Abstract
Little is known about how chaperones in the endoplasmic reticulum are organized into complexes to assist in the proper folding of secreted proteins. One notable exception is the complex of ERp57 and calnexin that functions as part the calnexin cycle to direct disulfide bond formation in N-glycoproteins. Here, we report three new complexes composed of the peptidyl prolyl cis-trans-isomerase cyclophilin B and any of the lectin chaperones: calnexin, calreticulin, or calmegin. The 1.7 Å crystal structure of cyclophilin with the proline-rich P-domain of calmegin reveals that binding is mediated by the same surface that binds ERp57. We used NMR titrations and mutagenesis to measure low micromolar binding of cyclophilin to all three lectin chaperones and identify essential interfacial residues. The immunosuppressant cyclosporin A did not affect complex formation, confirming the functional independence of the P-domain binding and proline isomerization sites of cyclophilin. Our results reveal the P-domain functions as a unique protein-protein interaction domain and implicate a peptidyl prolyl isomerase as a new element in the calnexin cycle.
Keywords: Chaperone Chaperonin, Lectin, NMR, Prolyl Isomerase, Protein Folding, X-ray Crystallography
Introduction
The endoplasmic reticulum (ER)2 is the cellular compartment responsible for the folding of secreted proteins. Molecular chaperones in the ER include general chaperones such as BiP, which promote folding by binding and stabilizing exposed hydrophobic stretches of partially folded protein, and more specialized chaperones such as ERp57 and cyclophilin B, which catalyze disulfide bond formation and prolyl peptide bond cis-trans isomerization (1). Several studies have used cross-linking and co-immunoprecipitation experiments to show the existence of large multiprotein complexes in the ER. These include a complex composed of GRP94, BiP, P5, PDI, ERdj3, cyclophilin B, ERp72, GRP170, UDP-glucose:glycoprotein glucosyltransferase (UGGT), SDF2-L1, and substoichiometric amounts of calreticulin (CRT) (2) and a complex of GRP94, ERp72, BiP, CRT, and cyclophilin B (3). Little is known about the nature of the protein-protein interactions that hold these complexes together and regulate their activity.
The calnexin cycle is a multichaperone system specific for the folding of N-glycosylated proteins (4). It involves enzymes that modify the N-glycan of nascent proteins and a family of lectin chaperones that specifically bind the monoglucosylated glycan Glc1Man9(GlcNAc)2 (5). These chaperones, calnexin (CNX), CRT, and calmegin (CMG), are composed of a globular lectin domain and a unique extended arm-like structure known as the P-domain due to an abundance of proline residues (6, 7). The tip of the P-domain of CNX and CRT binds the protein disulfide isomerase ERp57 to form a heterodimeric chaperone specific for N-glycosylated proteins (8–12).
Proteins synthesized on rough ER ribosomes enter the calnexin cycle by the co-translational addition of a Glc3Man9(GlcNAc)2 polysaccharide to asparagine side chains. The N-glycan is then trimmed by ER glucosidases to the monoglucosylated form that is recognized by CNX, CRT, and CMG (4). The complex of ERp57 with CNX, CRT, or CMG likely acts as a general chaperone in addition to its role in catalyzing proper disulfide bond formation through the isomerase activity of ERp57 (12). The engagement of the lectin chaperone·ERp57 complex is strictly regulated by the structure of the N-glycan. Once the terminal glucose residue is removed by glucosidase II, the trimmed glycoprotein no longer interacts with CNX/CRT/CMG. On the other hand, unfolded or misfolded glycoproteins that lack the terminal glucose can be rescued by the action of UDP-glucose:glycoprotein glucosyltransferase, which adds a glucose to the terminal mannose for renewed interaction with the folding machinery.
Cyclophilin B is one of a number of peptidyl prolyl cis-trans-isomerases (PPIases) found in the ER (13, 14). It is inhibited by cyclosporin A, which binds to its active site with high affinity (13). Treating cells with cyclosporin A interferes with the folding of collagen (15) and the maturation of transferrin (16), demonstrating the functional relevance of cyclophilin B in the ER. Cyclophilin B forms a complex with prolyl 3-hydroxylase and cartilage-associated protein (17). Recent studies showed that ER stress activates cyclophilin B expression, whereas its absence makes cells more sensitive to ER stress (18). Why the ER contains multiple PPIases and how they recruit substrates are open questions.
Important clues to the function of ER chaperones have come from the study of their interacting partners (1). The association of ERp57 with CNX enabled its function in N-glycoprotein folding to be established. Similarly, the association of ERdj5 with BiP and EDEM enabled elucidation of its role as a reductase involved in ER-associated decay (19). Here, we show that cyclophilin B binds directly to the P-domain of the lectin chaperones. The crystal structure of cyclophilin B in complex with the P-domain from CMG provides a mechanism for the specific recruitment of PPIase activity to unfolded N-glycoproteins and suggests that cyclophilin B functions as part of the calnexin cycle.
EXPERIMENTAL PROCEDURES
Protein Expression, Preparation, and Purification
Human cyclophilin B (residues 2–184 of the mature protein) was cloned into a pGEX-6P-1 vector (GE Healthcare, Uppsala, Sweden) and expressed in Escherichia coli BL21(DE3) in rich (LB) medium as a fusion with N-terminal GST tag. For NMR experiments, the recombinant protein was labeled by growth of E. coli BL21 in M9 minimal medium with [15N]ammonium sulfate as the sole source of nitrogen. Cells were harvested and broken in PBS, pH 7.4. The GST fusion protein was purified by affinity chromatography on glutathione-Sepharose resin, and the tag was removed by cleavage with PreScission protease (GE Healthcare), leaving a Gly-Pro-Leu-Gly-Ser N-terminal extension. The cleaved protein was additionally purified using ion-exchange chromatography (SP Sepharose) and dialyzed against NMR buffer (20 mm MES, 50 mm NaCl, pH 6.5).
The two-module fragment of canine CNX P-domain (residues 310–381) was expressed and purified as described earlier (11). The two-module (residues 211–261) and one-module (residues 221–256) fragments of mouse CRT P-domain and the one-module (residues 317–350) fragment of canine CMG P-domain were cloned into pET15b vector (GE Healthcare) and expressed in E. coli BL21(DE3) as a fusion with N-terminal His tag. The proteins were purified using a similar procedure to that for the calnexin P-domain. After cleavage with thrombin, the purified fragments contained an N-terminal Gly-Ser-His-Met extension and were exchanged into NMR buffer (20 mm MES, 50 mm NaCl, pH 6.5).
To obtain the D344K, D346K, D348K, E350K, and E352K CNX P-domain mutants and the K6A, K9A, K35A, K97A, and K183A cyclophilin B mutants, site-directed mutagenesis was performed on calnexin or cyclophilin B cDNA using mismatched primers and PCR with TaqPlus PrecisionTM polymerase (Stratagene, La Jolla, CA) following the manufacturer's protocol. Automated DNA sequencing was performed to verify all sequence modifications.
Pulldown Assays
0.5–2 mg of purified recombinant proteins (cyclophilin B, ERp57, ERp72) was coupled to 100 μl of NHS-activated Sepharose 4B (GE Healthcare) for 2 h at 4 °C in a total volume of 500 μl of HA buffer (115 mm potassium acetate, 20 mm HEPES, pH 7.0, 0.75 mm CaCl2, 0.1% Triton X-100) with rotation. After coupling, beads were blocked with 500 μl of 0.5 m NaCl, 0.5 m ethanolamine, pH 8.3 for 30 min with rocking at room temperature. Beads were washed four times with 500 μl of HA buffer and stored at 4 °C. Rat liver low density microsomes were prepared as described previously (20) and solubilized in ice-cold HA buffer at a concentration of 3–5 mg/ml. Insoluble portions were removed by centrifugation at 5500 × g for 10 min at 4 °C. 100 μl of the soluble fraction was applied to 10 μl of bait-coupled beads and incubated with gentle agitation at 4 °C for 2 h. Beads were washed with ice cold HA buffer and then resuspended in 50 μl of 2× Laemmli buffer containing 0.5 mm β-mercaptoethanol at 37 °C for 5 min. Supernatants were obtained by centrifugation at 4000 × g, and 10 μl of each eluate was analyzed by SDS-PAGE and Western blotting. Immunodetection was performed using anti-CRT from Daniel Tessier (National Research Council, Montreal, Canada) diluted 1:5000 in 5% skim milk TBS-T (0.1% Tween 20) and HRP-conjugated goat anti-rabbit IgG (Santa Cruz Biotechnology).
Crystallization
Initial crystallization conditions were identified utilizing hanging drop vapor diffusion with the PACT screen (Qiagen). The crystals of unliganded cyclophilin B were obtained by equilibrating a 0.6-μl drop of the cyclophilin B (7 mg/ml) in buffer (20 mm MES (pH 6.5), 50 mm NaCl), mixed with 0.6 μl of reservoir solution containing 25% (w/v) PEG 1500, and 0.1 m MMT buffer (pH 6.0) and suspended over 0.6 ml of reservoir solution. No additional cryoprotectant was added prior to flash-freezing the crystals in a N2 cold stream (Oxford Cryosystem). The crystals had a Matthews coefficient of 2.30 Å3 Da−1 and a solvent content of 46.6%.
The best cyclophilin·P-domain complex crystals were obtained by equilibrating a 1-μl drop of the cyclophilin·CMG (residues 317–350) mixture in a 1:3 molar ratio (8 mg/ml) in buffer (20 mm MES (pH 6.5), 50 mm NaCl), mixed with 1 μl of reservoir solution containing 22% (w/v) PEG 8000, 10 mm ZnCl2, and 0.1 m Tris buffer (pH 7.0) and suspended over 1 ml of reservoir solution. Crystals grew in 3–7 days at 22 °C. For data collection, crystals were cryoprotected by soaking in the reservoir solution supplemented with 20% (v/v) glycerol. The crystals had a Matthews coefficient of Vm = 2.12 Å3 Da−1 and a solvent content of 42.0%.
Structure Solution and Refinement
Diffraction data from single crystals of unliganded cyclophilin B and the cyclophilin· P-domain complex were collected on an ADSC Quantum-210 CCD detector (Area Detector Systems Corp.) at beamline A1 at the Cornell High Energy Synchrotron Source (CHESS) (see Table 1). Data processing and scaling were performed with HKL2000 (21). The structures were determined by molecular replacement with Phaser (22), using the coordinates of cyclophilin B (Protein Data Bank (PDB) entry 1CYN). The initial models obtained from Phaser were completed and adjusted with the program Coot (23) and were improved by several cycles of refinement, using the program REFMAC 5.2 (24) and model refitting. At the latest stage of refinement, we also applied the translation-libration-screw (TLS) option (25). The refinement statistics are given in Table 1. The final models have all residues in the allowed regions of Ramachandran plot according to the program PROCHECK.
TABLE 1.
Data collection and refinement statistics
| Cyclophilin B | Cyclophilin/P-domain | |
|---|---|---|
| Data collection | ||
| Space group | P32 | P1 |
| Cell dimensions | ||
| a, b, c (Å) | 64.67, 64.67, 39.46 | 43.41, 44.30, 55.43 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 89.94, 94.46, 114.03 |
| Resolution (Å) | 50-1.2 (1.22-1.20) | 50-1.7 (1.76-1.70)a |
| Rsym | 0.035 (0.105) | 0.087 (0.263) |
| I/σI | 49.8 (8.8) | 18.0 (4.6) |
| Completeness (%) | 100.0 (99.7) | 97.3 (95.0) |
| Redundancy | 5.3 (3.1) | 3.9 (3.4) |
| Refinement | ||
| Resolution (Å) | 55.99-1.20 | 55.22-1.70 |
| No. of reflections | 54608 | 37796 |
| Rwork/Rfree | 0.143/0.162 | 0.188/0.238 |
| No. of atoms | 1810 | 3535 |
| Protein | 1434 | 3017 |
| Water | 376 | 504 |
| Zn2+ | 2 | |
| MES | 12 | |
| B-factors | ||
| Protein | 7.64 | 13.92 |
| Water | 22.67 | 29.82 |
| Zn2+ | 16.53 | |
| MES | 45.14 | |
| r.m.s.b deviations | ||
| Bond lengths (Å) | 0.006 | 0.013 |
| Bond angles (°) | 1.12 | 1.39 |
a Highest resolution shell is shown in parentheses.
b r.m.s., root mean square.
NMR Spectroscopy
NMR resonance assignments of the two-module P-domain of calnexin were described earlier (11). Biological Magnetic Resonance Bank (BMRB) entries 15505 and 4878 were used to assign NMR signals for cyclophilin B and the CRT P-domain, respectively. NMR samples were exchanged in 20 mm MES, 50 mm NaCl, pH 6.5. For NMR titrations, unlabeled P-domain or cyclophilin B was added to 15N-labeled 0.2–0.4 mm titrated protein to the final molar ratio of 3:1. All NMR experiments were performed at 28 °C using a Bruker 600-MHz spectrometer. NMR spectra were processed using NMRPipe and analyzed with XEASY.
RESULTS
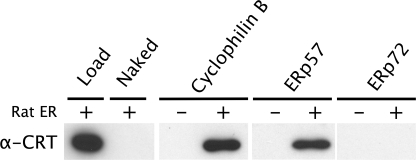
Based on previous reports of their association in cell extracts (2, 3), we tested the direct interaction of the lectin chaperone CRT with purified ER proteins. When covalently attached to beads, cyclophilin B was able to retain CRT to the same extent as ERp57, a known CRT-binding protein (Fig. 1). No retention of CRT was observed on beads coupled to ERp72 or on beads without protein.
FIGURE 1.
CRT binds to cyclophilin B and ERp57 but not ERp72. Lysed rat liver microsomes were incubated with Sepharose beads covalently loaded with the proteins indicated, and bound CRT was detected by Western blotting. Load, 20 μg of rat ER; Naked, beads blocked with ethanolamine; −, no microsomes added; +, microsomes added.
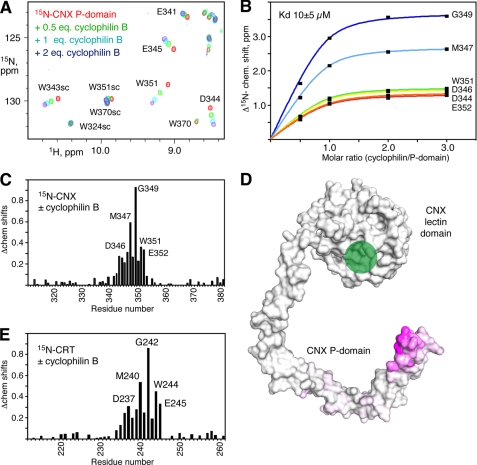
To identify the region of CNX/CRT responsible for binding cyclophilin B, we labeled a fragment of the CNX P-domain with 15N and monitored a titration with unlabeled cyclophilin B using NMR spectroscopy. We observed large chemical shift changes for a number of CNX amide signals, indicating specific binding between the two proteins (Fig. 2). Mapping of the chemical shift changes onto the crystal structure of CNX (PDB entry 1JHN) shows that the signals affected arise from the tip of the P-domain. Titration of the two-module P-domain from mouse CRT (residues 211–261) with cyclophilin produced very similar results (Fig. 2E and supplemental Fig. 1). A fit of the NMR chemical shift changes as a function of the amount of cyclophilin B added allowed an estimation of the binding affinity. The P-domain fragments of both CNX and CRT bound cyclophilin B with affinity on the order of 10 μm. This is very similar to the affinities of ERp57 binding to CNX (Kd of 6 μm) (10) and to CRT (7 μm) (9).
FIGURE 2.
Cyclophilin B interacts with the tip of the P-domain from CNX and CRT. A, downfield region of heteronuclear single quantum correlation spectra of 15N-labeled two-module P-domain (residues 310–381) titrated with increasing amounts of cyclophilin B. The spectra show specific chemical shift changes arising from residues at the tip of the P-domain. Backbone and side chain (sc) signal assignments are shown. B, Kd of the binding estimated from a fit of 5N chemical shift (chem. shift) changes for six residues in the P-domain. C, plot of weighted average 1H and 5N chemical shift changes in the 15N-labeled CNX P-domain upon the addition of unlabeled cyclophilin B. D, mapping of the chemical shifts measured in the P-domain on the x-ray crystal structure of CNX (7). Purple indicates a large chemical shift change; white indicates no change. The green circle marks the glycan binding site in the lectin domain. E, plot of chemical shift changes in the 15N-labeled single module P-domain of CRT (residues 211–261) upon the addition of unlabeled cyclophilin B.
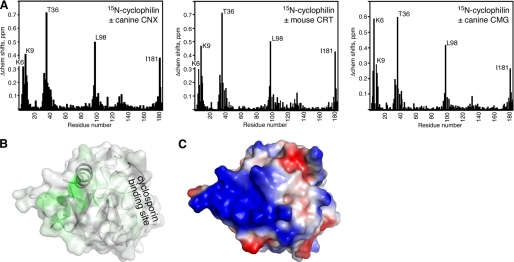
We carried out the reverse titration with 15N-labeled cyclophilin B to identify the cyclophilin residues involved in binding. Titration with various P-domain fragments led to large chemical shift changes in the N and C termini and two adjacent loops of cyclophilin (Fig. 3A). When mapped onto the cyclophilin B structure, the changes converge to a well defined surface opposite the cyclosporin A binding surface (26) (Fig. 3B). The affected surface is abundant in lysine residues and has a pronounced positively charged character (Fig. 3C). This is not surprising as the tip of a P-domain is rich in negatively charged aspartate and glutamate residues. Cyclophilin B binds heparin using this same surface, albeit the chemical shifts changes induced by heparin are much smaller (27). The binding affinity measured in the reverse titrations was the same (∼10 μm) as measured in titrations with 15N-labeled P-domains.
FIGURE 3.
Identification of the P-domain binding site on cyclophilin B. A, plot of chemical shift changes (Δchem shifts) in 15N-labeled cyclophilin B upon the addition of unlabeled P-domain fragments from CNX (residues 310–381), CRT (residues 211–261), and CMG (residues 317–350). Titration with a smaller fragment of CRT (residues 221–256) produced identical results. Cyclophilin residues showing large chemical shift changes are labeled. B, mapping of the chemical shifts from CNX addition onto the cyclophilin structure. Color is proportional to chemical shift changes from green (large change) to white (no change). C, electrostatic potential of cyclophilin showing the abundance of basic residues in the P-domain binding site. Positive charge is colored blue; negative charge is red.
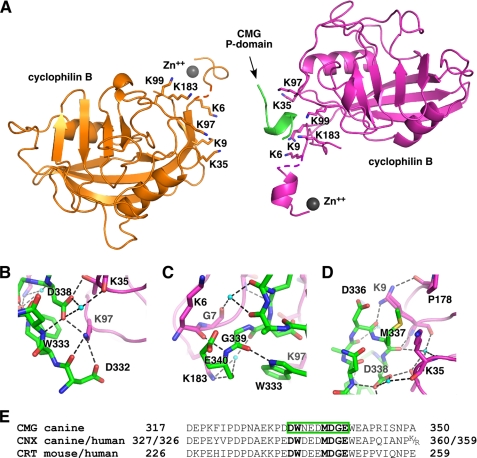
We next used x-ray crystallography to identify the molecular details of the P-domain-cyclophilin interaction. We first crystallized unliganded cyclophilin B and determined its structure to 1.2 Å resolution (Table 1). The structure is similar to the previous resolved structure with cyclosporin A bound (26) but includes six N-terminal residues absent in the prior structure (supplemental Fig. 2A). We then carried out crystallization screens of cyclophilin B with a number of two- and one-module P-domains. The best crystals were obtained with the CMG P-domain, and the complex structure was determined to 1.7 Å resolution (Table 1). The asymmetric unit contains two cyclophilin B molecules with the positively charged surfaces previously identified facing each other (Fig. 4A). Only a single P-domain fragment is present as there is insufficient space between the two cyclophilin molecules to accommodate two P-domains. We observed nearly complete molecules of cyclophilin B in the electron density maps, with the exception of Lys5 of chain A and Lys4–Lys5 of chain B. Crystal packing was mediated in part through the formation of an α-helix by the N-terminal cloning linker and the binding of a zinc atom (supplemental Fig. 3). Only nine bound residues of the tip of the P-domain are visible in the electron density map. The rest of the 38-residue fragment and the side chain of Glu335 appear to be disordered.
FIGURE 4.
Crystal structure of cyclophilin B bound to the CMG P-domain. A, ribbon representation of the complex structure. There are two molecules of cyclophilin (orange and magenta) in the asymmetric unit, but by happenstance, only one is in a complex with the CMG P-domain. The P-domain is green, and the two bound Zn2+ are shown in gray. Key lysine residues that contribute to P-domain binding are labeled. B, key intermolecular salt bridges and hydrogen bonds (dashed lines) involving cyclophilin Lys97 and the P-domain Asp338. C, close packing of Gly339 of the P-domain (green) with Gly7 of cyclophilin (magenta). D, the side chain of Met337 inserts between side chains of Lys9 and Lys35 of cyclophilin B, whereas the side chain of Lys9 adopts two alternative conformations forming intra- and intermolecular hydrogen bonds with the carbonyls of Pro178 and Asp336. Figures were made with PyMOL (35). E, sequence of the P-domain fragment crystallized and its alignment with other P-domains. CMG P-domain residues observed in the electron density map are boxed in green. Conserved residues making important contacts are shown in bold.
The crystal structure reveals the molecular complementarity of the P-domain and cyclophilin binding surfaces. The tip of the P-domain binds as an α-helical turn in which carbonyl groups of Asn334 and Glu335 make hydrogen bonds with amides of Asp338 and Gly339, respectively. The helical conformation is stabilized by hydrogen bonds between the imino proton of Trp333 with the carbonyl of Gly339 and between amide of Asn334 and the side chain of Asp338. Analysis of intermolecular contacts shows the importance of polar interactions mediated by Lys97 of cyclophilin B and Asp338 of the P-domain (Fig. 4B). The side chain of Lys97 forms salt bridges with the side chains of Asp338 and Asp332 and hydrogen-bonds with the carbonyl of Asp332. The side chain of Asp338 forms a salt bridge with the side chain of Thr36 and hydrogen-bonds with the carbonyl of Lys35 via an ordered water molecule. In addition, the carbonyl of Asp338 makes a hydrogen bond with the side chain of Lys183 via a water molecule. The side chain of Lys183 hydrogen-bonds with the carbonyl of Glu340, whereas the amide of Glu340 hydrogen-bonds with the carbonyl of Lys6, emphasizing the close approach of the backbone atoms (Fig. 4C). Nearby, the carbonyl of Gly7 employs an ordered water molecule to hydrogen-bond with the carbonyls of Glu335 and Asp336. The side chain of Lys9 makes polar contacts with the side chain and carbonyl of Asp336. Finally, the carbonyl of Met337 at the very tip of the P-domain hydrogen-bonds with the side chain of Thr36 directly and with amides of Lys35 and Thr36 and the carbonyl of Lys9 via a water molecule (Fig. 4D). Although the P-domain-interacting surface of cyclophilin B is abundant in lysine residues, only the side chains of Lys9, Lys97, and Lys183 are involved in the intermolecular salt bridges and hydrogen bonds.
The intermolecular contacts are stabilized by a few hydrophobic interactions. Thus, the side chain of Met337 faces away from the P-domain and inserts between the aliphatic part of the side chains of Lys9 and Lys35. The absence of a side chain for Gly339 allows packing of its Cα atom against the carbonyl of Gly7. It appears that no other residue could be tolerated at this position, explaining the conservation of this glycine in the CNX/CRT protein family (Fig. 4E). Additionally, the protruding edge of the Trp333 aromatic ring packs against carbonyl and Cα of Lys97 (Fig. 4C).
We performed site-directed mutagenesis of both cyclophilin B and the P-domain to verify the intermolecular surface and confirm the residues crucial for their interactions (Table 2). We expressed 15N-labeled mutants and used NMR to verify that all the mutants were correctly folded. We prepared the D344K, D346K, D348K, E350K, and E352K mutants of the CNX P-domain and tested their binding to cyclophilin B by NMR. Only the D348K mutation abolished binding to cyclophilin B, indicating that this residue is crucial for the complex formation. Likewise, among the cyclophilin B mutants K6A, K9A, K35A, K97A, and K183A, only the K97A mutant did not bind to the CNX P-domain. The K183A mutant showed impaired binding (supplemental Fig. 4).
TABLE 2.
Mutagenesis of cyclophilin B/P-domain interactions
| Cyclophilin | CNX P-domain | Bindinga |
|---|---|---|
| Wild type | Wild type | + |
| Wild type | D344K | + |
| Wild type | D346K | + |
| Wild type | D348K | − |
| Wild type | E350K | + |
| Wild type | E352K | + |
| K6A | Wild type | + |
| K9A | Wild type | + |
| K35A | Wild type | + |
| K97A | Wild type | − |
| K183A | Wild type | +/− |
a Determined by NMR chemical shift titrations.
Sequence alignment of the P-domains from CRT, CNX, and CMG shows that several residues are invariantly conserved in the tip region (Fig. 4E). Of these, the tryptophan residues are structurally important as they form a small hydrophobic core. Among the solvent-exposed residues, the aspartic acid and glycine residues following the conserved methionine at the tip of the P-domain are absolutely conserved and are crucial for the cyclophilin B binding. Interestingly, the binding region of the P-domain is strikingly similar to the region that interacts with ERp57 (10). Comparison of the chemical shift changes of the P-domain of CNX upon binding ERp57 and cyclophilin B shows that in both cases, binding is mediated by the extreme tip of the P-domain (10). Significantly, the CNX P-domain D348K mutation also abolishes binding to ERp57,3 and the homologous aspartic acid is required for CRT binding to ERp57 (28).
An overlay of our structure with the crystal structure of cyclophilin B in complex with cyclosporin A suggests that the prolyl isomerization and the P-domain binding sites are independent (supplemental Fig. 2B). To see whether binding to cyclosporin would affect the interactions of cyclophilin with a P-domain, we carried out NMR titrations in the presence of cyclosporin A. Binding of the P-domain of CNX was not affected by cyclosporin bound to the active site of cyclophilin B (supplemental Fig. 5), confirming that these binding events are functionally independent.
DISCUSSION
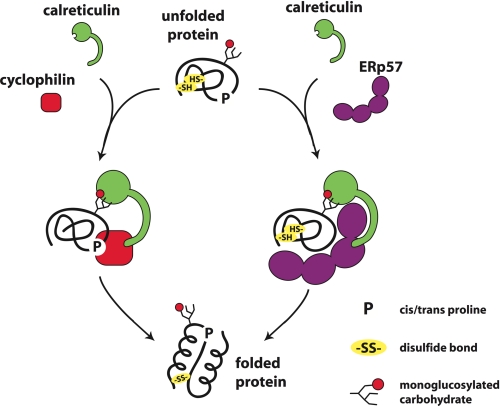
Protein maturation in the ER is a complex and highly ordered process involving enzymes that modify the covalent chemical structure of the proteins and chaperones that mediate the more subtle conformational changes associated with protein folding. Work from a number of groups, including our own, has shown that these processes are linked through the calnexin cycle wherein the trimming of the glycan of N-linked glycoproteins is tied to the folded state of the protein (reviewed in Ref. 4). Recruitment of the heterodimeric chaperone complex CNX-ERp57 is in turn regulated by the presence of a terminal glucose residue on the N-glycan. The complex of cyclophilin B with CNX and the other lectin chaperones suggests that cyclophilin B acts in the calnexin cycle in a fashion analogous to ERp57 (Fig. 5).
FIGURE 5.
Model of cyclophilin B action in concert with CRT and ERp57. An unfolded glycoprotein with cysteine residues (−SH) and proline residues (P) is engaged by CRT (CNX or CMG) through its lectin domain, which is specific for monoglucosylated N-glycans. The P-domain of lectin chaperone can recruit either cyclophilin B or ERp57 to promote respectively protein folding through prolyl peptide bond isomerization or disulfide bond formation.
Several lines of argument support the idea that CNX/CRT/CMG and cyclophilin B interact in vivo. Cyclophilin B and CNX/CRT/CMG co-localize in the ER and have been reported by two different groups to be associated in multichaperone ER complexes (2, 3). The specific interaction between cyclophilin and CRT has also been proposed to contribute to ER retention of cyclophilin B, which lacks other known ER retention signals (29). Finally, the interaction site on the lectin chaperones used to bind cyclophilin B is the same site used to bind ERp57 (9, 10). The P-domain appears to act as specialized protein-protein domain composed of a long semi-rigid linker with a binding site at its tip for either cyclophilin B or ERp57. The chemical shift changes observed in the lectins complexed with cyclophilin are essentially identical to those observed in the complex with ERp57; furthermore, both complexes have the same profile of sensitivity to amino acid substitutions in the P-domains. This suggests that the structure of the P-domain·ERp57 complex can be reliably modeled based on the P-domain·cyclophilin structure presented here.
The association of glycan binding activity with cyclophilin B provides a mechanism for the recruitment of PPIase activity in the ER to newly synthesized glycoproteins, such as the CH antibody heavy chain. The heavy chain CH1 domain possesses three cis-prolines in its native state, and its folding is markedly accelerated by cyclophilin B (30). As a component of the calnexin cycle, cyclophilin B would bind to the P-domains of the abundant lectin chaperones CNX, CRT, or CMG (Fig. 5). These would in turn bind nascent, unfolded monoglucosylated proteins to promote their folding through the action of cyclophilin B. Future work is required to test whether monoglucosylation affects the rate of proline isomerization of N-glycoproteins such as CH1.
The specific targeting of cyclophilin B provides insight into two long standing queries regarding the existence of multiple ER PPIases and the low activity of PPIases relative to the spontaneous isomerization rate. The ER contains seven PPIases, but it is unknown whether they are specific for different substrates or for different steps in folding (31, 32). Our results suggest that cyclophilin B functions primarily as a substrate-specific PPIase for monoglucosylated N-glycoproteins. The second question concerns the catalytic efficiency (kcat/Km) of PPIases relative to their cellular concentrations (33). As has been suggested by previous investigators, the low in vitro activities of PPIases may be due to the absence of other factors that increase their activity in vivo (34). In the case of cyclophilin B, the association with lectin chaperones should increase the catalytic efficiency through the recruitment of substrates and decrease in the effective Km.
In conclusion, the x-ray structure and NMR titrations presented in this study reveal that the lectin chaperone P-domains are highly specialized protein-protein interaction domains and implicate PPIase activity as a new element of the well characterized calnexin cycle.
Acknowledgments
Data acquisition at the Macromolecular Diffraction (MacCHESS) facility at the CHESS was supported by the National Science Foundation Award DMR 0225180 and the National Institutes of Health Award RR-01646. We thank Ali Fazel for liver microsomes and Daniel Tessier for anti-calreticulin serum.
This work was supported by Grants 81277 and 74567 from the Canadian Institutes of Heath Research (to D. Y. T. and K. G.).
This article was selected as a Paper of the Week.
The atomic coordinates and structure factors (codes 3ICH and 3ICI) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5.
G. Kozlov, unpublished data.
- ER
- endoplasmic reticulum
- CNX
- calnexin
- CRT
- calreticulin
- CMG
- calmegin
- PPIase
- peptidyl prolyl cis-trans-isomerase.
REFERENCES
- 1.Christis C., Lubsen N. H., Braakman I. (2008) FEBS J. 275, 4700–4727 [DOI] [PubMed] [Google Scholar]
- 2.Meunier L., Usherwood Y. K., Chung K. T., Hendershot L. M. (2002) Mol. Biol. Cell 13, 4456–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang L., Wu G., Tate C. G., Lookene A., Olivecrona G. (2003) J. Biol. Chem. 278, 29344–29351 [DOI] [PubMed] [Google Scholar]
- 4.Ellgaard L., Helenius A. (2001) Curr. Opin. Cell Biol. 13, 431–437 [DOI] [PubMed] [Google Scholar]
- 5.Zapun A., Petrescu S. M., Rudd P. M., Dwek R. A., Thomas D. Y., Bergeron J. J. (1997) Cell 88, 29–38 [DOI] [PubMed] [Google Scholar]
- 6.Ellgaard L., Bettendorff P., Braun D., Herrmann T., Fiorito F., Jelesarov I., Güntert P., Helenius A., Wüthrich K. (2002) J. Mol. Biol. 322, 773–784 [DOI] [PubMed] [Google Scholar]
- 7.Schrag J. D., Bergeron J. J., Li Y., Borisova S., Hahn M., Thomas D. Y., Cygler M. (2001) Mol. Cell 8, 633–644 [DOI] [PubMed] [Google Scholar]
- 8.Leach M. R., Cohen-Doyle M. F., Thomas D. Y., Williams D. B. (2002) J. Biol. Chem. 277, 29686–29697 [DOI] [PubMed] [Google Scholar]
- 9.Frickel E. M., Riek R., Jelesarov I., Helenius A., Wuthrich K., Ellgaard L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1954–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kozlov G., Maattanen P., Schrag J. D., Pollock S., Cygler M., Nagar B., Thomas D. Y., Gehring K. (2006) Structure 14, 1331–1339 [DOI] [PubMed] [Google Scholar]
- 11.Pollock S., Kozlov G., Pelletier M. F., Trempe J. F., Jansen G., Sitnikov D., Bergeron J. J., Gehring K., Ekiel I., Thomas D. Y. (2004) EMBO J. 23, 1020–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zapun A., Darby N. J., Tessier D. C., Michalak M., Bergeron J. J., Thomas D. Y. (1998) J. Biol. Chem. 273, 6009–6012 [DOI] [PubMed] [Google Scholar]
- 13.Price E. R., Zydowsky L. D., Jin M. J., Baker C. H., McKeon F. D., Walsh C. T. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 1903–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasel K. W., Glass J. R., Godbout M., Sutcliffe J. G. (1991) Mol. Cell. Biol. 11, 3484–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steinmann B., Bruckner P., Superti-Furga A. (1991) J. Biol. Chem. 266, 1299–1303 [PubMed] [Google Scholar]
- 16.Lodish H. F., Kong N. (1991) J. Biol. Chem. 266, 14835–14838 [PubMed] [Google Scholar]
- 17.Ishikawa Y., Wirz J., Vranka J. A., Nagata K., HP B. A. (2009) J. Biol. Chem. 6, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J., Choi T. G., Ding Y., Kim Y., Ha K. S., Lee K. H., Kang I., Ha J., Kaufman R. J., Lee J., Choe W., Kim S. S. (2008) J. Cell Sci. 121, 3636–3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ushioda R., Hoseki J., Araki K., Jansen G., Thomas D. Y., Nagata K. (2008) Science 321, 569–572 [DOI] [PubMed] [Google Scholar]
- 20.Lavoie C., Lanoix J., Kan F. W., Paiement J. (1996) J. Cell Sci. 109, 1415–1425 [DOI] [PubMed] [Google Scholar]
- 21.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 22.Read R. J. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 1373–1382 [DOI] [PubMed] [Google Scholar]
- 23.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 24.Murshudov G. N., Vagin A. A., Lebedev A., Wilson K. S., Dodson E. J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 247–255 [DOI] [PubMed] [Google Scholar]
- 25.Winn M. D., Murshudov G. N., Papiz M. Z. (2003) Methods Enzymol. 374, 300–321 [DOI] [PubMed] [Google Scholar]
- 26.Mikol V., Kallen J., Walkinshaw M. D. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 5183–5186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanoulle X., Melchior A., Sibille N., Parent B., Denys A., Wieruszeski J. M., Horvath D., Allain F., Lippens G., Landrieu I. (2007) J. Biol. Chem. 282, 34148–34158 [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y., Kozlov G., Pocanschi C. L., Brockmeier U., Ireland B. S., Maattanen P., Howe C., Elliott T., Gehring K., Williams D. B. (2009) J. Biol. Chem. 284, 10160–10173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arber S., Krause K. H., Caroni P. (1992) J. Cell Biol. 116, 113–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feige M. J., Groscurth S., Marcinowski M., Shimizu Y., Kessler H., Hendershot L. M., Buchner J. (2009) Mol. Cell 34, 569–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braakman I., Otsu M. (2008) Science 321, 499–500 [DOI] [PubMed] [Google Scholar]
- 32.Kay J. E. (1996) Biochem. J. 314, 361–385 [PMC free article] [PubMed] [Google Scholar]
- 33.Balbach J., Schmid F. X. (2000) in Mechanisms of Protein Folding (Pain R. H. ed), 2nd Ed., pp 212–249, Oxford University Press, Oxford [Google Scholar]
- 34.Harrison R. K., Stein R. L. (1990) Biochemistry 29, 3813–3816 [DOI] [PubMed] [Google Scholar]
- 35.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]