Abstract
Globotriaosylceramide (Gb3) is a well known receptor for Shiga toxin (Stx), produced by enterohemorrhagic Escherichia coli and Shigella dysenteriae. The expression of Gb3 also affects several diseases, including cancer metastasis and Fabry disease, which prompted us to look for factors involved in its metabolism. In the present study, we isolated two cDNAs that conferred resistance to Stx-induced cell death in HeLa cells by expression cloning: ganglioside GM3 synthase and the COOH terminus region of glutamate receptor, ionotropic, N-methyl-d-asparate-associated protein 1 (GRINA), a member of the transmembrane BAX inhibitor motif containing (TMBIM) family. Overexpression of the truncated form, named GRINA-C, and some members of the full-length TMBIM family, including FAS inhibitory molecule 2 (FAIM2), reduced Gb3, and lactosylceramide was accumulated instead. The change of glycolipid composition was restored by overexpression of Gb3 synthase, suggesting that the synthase is affected by GRINA-C and FAIM2. Interestingly, the mRNA level of Gb3 synthase was unchanged. Rather, localization of the synthase as well as TGN46, a trans-Golgi network marker, was perturbed to form punctate structures, and degradation of the synthase in lysosomes was enhanced. Furthermore, GRINA-C was associated with Gb3 synthase. These observations may demonstrate a new type of posttranscriptional regulation of glycosyltransferases.
Keywords: Glycolipids, Glycosylation, Golgi, Lipid Synthesis, Sphingolipid, Gb3, Gb3 Synthase, Shiga Toxin, TMBIM Family
Introduction
Glycosphingolipids (GSLs)2 are composed of an oligosaccharide chain linked to ceramide and are ubiquitously expressed in animal cells. One of the peculiarities of GSLs is their diversity, which is generated by various glycosyltransferases in a cell type-specific manner, and various GSLs have specific functions, such as cell adhesive molecules, cell signaling modulators, and receptors for microbial toxins (1). Among GSLs, globotriaosylceramide (Gb3) serves as a receptor for Shiga toxin (Stx), produced by enterohemorrhagic Escherichia coli and Shigella dysenteriae (2, 3). Stx inhibits protein synthesis after transportation into the cytosol and, as a result, causes diarrhea-associated hemolytic uremic syndrome (4). Gb3 is synthesized from lactosylceramide (LacCer) by α1,4-galactosyltransferase (Gb3/CD77 synthase) (5–7), and Gb3 synthase-deficient mice show loss of sensitivity to Stx, indicating that the expression of this enzyme is crucial for the pathogenesis of Stx (8).
Gb3 also has other biological significance, especially under pathological conditions. First, the lipid has been associated with human cancers and found to be elevated in several types of tumor cells, including Burkitt lymphoma (9, 10). Recent studies showed that expression of Gb3 is correlated with metastasis in colon cancers, and Gb3 knockdown inhibits cell invasiveness (11). Furthermore, tumor-specific HSP70 on the cell surface is determined by the expression of Gb3 (12). The second is Fabry disease, which is an X-linked lysosomal disease caused by α-galactosidase A deficiency (13). This enzyme is responsible for degradation of Gb3 and its deficiency results in the progressive accumulation of Gb3, causing serious manifestations in the cardiovascular system etc. Gb3 also affects HIV infection through binding to the HIV envelope protein, gp120, although it is still inconclusive whether Gb3 promotes infection or counters it (14, 15).
In the present study, we isolated two genes, the overexpression of which conferred Stx resistance to HeLa cells, by carrying out expression cloning. One gene was ganglioside GM3 synthase (ST3GalV), which competes with Gb3 synthase for the common precursor LacCer, and the other was a truncated cDNA of GRINA, glutamate receptor, ionotropic, N-methyl-d-asparate-associated protein 1, which encodes a C-terminal hydrophobic polypeptide of the hypothetical full-length, and the expression of the polypeptide reduces the biosynthesis of Gb3 posttranscriptionally.
EXPERIMENTAL PROCEDURES
Cell Culture, Antibodies, and Reagents
HeLa cells (ATCC CCL-2) were maintained in DMEM containing 10% fetal bovine serum (FBS). JEG-3 human choriocarcinoma cells (ATCC HTB-36) were maintained in minimal essential medium containing 10% FBS. Purchased antibodies were as follows: rat anti-HA IgG (Roche Diagnostics), mouse anti-GAPDH IgG (Millipore, Billerica, MA), rabbit anti-Gb3 synthase antibodies and mouse anti-transferrin receptor IgG (Sigma), mouse anti-GM130 IgG, mouse anti-BiP/Grp78 IgG, and mouse anti-IκB IgG (BD Transduction Laboratories, San Diego, CA), sheep anti-TGN46 antibodies (Serotech, Kidlington, UK), mouse anti-LacCer IgM (Ancell, Bayport, MN), and mouse anti-FAS IgM clone CH11 (MBL, Nagoya, Japan). Chicken anti-VAP-A antibodies were raised against the recombinant cytosolic domain of human VAP-A and affinity purified (Scrum Inc., Tokyo, Japan). Alexa-conjugated secondary antibodies were from Invitrogen.
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), puromycin, tunicamycin, and galactosylceramide type I containing α-hydroxy fatty acid and type II containing non-hydroxy fatty acid (GalCer (I) and (II)) were from Sigma. Deoxymannojirimycin was from Wako Chemicals (Tokyo, Japan). High performance thin layer chromatography plates (Silica Gel 60) and uridine 5′-diphosphate galactose (UDP-Gal) were from Merck (Darmstadt, Germany). d-[1-14C]Galactose (56 mCi/mmol) was from GE Healthcare. UDP-[6-3H]Gal was from American Radiolabeled Chemicals (St. Louis, MO). EXPRE35S[35S]-Protein Labeling Mix was from PerkinElmer Life Sciences. E-64d, pepstatin A, and lactacystin were from the Peptide Institute (Osaka, Japan). Geneticin was from Nacalai Tesque (Kyoto, Japan), hygromycin was from Invitrogen. Blasticidin-S was from Kaken Pharmaceutical (Tokyo, Japan). Endoglycosidase H and peptide:N-glycosidase F were from New England Biolabs (Ipswich, MA). Monogalactosyl diacylglycerol, glucosylceramide (GlcCer), LacCer, and Gb3 were from Matreya (Pleasant Gap, PA). Globotetrasylceramide (Gb4) and GM3 were generous gifts from Dr. Akemi Suzuki (Tokai University, Kanagawa, Japan).
Construction Vectors
Information on the vectors in this study is shown under supplemental “Experimental Procedures” and Fig. S7.
Preparation of Shiga Toxin 1 (Stx1) and Fluorescent Stx1 B Subunit
Stx1 derived from E. coli O157:H7 was purified previously (16). The pEF-Stx1 B (binding) subunit-His6 vector for recombinant histidine-tagged Stx1 B subunit (1BH) was constructed previously (16). Preparation of the fluorescent Stx1 B subunit (Alexa 555-Stx1B) is described under supplemental “Experimental Procedures”.
Transfection
FuGENE 6 (Roche Diagnostics) was used in both transient and stable transfections according to the manufacturer's instructions. In stable transfections, cells were transfected with linearized plasmids, and then subjected to geneticin (for pCXN2) or hygromycin (for pcDNA3.1 Hyg) selection at a concentrations of 800 or 150 μg/ml, respectively. Colonies were isolated by limiting dilution. All stable transfectants used in this study are listed under supplemental Fig. S8.
Retroviral Infection
As parent cells for retroviral infection, HeLa cells were stably transfected with linearized pcDNA3.1 Hyg/mCAT-1, the retroviral receptor. After hygromycin selection, one efficiently infectable clone was chosen, named HeLa-mCAT#8. Preparation of retroviruses and their infection of HeLa-mCAT#8 cells were performed using the Plat-E system, as described previously (17, 18). When pMXs-IP- and pMXs-IB-based retroviruses were used, the concentrations of puromycin and blasticidin-S for selection were 1 and 2.5 μg/ml, respectively.
Isolation of Stx-resistant Genes
HeLa-mCAT#8 cells (1 × 106 cells in a 10-cm plate) were infected with the pLIB HeLa cell cDNA-library packaged retroviral particles (Clontech, Mountain View, CA), as described previously (18, 19). After 1 week of infection, the infected cells (4.5 × 106 cells in three 15-cm plates) were cultured with 200 ng/ml of Stx1 for 1 week. Surviving colonies were then isolated with cloning cylinders, and 46 colonies were finally propagated. For identification of integrated genes, genomic polymerase chain reaction (PCR) was performed with the KOD Plus kit (TOYOBO, Osaka, Japan) and PrimeSTAR GXL kit (Takara Bio, Ohtsu, Japan), using the primers described previously (18, 19). Some colonies contained more than two inserted cDNAs. The amplified fragments were sequenced to identify the integrated cDNAs using a BLAST search. The fragments (12 cDNAs) were then re-cloned to pLIB, the retroviral vector used in the cDNA library, and retroviral particles were prepared again to examine whether the isolated genes showed resistance to Stx1. Two cDNAs, GM3 synthase with a long cytoplasmic region (NM_003896) and a truncated cDNA of GRINA (GRINA-C), were identified. Among 46 colonies, 16 colonies (from at least 4 different clones) contained GM3 synthase and 10 colonies contained GRINA-C, which were confirmed by PCR. As for the remaining 20 colonies, the responsible cDNAs could not be identified although most colonies showed the reduction of Gb3 (data not shown). The pLIB-GM3S encoding GM3 synthase with a long cytoplasmic region and a clone of its infectants (named rGM3S) was used in subsequent experiments.
Immunofluorescence Microscopy
Immunostaining was performed as described previously (20), and the specimens were visualized with a confocal laser-scanning microscope, LSM510 META (Carl Zeiss, Jena, Germany) equipped with a C-Apochromat ×63/1.2W Corr objective.
Lysate Preparation and Western Blot Analysis
Three methods were used to prepare lysates as follows. For method 1 the cells were sonicated in sonication buffer (10 mm Hepes/NaOH (pH 7.4) 1 mm EDTA, 0.25 m sucrose, protease inhibitor mixture) and then mixed with Laemmli SDS sample buffer. For method 2 the cells were sonicated as in method 1, mixed with 4 volumes of urea-containing buffer (50 mm Tris/HCl, pH 8.8, 7 m urea, 2 m thiourea, 2% CHAPS, 2% Triton X-100, 33 mm DTT, protease inhibitor mixture), and incubated for 1 h at 37 °C. The proteins were then alkylated with 100 mm iodoacetoamide to prevent the re-oxidation of SH residues. Lithium dodecyl sulfate was added to the samples at 2%. For method 3 the cells were lysed with RIPA buffer (50 mm Tris/HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA, 1% Triton, 0.1% deoxycholate, 0.1% SDS, protease inhibitor mixture) and subsequent treatment was the same as method 2. Protein concentrations were determined with the Pierce BCA assay reagent kit using BSA as the standard.
Proteins were resolved by SDS-PAGE, transferred to PVDF membranes using a wet transfer method, and probed with appropriate antibodies. Antigen signals were detected using SuperSignal West Femto Maximum Sensitivity Substrate (ThermoFisher Scientific) or Immobilon Western HRP (Millipore) and exposed to an x-ray film. Image J software was used for densitometric analysis. endoglycosidase H and peptide:N-glycosidase F treatment were performed according to the manufacturer's protocol.
In Western blot analysis of Gb3 synthase, cells (2 × 105 cells in 6-well plates, or 1 × 105 cells in 12-well plates) were cultured overnight at 37 °C and then incubated with or without inhibitors for another 24 h at 37 °C. The inhibitors used in this study are as follows: a lysosomal protease inhibitor mixture (50 μm E-64d and 10 μm pepstatin A) (21), a proteasome inhibitor (1 or 10 μm lactacystin), tunicamycin (1 μg/ml), and deoxymannojirimycin (1 mm).
In co-immunoprecipitation analysis, the supernatants of cell lysates in RIPA buffer were incubated with anti-HA-agarose (Sigma). After washing with RIPA buffer, the bound proteins were eluted with the urea-containing buffer as described above.
Metabolic Labeling of Glycolipids and TLC Analysis
In metabolic labeling, cells (1 × 106 cells in 6-cm plates) were cultured overnight at 37 °C, and then the medium was changed to Opti-MEM with 1% Neutridoma-SP (Roche Diagnostics) and the cells were labeled with 0.4 μCi of d-[1-14C]galactose for 15 h. The cells were lysed with 0.1% SDS, and lysates containing the same amount of proteins were then used for lipid extraction by the method of Bligh and Dyer (22). The lower fractions collected were dried under an N2 gas stream and separated on high performance thin layer chromatography plates with a solvent of chloroform, methanol, and 0.25% CaCl2 (60/35/8; v/v), except for supplemental Fig. S2B, in which the lipids were separated on TLC (Silica 60, Merck) with a solvent of methyl acetate, n-propyl alcohol, chloroform, methanol, and 0.25% KCl (50/50/50/20/8; v/v). The radioactive lipids were visualized and quantified using a BAS2500 Image Analyzer (Fujifilm, Kanagawa, Japan).
For alkali methanolysis to remove glycerolipids, the dried lipids were hydrolyzed with 0.1 n KOH in methanol for 1 h at 40 °C. After neutralization with HCl at 0.1 n, the methanol layer was washed with n-hexane twice, and the lipids were extracted by the method of Bligh and Dyer (22).
For the preparation of radiolabeled standards by in vitro galactose labeling, HeLa cells were sonicated in sonication buffer (10 mm Hepes/NaOH (pH 7.4), 1 mm EDTA, 15 mm MnCl2, 0.5% Triton X-100, 0.25 m sucrose, protease inhibitor mixture), which was used as an enzyme source. Twenty-five μg of the indicated lipids (substrates) and 300 μg of Triton X-100 were dissolved in chloroform. Chloroform was removed from the mixture by an N2 gas stream. The dried lipids were sonicated in 90 μl of reaction buffer (20 mm MES/NaOH (pH 6.4), 15 mm MnCl2, 6.25 μm UDP-Gal) and then the substrates were incubated with 12.5 pmol (0.25 μCi) of UDP-[6-3H]Gal and 10 μl of the prepared lysates for 1 h at 37 °C. The lipids in the reaction mixture were extracted by the method of Bligh and Dyer (22). The lipid standards were developed with metabolically labeled lipid samples on the same plate, and visualized using a BAS1800 Image Analyzer (prepared radiolabeled lipids) or stained with I2 vapor followed by scanning with an LAS1000 Image Analyzer (non-labeled lipids) (Fujifilm).
Pulse-Chase Labeling Analysis
Cells (1 × 105 cells in 12-well plates) were cultured for 1 day at 37 °C, incubated in 400 μl of methionine- and cysteine-free DMEM with 10% dialyzed FBS for 1 h, and labeled with 44.4 μCi of 35S-protein labeling mixture for 20 min. The labeled cells were washed with pre-warmed complete medium, and then incubated for the indicated chase time periods at 37 °C with or without the inhibitors. The cells were washed with cold PBS once, and lysed with RIPA buffer. After centrifugation at 20,000 × g, the supernatant, in which the protein concentration was adjusted to the same concentration, was incubated with Dynabeads protein G (Veritas, Tokyo, Japan) for 1 h at 4 °C. The subsequent procedure was performed according to the manufacturer's protocol. Method 3 as described above was used for sample elution and preparation. Comparisons were analyzed with a paired t test using Excel software, and p < 0.05 was considered significant.
RNA Isolation, RT-PCR, and Real Time PCR
Total RNA was isolated with TRIzol reagent (Invitrogen). The isolated RNA was treated with DNase I (Takara Bio) to remove the contaminated DNA, and then purified with TRIzol again. cDNA was synthesized from 2 μg of total RNA by reverse transcriptase (ReverTra Ace, Toyobo) and random hexamers (Roche) at 42 °C for 1 h and 95 °C for 5 min.
For simple RT-PCR, KOD Plus was used. PCR products were resolved on agarose gels and the band was scanned with an LAS1000 Image Analyzer. Primers used in these experiments were: hGb3S sense 1 (5′-GGCAACATCTTCTTCCTGGAGACTTC-3′) and hGb3S antisense 2 (5′-GGATGGAACACCACTTCTTGAAGACC-3′), actin sense 1 (5′-GATATGGAGAAGATTTGGCACC-3′) and actin antisense 1 (5′-CAACGTCACACTTCATGATGGA-3′).
For real time PCR, the LightCycler system with the LightCycler-FastStart DNA master SYBR Green I kit (Roche Diagnostics) was used according to the manufacturer's protocol (19). Primers used in these experiments were: hGb3S sense 1 described above and hGb3S antisense 1 (5′-CGAACTTCCACATGAGTGCGATCC-3′), GAPDH sense 1 (5′-GAGTCAACGGATTTGGTCGT-3′), and GAPDH antisense 1 (5′-TTGATTTTGGAGGGATCTCG-3′).
FACS Analysis
Trypsinized cells were washed with culture medium and wash buffer (1% BSA) in PBS at 4 °C. The cells were incubated with 10 μg/ml of Alexa 555 Stx1 B subunits for 45 min on ice. After washing with the wash buffer once, the cells were analyzed with FACSCalibur (BD Biosciences). In LacCer staining, the cells were incubated with mouse anti-LacCer IgM for 45 min on ice. After washing with the wash buffer, the cells were incubated with Alexa Fluor 488-conjugated anti-mouse IgG (H+L), which can recognize IgM.
To see the effect of transiently expressed proteins on the expression of Stx receptors, HeLa-mCAT#8 cells (2.5 × 104 cells in 12-well plates) were co-transfected with 0.25 μg of plasmids containing the objective genes and 0.05 μg of EGFP-N3 (Clontech, Mountain View, CA). After 3 days of transfection, the cells underwent FACS analysis. Spillover of EGFP fluorescence in the FL2 channel, and that of Alexa 555 in the FL1 channel were electronically compensated. After gating out debris and cell aggregates by FSC/SSC, the percentage of StxR lower cells in EGFP-positive cells was calculated by (cell number in lower right)/(cell number in upper and lower right) × 100.
MTT Assay
In the treatment with Stx1, cells (1.25 × 104 cells in 24-well plates) were cultured overnight at 37 °C, and then treated with Stx1 at the indicated concentrations. In the treatment with anti-FAS antibodies, cells (5 × 104 cells in 24-well plates) were cultured overnight at 37 °C, and then treated with anti-FAS IgM (100 ng/ml) and cycloheximide (10 μg/ml). After a 3-day incubation, the MTT assay was performed as described previously (23).
RESULTS
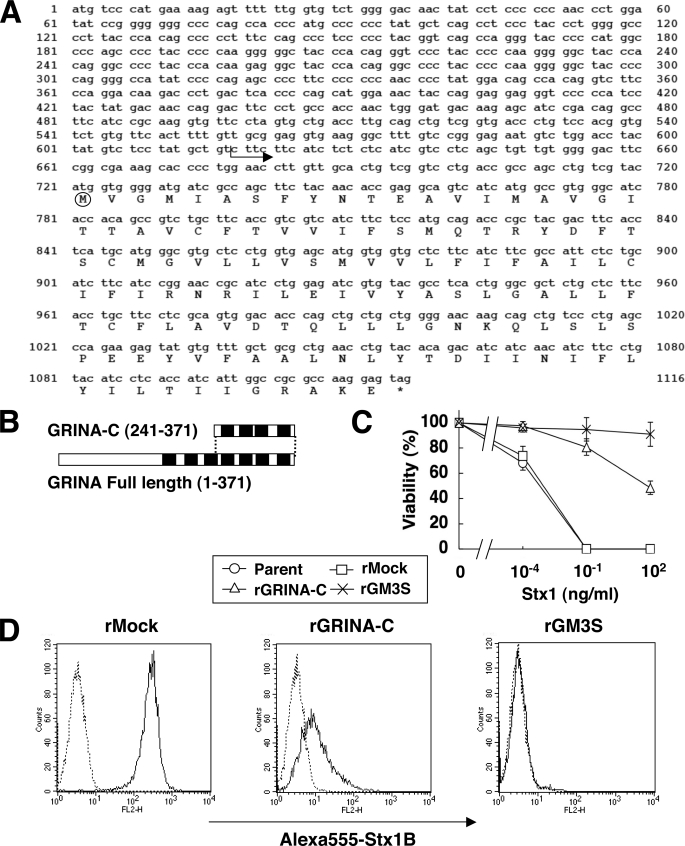
Isolation of cDNAs to Confer Resistance to Stx-induced Cell Death
To identify molecules that affect Gb3 metabolism, we attempted to isolate cDNAs, the overexpression of which confers resistance to Stx-induced cell death in HeLa cells, by the expression cloning method shown under “Experimental Procedures.” Forty-six Stx-resistant colonies were isolated, and finally two cDNAs were identified. One was ganglioside GM3 synthase (ST3GalV), which was inserted into 16 colonies. The inserted cDNA started on the 5′ side of the start codon and encoded the enzyme with the longest cytoplasmic region among several transcript variants (24). GM3 synthase-expressing cells (rGM3S) showed complete resistance to Stx1 even at higher concentrations (Fig. 1C). The other was a truncated cDNA of GRINA, glutamate receptor, ionotropic, N-methyl-d-asparate-associated protein 1 (10 colonies), which encodes a C-terminal hydrophobic polypeptide of the hypothetical full-length, as shown in Fig. 1, A and B. Truncated GRINA (named GRINA-C), with an influenza hemagglutinin (HA) tag attached at the C terminus, was retrovirally introduced to HeLa-mCAT#8 parent cells to prepare a new transfectant (rGRINA-C). The rGRINA-C cells showed moderate resistance to Stx1 as expected, whereas empty vector-infected cells (rMock) remained sensitive to Stx1 as the parent cells (Fig. 1C).
FIGURE 1.
Isolation of Stx-resistant genes and reduction of Stx receptors (StxRs) on the cell surface. A, an isolated Stx-resistant gene encoding the C-terminal human GRINA, named GRINA-C. The sequence shows the open reading frame of human GRINA (GenBankTM accession number NM_001009184), and the arrow indicates the position at which the isolated cDNA started. The circle indicates the predicted start codons of GRINA-C. B, schematics of GRINA-C and full-length GRINA. Black boxes show transmembrane domains. C, resistance to Stx1 in HeLa transfectants retrovirally expressing GRINA-C and GM3S. Stx1 was added at the indicated concentrations. Viability was estimated by MTT assay, and expressed as the percentage of the value (OD570) in the absence of Stx1: mean percentage ± S.D. obtained from three independent experiments. D, surface expression of StxRs on HeLa transfectants. Transfectants were stained with Alexa 555-Stx1 B subunit (solid line) or not (dotted line) and analyzed by FACS.
Stx Binding on the Cell Surface Is Reduced by Overexpression of GRINA-C
Next, the expression of Stx receptors (StxRs) on the cell surface was investigated using a fluorescent Stx1-B binding subunit (Alexa 555-Stx1B). FACS analysis showed that Stx binding was significantly detected in rMock cells, whereas the binding was much reduced in rGRINA-C cells and no Stx1B binding was detected in rGM3S cells (Fig. 1D). These results were consistent with the sensitivity to Stx shown in Fig. 1C, suggesting that reduction of StxRs on the cell surface reflected the sensitivity.
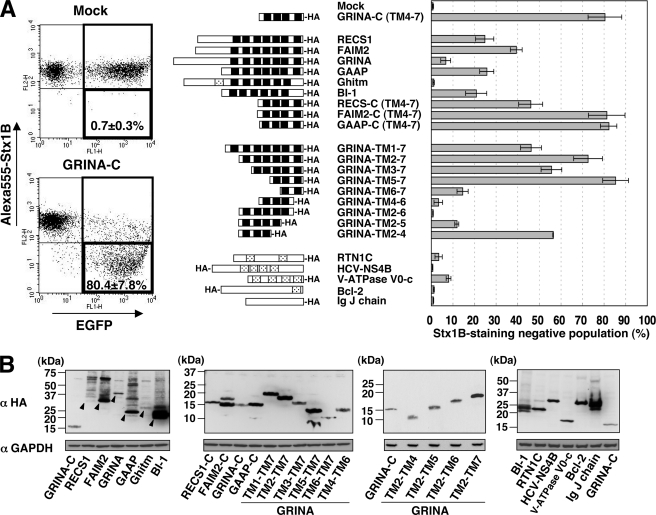
The Expression of TMBIM Family Reduces StxR Expression
GRINA is known as a member of the transmembrane BAX inhibitor motif-containing (TMBIM) family, which includes RECS1 (TMBIM1), FAIM2 (TMBIM2), GRINA (TMBIM3), GAAP (TMBIM4), Ghitm (TMBIM5), and BI-1 (TMBIM6), many of which are known as anti-apoptotic proteins (25–28). For example, FAIM2 was originally isolated as a molecule that inhibits FAS-mediated apoptosis (26). These members have a conserved hydrophobic domain (named UPF0005), including 6–8 transmembrane domains on the C-terminal side (Fig. 2, black boxes), and GRINA-C has a C-terminal half of the UPF0005 domain. PCR analysis showed that HeLa cells express all full-length TMBIM families except FAIM2 (data not shown). Then, to examine whether the action of GRINA-C has a dominant-negative effect against a member of the TMBIM family or a gain of function, full-length TMBIM families were transiently expressed in rGRINA-C cells to see whether the expression level of StxRs was restored. No restoration of StxRs was seen by expression of any TMBIM families, suggesting that the effect of GRINA-C is not dominant-negative (data not shown). Rather, overexpression of some TMBIM families, including RECS1, FAIM2, GAAP, and BI-1, reduced the expression of StxRs, although the degree of reduction was less than that by GRINA-C. The C-terminal regions of RECS-1, FAIM2, and GAAP (RECS1-C, FAIM2-C, GAAP-C), which are highly homologous to GRINA-C, greatly reduced StxRs as GRINA-C (Fig. 2A). Co-transfection of FAIM2 and GRINA-C showed an additive effect on the expression of Gb3, suggesting that at least part of the action mechanisms of these molecules on the Gb3 reduction are shared with each other (supplemental Fig. S1). The reduction of StxRs was much less by the expression of full-length GRINA, and no reduction was observed by expression of Ghitm, which is known as a mitochondrial protein with the UPF0005 domain.
FIGURE 2.
Effects of TMBIM family on the expression of StxRs. A, HeLa-mCAT#8 cells were transiently transfected with the indicated cDNAs and EGFP, and the transfectants were stained with Alexa 555-Stx1B. The Stx1B staining negative population was calculated as described under “Experimental Procedures:” mean percentage ± S.D. was obtained from at least three independent experiments. Representative dot plot data are shown on the left side. Black boxes indicate the transmembrane domains in UPF0005, and dotted boxes indicate other transmembrane domains. The number of TMs was referred to Uniprot. HA means HA-epitope tag. B, Western blot analysis of proteins in the transient transfectants shown above. In the cell lysate preparation, method 2 was used for the TMBIM family (membranes except the right one), and method 1 was used for other proteins (the right membrane), as described under “Experimental Procedures.” All proteins were tagged with the HA epitope. Arrowheads indicate the predicted monomeric proteins.
Full-length GRINA contains a longer hydrophilic region at the N terminus than others in the TMBIM family, and the expression level was also lower (Fig. 2B), which may be the reason for low activity. The removal of the N-terminal cytoplasmic region (GRINA-TM1–7) caused a significant reduction of StxRs as the other full-length TMBIM family (Fig. 2A), and the expression level also increased (Fig. 2B), suggesting that GRINA potentially has activity to reduce StxRs. Next, GRINA was sequentially deleted from the N terminus TM domain to see the effects (Fig. 2A). The reduction of StxRs was still observed by deletion to the 4th TM, containing the 5th to 7th TMs (GRINA-TM5–7). Deletion to the 5th TM, which contains only the 6th and 7th TMs, reduced the activity (GAINA-TM6–7). Next GRINA-C and GRINA-TM2–7, containing the 2nd to 7th TMs, were deleted from the C terminus. The deletion of the 7th TM lost activity (GRINA-TM4–6, TM2–6), which suggests that the 7th TM is required for activity; however, surprisingly, GRINA-TM2–4, which contains the 2nd to 4th TMs and hardly overlaps GRINA-TM5–7, also showed moderate activity to reduce StxR. These results suggest that this activity is not simply dependent on specific regions, but structurally related functions or molecular information of the C-terminal regions may be required for the reduction of StxRs.
Then we examined whether other proteins also reduce StxRs. The following five proteins were chosen to see the effect (the number of hydrophobic regions is shown in Fig. 2A): Reticulon-1C (RTN1C, a potential ER stress inducer (29)), NS4B polypeptide of hepatitis C virus (HCV-NS4B, a potential ER stress inducer (30)), V-ATPase V0-c subunit (V-ATPase V0-c), immunoglobulin J chain (IgJ chain, a component of secretory IgM (and A), an endoplasmic reticulum-associated degradation (ERAD) substrate due to unassembly (31)), and Bcl-2 (an anti-apoptotic protein). Among them, overexpression of RTN1C, HCV-NS4B, IgJ chain, and Bcl-2 showed less or no reduction of StxRs, indicating that the general anti-apoptotic activity and simple overexpression of secretory or membrane proteins are not the cause of Gb3 reduction (Fig. 2). GRINA-C-expressing cells did not show up-regulation of BiP/Grp78 (32) (supplemental Fig. S7) and down-regulation of IκB (33) (data not shown), both of which are observed in chronic as well as acute responses to ER stress. V-ATPase V0-c reduced it slightly, similar to full-length GRINA. V0-c is more hydrophobic than RTN1c and HCV-NS4B, which might suggest that the hydrophobicity of transmembrane proteins is involved in the phenomenon; however, the activity to reduce StxRs by GRINA-C, which also has the same number of TM domains, is much stronger than that by V0-c, which indicates that the TM domains of the TMBIM family have unique activity to reduce StxRs.
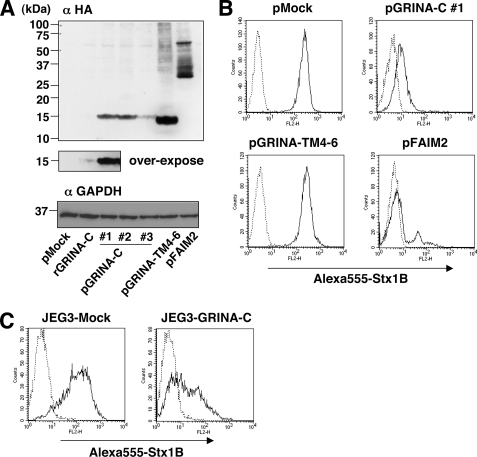
To confirm the above results, plasmid-based stable transfectants of GRINA-C, GRINA-TM4–6, and full-length FAIM2 were prepared (Fig. 3A). Three isolated clones expressing GRINA-C (pGRINA1, -2, and -3) showed the reduction of StxRs as expected, whereas empty vector-expressing cells (pMock) did not (Fig. 3B, and data not shown). The expression level of GRINA-C in these transfectants was higher than in the retrovirus-based transfectant (rGRINA-C < pGRINA-C #3 < pGRINA-C #1 = pGRINA-C #2), which means that only a small amount of GRINA-C is required for reduction of StxR (Fig. 3A). In FAIM2, the expression level is critical for reduction of StxR, and the retrovirus-based expression hardly reduces StxRs (data not shown). Some of the isolated clones to highly express FAIM2 showed the reduction of StxRs (representative clone: pFAIM2, Fig. 3, A and B), the tendency of which is consistent with the transient transfection experiment in Fig. 2A. The prepared pFAIM2 clone was completely resistant to FAS-mediated apoptosis, indicating that overexpressed FAIM2 is functional (data not shown). In contrast to pGRINA-C and pFAIM2 cells, the stable transfectant expressing GRINA-TM4–6 (pGRINA-TM4–6) did not reduce StxR, although its expression level is much higher than GRINA-C in pGRINA-C transfectants (Fig. 3A).
FIGURE 3.
Stable expression of GRINA-C, GRINA-TM4–6, and FAIM2. A, expression of HA-tagged GRINA-C, GRINA-TM4–6, and FAIM2 in plasmid-based stable HeLa transfectants. Cell lysate was prepared by method 2, as described under “Experimental Procedures.” B and C, surface expression of StxRs on the above HeLa transfectants (B), and JEG-3 cells stably expressing plasmid-based GRINA-C or Mock (C). The method was as described in the legend to Fig. 1D.
To investigate whether other cell lines show a similar phenomenon, human choriocarcinoma cell line JEG-3, which is Gb3-positive, was used. GRINA-C expressing JEG-3 showed the reduction of StxRs compared with Mock cells (Fig. 3C), suggesting that the phenomenon is shared among various types of cells.
The Synthesis of Gb3 Is Reduced in GRINA-C-expressing Cells
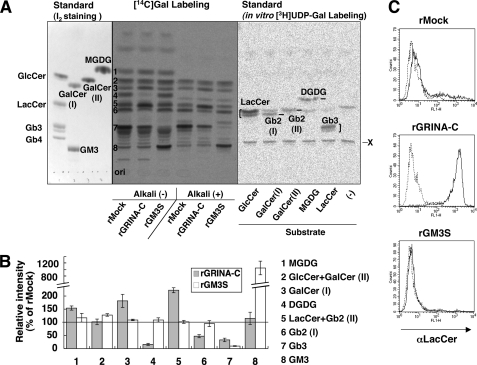
To investigate whether GSLs, especially the major Stx1 receptor Gb3, were affected by expression of the genes described above, metabolic labeling with [14C]galactose was carried out. TLC analysis showed that several galactose-labeled lipids were detected, among which the major bands were numbered 1 to 8, as shown in Fig. 4A. We identified these bands by the following experiments and information. (a) The samples were developed on the same plate with some glycolipid standards and the radioactive glycolipids prepared by in vitro galactose labeling using HeLa lysates and several lipid substrates. (b) mRNA of galactosylceramide (GalCer) synthase was expressed in HeLa cells used in this study (data not shown). (c) GalCer synthase synthesizes monogalactosyl diacylglycerol as well as GalCer (34, 35). (d) GalCer and monogalactosyl diacylglycerol were galactose-labeled in vitro, suggesting galabiosylceramide (Gb2; Galα1,4Gal-Cer) and digalactosyl diacylglycerol (Galα1,4Gal-DG, named DGDG) are potentially expressed in HeLa cells. Actually, HeLa cells are known to express Gb2 (36), and Gb3 synthase can use GalCer as a substrate to synthesize Gb2 (5, 6). (e) Alkali lysis was performed to discriminate sphingolipids from glycerolipids, and bands 1 and 4 are glycerolipids. (f) The band of Gb2 (I) may be overlapped with the lower band of LacCer as shown in the galactose labeling standard prepared in vitro; however, in glycolipids of HeLa cells, the upper band looks dominant as shown in band 7 (Gb3), suggesting that most LacCer is included in band 5 as the upper band and band 6 does not contain as much LacCer. (g) Knockdown of Gb3 synthase (shGb3S cells) showed reduction of bands 4, 6, and 7 (supplemental Fig. S2A). From the above information, the bands were identified as shown in Fig. 4B. The patterns of [14C]galactose-labeled lipids were quite different among rMock, rGRINA-C, and rGM3S. As expected, rGM3S cells showed a considerable increase in GM3 (number 8, 1052.9 ± 207.6%) and instead a reduction of Gb3 (number 7, 9.8 ± 1.6%) compared with rMock cells (Fig. 4, A and B). On the other hand, rGRINA-C showed a moderate reduction of Gb3 (number 7, 33.0 ± 6.7%) as well as DGDG (number 4, 15.8 ± 3.8%) and Gb2 (I) (number 6, 47.1 ± 5.9%), and instead an increase in LacCer (number 5, 222.1% ± 9.2%). The reduction of Gb3, Gb2, and DGDG in rGRINA-C cells was also observed in a different TLC development system (supplemental Fig. S2B). The accumulation of LacCer was detected in FACS analysis using anti-LacCer antibodies in rGRINA-C cells (Fig. 4C). The synthesis of GM3 did not change by the expression of GRINA-C (number 8, 115.0 ± 23.4%). The pattern of GSL biosynthesis in rGRINA-C cells was quite similar to that in shGb3S cells (supplemental Fig. S2A). These data suggest that GRINA-C inhibits the Gb3 synthase step, which synthesizes Gb3 as well as Gb2 and DGDG. Similar patterns of GSL biosynthesis were also seen in pGRINA-C and pFAIM2 cells, but not in pGRINA-TM4–6 cells (data not shown), consistent with the result in Fig. 2.
FIGURE 4.
Reduction of Gb3 and increase of LacCer in GRINA-C-expressing transfectants. A, galactose metabolic labeling analysis. HeLa transfectants (rMock, rGRINA-C, rGM3S) were labeled with [14C]galactose, and the labeled lipids, with or without treatment with mild alkali-catalyzed methanolysis, were separated on a high performance thin layer chromatography plate. The major bands were numbered from 1 to 8. Radiolabeled or non-radiolabeled lipid standards are shown on both sides. X is an unidentified product. B, quantification of the labeling experiments, as shown in A. The amounts of each [14C]galactose-labeled lipid are expressed as the percentage of band intensity in rMock cells: mean percentage ± S.D. obtained from three independent experiments. C, surface expression of LacCer on HeLa transfectants. Transfectants were stained with mouse anti-LacCer IgM (solid line) or not (dotted line) followed by Alexa Fluor 488-conjugated anti-mouse IgG (H+L).
Overexpression of Gb3 Synthase Restores the Expression of Gb3 in GRINA-C-expressed Cells
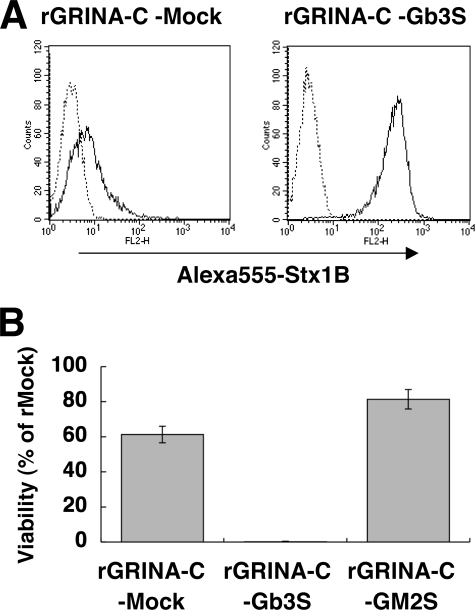
The effect of GRINA-C was assumed to be through the inhibition of Gb3 synthesis; therefore, Gb3 synthase was then retrovirally overexpressed in rGRINA-C cells (rGRINA-C-Gb3S) to see the effects. As shown in Fig. 5, A and B, rGRINA-C-Gb3S cells restored surface expression of StxRs and Stx sensitivity, whereas expression of empty or GM2 synthase had no effect (rGRINA-Mock or rGRINA-GM2syn). The glycolipid pattern of rGRINA-C-Gb3S cells, including Gb3, Gb2, DGDG, and LacCer, also returned to that of rMock cells (supplemental Fig. S2C). These data indicate that reduction of Gb3 is the cause of the resistance to Stx by GRINA-C.
FIGURE 5.
Restoration from GRINA-C-induced reduction of StxRs by overexpression of Gb3 synthase. A, surface expression of StxRs on rGRINA-C-Gb3S cells and rGRINA-C-Mock cells. Transfectants were stained with Alexa 555-Stx1 B subunit (solid line) or not (dotted line). B, restoration of susceptibility to Stx1 by overexpression of Gb3 synthase. The above HeLa transfectants were treated with 100 ng/ml of Stx1. Viability was estimated as described in the legend to Fig. 1C: mean percentage ± S.D. (n = 3).
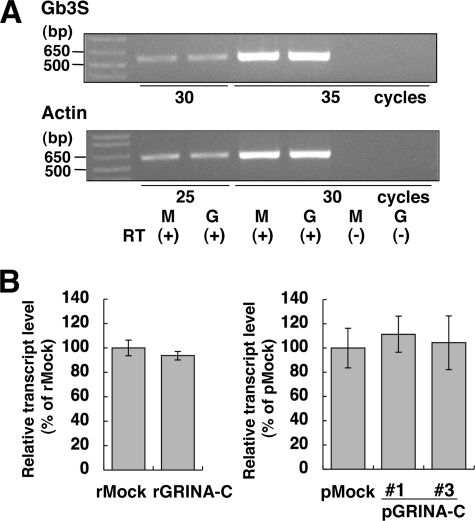
The mRNA Level of Gb3 Synthase Is Not Changed by Expression of GRINA-C
Next, the effect of GRINA-C on the mRNA level of Gb3 synthase was investigated. RT-PCR analysis showed that the amount of Gb3 synthase mRNA in rGRINA-C cells was almost the same as that in rMock cells (Fig. 6A). To confirm the result, quantitative PCR was also carried out using two combinations of transfectants (retrovirus series and plasmid series) (Fig. 6B). The relative amount of Gb3 synthase mRNA in GRINA-C-expressing cells was almost the same as that in control cells (rGRINA-C versus rMock, and pGRINA-C numbers 1 and 3 versus pMock), ruling out the possibility that reduction of Gb3 by GRINA-C was the result of alteration of the transcriptional level of Gb3 synthase.
FIGURE 6.
No alteration in the mRNA level of Gb3 synthase by the expression of GRINA-C. A, RT-PCR of Gb3 synthase mRNA from rGRINA-C and rMock cells. Amplification was performed for the different cycles indicated. RT(−) means the PCR template without reverse transcription. Actin was the reference control. B, quantitative real time PCR of Gb3 synthase mRNA. The mRNA from rGRINA-C cells was compared with that from rMock cells, and mRNAs from pGRINA-C#1 and -#3 were compared with that from pMock. Relative mRNA levels of Gb3 synthase were expressed as the percentage of the value in each control cell line: mean percentage ± S.D. obtained from at least three independent experiments.
GRINA-C Reduces the Exogenously Expressed Gb3 Synthase
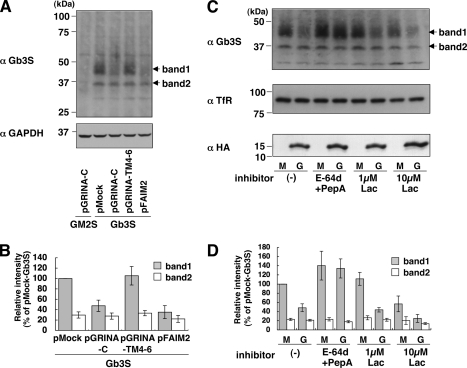
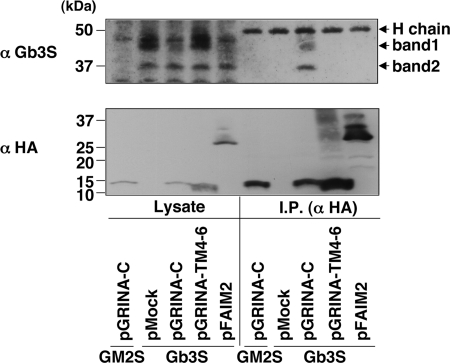
We attempted to compare the amount of Gb3 synthase at a translational level. An antibody against Gb3 synthase was able to detect Gb3 synthase by Western blot, but only in overexpression, and failed to detect the endogenous Gb3 synthase, likely because of its low expression level. Gb3 synthase was then retrovirally expressed in the 4 transfectants described above (pMock, pGRINA-C#1, pGRINA-TM4–6, and pFAIM2) to see the expression levels of exogenous Gb3 synthase from an alternative view. All transfectants (pMock-Gb3S, pGRINA-C-Gb3S, pGRINA-TM4–6-Gb3S, and pFAIM2-Gb3S) showed a similar mRNA level of Gb3 synthase, which means that these transfectants can be compared to determine the protein level of Gb3 synthase without an effect on transcription, although Gb3 was restored by the overexpression (supplemental Fig. S3, A and B). Western blot analysis showed that two bands derived from overexpressed Gb3 synthase were detected (named as band 1 and 2), but were not detected in pGRINA-C-GM2S cells as a negative control (Fig. 7A). The pGRINA-C-Gb3S and pFAIM2-Gb3S cells apparently showed the reduction of band 1 but not band 2 in comparison with pMock-Gb3S. From endoglycosidase H and peptide:N-glycosidase F digestion analyses, band 1 was thought to be a mature form of Gb3 synthase, and band 2 was an immature high-mannose form (supplemental Fig. S4A). Quantitative analysis showed that the mature form of pGRINA-C-Gb3S cells was reduced to 47% and that of pFAIM2-Gb3S cells to 35% (Fig. 7B). In contrast, GRINA-TM4–6 did not change any bands, consistent with this plasmid not affecting the level of Gb3. The reduction was not observed in B4GalT6, another galactosyltransferase, suggesting that not all glycosyltransferases are affected by the expression of GRINA-C (supplemental Fig. S4B).
FIGURE 7.
Reduction of Gb3 synthase by the expression of GRINA-C and FAIM2 in a lysosomal protease-dependent manner. A, comparison of the protein levels of Gb3 synthase among 4 HeLa transfectants (pMock-, pGRINA-C-, pGRINA-TM4–6-, and pFAIM2-Gb3S), in which Gb3 synthase was retrovirally expressed. These transfectants had a similar amount of Gb3 synthase mRNA, as shown under supplemental Fig. S3A. pGRINA-C-GM2S was a negative control of Gb3 synthase. Cell lysate was prepared by method 3, as described under “Experimental Procedures.” Band 1 is a mature form and band 2 is an immature form as shown under supplemental Fig. S4A. C, effects of protease inhibitors on the reduction of Gb3 synthase by GRINA-C. pGRINA-C-Gb3S (G) cells and pMock-Gb3S (M) cells were incubated with a lysosomal protease inhibitor mixture (50 μm E-64d and 10 μm pepstatin A (PepA)) or a proteasome inhibitor (1 or 10 μm lactacystin (Lac)) for 24 h. B and D, quantification of A and C. The amounts of Gb3 synthases (bands 1 and 2) were expressed as a percentage of the intensity of band 1 in pMock-Gb3S cells: mean percentage ± S.D. obtained from three independent experiments.
The Reduction of Gb3 Synthase Was Restored by the Addition of Lysosome Inhibitors
The reduction of only the mature form of Gb3 synthase by the expression of GRINA-C raised the possibility of enhanced degradation of Gb3 synthase. Two major degradation pathways, lysosome and proteasome, were inhibited by inhibitors to see the effect on the reduction. The mixture of E-64d and pepstatin A, a cell-permeable cysteine protease inhibitor and an aspartic protease inhibitor, respectively, to inhibit lysosomal proteases, effectively restored the amount of the mature form, whereas lactacystin, a proteasome inhibitor, did not restore it even at 10 μm (Fig. 7, C and D). The immature form of Gb3 synthase was not changed under any conditions tested. The increased mature form by treatment with lysosome inhibitors was endoglycosidase H resistant, suggesting that Gb3 synthase was degraded in lysosomes after maturation (supplemental Fig. S4C).
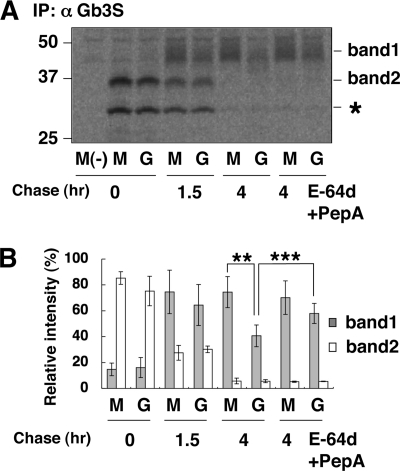
To see the effect of GRINA-C on translation and degradation of Gb3 synthase more accurately, a pulse-chase experiment was performed. As shown in Fig. 8, A and B, there was no significant difference in the newly synthesized Gb3 synthase level (band 2) between pMock-Gb3S and pGRINA-C-Gb3S cells in the 20-min pulse, and during the chase period, the level of the immature form of Gb3 synthase was similarly changed between the two cell types; however, the metabolism of the mature form was different between them. In pMock-Gb3S cells, the level of matured Gb3 synthase was maintained at least until 4 h of chase, whereas the level at the 4-h chase was significantly reduced in pGRINA-C-Gb3S cells compared with that at 1.5 h of chase. The reduction was partially restored by treatment with lysosome inhibitors during the chase. These results support the hypothesis that GRINA-C promotes the degradation of Gb3 synthase after maturation.
FIGURE 8.
Pulse-chase analysis to see the degradation of Gb3 synthase. A and B, pulse-chase analysis of Gb3 synthase in pGRINA-C-Gb3S (G) cells and pMock-C Gb3S cells (M). The amounts of synthesized Gb3 synthases (band 1 and 2) are expressed as a percentage of the total intensity of bands 1 and 2 in pMock-Gb3S cells at 0 h: mean percentage ± S.D. obtained from four independent experiments. Paired t test was used for statistical analysis. **, p < 0.01. ***, p < 0.001. The single asterisk indicates an unidentified band.
GRINA-C Was Co-immunoprecipitated with Gb3 Synthase
To see the interaction between Gb3 synthase and GRINA-C or FAIM2, co-immunoprecipitation experiments were carried out (Fig. 9). The binding of GRINA-C to both mature and immature forms of Gb3 synthase was detected even in RIPA buffer containing 0.1% SDS and 0.1% deoxycholate. This interaction is specific because GRINA-TM4–6 did not co-immunoprecipitate with the Gb3 synthase. This result suggests that GRINA-C inhibits Gb3 synthase through their interaction directly or indirectly. FAIM2 did not co-immunoprecipitate with Gb3 synthase in this condition. This might be correlated with the fact that GRINA-C reduces Gb3 much more effectively than the full-length TMBIM family molecules including FAIM2.
FIGURE 9.
Co-immunoprecipitation of GRINA-C with Gb3 synthase. pMock-, pGRINA-C-, pGRINA-TM4–6-, and pFAIM2-Gb3S cells were lysed with RIPA buffer and immunoprecipitated with anti-HA-agarose. The immunoprecipitated proteins (I.P.) and cell lysates equivalent to 1/34 of the applied immunoprecipitates were subjected to SDS-PAGE and Western blot with the indicated antibodies.
Localization of GRINA-C and FAIM2 and Their Effects on the Distribution of TGN
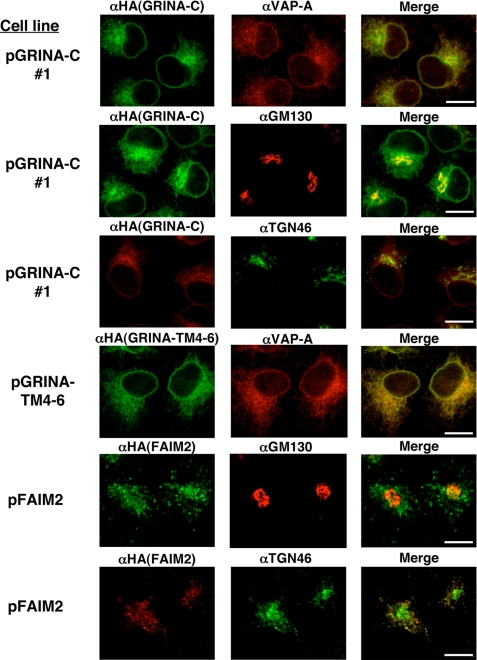
Next, intracellular localizations of GRINA-C and FAIM2 were investigated with a confocal microscope. As shown in Fig. 10, the staining pattern of GRINA-C was similar to that of VAP-A, an ER membrane protein, but GRINA-C was more concentrated in the perinuclear region of the cell than VAP-A. Concentrated staining was colocalized with GM130, a cis-Golgi marker protein, and TGN46, a trans-Golgi network (TGN) marker. In contrast, GRINA-TM4–6, which has no activity to reduce Gb3, was localized only to the ER (Fig. 10), although the expression level of GRINA-TM4–6 in the transfectant was higher than that of GRINA-C (Fig. 3). These results imply that localization of the perinuclar region is important for GRINA-C to reduce Gb3. Unlike GRINA-C, the staining pattern of FAIM2 was more punctate and less merged with GM130. The punctate structures of FAIM2 were completely merged with the dispersed dots in TGN46, suggesting that FAIM2 is localized at the TGN and the following vesicles (endosome etc.). From Fig. 10 we noticed that a lot of the punctate pattern of TGN46 was observed not only in pFAIM2 cells but also in pGRINA-C cells; therefore, colocalization of TGN46 and GM130 in these cells was compared with that in control cells. In pMock cells, TGN46 was colocalized or at least aligned with GM130, and almost all cells (90%) had less punctate staining of TGN46 (supplemental Fig. S5). In contrast, about half of pGRINA-C and pFAIM2 cells had punctate staining of TGN46, which was not merged with GM130 (supplemental Fig. S5). These results suggest that expression of GRINA-C and FAIM2 affects the distribution of TGN, although localization patterns of GRINA-C and FAIM2 were different.
FIGURE 10.
Intracellular localization of GRINA-C and FAIM2. HeLa transfectants were stained with rat anti-HA antibodies and the indicated antibodies (chicken anti-VAP-A (ER), mouse anti-GM130 (cis-Golgi apparatus), sheep anti-TGN46 (trans-Golgi network)), followed by Alexa Fluor 488 (green) and Alexa Fluor 594 (red) IgG conjugates. Scale bars, 10 μm.
Localization of Gb3 Synthase Was Also Perturbed by the Expression of GRINA-C
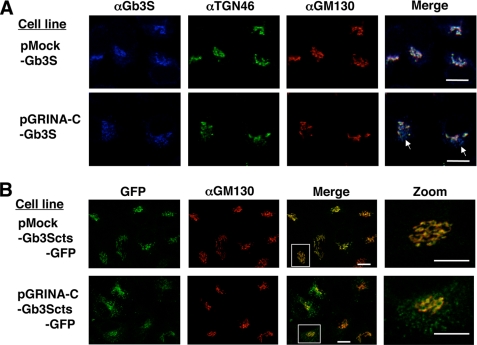
Next, the localization of exogenous Gb3 synthase was investigated. The signal intensity of Gb3 synthase was uneven in both pMock-Gb3S and pGRINA-C-Gb3S cells, and many cells reduced the signal to the background level, especially in pGRINA-C-Gb3S cells, probably because the expression level was reduced; therefore, only cells with clear staining of Gb3 synthase were observed. Fig. 11A shows that Gb3 synthase was colocalized with both TGN46 and GM130 in control cells (pMock-Gb3S), whereas puctate staining of Gb3 synthase, colocalized with TGN46 but not GM130, was observed in more than half of the clearly stained pGRINA-C-Gb3S cells. To confirm the abnormal distribution of Gb3 synthase, we then prepared a probe that contained the cytoplasmic, transmembrane, and stem regions of Gb3 synthase fused with the GFP fluorescent protein at the C terminus (Gb3Scts-GFP), and expressed it in pMock and pGRINA-C cells. The probe was colocalized with GM130 in pMock-Gb3Scts-GFP cells, whereas punctate staining, unmerged with GM130, was observed in pGRINA-C-Gb3Scts-GFP cells, consistent with the staining of full-length Gb3 synthase (Fig. 11B). These results indicate that expression of GRINA-C perturbs the localization of Gb3 synthase as well as TGN46.
FIGURE 11.
Dispersed dot-like structures of Gb3 synthase and TGN46. A, HeLa transfectants were stained with rabbit anti-Gb3 synthase, sheep anti-TGN46, and mouse anti-GM130, followed by Alexa Fluor 350-conjugated anti-rabbit IgG (blue), Alexa Fluor 488 anti-sheep IgG (green), and Alexa Fluor 594 anti-mouse IgG (red). Arrows indicate dispersed dot-like structures. B, HeLa transfectants expressing Gb3Scts-EGFP was stained with mouse anti-GM130, followed by Alexa Fluor 594 anti-mouse IgG. Scale bars, 10 μm. White boxes are zoomed on the right side.
DISCUSSION
GSL biosynthesis is regulated by various factors, but in many cases the transcriptional levels of glycosyltransferases have been mainly investigated to explain the change in lipid composition. By functional screening using Stx, we isolated an unexpected factor, the C terminus of GRINA (GRINA-C), which reduced Gb3 biosynthesis without changing the amount of Gb3 synthase mRNA. Importantly, not only GRINA-C but also some of the full-length TMBIM family, including FAIM2, showed the same activity, suggesting that these molecules have shared biological functions for GSL biosynthesis.
GRINA was originally identified as a glutamate-binding subunit of an NMDA receptor-associated complex (37), but the biological function remains unclear. From sequence similarity, the TMBIM family consists of at least 6 genes, including GRINA, and most members have anti-apoptotic activity. For example, FAIM2 was identified as Fas inhibitory molecule 2 (26), and BI-1 was first identified as BAX inhibitor-1 (25). BI-1 and GAAP have been reported to modulate the intracellular calcium flux, which inhibits apoptosis by several stimuli (38, 39). An interesting feature of the family is their highly conserved multispanning transmembrane domains named UPF0005; however, the biological activity of the domain is still unknown. In this study, overexpression of molecules containing whole or part of UPF0005 resulted in reduction of Gb3. In particular, GRINA-C, the C-terminal half of UPF0005, is much conserved with the equivalent sequences of RECS-1, FAIM2, and GAAP (40), all of which greatly reduce Gb3. These results indicate that the conserved domain has specific biological information.
How do GRINA-C and FAIM2 reduce Gb3? In this study, three cellular phenomena were observed by overexpression of GRINA-C and/or FAIM2 along with the reduction of Gb3: the co-immunoprecipitation of GRINA-C with Gb3 synthase, the promotion of Gb3 synthase degradation in the lysosome, and the perturbed localization of Gb3 synthase and TGN46. The relationship among the three cellular phenomena and their dominance in Gb3 biosynthesis still remains unclear. However, among these phenomena, the interaction of GRINA-C with Gb3 synthase is a strong piece of evidence that GRINA-C directly inhibits Gb3 synthase to reduce Gb3. The interaction of FAIM2 with Gb3 synthase could not be observed in this study; however, we do not rule out the possibility that the weak or transient interaction occurs between FAIM2 and Gb3 synthase in cells, or a small amount of intracellular degradation products of FAIM2 interacts with Gb3 synthase.
Overexpression of GRINA-C and FAIM2 also showed the promotion of Gb3 synthase degradation in a lysosomal protease-dependent manner. Many factors are thought to be involved in protein degradation in cells, including glycosylation and chaperones. Treatment with tunicamycin, an N-glycan inhibitor, promotes degradation of some glycoproteins (41, 42). However, N-glycosylation is unlikely to be involved in degradation of the Gb3 synthase by GRINA-C because of the following results. 1) In pMock-Gb3S cells, treatment with tunicamycin and deoxymannojirimycin, a specific inhibitor of the cis-Golgi processing enzyme α-mannosidase I, still showed the accumulated Gb3 synthase that was non-glycosylated or immature, although the mature form was reduced. Such an accumulation of the intermediates was not observed in pGRINA-C-Gb3S cells (supplemental Fig. S6). 2) Gb3 synthase was degraded by GRINA-C after maturation as shown in Fig. 8. The possibility cannot be ruled out that the difference in terminal glycosylation, including mannose 6-phosphate and sialic acid, is involved in this phenomena, and should be clarified in the future. Chaperones are also important factors to understand protein stability. For example, a molecular chaperone Cosmc is required for activity of the core 1 β3-galactosyltransferase, which is involved in O-glycoprotein synthesis (43). If Gb3 synthase requires a specific chaperone, the interaction of GRINA-C and possibly FAIM2 may inhibit interaction of the factor with Gb3 synthase to promote degradation. As a similar example, overexpression of the V-ATPase V0-c subunit has been known to reduce expression of the β1 integrin through their bindings. Interestingly, the reduced level of β1 integrin was not restored by treatment with MG132, a proteasome inhibitor (44), which is similar to the effect of GRINA-C on Gb3 synthase as shown in Fig. 7C.
We also observed that overexpression of GRINA-C and FAIM2 affects distribution of the Gb3 synthase and TGN46. The perturbed distribution of Gb3 synthase might also affect Gb3 biosynthesis in addition to the direct interaction described above because if Gb3 synthase is mislocalized and unable to encounter LacCer, Gb3 biosynthesis should be suppressed. However, the perturbed distribution does not seem to be the principal reason for reduction of Gb3 by overexpression of GRINA-C, because the rGRINA-C cells, in which the expression level of GRINA-C is much lower than in pGRINA-C#1 cells but Gb3 is reduced, showed no detectable alteration of TGN46 distribution unlike pGRINA-C#1 cells (data not shown). This evidence suggests that interaction of GRINA-C with the Gb3 synthase might more dominantly affect Gb3 biosynthesis than the TGN perturbation. Then, how do GRINA-C and FAIM2 perturb TGN? Intriguingly, the similar effect of GRINA-C and FAIM2 on localization of TGN46 was observed although localizations of GRINA-C and FAIM2 were quite different. FAIM2 was localized in dispersed dot structures that were co-stained with TGN46, whereas GRINA-C was mainly localized in the ER. However, GRINA-C was also localized at the perinuclear region, probably the Golgi apparatus; therefore, a small amount of GRINA-C in the Golgi might affect distribution of TGN molecules. Identification of other affected molecules and elucidation of molecular activity in the shared hydrophobic regions will help to solve the meaning of the TGN perturbation. For example, BI-1 functions as a leaky calcium channel on ER (38). It would be interesting if the UPF0005 domain had a shared function of ion channels, and influence on membrane trafficking.
Does the phenomenon physiologically occur? Several possibilities can be considered. Case 1: GRINA-C or other short forms of the TMBIM family might be synthesized as splicing variants. It is still unknown whether transcripts equivalent to GRINA-C cDNA are expressed. As shown in Fig. 2, however, various deletion mutants show higher activity to reduce Gb3 despite the different number and range of TM domains, and those of different molecules. Furthermore, GRINA-C and, probably other mutants, require only a small amount of protein for activity, as shown in Fig. 3A; therefore, these molecules are likely to function at physiological expression levels if their transcripts exist. Another possibility is that it is synthesized by limited proteolysis in some situations. Case 2: the full-length TMBIM family might be overexpressed, affecting glycolipid biosynthesis. There are several reports that a member of the TMBIM family is up-regulated in some cases; for example, rat GRINA is up-regulated after chronic exposure to ethanol or ischemia-reperfusion injury (45, 46). RECS1 was originally identified as a molecule up-regulated after exposure to shear stress in human umbilical vein endothelial cells (47). Human BI-1, FAIM-2, and GAAP were up-regulated in some tumors (48–52). However, it should be considered whether physiological or pathological up-regulation is enough to reduce Gb3 because retrovirus-based expression of the full-length TMBIM family hardly reduced the Gb3. Case 3: infection of microorganisms containing similar genes to the TMBIM family might cause the reduction of Gb3. Not only higher eukaryotes, including both animals and plants, but also protista, yeast, bacteria, and even some viruses code for proteins with UPF0005 or a BI-1-like domain (40). For example, orthopox viruses, including vaccinia, camelpox, cowpox, and monkeypox, code for full-length or part of GAAP, the sequence of which is more than 70% similar to human GAAP (27). Human cytomegalovirus (HHV-5) has a set of 10 contiguous genes (US12–21), which encode proteins that are similar to the mammalian TMBIM family, especially US21 (53). If these viruses infect host cells in which many of the proteins are expressed, the reduction of intracellular Gb3 might occur.
Together with the reduction of Gb3, the accumulation and surface expression of LacCer are also physiologically notable points, because LacCer has been known to function as a signal mediator in some cases. A type of fungal β-glucans, derived from Candida albicans etc., specifically bound to LacCer and enhanced neutrophil functions, including chemotaxis (54). LPS and IFN-γ treatment increased intracellular LacCer, which mediated iNOS gene expression in primary rat astrocytes (55). Furthermore, vascular endothelial growth factor also increased LacCer, which up-regulated platelet endothelial cell adhesion molecule expression and stimulated angiogenesis in human endothelial cells (56). If the TMBIM family or related proteins are overexpressed in Gb3-positive cells, endothelial cells etc., sensitivity to the above stimulators might be up-regulated along with an increase in LacCer.
Overall, we found the TMBIM family to be potential molecules affecting Gb3 synthesis and TGN homeostasis. Physiological verification of this phenomenon is expected to elucidate the organization of glycosyltransferases in TGN and the functions of the TMBIM family in detail.
Supplementary Material
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology of Japan Grant-in-aid for Young Scientists (B) No. 20770114 (to T. Y.) and Japan Society for the Promotion of Science Grant-in-aid for Scientific Research (B) No. 22370054 (to K. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures” and Figs. S1–S8.
- GSL
- glycosphingolipid
- GRINA
- glutamate receptor, ionotropic, N-methyl-d-asparate-associated protein 1
- GRINA-C
- COOH-terminal polypeptide of GRINA
- TM
- transmembrane
- TMBIM
- transmembrane BAX inhibitor motif containing
- FAIM2
- FAS inhibitory molecule 2
- ER
- endoplasmic reticulum
- TGN
- trans-Golgi network
- Gb3
- globotriaosylceramide
- LacCer
- lactosylceramide
- GalCer
- galactosylceramide
- DGDG
- digalactosyl diacylglycerol
- Gb3S
- Gb3 synthase
- GM3S
- GM3 synthase
- Stx1
- Shiga toxin 1
- Stx1B
- Stx1 B subunit
- StxRs
- Stx receptors
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
REFERENCES
- 1.Hakomori S. I. (2008) Biochim. Biophys. Acta 1780, 325–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacewicz M., Clausen H., Nudelman E., Donohue-Rolfe A., Keusch G. T. (1986) J. Exp. Med. 163, 1391–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lingwood C. A., Law H., Richardson S., Petric M., Brunton J. L., De Grandis S., Karmali M. (1987) J. Biol. Chem. 262, 8834–8839 [PubMed] [Google Scholar]
- 4.Noris M., Remuzzi G. (2005) J. Am. Soc. Nephrol. 16, 1035–1050 [DOI] [PubMed] [Google Scholar]
- 5.Keusch J. J., Manzella S. M., Nyame K. A., Cummings R. D., Baenziger J. U. (2000) J. Biol. Chem. 275, 25315–25321 [DOI] [PubMed] [Google Scholar]
- 6.Kojima Y., Fukumoto S., Furukawa K., Okajima T., Wiels J., Yokoyama K., Suzuki Y., Urano T., Ohta M., Furukawa K. (2000) J. Biol. Chem. 275, 15152–15156 [DOI] [PubMed] [Google Scholar]
- 7.Steffensen R., Carlier K., Wiels J., Levery S. B., Stroud M., Cedergren B., Nilsson Sojka B., Bennett E. P., Jersild C., Clausen H. (2000) J. Biol. Chem. 275, 16723–16729 [DOI] [PubMed] [Google Scholar]
- 8.Okuda T., Tokuda N., Numata S., Ito M., Ohta M., Kawamura K., Wiels J., Urano T., Tajima O., Furukawa K., Furukawa K. (2006) J. Biol. Chem. 281, 10230–10235 [DOI] [PubMed] [Google Scholar]
- 9.Nudelman E., Kannagi R., Hakomori S., Parsons M., Lipinski M., Wiels J., Fellous M., Tursz T. (1983) Science 220, 509–511 [DOI] [PubMed] [Google Scholar]
- 10.Farkas-Himsley H., Hill R., Rosen B., Arab S., Lingwood C. A. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 6996–7000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kovbasnjuk O., Mourtazina R., Baibakov B., Wang T., Elowsky C., Choti M. A., Kane A., Donowitz M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 19087–19092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gehrmann M., Liebisch G., Schmitz G., Anderson R., Steinem C., De Maio A., Pockley G., Multhoff G. (2008) PLoS One 3, e1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clarke J. T. (2007) Ann. Intern. Med. 146, 425–433 [DOI] [PubMed] [Google Scholar]
- 14.Puri A., Hug P., Jernigan K., Barchi J., Kim H. Y., Hamilton J., Wiels J., Murray G. J., Brady R. O., Blumenthal R. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14435–14440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lingwood C. A., Binnington B., Manis A., Branch D. R. (2010) FEBS Lett. 584, 1879–1886 [DOI] [PubMed] [Google Scholar]
- 16.Watanabe M., Matsuoka K., Kita E., Igai K., Higashi N., Miyagawa A., Watanabe T., Yanoshita R., Samejima Y., Terunuma D., Natori Y., Nishikawa K. (2004) J. Infect. Dis. 189, 360–368 [DOI] [PubMed] [Google Scholar]
- 17.Morita S., Kojima T., Kitamura T. (2000) Gene Ther. 7, 1063–1066 [DOI] [PubMed] [Google Scholar]
- 18.Hanada K., Kumagai K., Yasuda S., Miura Y., Kawano M., Fukasawa M., Nishijima M. (2003) Nature 426, 803–809 [DOI] [PubMed] [Google Scholar]
- 19.Tomishige N., Kumagai K., Kusuda J., Nishijima M., Hanada K. (2009) Mol. Biol. Cell 20, 348–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawano M., Kumagai K., Nishijima M., Hanada K. (2006) J. Biol. Chem. 281, 30279–30288 [DOI] [PubMed] [Google Scholar]
- 21.Okemoto-Nakamura Y., Yamakawa Y., Hanada K., Tanaka K., Miura M., Tanida I., Nishijima M., Hagiwara K. (2008) Microbiol. Immunol. 52, 357–365 [DOI] [PubMed] [Google Scholar]
- 22.Bligh E. G., Dyer W. J. (1959) Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 23.Hanada K., Hara T., Fukasawa M., Yamaji A., Umeda M., Nishijima M. (1998) J. Biol. Chem. 273, 33787–33794 [DOI] [PubMed] [Google Scholar]
- 24.Uemura S., Yoshida S., Shishido F., Inokuchi J. (2009) Mol. Biol. Cell 20, 3088–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Q., Reed J. C. (1998) Mol. Cell 1, 337–346 [DOI] [PubMed] [Google Scholar]
- 26.Somia N. V., Schmitt M. J., Vetter D. E., Van Antwerp D., Heinemann S. F., Verma I. M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 12667–12672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gubser C., Bergamaschi D., Hollinshead M., Lu X., van Kuppeveld F. J., Smith G. L. (2007) PLoS Pathog. 3, e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oka T., Sayano T., Tamai S., Yokota S., Kato H., Fujii G., Mihara K. (2008) Mol. Biol. Cell 19, 2597–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Di Sano F., Fazi B., Tufi R., Nardacci R., Piacentini M. (2007) J. Neurochem. 102, 345–353 [DOI] [PubMed] [Google Scholar]
- 30.Zheng Y., Gao B., Ye L., Kong L., Jing W., Yang X., Wu Z., Ye L. (2005) J. Microbiol. 43, 529–536 [PubMed] [Google Scholar]
- 31.Ushioda R., Hoseki J., Araki K., Jansen G., Thomas D. Y., Nagata K. (2008) Science 321, 569–572 [DOI] [PubMed] [Google Scholar]
- 32.Rutkowski D. T., Kaufman R. J. (2007) Trends Biochem. Sci. 32, 469–476 [DOI] [PubMed] [Google Scholar]
- 33.Bridges J. P., Xu Y., Na C. L., Wong H. R., Weaver T. E. (2006) J. Cell Biol. 172, 395–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Bijl P., Strous G. J., Lopes-Cardozo M., Thomas-Oates J., van Meer G. (1996) Biochem. J. 317, 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujimoto H., Tadano-Aritomi K., Tokumasu A., Ito K., Hikita T., Suzuki K., Ishizuka I. (2000) J. Biol. Chem. 275, 22623–22626 [DOI] [PubMed] [Google Scholar]
- 36.Samuel J. E., Perera L. P., Ward S., O'Brien A. D., Ginsburg V., Krivan H. C. (1990) Infect. Immun. 58, 611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar K. N., Tilakaratne N., Johnson P. S., Allen A. E., Michaelis E. K. (1991) Nature 354, 70–73 [DOI] [PubMed] [Google Scholar]
- 38.Kim H. R., Lee G. H., Ha K. C., Ahn T., Moon J. Y., Lee B. J., Cho S. G., Kim S., Seo Y. R., Shin Y. J., Chae S. W., Reed J. C., Chae H. J. (2008) J. Biol. Chem. 283, 15946–15955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Mattia F., Gubser C., van Dommelen M. M., Visch H. J., Distelmaier F., Postigo A., Luyten T., Parys J. B., de Smedt H., Smith G. L., Willems P. H., van Kuppeveld F. J. (2009) Mol. Biol. Cell 20, 3638–3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu L., Smith T. F., Goldberger G. (2009) Apoptosis 14, 1255–1265 [DOI] [PubMed] [Google Scholar]
- 41.Olden K., Pratt R. M., Yamada K. M. (1978) Cell 13, 461–473 [DOI] [PubMed] [Google Scholar]
- 42.Ciccarelli E., Alonso M. A., Cresteil D., Bollen A., Jacobs P., Alvarez F. (1993) Eur. J. Biochem. 213, 271–276 [DOI] [PubMed] [Google Scholar]
- 43.Ju T., Cummings R. D. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 16613–16618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee I., Skinner M. A., Guo H. B., Sujan A., Pierce M. (2004) J. Biol. Chem. 279, 53007–53014 [DOI] [PubMed] [Google Scholar]
- 45.Bao X., Hui D., Näassila M., Michaelis E. K. (2001) Neurosci. Lett. 315, 5–8 [DOI] [PubMed] [Google Scholar]
- 46.Aikawa H., Tomita H., Ishiguro S., Nishikawa S., Sugano E., Tamai M. (2003) Tohoku J. Exp. Med. 199, 25–33 [DOI] [PubMed] [Google Scholar]
- 47.Yoshisue H., Suzuki K., Kawabata A., Ohya T., Zhao H., Sakurada K., Taba Y., Sasaguri T., Sakai N., Yamashita S., Matsuzawa Y., Nojima H. (2002) Atherosclerosis 162, 323–334 [DOI] [PubMed] [Google Scholar]
- 48.Grzmil M., Kaulfuss S., Thelen P., Hemmerlein B., Schweyer S., Obenauer S., Kang T. W., Burfeind P. (2006) J. Pathol. 208, 340–349 [DOI] [PubMed] [Google Scholar]
- 49.Harima Y., Togashi A., Horikoshi K., Imamura M., Sougawa M., Sawada S., Tsunoda T., Nakamura Y., Katagiri T. (2004) Int. J. Radiat. Oncol. Biol. Phys. 60, 237–248 [DOI] [PubMed] [Google Scholar]
- 50.Igney F. H., Krammer P. H. (2002) Nat. Rev. Cancer 2, 277–288 [DOI] [PubMed] [Google Scholar]
- 51.Bucan V., Reimers K., Choi C. Y., Eddy M. T., Vogt P. M. (2010) Cell Mol. Biol. Lett. 15, 296–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van't Veer L. J., Dai H., van de Vijver M. J., He Y. D., Hart A. A., Mao M., Peterse H. L., van der Kooy K., Marton M. J., Witteveen A. T., Schreiber G. J., Kerkhoven R. M., Roberts C., Linsley P. S., Bernards R., Friend S. H. (2002) Nature 415, 530–536 [DOI] [PubMed] [Google Scholar]
- 53.Lesniewski M., Das S., Skomorovska-Prokvolit Y., Wang F. Z., Pellett P. E. (2006) Virology 354, 286–298 [DOI] [PubMed] [Google Scholar]
- 54.Yoshizaki F., Nakayama H., Iwahara C., Takamori K., Ogawa H., Iwabuchi K. (2008) Biochim. Biophys. Acta 1780, 383–392 [DOI] [PubMed] [Google Scholar]
- 55.Pannu R., Won J. S., Khan M., Singh A. K., Singh I. (2004) J. Neurosci. 24, 5942–5954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rajesh M., Kolmakova A., Chatterjee S. (2005) Circ. Res. 97, 796–804 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.