α5 (supplemental Fig. S4), suggesting that despite the focal adhesion-like appearance, these adhesion junctions are mediated by E-cadherin.
FIGURE 3.
Actin assembly at cadherin-mediated adhesion sites depends on zyxin. A, E-cadherin-decorated adhesion plaques and the leading edge of wild type (WT) cells on E-cadherin-coated substrate. In zyxin-knockdown cells, E-cadherin-positive plaques were disorganized, and E-cadherin puncta were not found at the leading edge. B, wild type and zyxin-knockdown cells plated on E-cadherin substrates permeabilized with saponin and treated with Alexa Fluor 568-labeled actin monomers, followed by fixation and phalloidin staining. Fluorescently labeled exogenous actin decorated actin fibers, tips of actin bundles, and the leading lamellipodial edge in saponin-permeabilized wild type cells. For zyxin-knockdown cells, exogenous actin weakly decorated actin fibers but not actin bundle tips or the leading edge. C, VASP localized to the tips of actin bundles and the leading edge in normal cells. In zyxin-knockdown cells, VASP was not found at the tips of actin bundles, but it remained localized to the leading edge. Scale bars, 10 μm.
Similar to focal adhesions in molecular composition, E-cadherin-positive plaques co-localized with zyxin, actin, VASP, and vinculin, but also recruited α-catenin and β-catenin (supplemental Fig. S3). In addition, the E-cadherin puncta at the leading edge contained zyxin, VASP, and vinculin (supplemental Fig. S3). Time lapse imaging of GFP-tagged zyxin-expressing cells showed these lamellipodial spots elongating to form adhesion plaques (supplemental Fig. S5 and supplemental Movie S1), suggesting that these zyxin-containing cadherin puncta are precursors for cadherin plaques.
In zyxin-knockdown cells, E-cadherin plaques were sparsely distributed across the cell body (Fig. 3A). In addition, the small E-cadherin puncta at the leading edge were mostly absent in zyxin-knockdown cells (Fig. 3A). Actin bundles in zyxin-knockdown cells were loosely organized compared with those in wild type cells (Fig. 3, B and C). This suggests that the loss of zyxin disrupted the formation of cadherin-mediated nascent adhesion and the organization of actin networks.
In saponin-permeabilized wild type cells, exogenous actin incorporated into both E-cadherin puncta and plaques, suggesting that these regions are the sites of rapid actin assembly (Fig. 3B). In saponin-permeabilized zyxin-knockdown cells, actin assembly was diminished at both E-cadherin puncta and plaques (Fig. 3B). Interestingly, in zyxin-knockdown cells, VASP was absent from cadherin plaques but remained localized to the leading edge (Fig. 3C). Because the leading edge of zyxin-knockdown cells lacked E-cadherin puncta (Fig. 3A), this VASP recruitment is independent of E-cadherin adhesion. As was the case at focal adhesions, the absence of VASP at cadherin plaques coincided with reduced actin assembly. However, despite the presence of VASP at the leading edge in zyxin-knockdown cells, actin assembly was reduced at the lamellipodia (Fig. 3B). This is consistent with reduced actin assembly at VASP positive cell-cell contacts (see below, Fig. 4). These results suggest that zyxin facilitates actin assembly at both E-cadherin plaques (VASP-dependent) and puncta (VASP-independent).
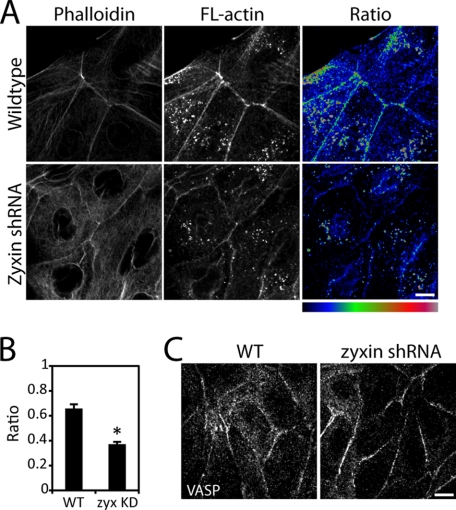
FIGURE 4.
Actin assembly at cell-cell contacts depends on zyxin. A, wild type (WT) and zyxin-knockdown cells were permeabilized with saponin and treated with Alexa Fluor 568-labeled actin monomers, followed by fixation and phalloidin staining. Labeled actin is incorporated into actin network at cell-cell adhesion in wild type cells, but not zyxin-knockdown cells. B, ratio of labeled actin to phalloidin intensity at cell-cell adhesion is higher in wild type cells than in zyxin-knockdown cells. Data represent the average ratio ± S.E. (error bars). n = 25 cells. *, p < 0.001. C, VASP is localized to cell-cell contacts in normal and zyxin-knockdown cells. Scale bar, 10 μm.
Zyxin Promotes Actin Assembly at Cell-Cell Contacts
We analyzed whether suppressing zyxin affects actin polymerization at the sites of cell-cell adhesion. When cells in colonies were saponin-permeabilized, exogenous actin incorporated into the endogenous actin network at cell-cell contacts (Fig. 4A). To determine the relative amount of actin assembly, we generated ratio images that correspond to the fluorescence intensity ratio of exogenous actin to total actin filaments labeled by phalloidin (Fig. 4A, Ratio). The region with a high ratio indicates more exogenous actin assembled with endogenous actin relative to the region with a low ratio. In wild type cells, the ratio was highest at zyxin-rich multicellular junctions (Fig. 4, A and B). The average ratio of exogenous actin to phalloidin was greater for wild type cells compared with zyxin-knockdown cells (Fig. 4B). In addition to the strong correlation between zyxin localization and actin assembly in wild type cells, low exogenous actin incorporation at cell-cell contacts of zyxin-knockdown cells suggests that zyxin promotes actin assembly at sites of cell-cell adhesion.
We suspected that the reduced actin assembly along cell-cell contacts in zyxin-deficient cells is due to the absence of VASP, as was the case for focal adhesions (supplemental Fig. S1B) (5) and cadherin plaques (Fig. 3, B and C). Surprisingly, VASP remained localized along cell-cell contacts in zyxin-knockdown cells (Fig. 4C). These data suggest that VASP localization alone is not sufficient to promote actin assembly at these cell-cell contacts.
Efficient Wound Closure Requires Zyxin
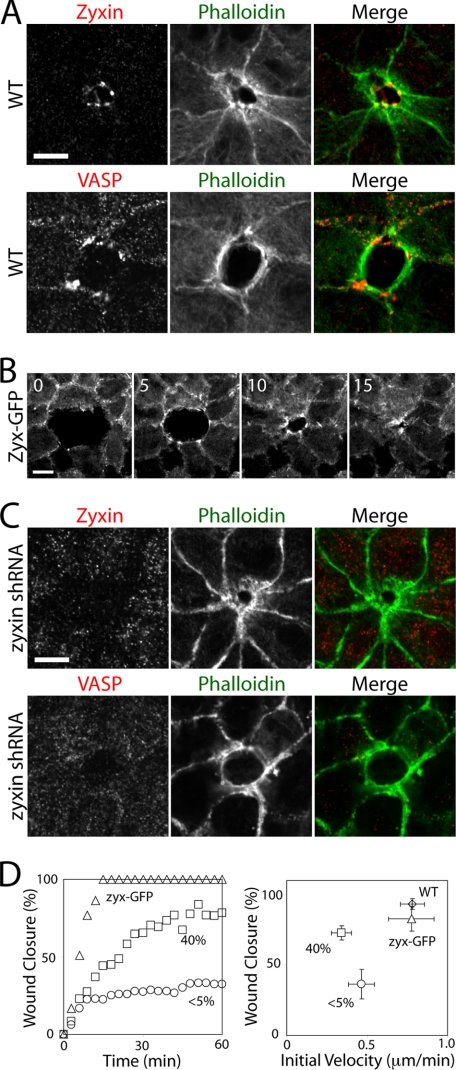
Because zyxin is a mechano-sensing protein that localizes to the sites of force-bearing cell adhesion, we sought to analyze how zyxin affects cell-cell contacts that are under tension. We ablated a single cell in a cell monolayer of normal or zyxin-deficient cells and analyzed collective cell movement that requires contractile actin bundles. After the ablation of a single cell in a normal cell monolayer, an actin ring formed along the lining of wound edge (Fig. 5A). As the actin ring decreased in size, the wound circumference also decreased, suggesting that the contractile circumferential actin bundles are exerting forces at cell-cell junctions to close the opening and extrude out the dead cell. Zyxin and VASP-positive puncta localized to cell-cell junctions at the wound edge where the tips of actin bundles interconnect the neighboring cells (Fig. 5A). When a cell monolayer of GFP-tagged zyxin-expressing cells was wounded, zyxin-GFP rapidly accumulated to cell-cell junctions at the wound edge, and the zyxin puncta moved with the leading edge of closing wound (Fig. 5B and supplemental Movie S2), suggesting that zyxin, together with VASP, may play a role in the actin contraction between neighboring cells during the wound closure.
FIGURE 5.
Zyxin is required for efficient wound healing. A, formation of purse-string actin network and the localization of zyxin-VASP puncta during wound healing of normal cells. Single cell-sized wound in a cell monolayer was generated using a laser ablation system. B, time lapse montage of wound healing by GFP-tagged zyxin cells. The accumulation of zyxin at the wound edge is rapid and increases during wound closure. Time in minutes. See also supplemental Movie S2. C, absence of zyxin and VASP at the cell-cell junctions during wound healing of zyxin-deficient cells. D, quantification of the rate and degree of wound closure after 1 h. Scale bars, 10 μm.
In zyxin-deficient cells, a similar actin ring formed, but with no zyxin or VASP accumulation at the edge of cell-cell contacts facing the wound (Fig. 5C). Furthermore, the wound closure rate of zyxin-deficient cells was slower than wild type cells, and the wound closure was often incomplete (Fig. 5D). The partial zyxin-knockdown cells (∼40% of wild type zyxin levels (supplemental Fig. S3)) also exhibited a wound closure rate similar to the complete knockdown cells, albeit with a more complete wound closure (Fig. 5D). These results suggest that zyxin, together with VASP, regulates actin contraction at cell-cell junctions during wound closure.
DISCUSSION
In addition to the roles of zyxin at focal adhesions (supplemental Fig. S1) (5, 6, 10, 11), our results show that zyxin also regulates actin assembly at sites of cell-cell adhesion. Actin incorporation at cell-cell contacts was significantly reduced in zyxin-knockdown cells, especially at multicellular junctions where zyxin is often enriched (Fig. 4B). Unlike at focal adhesions, VASP accumulated along cell-cell contacts in zyxin-deficient cells. Therefore, zyxin-mediated actin assembly along the sites of cell-cell adhesion is independent of VASP localization.
On E-cadherin-coated substrates, actin assembly is attenuated at adhesion plaques and the lamellipodia in zyxin-knockdown cells (Fig. 3A). Interestingly, VASP is still localized to the tips of cadherin-based lamellipodia (Fig. 3C). Despite the absence of zyxin, VASP can interact with vinculin (20, 21), lipoma-preferred partner (22), and Wiskott-Aldrich syndrome protein (23). In zyxin-deficient cells, these proteins may recruit VASP to sites of cell-cell contacts. However, the defect in actin assembly at leading edges is not dependent on VASP localization, consistent with our observations at cell-cell adhesion (Fig. 4). Because actin polymerization at the lamellipodia is necessary for the formation of nascent cell adhesion on extracellular matrix (24, 25), the reduced actin polymerization at the leading edge, rather than the reduction in E-cadherin level, is likely responsible for the slow cell spreading on E-cadherin substrate. This also suggests that actin assembly may be important for the early stage of cell-cell adhesion as observed in hanging drop assays. Our data show that zyxin plays a role in the formation of cadherin-mediated adhesion in a VASP-independent manner.
Although VASP-positive junctions are often sites of actin assembly, it is not clear why the presence of VASP does not enhance actin assembly at cell-cell contacts of zyxin-knockdown cells. It is possible that the localization of VASP is not sufficient and VASP needs to be activated to promote actin assembly. Based on the analysis of overexpressed zyxin mutants and detergent insolubility as a measure of VASP activation, the LIM domains of zyxin have been suggested to negatively regulate the ability of the ActA domain to activate VASP at cell-cell adhesion sites (13). This may explain how the VASP-zyxin interaction regulates zyxin-bound VASP, but it does not explain how the rest of VASP may be regulated at cell-cell contacts.
Previous studies have shown that VASP has an intrinsic ability to regulate actin assembly. For example, the recruitment of VASP via the conserved proline-rich domain of ActA is sufficient to promote Listeria motility in a system with purified proteins (26). Furthermore, anticapping activity of VASP does not require any association with ActA (27), suggesting that VASP is constitutively active. However, VASP is not an actin nucleator. Instead, VASP activity is essential for the organization of branched versus linear actin network (28) and may not be essential for actin assembly at cell-cell adhesion sites. Actin assembly at sites of cell-cell adhesion may be regulated by other actin nucleators. For example, the Arp2/3 complex (29–31), formin-1 (32), and Dia1 (33) have been shown to localize to cell-cell contacts. Interestingly, VASP localization at cell-cell adhesion depends on Dia1 (33). The relationship between zyxin and other actin nucleators remains unclear.
Although VASP localization is independent of zyxin at cell-cell contacts (Fig. 4C) and at the tips of E-cadherin mediated lamellipodia (Fig. 3C), our study also demonstrates that VASP accumulation strongly depends on zyxin at the E-cadherin plaques (Fig. 3C) and at cell-cell junctions of wound-bordering cells (Fig. 5A). Interestingly, both junctions are located at the ends of prominent contractile actin bundles, suggesting that these junctions are under high mechanical load and that zyxin, a mechano-sensing protein, may be involved in force-induced VASP recruitment.
During wound closure, the cadherin junctions of wound-healing cells are under constant tension from an actin-myosin contractile “purse-string” that shortens to seal the wound (34). The circumferential actin bundles are enriched with myosin II, and together, they generate contractile forces (34, 35). The wound healing of a small incision depends on cadherin junctions since the addition of anti-cadherin antibody inhibits wound closure (36). Furthermore, the force-induced α-catenin-vinculin interaction is shown to occur at cell-cell junctions of wound-bordering cells (37). In addition, we found that, unlike VASP localization at quiescent cell-cell junctions (Fig. 4C), VASP accumulation at the edges of cell-cell junctions of wound-bordering cells strongly depends on zyxin, and the absence of zyxin and VASP at these cadherin junction sites decreases the rate of wound closure (Fig. 5). Therefore, our data suggest that the purse-string contraction of wound-healing cells requires actin assembly mediated by the zyxin·VASP complex.
Collectively, these data suggest that the mechano-sensing ability of zyxin is critical for VASP recruitment and the regulation of actin dynamics at force-bearing cell-extracellular matrix and cell-cell junctions. Previous studies have shown that the actin network at sites of cell-cell adhesion is highly dynamic in diverse systems (38, 39), yet it remains unclear how the mechanical forces affect the actin dynamics. Our study identifies zyxin as a key regulator of actin assembly at the sites of force-bearing cell adhesion in simple epithelial cells.
Supplementary Material
Acknowledgments
We thank Dr. Mary Beckerle (University of Utah) for sharing the zyxin B71 antibody, Dr. Juergen Wehland (Gesellschaft für Biotechnologische Forschung, Germany) for the zyxin-GFP plasmid, and Mimi Zhu for the analysis of the cell-spreading assay.
This work was supported by a Beckman Young Investigator Award (to S. Y.), a Hellman Family New Faculty Award (to S. Y.), and by the University of California Cancer Research Coordinating Committee (S. Y.), the Achievement Rewards for College Scientists Award (to T. N. N.), and Training Program in Biomolecular Technology Fellowship T32-GM08799 (to T. N. N.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and Movies S1 and S2.
- Ena
- Enabled
- VASP
- vasodilator-stimulated phospho-protein.
REFERENCES
- 1.Crawford A. W., Beckerle M. C. (1991) J. Biol. Chem. 266, 5847–5853 [PubMed] [Google Scholar]
- 2.Beckerle M. C. (1997) Bioessays 19, 949–957 [DOI] [PubMed] [Google Scholar]
- 3.Yoshigi M., Hoffman L. M., Jensen C. C., Yost H. J., Beckerle M. C. (2005) J. Cell Biol. 171, 209–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colombelli J., Besser A., Kress H., Reynaud E. G., Girard P., Caussinus E., Haselmann U., Small J. V., Schwarz U. S., Stelzer E. H. (2009) J. Cell Sci. 122, 1665–1679 [DOI] [PubMed] [Google Scholar]
- 5.Hirata H., Tatsumi H., Sokabe M. (2008) J. Cell Sci. 121, 2795–2804 [DOI] [PubMed] [Google Scholar]
- 6.Drees B., Friederich E., Fradelizi J., Louvard D., Beckerle M. C., Golsteyn R. M. (2000) J. Biol. Chem. 275, 22503–22511 [DOI] [PubMed] [Google Scholar]
- 7.Crawford A. W., Michelsen J. W., Beckerle M. C. (1992) J. Cell Biol. 116, 1381–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reinhard M., Jouvenal K., Tripier D., Walter U. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 7956–7960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nix D. A., Fradelizi J., Bockholt S., Menichi B., Louvard D., Friederich E., Beckerle M. C. (2001) J. Biol. Chem. 276, 34759–34767 [DOI] [PubMed] [Google Scholar]
- 10.Hoffman L. M., Jensen C. C., Kloeker S., Wang C. L., Yoshigi M., Beckerle M. C. (2006) J. Cell Biol. 172, 771–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fradelizi J., Noireaux V., Plastino J., Menichi B., Louvard D., Sykes C., Golsteyn R. M., Friederich E. (2001) Nat. Cell Biol. 3, 699–707 [DOI] [PubMed] [Google Scholar]
- 12.Bear J. E., Gertler F. B. (2009) J. Cell Sci. 122, 1947–1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen M. D., Beckerle M. C. (2006) J. Biol. Chem. 281, 16178–16188 [DOI] [PubMed] [Google Scholar]
- 14.Furman C., Sieminski A. L., Kwiatkowski A. V., Rubinson D. A., Vasile E., Bronson R. T., Fässler R., Gertler F. B. (2007) J. Cell Biol. 179, 761–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vasioukhin V., Bauer C., Yin M., Fuchs E. (2000) Cell 100, 209–219 [DOI] [PubMed] [Google Scholar]
- 16.Scott J. A., Shewan A. M., den Elzen N. R., Loureiro J. J., Gertler F. B., Yap A. S. (2006) Mol. Biol. Cell 17, 1085–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffman L. M., Nix D. A., Benson B., Boot-Hanford R., Gustafsson E., Jamora C., Menzies A. S., Goh K. L., Jensen C. C., Gertler F. B., Fuchs E., Fässler R., Beckerle M. C. (2003) Mol. Cell. Biol. 23, 70–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rottner K., Krause M., Gimona M., Small J. V., Wehland J. (2001) Mol. Biol. Cell 12, 3103–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lambert M., Thoumine O., Brevier J., Choquet D., Riveline D., Mège R. M. (2007) Exp. Cell Res. 313, 4025–4040 [DOI] [PubMed] [Google Scholar]
- 20.Brindle N. P., Holt M. R., Davies J. E., Price C. J., Critchley D. R. (1996) Biochem. J. 318, 753–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reinhard M., Rüdiger M., Jockusch B. M., Walter U. (1996) FEBS Lett. 399, 103–107 [DOI] [PubMed] [Google Scholar]
- 22.Petit M. M., Fradelizi J., Golsteyn R. M., Ayoubi T. A., Menichi B., Louvard D., Van de Ven W. J., Friederich E. (2000) Mol. Biol. Cell 11, 117–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castellano F., Le Clainche C., Patin D., Carlier M. F., Chavrier P. (2001) EMBO J. 20, 5603–5614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galbraith C. G., Yamada K. M., Galbraith J. A. (2007) Science 315, 992–995 [DOI] [PubMed] [Google Scholar]
- 25.Choi C. K., Vicente-Manzanares M., Zareno J., Whitmore L. A., Mogilner A., Horwitz A. R. (2008) Nat. Cell Biol. 10, 1039–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loisel T. P., Boujemaa R., Pantaloni D., Carlier M. F. (1999) Nature 401, 613–616 [DOI] [PubMed] [Google Scholar]
- 27.Pasic L., Kotova T., Schafer D. A. (2008) J. Biol. Chem. 283, 9814–9819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bear J. E., Svitkina T. M., Krause M., Schafer D. A., Loureiro J. J., Strasser G. A., Maly I. V., Chaga O. Y., Cooper J. A., Borisy G. G., Gertler F. B. (2002) Cell 109, 509–521 [DOI] [PubMed] [Google Scholar]
- 29.Yamada S., Nelson W. J. (2007) J. Cell Biol. 178, 517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verma S., Shewan A. M., Scott J. A., Helwani F. M., den Elzen N. R., Miki H., Takenawa T., Yap A. S. (2004) J. Biol. Chem. 279, 34062–34070 [DOI] [PubMed] [Google Scholar]
- 31.Kovacs E. M., Goodwin M., Ali R. G., Paterson A. D., Yap A. S. (2002) Curr. Biol. 12, 379–382 [DOI] [PubMed] [Google Scholar]
- 32.Kobielak A., Pasolli H. A., Fuchs E. (2004) Nat. Cell Biol. 6, 21–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carramusa L., Ballestrem C., Zilberman Y., Bershadsky A. D. (2007) J. Cell Sci. 120, 3870–3882 [DOI] [PubMed] [Google Scholar]
- 34.Jacinto A., Martinez-Arias A., Martin P. (2001) Nat. Cell Biol. 3, E117–E123 [DOI] [PubMed] [Google Scholar]
- 35.Bement W. M., Forscher P., Mooseker M. S. (1993) J. Cell Biol. 121, 565–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Danjo Y., Gipson I. K. (1998) J. Cell Sci. 111, 3323–3332 [DOI] [PubMed] [Google Scholar]
- 37.Yonemura S., Wada Y., Watanabe T., Nagafuchi A., Shibata M. (2010) Nat. Cell Biol. 12, 533–542 [DOI] [PubMed] [Google Scholar]
- 38.Cavey M., Rauzi M., Lenne P. F., Lecuit T. (2008) Nature 453, 751–756 [DOI] [PubMed] [Google Scholar]
- 39.Yamada S., Pokutta S., Drees F., Weis W. I., Nelson W. J. (2005) Cell 123, 889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.