Abstract
Most mammalian cell types depend on multiple Src family kinases (SFKs) to regulate diverse signaling pathways. Strict control of SFK activity is essential for normal cellular function, and loss of kinase regulation contributes to several forms of cancer and other diseases. Previous x-ray crystal structures of the SFKs c-Src and Hck revealed that intramolecular association of their Src homology (SH) 3 domains and SH2 kinase linker regions has a key role in down-regulation of kinase activity. However, the amino acid sequence of the Hck linker represents a suboptimal ligand for the isolated SH3 domain, suggesting that it may form the polyproline type II helical conformation required for SH3 docking only in the context of the intact structure. To test this hypothesis directly, we determined the crystal structure of a truncated Hck protein consisting of the SH2 and SH3 domains plus the linker. Despite the absence of the kinase domain, the structures and relative orientations of the SH2 and SH3 domains in this shorter protein were very similar to those observed in near full-length, down-regulated Hck. However, the SH2 kinase linker adopted a modified topology and failed to engage the SH3 domain. This new structure supports the idea that these noncatalytic regions work together as a “conformational switch” that modulates kinase activity in a manner unique to the SH3 domain and linker topologies present in the intact Hck protein. Our results also provide fresh structural insight into the facile induction of Hck activity by HIV-1 Nef and other Hck SH3 domain binding proteins and implicate the existence of innate conformational states unique to individual Src family members that “fine-tune” their sensitivities to activation by SH3-based ligands.
Keywords: Crystal Structure, Crystallography, Protein Kinases, Protein Structure, SH2 Domains, SH3 Domains, Src, Tyrosine Kinase, Hck, Src Family Kinase
Introduction
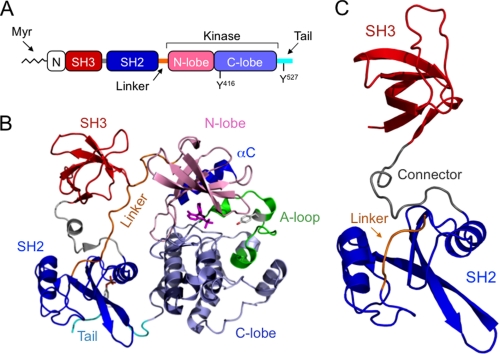
The human kinome encodes 11 Src-related kinase sequences (1). Nearly all mammalian cell types express multiple Src family kinases (SFKs),2 which regulate diverse pathways involved in cell growth, survival, differentiation, adhesion, and migration (2). Strict control of SFK activity is essential to normal cellular function, and loss of kinase regulation contributes to several forms of cancer and other diseases (3). The conserved structural organization of SFKs includes a myristoylated (and in most cases palmitoylated) N-terminal region, modular SH3 and SH2 domains joined by a short connector, an SH2 kinase linker, a bilobed kinase domain, and a C-terminal negative regulatory tail (Fig. 1A) (2). The crystal structures of several near full-length SFKs, consisting of SH3, SH2, and kinase domains plus a tyrosine-phosphorylated C-tail, demonstrate that intramolecular interactions involving the SH3 and SH2 domains both contribute to negative regulation of kinase activity (4–6). These noncatalytic domains pack in tandem against the back surface of the kinase domain, stabilizing it in a closed, inactive conformation (Fig. 1B). The crystal structures revealed that the SH3 domain engages the SH2 kinase linker, which adopts a polyproline type II (PPII) helical conformation in the context of the overall structure, typical of most SH3 ligands (Fig. 1B). The SH2 domain binds to the C-terminal tail, which is phosphorylated on a conserved tyrosine residue by the independent regulatory kinases, Csk and Chk (7).
FIGURE 1.
X-ray crystal structure of the Hck regulatory region. A, domain organization of Src family kinases. See text for descriptions. B, crystal structure of near full-length, down-regulated Hck (PDB code 1QCF). Note that the SH3 domain (red) engages the SH2 kinase linker (orange), and the SH2 domain (blue) binds to the tyrosine-phosphorylated tail (cyan). Together, these interactions pack against the back of the kinase domain away from the active site to stabilize the inactive conformation. C, crystal structure of the Hck SH3-SH2-linker region (Hck32L). A and B were adapted from a review article by Engen et al. (2) for comparative purposes.
Intramolecular interactions maintain the kinase in an inactivated state, and disruption of these regulatory interactions often leads to SFK activation. In the case of Hck, a member of the Src kinase family that is expressed in macrophages and other myeloid hematopoietic cells (8), mutations within the SH3 domain induce kinase activation presumably by disrupting interaction with the SH2 kinase linker. Complementary mutations in the SH2 kinase linker region, distal to the kinase domain, also are sufficient to induce kinase activation. Replacement of the tyrosine residue in the negative regulatory tail with phenylalanine is also sufficient to activate the kinase domain by releasing the tail from the SH2 domain (9, 10).
Intermolecular interactions with partner proteins can also induce kinase activation, by displacing the SH3 domain, the SH2 domain, or both, from their regulatory positions on the back of the kinase domain. For example, the Nef protein of HIV-1 binds tightly to the SH3 domain of Hck, inducing constitutive kinase activation both in vitro and in cells (11–14). In the case of c-Src, one mode of activation involves bipartite interaction with focal adhesion kinase, which causes displacement of both the SH3 and SH2 domains from their respective internal ligand sites (15). These observations suggest that different members of the Src kinase family have distinct requirements for activation, which may be dictated in part by the affinities of their respective SH2 and SH3 domains for their intramolecular binding sites.
The two distinct modes regulating SFK activity, one involving SH3-linker interaction and the other SH2-tail engagement, has raised the question of whether disruption of either interaction alone is sufficient for activation or whether displacement of both is required (2). Previous work has shown that tightening the Hck SH2-tail interaction does not impair kinase activation through SH3 displacement by Nef in cells (12) or in vitro (14). These observations suggest that the SH2 kinase linker may function as an “activation switch,” with perturbation of its interaction with the SH3 domain sufficient for kinase activation at least in the context of Hck. An SH3-dominant mode of activation such as this predicts that the SH2 kinase linker may represent a suboptimal ligand for the SH3 domain in the context of the down-regulated Hck structure. To approach this question from a structural perspective, we solved the x-ray crystal structure of the complete Hck regulatory region (SH3-SH2 linker) in the absence of the kinase domain. We observed that the structures and relative orientations of the SH2 and SH3 domains in this shorter protein were very similar to those observed in near full-length, down-regulated Hck. However, the SH2 kinase linker adopted a modified structure in the absence of the kinase domain and failed to engage the SH3 domain. This new structure supports the idea that the linker region exists as a PPII helix only in the context of the overall down-regulated structure. We also observed that a high affinity peptide ligand for SFK SH3 domains readily induced activation of a form of Hck in which the SH2-tail interaction is locked. In contrast, the same peptide had no effect on the activity of a similar down-regulated form of c-Src. Our data provide a structural basis for the observation that Hck can be readily activated by Nef and other SH3-binding proteins (10, 12). These data imply that innate conformational differences exist between Src family members that “fine-tune” their sensitivities to activation by SH3-based ligands and indicate isoform-specific mechanisms may modulate kinase activities.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of Recombinant Hck32L
The coding region for the human Hck SH3 and SH2 domains plus the SH2 kinase linker (residues 72–256, Hck32L; Hck numbering) was amplified by PCR and subcloned into the pET21a bacterial expression vector (EMD Chemicals Inc., San Diego) using NdeI and XhoI restriction sites. The nucleotide sequence was verified for the entire coding region. The Hck32L protein was expressed with the vector-encoded C-terminal His tag in Escherichia coli Rosetta 2 (DE3) cells. Cultures were grown in Luria-Bertani medium, and protein expression was induced with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside at 18 °C for 16 h. Soluble Hck32L protein was extracted using a microfluidizer (Microfluidics, Newton, MA) and purified using a nickel-nitrilotriacetic acid column (GE Healthcare). Soluble aggregates were removed by gel filtration chromatography using a Hi-Load Superdex 75 16/60 column (GE Healthcare) equilibrated with 25 mm sodium phosphate, pH 7.5, 150 mm NaCl, 10% glycerol, 1 mm DTT, and 0.02% sodium azide. The peak containing Hck32L was further purified over Hi-Trap QP (GE Healthcare) at pH 7.5 and a 0–1.0 m NaCl gradient.
Expression and Purification of Recombinant SFKs and in Vitro Kinase Assay
Recombinant Hck-YEEI and Src-YEEI proteins were expressed in Sf-9 insect cells and purified by ion exchange and immobilized metal affinity chromatography as described elsewhere (14). The SH3 domain binding peptide VSL12 (VSLARRPLPPLP) (16) was synthesized by the University of Pittsburgh Genomics and Proteomics Core Laboratory. VSL12 mass and purity were verified by mass spectrometry. In vitro kinase assays were performed using the Tyr-2 FRET peptide substrate and the Z'-LyteTM assay (Invitrogen) as described previously (14, 17).
Crystallization
Diffraction quality crystals were grown by sitting-drop vapor diffusion at 20 °C by mixing Hck32L (6.5 mg/ml), 25% (v/v) polyethylene glycol 3,350, 0.15 m KSCN, 0.05 m NaCl, and a 1:100 dilution of Hck32L crystal seeds (grown in 25% v/v polyethylene glycol 3,350, 0.1 m Tris, pH 8.5, 0.2 m NaCl). Crystals formed in 7–10 days with approximate dimensions of 0.3 × 0.05 × 0.05 mm. Prior to data collection, crystals were cryoprotected by stepwise transfer to mother liquor supplemented with a final concentration of ethylene glycol to 10% (v/v) and flash-cooled in liquid nitrogen.
Data Collection and Processing
X-ray diffraction data (Table 1) were collected to a resolution of 2.6 Å from a single Hck32L crystal. All data were collected at the University of Pittsburgh X-ray Facility using x-rays of CuKα wavelength and Rigaku Saturn 944 CCD detector. Data integration and scaling were carried out using HKL2000 (18). Diffraction from the Hck32L crystal is consistent with the trigonal P31 space group with one molecule in the asymmetric unit.
TABLE 1.
X-ray data collection and structure refinement statistics for Hck32L
| Data collectiona | |
| Space group | P31 |
| a, b, c | 60.71, 60.71, 49.58 Å |
| α, β, γ | 90.0, 90.0, 120.0° |
| Resolution | 50.00 to 2.61 Å (2.66 to 2.61 Å) |
| Rmerge | 0.056 (0.278) |
| Average I/σI | 15.0 (2.6) |
| Completeness | 98.2% (78.9%) |
| Redundancy | 3.4 (2.3) |
| Model refinementb | |
| Resolution | 25.89 to 2.61 Å |
| No. of reflections | 5,920 |
| Rwork/Rfree | 0.12/0.17 |
| Non-H atoms | |
| Protein | 1,314 |
| Solvent | 35 |
| Average B factors | |
| Protein | 43.11 Å2 |
| Solvent | 39.94 Å2 |
| Root mean square deviations | |
| Bond lengths | 0.007 Å |
| Bond angles | 1.062° |
| Ramachandran | |
| Outliers | 0.0% |
| Generous | 14.9% |
| Favored | 85.1% |
a Values in parentheses are for highest resolution shell.
b Data cutoff for refinement was F/σF ≥ 2.
Structure Determination and Refinement
The structure factor data were analyzed using the program phenix.xtriage (19), and pseudo-merohedral twinning was detected possessing the twin-law operator (-h, -k, l) and twin fraction (0.48). Structure solution of Hck32L was determined by molecular replacement with the program PHASER (19), using the structure coordinates of the human Hck kinase (PDB code 1QCF) SH3 and SH2 domains as independent search models. The molecular replacement solutions were refined using rigid-body, individual coordinate, individual isotropic b-factor, occupancy, simulated annealing, and TLS refinement using the program phenix. refine (19). After the last cycle of refinement and model building, water molecules were added using phenix.refine. A final round of refinement using all atoms was then conducted, and final refinement statistics are summarized in Table 1. Model building was conducted using the program Coot (20). The stereochemical quality of the final refined model was evaluated using Procheck (21) and MolProbity (22). Figures of the x-ray structure and electron density were produced using PyMol (23). The program ESPript (24) was used to produce sequence alignment shown in Fig. 2C.
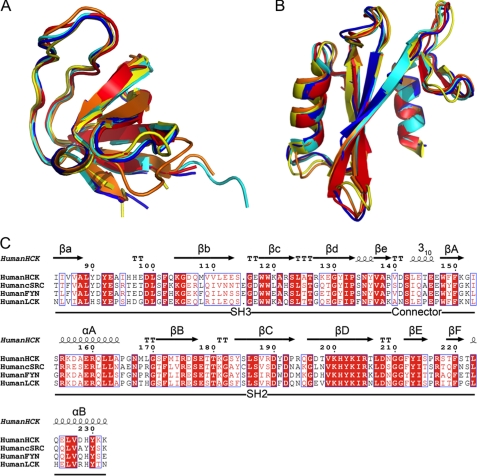
FIGURE 2.
Superposition of Src family kinase SH3 and SH2 domains. A, superposition of SH3 domains from Hck32L (red), Fyn32 (yellow; PDB code 1G83), Lck32 (blue; PDB code 1LCK), down-regulated near full-length Hck (cyan; PDB code 1QCF), and down-regulated near full-length c-Src (orange; PDB code 2SRC). B, superposition of the SH2 domains from the same structures in A. C, structure-based sequence alignment of SH3 and SH2 domains of Hck, c-Src, Fyn, and Lck (all residue numbers are based on the human c-Src crystal structure; PDB code 2SRC).
RESULTS AND DISCUSSION
Conservation of SH2 and SH3 Domain Architecture
Previous structural analyses of the down-regulated conformations of Hck and c-Src revealed intramolecular interactions between the SH3 domain and the SH2 kinase linker (see Introduction). However, subsequent studies have shown that the amino acid sequence of the linker represents a suboptimal ligand for the isolated SH3 domain (25, 26), suggesting that it may form the PPII helical conformation required for SH3 docking only in the context of the overall structure. To test this hypothesis directly, we determined the x-ray crystal structure of the regulatory region of Hck to 2.6 Å. This protein encompasses the SH3 and SH2 domains as well as the SH2 kinase linker (referred to hereafter as Hck32L) (Fig. 1C). Consistent with the structures of other SH3 and SH2 domains, the Hck32L SH3 domain folds as a β-sandwich composed of five β-strands (βa, βb, βc, βd, and βe), and the SH2 domain is composed of a central six-stranded β-sheet (βA, βB, βC, βD, βE, and βF) flanked by two α-helices (αA and αB) (Fig. 1C) (5, 6, 27–31). Superposition of the individual SH3 and SH2 domains in the Hck32L fragment with those from the structures of down-regulated near full-length human Hck and c-Src, as well as SH3-SH2 structures of human Lck and Fyn, demonstrates strong structural conservation among these domains despite varying degrees of sequence identity (Fig. 2 and Table 2).
TABLE 2.
Sequence and Structural Comparison of Hck32L domains with other SFKs
Sequence identity and superposition root mean square deviations (r.m.s.d.) were calculated using Fyn32 (PDB code 1G83), Lck32 (PDB code 1LCK), near full-length Hck (PDB code 1QCF), and near full-length c-Src (PDB code 2SRC) structures with the indicated number of α-carbon atoms aligned. Superpositions were calculated using secondary structure matching in Coot (20).
| Hck32L | SH3 domain |
SH2 domain |
||||
|---|---|---|---|---|---|---|
| Sequence identity | α-Carbon RMSD | α-Carbon atoms | Sequence identity | α-Carbon RMSD | α-Carbon atoms | |
| % | Å | % | Å | |||
| Fyn32 | 60 | 0.9 | 57 | 62 | 1.3 | 98 |
| Lck32 | 56 | 1.1 | 59 | 71 | 0.9 | 100 |
| Hck | 100 | 0.7 | 57 | 100 | 0.6 | 100 |
| c-Src | 53 | 1.2 | 57 | 54 | 1.1 | 100 |
Relationship of SH3 and SH2 Domains within the Hck32L Crystal Structure
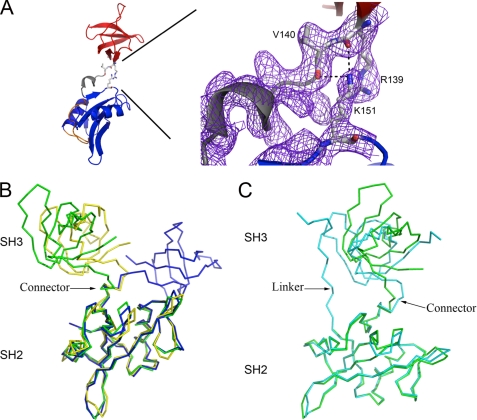
The SH3 and SH2 domains in the Hck32L crystal structure make minimal interdomain contacts, consistent with previous dynamics studies (25). One exception involves the main-chain carbonyls of Arg-139 and Val-140 (SH3 domain), which form salt bridges with the side-chain Nζ atom of Lys-151 (SH2 domain) (Fig. 3A). Limited SH3-SH2 inter-domain contacts are found in other tandem SH3-SH2 domain arrangements, including those in the Fyn32, Lck32, down-regulated c-Src and Hck, as well as the c-Abl core, which closely resembles c-Src and Hck in terms of regulatory domain orientation (5, 6, 27–33). A connector of ∼10 amino acids joins the SH3 and SH2 domains in the Hck32L structure (residues 140–147 in Hck; all numbering based on the human c-Src crystal structure; PDB code 2SRC). This connector forms a 310-helix (Fig. 1C), a structural feature observed in other SFKs (5, 6, 27–31).
FIGURE 3.
Comparison of Src family kinase SH3-SH2 domain architecture. A, interdomain contacts in Hck32L involve numerous salt bridges (denoted by black dashes) between residues Arg-139/Val-140 (depicted as balls-and-sticks) in the SH3 and Lys-151 in the SH2 domains. These residues have well defined conformations as shown by the 2Fo − Fc electron density map (purple mesh), generated in Coot (20), and contoured at 1σ. B, comparisons of the tandem SH3-SH2 domain organizations in Hck32L (green), Fyn32 (yellow), and Lck32 (blue). C, comparison of the SH3-SH2 domain organization in Hck32L (green) to the down-regulated near full-length Hck (cyan) (PDB identifiers as indicated in Fig. 2).
We next superimposed the SH2 domain of Hck32L with that of the Fyn32 and Lck32 structures and compared the relative orientations of the SH3 domains (Fig. 3B). This analysis shows that the relative orientations of the SH3 and SH2 domains are very similar between the Hck and Fyn structures, with the SH3 domains rotated away from each other by only 17° (angle between axes passing through the center of mass of each domain). However, the Lck SH3 domain orientation significantly differs from Hck and is rotated away from the SH2 domain by 100° relative to the Hck SH3 domain. This comparison demonstrates inter-domain flexibility across SFKs even in the crystal state and is consistent with 15N NMR relaxation data that showed differences in correlation times between the SH3-SH2 domains of Fyn, Lck, and c-Abl (27, 34, 35).
In near full-length down-regulated Hck, hydrophobic residues on the surface of SH3 (Ile-85, Val-87, and Leu-89) and SH2 (Trp-148, Phe-149, Tyr-184, and Leu-223) domains flank the connector 310-helix, potentially allowing some rotational movement of the domains with respect to each other but preventing gross reorientation that would expose these surfaces to an aqueous environment. To determine the structural constraints imparted by the kinase domain, the SH2 domain of our new Hck32L structure was superimposed onto that of near full-length down-regulated Hck, resulting in a root mean square deviation of 0.7 Å (over all α-carbons, 86 atoms) (Fig. 3C). This analysis reveals that the SH3 domain orientation of Hck32L deviates to some extent from the position of the SH3 domain in this near full-length Hck structure. The SH3 domains of Hck32L and near full-length Hck are oriented away from each other by 55° and deviate by 0.8 Å over 55 α-carbon atoms. The Fyn32 and Lck32 SH3 domains are oriented away and deviate from the SH3 domain of down-regulated near full-length Hck by 83°/0.8 Å and 96°/2.4 Å, respectively (superpositions not shown). The large deviation of the Lck SH3 domain may be attributed to the composition of the Lck inter-domain connector, which has a Pro-Xaa-Pro motif not found in Fyn, Hck, or c-Src. A conserved salt bridge (found in Fyn and c-Src) between a Lys (Lys-151) in the SH3 domain and a Glu (Glu-147) in the SH3-SH2 connector is absent in Lck due to the replacement of Glu with Pro in the Pro-Xaa-Pro motif. Loss of this SH3-connector interaction and the conserved β-turn that precedes the 310-helix in the connector, may destabilize a conformation compatible with the SH3 domain orientation found in the down-regulated states of Hck and c-Src. The SH3 domain orientation found in Lck32 may be stabilized by formation of a salt bridge between the main-chain amide of Lys-139 (SH3 domain) and the side-chain carboxylate of Glu-225 (SH2 domain).
The altered SH3 domain orientation observed in our Hck32L structure in comparison with the larger down-regulated Hck structure may be attributed to the loss of the conserved β-turn that precedes the 310-helix in the connector, and formation of inter-domain salt bridges between the main-chain carbonyl oxygen atoms of Arg-139 and Val-140 and a side-chain nitrogen of Lys-151 as described above (Fig. 3A). It is noteworthy that molecular dynamics simulations on near full-length down-regulated Hck and c-Src, which modeled the conformational changes involved in inducing the activated state, suggest that coupling between the SH3 and SH2 domains in the down-regulated state is dependent on the connector region. However, upon activation, the β-turn and 310-helix are structurally modified, resulting in adoption of a more flexible connector conformation (36). In addition, replacement of c-Src connector residues with glycines induced kinase activation, supporting a role for the connector in suppressing activation (36). Consequently, the conformation of the connector region observed in Hck32L may reflect its structure in an activated state, with the connector functioning as a conformational “switch” to regulate the activity of the kinase. Although the SH3 domain orientation in Hck32L may result from unstructuring of the β-turn preceding the connector and crystal contacts, the domain organization described here is also observed in a solution NMR analysis of an identical Hck32L protein.3 These observations suggest that activation of Hck by SH3 and/or SH2 domain displacement may lead to a similar reorganization of the SH3-SH2 region initiated by the connector 310-helical switch.
SH2 Kinase Linker Is Largely Unstructured in Hck32L
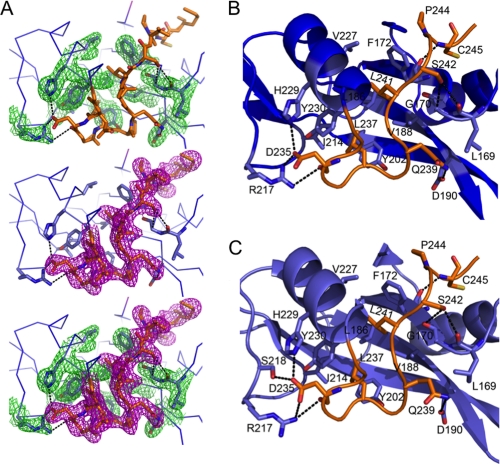
The complete SH2 kinase linker (residues Gly-233 to Glu-261) is present in the Hck32L protein, and electron density for the SH2-proximal linker residues Gly-233 to Met-246 was visible (Fig. 4A). As shown in Fig. 4B, these residues form hydrogen bonds, hydrophobic interactions, and salt bridges with residues in the SH2 domain. Hydrophobic interactions are observed between linker residue Leu-237 and SH2 Tyr-202, Ile-214, and Tyr-230; linker Leu-241 and SH2 Phe-172, Leu-186, Val-188, and Val-227; and linker Pro-244 and SH2 Phe-172. Flanking these hydrophobic interactions are hydrogen bonding and ionic contacts between linker residue Asp-235 and SH2 Arg-217, Ser-218, and His-229; linker Gln-239 and SH2 Asp-190; linker Ser-242 and SH2 Gly-170; and linker Cys-245 and SH2 Gln-161 and Phe-172. Very similar interactions are conserved in the down-regulated form of near full-length Hck (Fig. 4C) and may help to anchor the N-terminal end of the linker in the proper orientation for SH3 domain engagement in the full-length structure.
FIGURE 4.
Conservation of SH2 contacts with the proximal end of the linker in Hck32L and near full-length Hck. A, extensive hydrogen bonding and salt bridges (both indicated by black dashed lines) between side chains stabilize the conformation of the linker adjacent to the SH2 domain in Hck32L. These interactions are evident from the well ordered and defined electron density maps, generated in Coot (20), with 2Fo − Fc map coefficients, contoured at 1σ showing the SH2 domain residues (top, green mesh), linker residues (middle, magenta mesh), and both SH2 domain/linker residues (bottom, green/magenta mesh). B, interactions between the SH2 domain (blue) and SH2 kinase linker (orange) in Hck32L. C, interactions between the SH2 domain (light blue) and SH2 kinase linker (orange) within near full-length, down-regulated Hck (PDB code 1QCF).
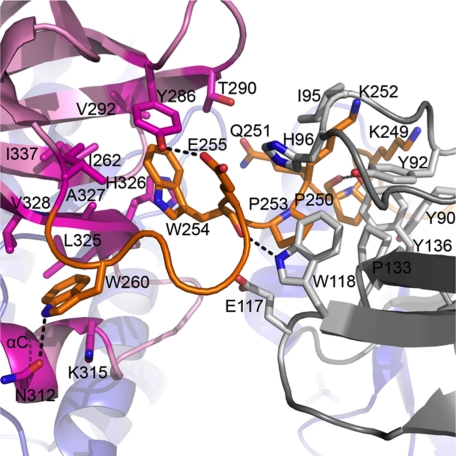
In the context of the near full-length down-regulated structure of Hck, the SH2 kinase linker makes multiple contacts with both the SH3 and kinase domains that contribute, along with SH2-tail interaction, to maintenance of the down-regulated state (5, 6, 13, 29, 31). SH2-tail interaction orients the linker to permit intercalation between the SH3 and kinase domains, positioning the linker PPII helix for interaction with the SH3 domain. In addition, the linker makes two contacts with the N-lobe of the kinase domain. These contacts involve Trp-254 and Trp-260, which insert into a hydrophobic pocket that is formed by the β-sheet and αC helix of the N-lobe (modeled in Fig. 5). The SH2 kinase linker sequence retained in the Hck32L protein possesses all of the determinants required for PPII helix formation and SH3 engagement observed in the down-regulated structure of near full-length Hck (5, 29). However, analysis of Hck32L crystal packing shows that crystal contacts are formed instead between the SH2 domain and the PPII helix-binding site of the SH3 domain. These crystal contacts are consistent with the occlusion of the linker from the SH3-binding site. In addition, linker disorder in Hck32L may also be due to the loss of the Trp-254 and Trp-260 interactions, which require the presence of the N-lobe of the kinase domain. These observations provide direct structural evidence that without the kinase domain, the linker cannot adopt the PPII helical conformation essential for SH3 binding. Our structural findings are supported by previous dynamic analysis of an identical Hck32L construct using hydrogen-deuterium exchange coupled to mass spectrometry, which also failed to detect SH3-linker interaction (25). In addition, this study showed that a peptide based on the linker sequence failed to interact with a tandem Hck SH3-SH2 protein in trans, supporting the idea that the linker is intrinsically unstructured (25). Taken together, these results strongly suggest that the presence of the kinase domain is essential to structure the linker in a PPII helical conformation compatible with SH3 binding and thus plays an important role in driving its own down-regulation.
FIGURE 5.
SH2 kinase linker interacts with the SH3 and kinase domains in near full-length Hck. Side chains of interacting residues of the SH2 kinase linker (orange), SH3 domain (gray), and kinase domain (pink) are shown as sticks, and hydrogen bonding is indicated by the black dashed lines (PDB code 1QCF).
Activation of Hck-YEEI but Not Src-YEEI by an SH3 Domain Peptide Ligand
The finding that the linker in Hck32L is unstructured and does not interact with the SH3 domain suggests that the native linker sequence is a poor ligand for the SH3 domain, even in the context of full-length down-regulated Hck. In contrast, a recent crystal structure of c-Src, in which the SH2 domain is released from the tail and the kinase adopts an active conformation (37), reveals that the linker remains bound to the SH3 domain despite the loss of almost all kinase domain contacts (modeled in Fig. 6A). This difference suggests that Hck may be more susceptible than c-Src to activation by SH3-directed ligands. To test this hypothesis, we expressed and purified both Hck and c-Src in their near full-length, down-regulated forms. Both proteins also have modified C-terminal tails, in which the natural sequence following the conserved regulatory tyrosine is replaced with pTyr-Glu-Glu-Ile (pYEEI) to enhance SH2-tail interaction (5, 14, 38). This modification allowed us to address the impact of an SH3-directed peptide ligand on kinase activation through the SH3 domain, while keeping the SH2-tail interaction intact.
FIGURE 6.
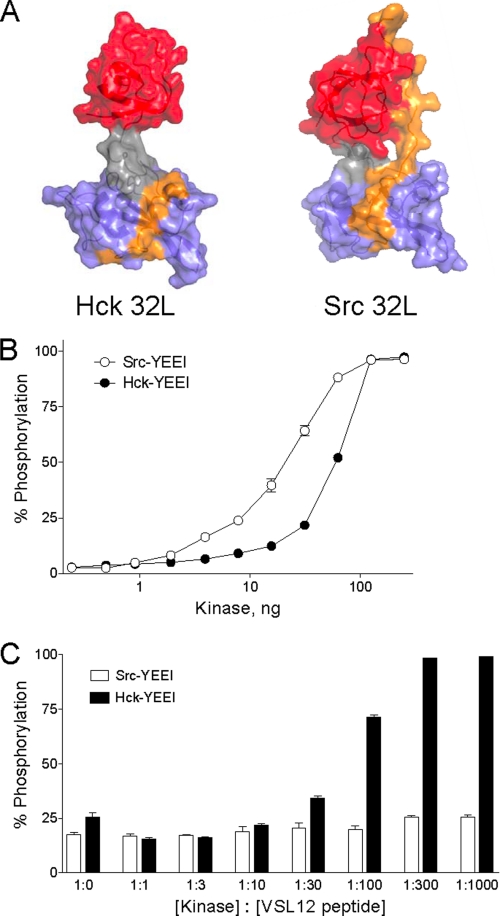
Activation of Hck but not c-Src by an SH3-binding peptide. A, comparison of the Hck32L crystal structure (this study) and the corresponding SH3-SH2 linker region from an active c-Src structure (PDB code 1Y57). In the c-Src structure, the SH3-SH2 linker region is uncoupled from the kinase domain (see text for details). The SH3 domain is rendered in red; the SH3-SH2 connector in gray; the SH2 domain in blue, and the linker in orange. Note that the linker remains bound to the SH3 domain in the c-Src structure but is disordered and therefore not observed in the Hck32L structure with the exception of SH2 contacts. B, Src-YEEI and Hck-YEEI are active in vitro. Recombinant purified Src-YEEI and Hck-YEEI were titrated into the Z'-LyteTM assay as shown. Each reaction was monitored in triplicate, and data are expressed as percent of peptide phosphorylation relative to a control phosphopeptide ± S.D. C, SH3-binding peptide VSL12 activates Hck-YEEI but not Src-YEEI. In vitro kinase assays were performed using equivalent amounts of Src-YEEI and Hck-YEEI that gave ∼20–25% maximal activity in the absence of the VSL12 peptide. Reactions were run in the presence of increasing kinase: VSL12 peptide ratios as shown. Each reaction was monitored in triplicate, and data are expressed as percent of peptide phosphorylation relative to a control phosphopeptide ± S.D.
We first determined the specific activity of each recombinant SFK in vitro with a FRET-peptide substrate (see “Experimental Procedures”). Each kinase was titrated into the assay, and maximal activity was observed with about 100 ng of input kinase per well (Fig. 6B). Under these conditions, kinase activation most likely results from random collisions of kinase molecules, the frequency of which rises as a function of the amount of kinase protein present. Based on these kinase titration curves, we then chose amounts of Hck-YEEI and c-Src-YEEI that gave similar sub-maximal levels of kinase activity to test the effect of a peptide ligand for the SH3 domain on kinase activation. The peptide ligand used in these studies is known as VSL12, a 12-mer with the sequence VSLARRPLPPLP that has similar affinities for the isolated Src and Hck SH3 domains (0.5–1.0 μm as determined by surface plasmon resonance) (16, 26). As shown in Fig. 6C, addition of the VSL12 peptide ligand induced complete activation of Hck-YEEI. The EC50 for half-maximal Hck activation was observed at a kinase:peptide molar ratio between 1:30 and 1:100. In striking contrast, VSL12 had no effect on c-Src-YEEI activity, even at a kinase:peptide ratio of 1:1000. This observation suggests that within full-length Hck, intra-molecular SH3-linker interaction is relatively weak, and the kinase domain is therefore susceptible to activation by SH3-directed ligands that perturb this negative regulatory interaction. In contrast, the inability of the peptide to stimulate c-Src-YEEI suggests that regulation of this kinase may also require SH2-tail displacement. Indeed, previous work has shown that peptides based on focal adhesion kinase, a physiological signaling partner for c-Src, require dual engagement of both the SH3 and SH2 domains for kinase activation in vitro (15). Together with the new Hck32L crystal structure presented here, our data strongly support the idea that individual Src family members may have inherently different sensitivities to SH3- and SH2-based inputs despite the overall similarity in the domain organization of their down-regulated conformations. These differences may help to explain their ability to respond in unique ways to specific cellular environments and to serve nonredundant functions even when co-expressed in the same cellular context.
Acknowledgments
We thank Drs. Jinwoo Ahn, Hyun Lee, and Inja Byeon for their valuable assistance with protein preparations.
This work was supported in part by National Institutes of Health Grants AI077444 and GM082251 (Pittsburgh Center for HIV-Protein Interactions).
The atomic coordinates and structure factors (code 3NHN) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
I. Byeon and A. Gronenborn, unpublished data.
- SFK
- Src family kinase
- SH
- Src homology
- PDB
- Protein Data Bank
- PPII
- polyproline type II.
REFERENCES
- 1.Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. (2002) Science 298, 1912–1934 [DOI] [PubMed] [Google Scholar]
- 2.Engen J. R., Wales T. E., Hochrein J. M., Meyn M. A., 3rd, Banu Ozkan S., Bahar I., Smithgall T. E. (2008) Cell. Mol. Life Sci. 65, 3058–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim L. C., Song L., Haura E. B. (2009) Nat. Rev. Clin. Oncol. 6, 587–595 [DOI] [PubMed] [Google Scholar]
- 4.Boggon T. J., Eck M. J. (2004) Oncogene 23, 7918–7927 [DOI] [PubMed] [Google Scholar]
- 5.Schindler T., Sicheri F., Pico A., Gazit A., Levitzki A., Kuriyan J. (1999) Mol. Cell 3, 639–648 [DOI] [PubMed] [Google Scholar]
- 6.Xu W., Doshi A., Lei M., Eck M. J., Harrison S. C. (1999) Mol. Cell 3, 629–638 [DOI] [PubMed] [Google Scholar]
- 7.Chong Y. P., Ia K. K., Mulhern T. D., Cheng H. C. (2005) Biochim Biophys. Acta 1754, 210–220 [DOI] [PubMed] [Google Scholar]
- 8.Guiet R., Poincloux R., Castandet J., Marois L., Labrousse A., Le Cabec V., Maridonneau-Parini I. (2008) Eur. J. Cell Biol. 87, 527–542 [DOI] [PubMed] [Google Scholar]
- 9.Briggs S. D., Smithgall T. E. (1999) J. Biol. Chem. 274, 26579–26583 [DOI] [PubMed] [Google Scholar]
- 10.Schreiner S. J., Schiavone A. P., Smithgall T. E. (2002) J. Biol. Chem. 277, 45680–45687 [DOI] [PubMed] [Google Scholar]
- 11.Briggs S. D., Sharkey M., Stevenson M., Smithgall T. E. (1997) J. Biol. Chem. 272, 17899–17902 [DOI] [PubMed] [Google Scholar]
- 12.Lerner E. C., Smithgall T. E. (2002) Nat. Struct. Biol. 9, 365–369 [DOI] [PubMed] [Google Scholar]
- 13.Moarefi I., LaFevre-Bernt M., Sicheri F., Huse M., Lee C. H., Kuriyan J., Miller W. T. (1997) Nature 385, 650–653 [DOI] [PubMed] [Google Scholar]
- 14.Trible R. P., Emert-Sedlak L., Smithgall T. E. (2006) J. Biol. Chem. 281, 27029–27038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas J. W., Ellis B., Boerner R. J., Knight W. B., White G. C., 2nd, Schaller M. D. (1998) J. Biol. Chem. 273, 577–583 [DOI] [PubMed] [Google Scholar]
- 16.Rickles R. J., Botfield M. C., Zhou X. M., Henry P. A., Brugge J. S., Zoller M. J. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 10909–10913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emert-Sedlak L., Kodama T., Lerner E. C., Dai W., Foster C., Day B. W., Lazo J. S., Smithgall T. E. (2009) ACS Chem. Biol. 4, 939–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otwinowski Z., Minor W., Carter C. W., Jr. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 19.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 21.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 22.Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 23.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, Palo Alto, CA [Google Scholar]
- 24.Gouet P., Robert X., Courcelle E. (2003) Nucleic Acids Res. 31, 3320–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hochrein J. M., Lerner E. C., Schiavone A. P., Smithgall T. E., Engen J. R. (2006) Protein Sci. 15, 65–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lerner E. C., Trible R. P., Schiavone A. P., Hochrein J. M., Engen J. R., Smithgall T. E. (2005) J. Biol. Chem. 280, 40832–40837 [DOI] [PubMed] [Google Scholar]
- 27.Arold S. T., Ulmer T. S., Mulhern T. D., Werner J. M., Ladbury J. E., Campbell I. D., Noble M. E. (2001) J. Biol. Chem. 276, 17199–17205 [DOI] [PubMed] [Google Scholar]
- 28.Eck M. J., Atwell S. K., Shoelson S. E., Harrison S. C. (1994) Nature 368, 764–769 [DOI] [PubMed] [Google Scholar]
- 29.Sicheri F., Moarefi I., Kuriyan J. (1997) Nature 385, 602–609 [DOI] [PubMed] [Google Scholar]
- 30.Williams J. C., Weijland A., Gonfloni S., Thompson A., Courtneidge S. A., Superti-Furga G., Wierenga R. K. (1997) J. Mol. Biol. 274, 757–775 [DOI] [PubMed] [Google Scholar]
- 31.Xu W., Harrison S. C., Eck M. J. (1997) Nature 385, 595–602 [DOI] [PubMed] [Google Scholar]
- 32.Nagar B., Hantschel O., Seeliger M., Davies J. M., Weis W. I., Superti-Furga G., Kuriyan J. (2006) Mol. Cell 21, 787–798 [DOI] [PubMed] [Google Scholar]
- 33.Nagar B., Hantschel O., Young M. A., Scheffzek K., Veach D., Bornmann W., Clarkson B., Superti-Furga G., Kuriyan J. (2003) Cell 112, 859–871 [DOI] [PubMed] [Google Scholar]
- 34.Fushman D., Xu R., Cowburn D. (1999) Biochemistry 38, 10225–10230 [DOI] [PubMed] [Google Scholar]
- 35.Hofmann G., Schweimer K., Kiessling A., Hofinger E., Bauer F., Hoffmann S., Rösch P., Campbell I. D., Werner J. M., Sticht H. (2005) Biochemistry 44, 13043–13050 [DOI] [PubMed] [Google Scholar]
- 36.Young M. A., Gonfloni S., Superti-Furga G., Roux B., Kuriyan J. (2001) Cell 105, 115–126 [DOI] [PubMed] [Google Scholar]
- 37.Cowan-Jacob S. W., Fendrich G., Manley P. W., Jahnke W., Fabbro D., Liebetanz J., Meyer T. (2005) Structure 13, 861–871 [DOI] [PubMed] [Google Scholar]
- 38.Porter M., Schindler T., Kuriyan J., Miller W. T. (2000) J. Biol. Chem. 275, 2721–2726 [DOI] [PubMed] [Google Scholar]