Abstract
Polyglutamine expansion within the androgen receptor (AR) causes spinal and bulbar muscular atrophy (SBMA) and is associated with misfolded and aggregated species of the mutant AR. We showed previously that nuclear localization of the mutant AR was necessary but not sufficient for SBMA. Here we show that an interdomain interaction of the AR that is central to its function within the nucleus is required for AR aggregation and toxicity. Ligands that prevent the interaction between the amino-terminal FXXLF motif and carboxyl-terminal AF-2 domain (N/C interaction) prevented toxicity and AR aggregation in an SBMA cell model and rescued primary SBMA motor neurons from 5α-dihydrotestosterone-induced toxicity. Moreover, genetic mutation of the FXXLF motif prevented AR aggregation and 5α-dihydrotestosterone toxicity. Finally, selective androgen receptor modulators, which prevent the N/C interaction, ameliorated AR aggregation and toxicity while maintaining AR function, highlighting a novel therapeutic strategy to prevent the SBMA phenotype while retaining AR transcriptional function.
Keywords: Neurodegeneration, Polyglutamine Disease, Protein Folding, Protein Motifs, Protein Phosphorylation, Androgen Receptor, Motor Neuron Disease
Introduction
Spinal and bulbar muscular atrophy (SBMA)2 is an X-linked, slowly progressive, adult onset, neurodegenerative disease that primarily affects males. Symptoms include proximal limb weakness and muscle atrophy, fasciculations, tremor, dysphagia, and dysarthria (1). SBMA results from expansion of a polymorphic trinucleotide (CAG) repeat that encodes a polyglutamine (polyQ) tract near the amino terminus of the androgen receptor (AR) (2). Although many patients exhibit signs of androgen insensitivity, including gynecomastia, testicular atrophy, and infertility, loss of AR function is not the primary mechanistic basis for disease in SBMA (3). Rather, the presence of an expanded polyglutamine tract within the AR leads to its misfolding, aberrant cleavage, and accumulation (4, 5), although how these phenomena lead to neuronal dysfunction and death is unclear.
SBMA is a member of a family of nine neurodegenerative diseases caused by an expanded polyglutamine tract; these include spinocerebellar ataxia types 1, 2, 3, 6, 7, and 17, dentatorubral pallidoluysian atrophy, and Huntington disease (reviewed in Ref. 6). In addition to neuronal loss, a common pathogenic feature of this family of neurodegenerative diseases is the accumulation of misfolded and aggregated mutant protein in the affected neuronal populations. SBMA is characterized by the specific loss of lower motor neurons from the brainstem and anterior horn of the spinal cord (7). This neuronal loss, the formation of nuclear inclusions, and the subsequent symptoms of the disease depend on the binding of the polyglutamine-expanded AR to its ligands testosterone or 5α-dihydrotestosterone (DHT) (8–11).
The AR is a transcription factor and a member of the nuclear receptor superfamily that is regulated by hormone binding. Unliganded AR is found in the cytoplasm in an aporeceptor complex that contains heat shock proteins (Hsc70, Hsp40, and Hsp90), p23, and immunophilins (12). Androgens such as testosterone and DHT bind with high affinity to the AR, causing its release from the aporeceptor complex. Upon release, the AR undergoes several conformational changes; one such change is the interdomain interaction between the 23FQNLF27 motif near the amino terminus and the AF2 domain near the ligand-bound carboxyl terminus (13–18). Of note, this 23FQNLF28 motif lies in close proximity to the polyglutamine tract, which begins at amino acid 58. The amino/carboxyl (N/C) interaction stabilizes the AR, enhances hormone binding (14, 19), and provides a binding face for coactivator recruitment (19).
The cellular pathways and molecular mechanisms that give rise to neuronal dysfunction and cell death in polyQ diseases remain poorly understood. Polyglutamine length has been shown to determine the rate of inclusion formation (20) and, in the case of AR, modulates the folding and structure of the amino-terminal domain (21). Generally, there appears to be a pathological threshold of 36–40 glutamines that favors a conformational change from a random coil to a β-structure (22). Studies indicate that small monomeric or oligomeric species, consisting largely of β-sheet structures, are cytotoxic to neurons and lead to neuronal dysfunction (23). It is unclear whether a single misfolded polyglutamine monomer can provide a “seed” to induce aggregation through its ability to self-associate or whether sequences outside of the glutamine tract are required to promote this interaction. An expanded polyQ ataxin-1 deletion of a self-association domain outside of the polyglutamine tract inhibited aggregation but did not ameliorate disease (24). Moreover, recent studies reveal a role for the amino-terminal 17 amino acids of the Huntingtin protein in its aggregation (25, 26). Finally, although α-synuclein does not have an expanded polyglutamine tract, it self-associates through an intramolecular interaction (27). This interaction stabilizes the protein conformation of α-synuclein by masking one of its own domains, the highly amyloidogenic NAC (non-Aβ component of Alzheimer disease amyloid) domain, from promoting aggregation (27, 28).
We sought to determine whether a similar interaction of expanded polyQ AR plays a role in toxicity and aggregation. We utilized various ligands of AR that inhibit the N/C interaction induced by testosterone or DHT, as well as genetic mutations of the 23FQNLF27 motif that inhibit the N/C interaction, to determine whether the N/C interaction was required for the altered metabolism and toxic effects of the expanded polyQ AR. Our results demonstrate that both pharmacologic and genetic disruption of the N/C interaction of expanded polyQ AR prevent inclusion formation and toxicity of mutant AR in models of SBMA. Moreover, we show that phosphorylation of the AR at both Ser-81 and Ser-308 is dependent upon this interdomain interaction, demonstrating the utility of using this phosphorylation status to mark this disease-associated structural feature.
EXPERIMENTAL PROCEDURES
Construction of Mutant AR Cell Lines
FXXLF mutant AR-expressing PC12 cells were constructed by excision of pCMVhAR-F23A and pCMVhAR-L26A/F27A plasmids with NheI/NarI and ligation of each mutated fragment into both pTREAR10Q and pTREAR112Q plasmids. Mutation of Ser-81 and Ser-308 to alanine was performed by sequential site-directed mutagenesis (QuikChange II, Stratagene) of pTREAR112Q. All mutations and CAG lengths were sequence-verified. Transfection of tet-On PC12 cells (Clontech, Mountainview, CA) was performed with mutant pTREAR plasmids and the hygromycin-resistant plasmid pTK-Hygro using calcium phosphate. Transfected cells were selected by the addition of 200 μg/ml hygromycin (Invitrogen). Single colonies were isolated, expanded, and screened for AR protein expression by inducing AR with 1 μg/ml doxycycline (Clontech); positive clones were verified via slot-blot and immunoblotting with the antibody ARN20 (Santa Cruz Biotechnology, Santa Cruz, CA). Genomic DNA was isolated from positive clones, and both the mutation(s) and CAG repeat length were verified by PCR and sequencing. The amount of doxycycline required to induce a level of AR protein expression comparable with that in the pTREAR112Q cell line was determined.
Cell Culture and Reagents
Stable PC12 cells inducibly expressing AR were maintained as described (11). All experiments were performed in media that contained charcoal-stripped serum (stripping protocol from Sigma). Cells containing AR10Q and AR112Q were induced with 10 or 500 ng/ml doxycycline (Clontech). All other cell lines were induced to express equivalent amounts of AR to AR10Q and AR112Q. DHT (Sigma) and bicalutamide (Toronto Research Chemicals, North York, Ontario, Canada) were resuspended in EtOH.
Primary Motor Neuron Cultures
Dissociated spinal cord cultures were established according to Roy et al. (29). Spinal cords were held on ice during rapid PCR genotyping, and spinal cords of nontransgenic or AR112Q were then pooled, plated on poly-d-lysine-coated glass coverslips in 24-well plates, and cultured for 3 weeks in glia-conditioned media containing minimal essential medium, 3% charcoal-stripped horse serum, 35 mm NaHCO3, 0.5% dextrose, 1% N3, and 10 nm 2.5S nerve growth factor. After 3 weeks, cultures were treated with DHT or other AR ligands as indicated. After 7 days, cells were fixed and immunostained for neurofilament heavy chain with SMI32 as described below, and 10 random fields of motor neurons (determined by size and morphology) were counted. A two-way ANOVA/post-hoc Tukey test was performed using SigmaStat software (SPSS, Inc., San Jose, CA).
Biochemical Analysis
Cells were harvested on ice with cold PBS, and cell pellets were lysed with immunoprecipitation assay buffer or Triton-DOC buffer (containing 100 μg/ml PMSF, 0.2 mm sodium orthovanadate, 2 nm okadaic acid (Calbiochem), and phosphatase substrate (Sigma)) and sonicated. Western analysis was performed using a standard protocol for antibodies ARN20, AR441, ARH280, GAPDH (Santa Cruz Biotechnology), and NCL-AR-318 (NovoCastra Laboratories Ltd., Newcastle Upon Tyne, United Kingdom) or ARα(p)Ser81 (Millipore, Billerica, MA) and ARα(p)Ser308 (Santa Cruz Biotechnology). Filter trap analysis was performed as described (30).
Immunofluoresence
PC12 cells and primary neuron cultures were fixed and stained as described previously (31). Antibodies included ARN20, ARH280, AR441 (1:100) (Santa Cruz Biotechnology), and SMI32 (1:1000) (Sternberger Monoclonals Inc., Baltimore). Cells were visualized with a Leica microscope (Leica Microsystems GmbH, Wetzler, Germany), and images were captured with a Leica camera and analyzed with iVision-MacTM software (BioVision Technologies, Exton, PA). In PC12 cells, the percentage of inclusions was determined by counting at least 1000 cells averaged from three independent wells; statistical analysis was performed with SigmaStat software using a one-way ANOVA/post-hoc Tukey test. Experiments were repeated at least three times.
PC12 Cell Toxicity Assay
The toxicity of PC12 cell lines (AR10Q, AR112Q, AR110Q-F23A, and AR110Q-L26A/F27A) was analyzed as described previously (31). Three independent experiments were performed in triplicate, and statistical significance was determined by SigmaStat using a one-way ANOVA/post-hoc Tukey test.
Protein Stability Assays
AR expression was induced with doxycycline in PC12 cell lines (AR112Q, AR110Q-F23A, and AR110Q-L26A/F27A) for 48 h. Cells were then washed three times with PBS to remove media containing doxycycline and treated with media containing 30 μg/ml cycloheximide and EtOH or 10 nm DHT. Cells were harvested at the given times after media replacement. AR levels were determined by Western analysis. Films were scanned, and densitometry was calculated using Kodak 1D image analysis software (Eastman Kodak Co.).
Mammalian Two-hybrid Assay
HEK293 cells were seeded at 1.6 × 105 in 12-well plates in antibiotic-free DMEM supplemented with 10% fetal bovine serum and 2 mm l-glutamine. Cells were transfected using Lipofectamine 2000 (Invitrogen) in Opti-MEM with fusion constructs consisting of VP16-AR111Q (amino acids 12–660) and GAL4DBD-AR (amino acids 624–919). The interaction between the two encoded proteins activates the co-transfected GAL4-driven luciferase reporter, and expression was monitored in a dual-luciferase system (Promega, Madison, WI). A co-transfected construct encoding Renilla luciferase was used to normalize transfection efficiency. The cells were washed 12 h after transfection, and the medium was replaced with charcoal serum-stripped DMEM containing EtOH, DHT, or bicalutamide (performed in triplicate). After a 48-h incubation, cells were lysed and luciferase activity was measured on a luminometer according to the manufacturer's instructions. The ratios of firefly luciferase to Renilla luciferase activity from quadruplicate samples were averaged and normalized to DHT-treatment, which was set to 100. Significance was assessed with SigmaStat using a one-way ANOVA/post-hoc Tukey test.
RESULTS
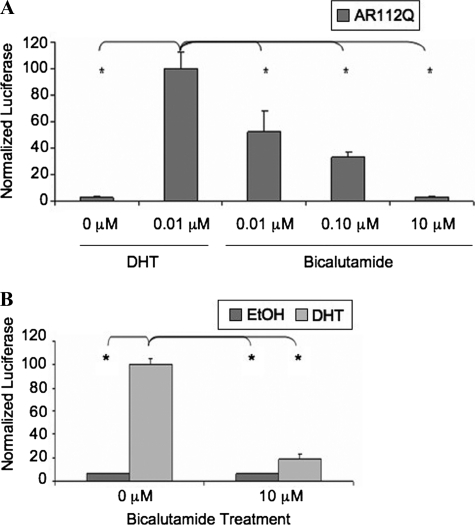
Bicalutamide Alters Nuclear Inclusion Formation and Prevents DHT-dependent Toxicity in PC12 Cells
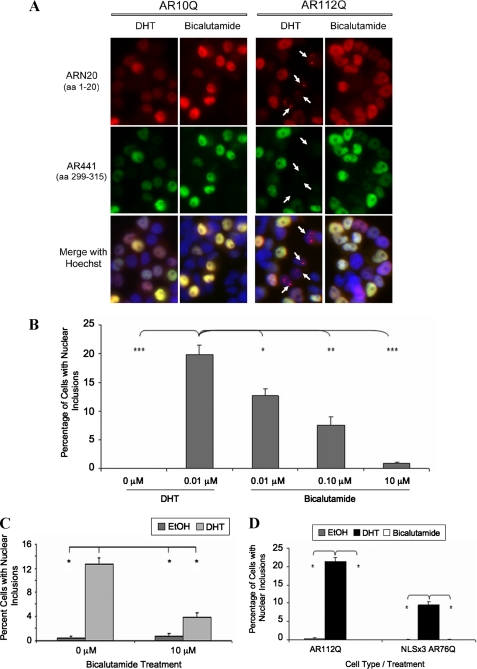
Using rat adrenal gland-derived PC12 cells, we previously created an inducible cell model of SBMA that, upon addition of doxycycline, expresses full-length human AR with an expanded polyglutamine tract (112Q) and, in response to DHT, forms nuclear inclusions of AR (11). This cell model recapitulates the histopathology of SBMA motor neurons, in which intranuclear inclusions consist primarily of proteolyzed amino-terminal fragments of AR3 (5, 32) (Fig. 1A). Using this SBMA cell model, we wanted to assess the effects that other AR ligands might have on the altered metabolism of expanded polyQ AR. We treated AR10Q- or AR112Q-expressing cells with DHT or increasing amounts of bicalutamide for 4 days. Immunofluorescence analysis revealed that DHT caused the formation of nuclear inclusions containing only amino-terminal AR epitopes (ARN20-positive, AR441-negative) in AR112Q but not AR10Q-expressing cells, as expected (Fig. 1A). High concentrations (10 μm) of the nonsteroidal compound bicalutamide, a transcriptional antagonist of the AR, failed to induce the formation of nuclear inclusions in AR112Q-expressing cells, despite nuclear AR localization (Fig. 1A), whereas comparably high concentrations of DHT induced substantial aggregation (Fig. 6B and data not shown) similar to that induced by low concentrations of DHT. Lower concentrations (0.01 and 0.1 μm) of bicalutamide induced inclusions at low frequency (Fig. 1B). In addition, bicalutamide reduced AR inclusions even in the presence of DHT (Fig. 1C). Biochemical analysis also revealed a reduction of high molecular weight species when cells were exposed to bicalutamide, either alone or in the presence of DHT (Fig. 2B), supporting an effect of bicalutamide on aggregation.
FIGURE 1.
Bicalutamide prevents nuclear inclusions in a cell model of SBMA. Stably transfected PC12 cells were induced with 10 ng/ml doxycycline to express AR10Q or AR112Q for 96 h. Cells were treated with EtOH, 0.01 μm DHT, or 0.01, 0.1, or 10 μm bicalutamide. A, cells were fixed and immunostained with an antibody to the amino terminus of AR (AR-N20), an antibody (AR-441) detecting a more internal epitope of AR (amino acids 299–315), and Hoechst to reveal nuclei. Panels depict cells treated with 0.01 μm DHT or 10 μm bicalutamide. B, 1000 cells were counted for each treatment, in triplicate. The number of cells with ARN20-staining inclusions was averaged, expressed as a percentage of total cells, and graphed. One-way ANOVA was performed (***, p < 0.001; **, p < 0.01; *, p < 0.05). C, AR112Q cells were treated alone or in combination with EtOH, 0.01 μm DHT, and 10 μm bicalutamide for 96 h. Inclusions were counted as in B. One-way ANOVA was performed: *, p < 0.001. These findings are representative of three independent experiments. D, bicalutamide does not induce inclusions of a constitutively nuclear expanded polyQ-AR. AR112Q or NLSx3-AR76Q-expressing cells were treated for 48 h with EtOH, 0.01 mm DHT, or 10 mm bicalutamide. Over 1000 cells were counted/well, and the percentage of cells containing inclusions was averaged and graphed. One-way ANOVA was performed (*, p < 0.001).
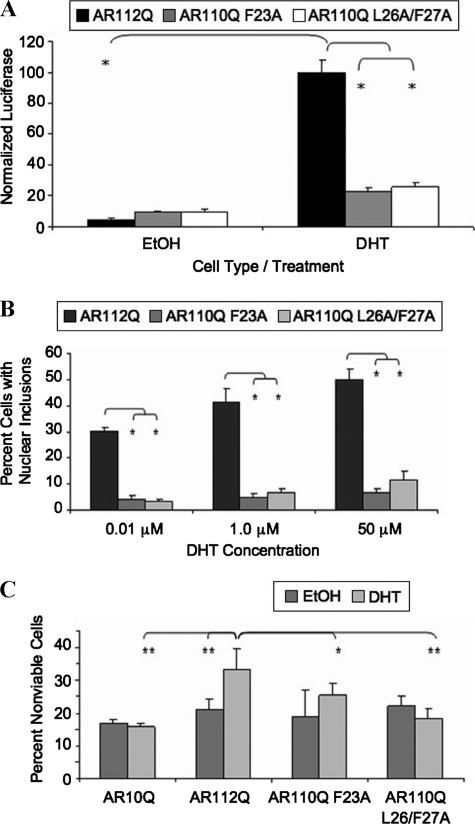
FIGURE 6.
Mutation of the FXXLF motif of AR polyQ-expanded AR prevents the N/C interaction, nuclear inclusions, and toxicity in an SBMA cell model. A, mammalian two-hybrid assay was performed as described in the legend for Fig. 5, including the AR111Q-F23A or ARQ111-L26A/F27A mutant fused to a VP16 activation domain. Student's t test was performed (*, p < 0.01). B, stably transfected PC12 cells were induced to express equivalent levels of AR10Q, AR112Q, AR110Q-F23A, or AR110Q-L26A/F27A. Cells were treated with increasing amounts of DHT (0.01, 1.0, and 50 μm) for 48 h, fixed, and immunostained as in Fig. 1A. Over 1000 cells were counted, and the percentage of cells containing nuclear inclusions graphed. Student's t test was performed (*, p < 0.02). C, toxicity was assessed as in Fig. 2A. Two-way ANOVA was performed (**, p < 0.001;*, p < 0.01). The findings are representative of three independent experiments.
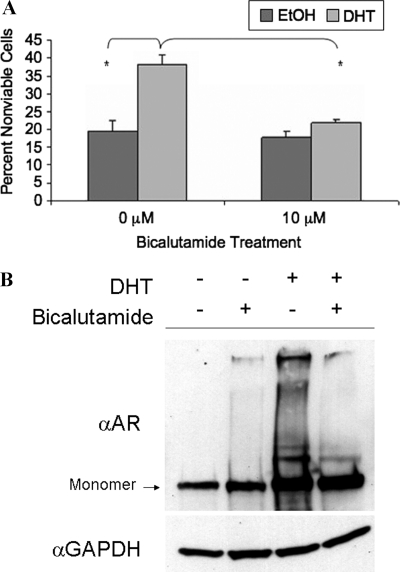
FIGURE 2.
Bicalutamide prevents DHT-induced toxicity in an SBMA cell model. A, stably transfected PC12 cells were induced with 500 ng/ml doxycycline to express AR112Q for 12 days. Cells were treated with EtOH, 0.01 μm DHT, 10 μm bicalutamide, or bicalutamide in the presence of DHT. Cells were harvested and stained with trypan blue, and at least 300 cells were counted in triplicate. Two-way ANOVA was performed (*, p < 0.001). B, Western analysis was performed on protein lysates of AR112Q induced with 500 ng/ml doxycycline and treated with EtOH or 0.01 μm DHT in the presence or absence of 10 μm bicalutamide for 48 h. Comparable results were obtained from cells treated for 12 days (data not shown).
DHT treatment of AR112Q-expressing cells not only caused the formation of nuclear inclusions (Fig. 1A) but also resulted in significant and substantial cell death (Fig. 2A). We then determined whether bicalutamide would cause toxicity and whether it could prevent the DHT-induced toxicity caused by expanded polyQ-AR. Bicalutamide failed, when used alone, to cause toxicity in these cells (Fig. 2A). Moreover, combined treatment with bicalutamide and DHT resulted in substantial and significant protection from DHT-induced toxicity. These data indicate that bicalutamide is capable of rescuing AR112Q-expressing PC12 cells from the toxic effects of DHT.
We next evaluated AR protein levels in cells treated with bicalutamide. Biochemical analysis revealed that although bicalutamide alone did not stabilize the AR, it did not alter AR stabilization by DHT (Fig. 2B), indicating that the mechanism by which bicalutamide prevented DHT-induced toxicity was not through loss of AR protein stability. Moreover, the lack of stabilization of AR112Q was observed only with the highest concentration of bicalutamide when used alone; low levels of bicalutamide, which partially stabilized AR112Q (data not shown), also decreased inclusions (Fig. 1B) and toxicity (data not shown). Thus, these data support the conclusion that bicalutamide impacts AR aggregation and toxicity independently of effects on AR stability.
Although immunostaining of PC12 cells expressing AR in the presence of bicalutamide indicated that this ligand induced nuclear localization of expanded polyQ AR, we sought to rule out the possibility that bicalutamide reduced nuclear inclusions by delaying the transit of AR into the nucleus. To this end, we utilized PC12 cells expressing expanded AR (76Q) with an exogenous nuclear localization signal (NLSX3) fused to the amino terminus; in these cells, both unliganded and liganded AR is confined to the nuclear compartment (31). Treatment of NLSX3-AR76Q-expressing cells with bicalutamide resulted in a substantial reduction of the number of cells with nuclear inclusions compared to treatment with DHT (Fig. 1D), indicating that bicalutamide inhibits mutant AR aggregation and toxicity independently of any potential effect on nuclear translocation.
Bicalutamide Prevents DHT-dependent Toxicity in Primary SBMA Motor Neurons
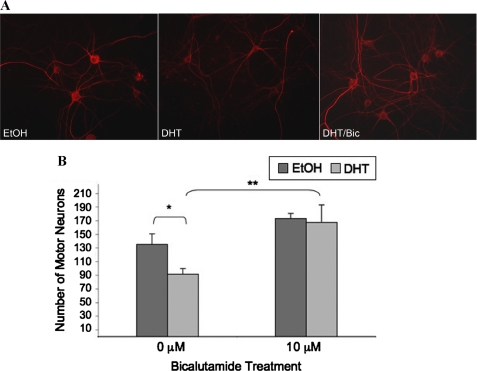
We next investigated the effect of bicalutamide on SBMA motor neuron viability. Motor neuron toxicity assays were performed on dissociated spinal cord cultures derived from transgenic SBMA mouse embryos (Fig. 3A). This transgenic mouse model of SBMA expresses full-length AR with a polyglutamine tract 112 amino acids in length and recapitulates the gender-specific motor deficits and intranuclear inclusions of SBMA (8). Motor neuron cultures derived from these mice reproduce the hormone-dependent AR aggregation and toxicity observed in SBMA (31, 33). Treatment of 3-week-old differentiated spinal cord cultures with DHT resulted in the loss of ∼35% of the motor neurons (Fig. 3B) as observed previously (31, 33). In contrast, bicalutamide not only failed to induce motor neuron death but also prevented DHT-induced toxicity (Fig. 3B). Moreover, qualitative comparisons of neurofilament heavy chain staining revealed that treatment with DHT, but not bicalutamide, resulted in reduced unphosphorylated NF-H immunostaining (Fig. 3A), as seen in vivo (8). In addition, DHT-treated axons appeared thinner and shorter, suggesting reduced neuronal health. Bicalutamide appeared to produce a qualitative improvement in these features (Fig. 3A, right panel).
FIGURE 3.
Bicalutamide prevents DHT-induced toxicity in primary motor neuron cultures of SBMA mice. After 3 weeks of differentiation, dissociated spinal cord cultures were treated for 7 days with EtOH or 10 μm DHT in the presence or absence of 10 μm bicalutamide (Bic). A, cells were fixed and immunostained with the antibody SMI-32 (unphosphorylated neurofilament heavy chain) and then stained with Hoechst. B, counts were obtained from 10 random fields at ×20 under a Leica microscope from three separate wells of each treatment of motor neurons and then graphed. Two-way ANOVA was performed with post-hoc Tukey test (*, p < 0.001).
Bicalutamide Alters Phosphorylation of Expanded PolyQ AR
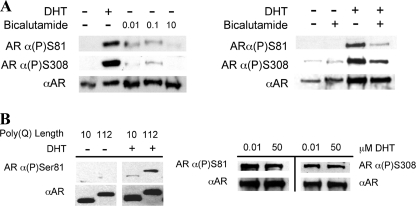
Having established that bicalutamide prevented both AR aggregation and toxicity in our PC12 cell model of SBMA as well as in transgenic SBMA motor neurons, we next explored the possible mechanism for these effects. It is known that DHT binding to the AR induces a conformational change that results in the phosphorylation of numerous residues within the AR (34–37). It has been shown previously that bicalutamide prevents phosphorylation of normal polyQ AR at Ser-81 (38). We therefore examined whether bicalutamide would alter the phosphorylation of Ser-81 on expanded polyQ AR. Western analysis of AR112Q-expressing PC12 cells revealed that bicalutamide failed to induce, to the same level as DHT, the phosphorylation of both Ser-81 and Ser-308 (Fig. 4A, left), and it reduced the phosphorylation of both residues even in the presence of DHT (Fig. 4A, right). Moreover, we observed that phosphorylation of Ser-81 was enhanced by polyglutamine expansion (Fig. 4B). We therefore reasoned that phosphorylation of one or both of these residues might be linked to the ability of the mutant AR to form inclusions. To test this hypothesis, we mutated Ser-81 to alanine to prevent its phosphorylation, either alone (S81A) or in combination with a mutation of Ser-308 (S81A/S308A). The phosphorylation-null mutant neither of Ser-81 nor of Ser-81/Ser-308 had any effect on the frequency of nuclear inclusion formation (data not shown), demonstrating that alteration of phosphorylation at these sites is not the primary mechanism by which bicalutamide prevents inclusions.
FIGURE 4.
Bicalutamide and polyQ length alter phosphorylation of AR. A, left, Western analysis was performed on protein lysates obtained from AR112Q-expressing cells treated in the presence or absence of 0.01 μm DHT or 0.01, 0.1, or 10 μm bicalutamide. Protein lysates were loaded to achieve equivalent AR levels. Right, bicalutamide reduced phosphorylation of Ser-81 and Ser-308 even in the presence of DHT. B, left, Western analysis was performed on protein lysates obtained from AR10Q- and AR112Q-expressing cells treated with EtOH or 50 μm DHT. The panels of AR10Q and AR112Q came from the same Western blot. Right, the Ser-81 phosphorylation state of the AR was unaffected by DHT concentration. Western analysis was performed on protein lysates obtained from AR112Q-expressing cells treated in the presence of either 0.01 or 50 μm DHT for 96 h. The membranes were probed with antibodies for phosphorylated Ser-81 (AR α(P)Ser81) or phosphorylated Ser-308 (ARα(P)Ser308) and pan-AR (α-AR) (AR-H280 (A) or AR-N20 (B)).
Bicalutamide Disrupts the N/C Interaction of Polyglutamine-expanded AR
Bicalutamide has been shown to prevent the interdomain interaction between the amino and carboxyl AR termini (39, 40) that occurs upon agonistic androgen binding (13). To confirm that this is also the case for an expanded polyQ-AR, we monitored the interaction of the amino and carboxyl termini of the polyglutamine-expanded AR (111Q) using a mammalian two-hybrid assay. Cells were transfected with a construct encoding the VP16 activation domain fused to the amino terminus of AR (amino acids 12–660) and a construct encoding the GAL4 DNA-binding domain fused to the carboxyl terminus of AR (amino acids 624–919). The interaction of these fusion proteins was monitored by their ability to activate a luciferase reporter under the control of Gal4 regulatory sequences. Co-transfection with a Renilla luciferase plasmid was used to control for transfection efficiency. As expected, in contrast to the substantial N/C interaction of expanded polyQ AR upon DHT binding, there was minimal N/C interaction in both the absence of ligand and the presence of bicalutamide (Fig. 5A). An examination of the effect of bicalutamide on expanded polyQ AR N/C interaction in the mammalian two-hybrid system revealed that low levels of bicalutamide induced a significant N/C interaction (albeit still lower than that caused by DHT binding) (Fig. 5A). The extent of N/C interaction induced by bicalutamide was correlated with both the frequency of inclusions (Fig. 1B) and the level of AR phosphorylation at serines 81 and 308 (Fig. 4A), consistent with a role for the N/C interaction in these events. We next examined the effect of bicalutamide in the presence of DHT on the N/C interaction of AR. Bicalutamide substantially prevented the ability of AR to form an N/C interaction in the presence of DHT (Fig. 5B). This correlated with the reduction of inclusions (Fig. 1C) and the prevention of DHT-induced toxicity (Fig. 2A).
FIGURE 5.
Bicalutamide prevents the N/C interaction of polyQ-expanded AR. Mammalian two-hybrid assay was performed by transfecting HEK293 cells with the N-terminal AR111Q (amino acids 12–660) fused to a VP16 activation domain, the C-terminal AR (amino acids 624–919) fused to the Gal4 DNA-binding domain, the Gal4-luciferase reporter, and the Renilla-luciferase transfection control plasmid. Dual-luciferase activity was determined from cell lysates. A, cells were treated in triplicate with EtOH, 0.01 μm DHT, or increasing amounts of bicalutamide (0.01, 0.1, or 10 μm) for 48 h. The values obtained from the ratios of Gal4-Luc/Renilla-Luc were normalized to the value of DHT-treated AR111Q set to 100. One-way ANOVA was performed (*, p < 0.001). The findings are representative of three independent experiments. B, cells were treated with EtOH or 0.01 μm DHT alone or in combination with 10 μm bicalutamide for 48 h. The values obtained from the ratios of Gal4-Luc/Renilla-Luc were normalized to the value of DHT-treated AR111Q set to 100. One-way ANOVA was performed (*, p < 0.001). The findings are representative of three independent experiments.
Genetic Disruption of the N/C Interaction of AR Prevents Nuclear Inclusions and Toxicity
To directly test the hypothesis that the N/C interaction plays a role in the altered metabolism of expanded polyQ AR, we utilized two separate mutations of the 23FQNLF27 motif at the amino terminus of the AR, previously shown to disrupt the interdomain interaction (14). We confirmed the effect of FXXLF mutation on the N/C interaction of expanded polyQ AR by utilizing the mammalian two-hybrid assay. Two distinct mutations of the 23FQNLF27 motif (F23A and L26A/F27A) disrupted the N/C interaction of an expanded polyQ AR in the presence of DHT (Fig. 6A). To test the effect of these mutations on expanded AR aggregation and toxicity, we made stable inducible PC12 cell lines containing either the single (F23A) or the double (L26A/F27A) FXXLF mutation in expanded polyQ AR. Cells expressing either of the FXXLF mutant AR proteins showed nuclear localization of the mutant AR in response to DHT (data not shown). Immunofluorescence analysis of these cell lines following DHT treatment showed a substantial reduction in nuclear inclusions (Fig. 6B), despite comparable AR protein levels (data not shown). Earlier studies indicated that the N/C interaction contributes to the stabilization of ligand binding (13). To rule out the possibility that the effect of FXXLF mutation on inclusion frequency was secondary to changes in ligand binding, we tested the effects of the FXXLF mutations using higher DHT concentrations. Regardless of DHT concentration (up to 50 μm), mutation of the FXXLF motif substantially abrogated the formation of AR nuclear inclusions (Fig. 6B).
We next sought to determine whether the FXXLF mutations could prevent DHT-dependent toxicity of expanded polyQ AR. As expected, we observed DHT-dependent toxicity of expanded AR compared with a normal polyQ AR (Fig. 6C). However, DHT failed to induce toxicity of both F23A- and L26A/F27A-expressing cells (Fig. 6C), indicating that an intact FXXLF sequence is required for DHT-dependent, polyQ-dependent toxicity.
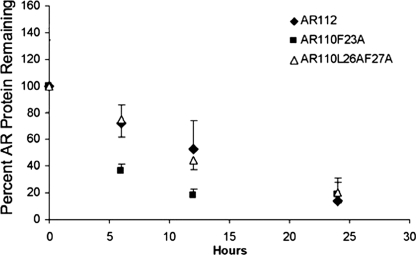
It was previously shown that the FXXLF motif may contribute to the regulation of AR degradation (41, 42). Although we induced AR expression in the mutant cell lines to produce levels of FXXLF mutant AR equivalent to that of AR112Q (wild type for the FXXLF motif), we questioned whether the mutation altered the stabilization of the AR by hormone. Treatment with epoxomicin to inhibit the proteasome revealed the accumulation of both AR mutant proteins, indicating that these proteins continued to be degraded by the proteasome (data not shown). We also carried out half-life analysis of AR protein, quantifying AR protein at time points following washout of doxycycline and in the presence of cycloheximide. This analysis indicated that, although the mutation of Leu-26/Phe-27 decreased the turnover of both unliganded and liganded expanded AR, mutation of Phe-23 had no effect on unliganded AR turnover and a modest, destabilizing effect on liganded AR (Fig. 7). Given the similar effects of these mutations on AR aggregation and toxicity, it is unlikely that turnover, as regulated by the FXXLF motif of the mutant AR, plays a major role in the cytoprotective effects of these mutations. Taken together, these results suggest that FXXLF mutation abrogates expanded polyQ AR aggregation and toxicity largely because of disruption of the interdomain N/C interaction.
FIGURE 7.
Effect of FXXLF mutation on AR protein half-life. Stably transfected PC12 cells were induced to express equivalent levels of AR112Q, AR110Q-F23A, or AR110Q-L26A/F27A for 48 h. After 48 h of doxycycline induction and EtOH or 0.01 μm DHT treatment, the cells were washed with PBS to remove doxycycline, and 30 μg/ml cycloheximide with EtOH or DHT was added. Cells were harvested at various time points, and Western analysis was performed on protein lysates. Blots were probed with pan-AR (AR-H280) and GAPDH antibodies. Densitometry was performed on the film, and the ratio of AR/GAPDH was graphed for each cell line. The findings are representative of three independent experiments.
Selective AR Modulators (SARMs) That Prevent N/C Interaction Also Prevent AR Aggregation and Toxicity
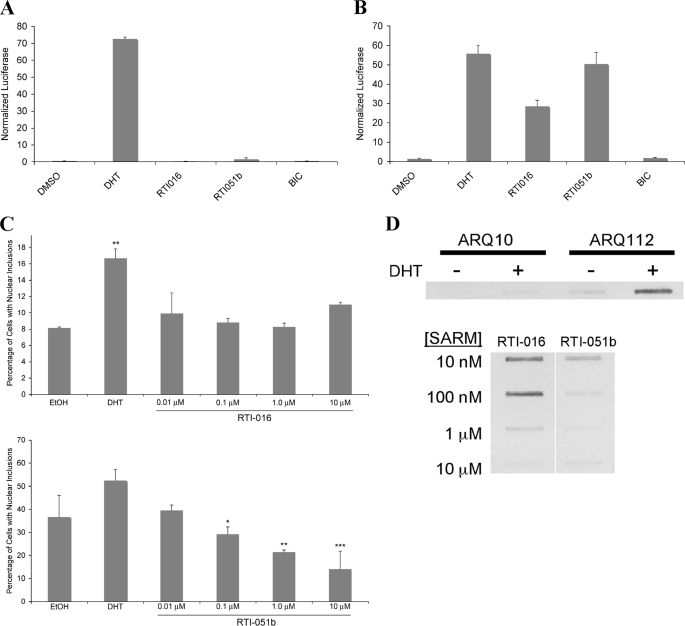
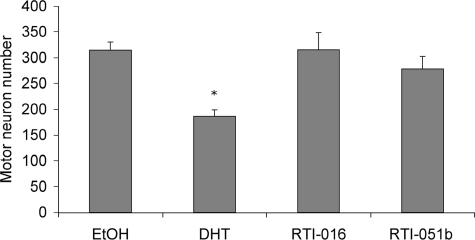
We next sought to determine whether additional AR ligands, selected to inhibit the N/C interaction, could prevent AR aggregation and toxicity. Two such ligands, derived from the chemical backbone of RU486 (40), failed to induce the AR N/C interaction (Fig. 8A). Despite the lack of N/C interaction, these ligands stimulated AR transcription on an MMTV promoter (Fig. 8B). Although the MMTV promoter shows little dependence on the N/C interaction for activity (43, 44), SARMs of the RTI class have been shown to exhibit a range of partial to complete agonist activities on a non-MMTV reporter (40) and on endogenous gene promoters (44). Both RTI-016 and RTI-051b promoted the nuclear translocation of AR, as expected (data not shown). However, both SARMs failed to induce AR inclusions to the extent seen in the presence of DHT (Fig. 8C). Moreover, filter trap analysis revealed that, although DHT induced substantial filter-trapped aggregated AR, the two alternative ligands shown here failed to induce AR aggregation to a similar extent (Fig. 8D). Finally, we tested the ability of these two selected SARMs to induce AR112Q toxicity in SBMA motor neurons. Unlike DHT, both RTI-016 and RTI-051b failed to induce toxicity (Fig. 9). Thus, not only does the AR antagonist bicalutamide fail to induce mutant AR aggregation and toxicity, but two structurally unrelated AR ligands also fail to induce AR aggregation and toxicity, further implicating the importance of the N/C interaction in both aspects of SBMA pathology. Moreover, the identification of AR ligands that prevent toxicity even while functioning as partial or complete transcriptional agonists provides a novel approach to the development of therapeutic compounds for SBMA.
FIGURE 8.
SARMs that do not reduce the AR N/C interaction but promote AR transcriptional activity fail to induce aggregation of expanded polyQ AR in an SBMA cell model. A and B, HepG2 cells were transfected with plasmids for mammalian two-hybrid analysis (A) and transcriptional activity (B). For AR mammalian two-hybrid assays, the DNA transfected into the cells consisted of pcDNA-AR1-660, VP16-AR507-919, MMTV-Luc, and pCMV-βGal. For AR transcriptional assay, the DNA consisted of pSG5-AR, MMTV-Luc, and pCMV-βGal. Cells were treated with hormone for 48 hr, lysed, and firefly luciferase (reporter) and β-galactosidase (transfection normalization) assays performed. C and D, stably- transfected PC12 cells were induced with 1 μg/ml of doxycycline to express AR112Q for 48 hr. Cells were treated with EtOH, DHT (50 μm) or various concentrations of RTI-016 and RTI-051b. C, cells were fixed and immunostained with an antibody to the N terminus of AR (AR-H280), an antibody (AR-441) detecting a more internal epitope of AR (amino acids 299-315), and Hoechst. One thousand cells were counted for each treatment in triplicate. The number of ARN20-staining inclusions was averaged, expressed as a percentage of total cells, and graphed. Student's t test was performed. * = p < 0.05; ** = p < 0.01; *** = p < .005. D, insoluble androgen receptor protein was detected using filter trap analysis. Protein lysates of treated cells were pulled by vaccum through cellulose acetate. The membrane was probed with an antibody for the N terminus of AR (AR-H280).
FIGURE 9.
SARMs fail to induce toxicity in primary motor neurons of SBMA mice. After 3 weeks of differentiation, dissociated spinal cord cultures were treated for 7 days with EtOH, DHT, RTI-016, or RTI-051b. Ligands were used at 10 μm concentration. Counts were obtained from analysis using 200× magnification of 10 random fields from three separate wells of each treatment of motor neurons. One-way ANOVA/post-hoc Tukey test was performed (*, p < 0.001, compared with all other conditions).
Phosphorylation Is a Marker for the N/C Interaction of AR
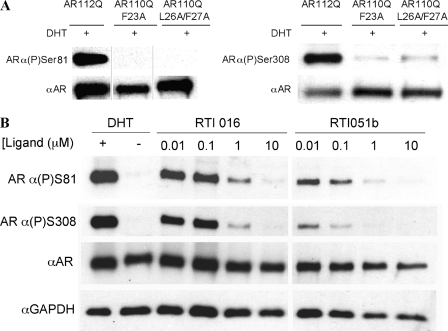
We showed that bicalutamide failed to induce substantial phosphorylation of both Ser-81 and Ser-308 (Fig. 4A). To determine whether this result was specific to bicalutamide-bound AR or resulted from the disruption of the N/C interaction of AR, we performed Western analysis of phosphorylation of the genetic mutants AR110Q-F23A and AR110Q-L26A/F27A. We found that phosphorylation of both Ser-81 and Ser-308 was absent when the N/C interaction was genetically disrupted (Fig. 10A), demonstrating that an intact FQNLF motif is required for phosphorylation at these residues. Finally, we assayed phosphorylation of these sites upon treatment of AR112Q cells expressing the expanded polyQ AR with SARMs known to disrupt the N/C interaction. Consistent with our previous results, we found that treatment with SARMs that prevent the N/C interaction disrupted the phosphorylation of serines 81 and 308 (Fig. 10B). These data indicate that the interdomain N/C interaction is required for phosphorylation of serines 81 and 308 and that these post-translational modifications can be used to report on the N/C interaction.
FIGURE 10.
Disruption of the AR N/C interaction prevents phosphorylation of serine 81 and serine 308. A, Western analysis was performed on cell lysates from stably transfected PC12 cells induced to express equivalent levels of AR112Q, AR110Q-F23A, and AR110Q-L26A/F27A and treated with EtOH or 0.01 μm DHT for 48 h. Membranes were probed with antibodies against phosphorylated Ser-81 (ARα(P)Ser81) (left) or phosphorylated Ser-308 (ARα(P)Ser308) (right). Note that the lanes on each Western blot are from the same blot; those shown on the left are noncontiguous. Membranes were reprobed for pan-AR (AR-N20 or AR-H280) and GAPDH (not shown). B, AR112Q cells were treated with EtOH, 0.01 μm DHT, 0.01, 0.1, 1, or 10 μm RTI-016, or RTI-051b for 48 h. Cell lysates were probed with antibodies detecting AR(P)Ser81, AR(P)Ser308, pan-AR (AR-H280), and GAPDH.
DISCUSSION
The roles of both hormone binding and nuclear localization of the mutant, polyglutamine-expanded AR have been established as essential components of the pathogenic process in SBMA (8–11, 31). However, additional details of the pathogenic metabolism of the AR that leads to its toxicity remain obscure. Our studies presented here focused on this question, in an effort to identify upstream steps in the pathogenic process that can be targeted for therapeutic benefit. To this end, we have identified a step in the normal metabolism of AR, a conformational change involving the N/C interaction, as a critical step in mutant AR aggregation and toxicity.
In our analysis of the effect of AR antagonistic ligands, we found that the nonsteroidal AR antagonist bicalutamide reduced mutant AR nuclear inclusion formation and DHT-dependent toxicity in a PC12 cell model of SBMA. Moreover, bicalutamide treatment prevented DHT-induced toxicity in primary motor neurons derived from SBMA mice. Bicalutamide affects AR metabolism in multiple ways, including changes in AR phosphorylation, turnover, DNA binding, recruitment of transcriptional coregulators and the interaction between the amino and carboxyl termini of AR.
Bicalutamide was shown previously to prevent phosphorylation of the AR at Ser-81 (38). This observation, coupled with the ability of bicalutamide to ameliorate inclusions and toxicity of expanded polyQ AR, suggested that phosphorylation at this site might play a role in SBMA pathogenesis. However, our finding that the phosphorylation-null mutations of Ser-81 and of Ser-308 failed to impact expanded AR aggregation indicates not only that phosphorylation of Ser-81 and Ser-308 are not essential components of the pathogenic pathway in SBMA but also that prevention of phosphorylation of these sites is not the mechanism by which bicalutamide ameliorates the abnormal metabolism of expanded polyQ AR.
We also observed that a high dose of bicalutamide reduced AR protein levels, suggesting that reductions in AR protein were responsible for the decrease in both nuclear inclusions and AR toxicity. Moreover, a previous study had shown, in contrast to our results, that bicalutamide induced toxicity in photoreceptor neurons of a fly model of SBMA without reducing AR levels (45). It may be that the effective dose in fly photoreceptors was substantially lower than the dose causing increased AR turnover in our studies. However, we found an amelioration of toxicity and AR aggregation even at lower doses of bicalutamide. Thus, whether the different effects of bicalutamide in that study (45), compared with the studies reported here, reflect only a potential difference in dose or are also related to differences in AR metabolism between the invertebrate model and our mammalian models is currently unknown. Nonetheless, in our studies, bicalutamide also decreased AR aggregation (Fig. 1C) and toxicity (Fig. 2A) in the presence of DHT without altering AR protein levels (Fig. 2B), indicating that effects on AR turnover are not solely responsible for the bicalutamide rescue of DHT-dependent AR toxicity. Moreover, lower concentrations of bicalutamide, which similarly had no effect on AR protein levels, also decreased expanded AR toxicity in the presence of DHT (data not shown).
An earlier study (46) of the effect of androgen ablation on disease in mutant AR transgenic mice included an analysis of flutamide, a bicalutamide-related AR antagonist that also prevents the N/C interaction, albeit with lower AR affinity and potency. Flutamide failed to impact disease onset or progression in these mice and had no effect on AR aggregation (46). However, whether the dose used in that study allowed effective levels to be achieved in the nervous system is unknown.
The binding of androgens to the AR initiates a set of conformational changes within the AR that result in interdomain interactions as well as translocation of AR to the nucleus. One interdomain interaction that is key to both the stabilization of the AR and its activation to a fully functional transcription factor occurs between the FXXLF motif at the amino terminus and the ligand-bound AF-2 motif at the carboxyl terminus (14, 41). This interaction, in concert with androgen binding, stabilizes the androgen receptor and recruits coactivators that are important for the transcriptional activation function of the AR. The binding of the AR by the antagonist bicalutamide prevents this interaction. Moreover, the novel agonistic ligands (SARMs) used in this study also prevent the N/C interaction. Thus, our results, using both pharmacologic and genetic approaches, demonstrate that the interaction between the amino and carboxyl termini of expanded polyQ AR is a critical and early event essential for the abnormal metabolism and toxicity of the mutant AR.
Although our results showed that bicalutamide prevented DHT-dependent AR toxicity without impacting AR protein levels, we cannot rule out a role for protein degradation in the mechanism by which the N/C interaction contributes to disease. It is well known that DHT binding stabilizes the AR (47), unlike the effect of hormone binding by several other steroid hormone receptors. Earlier studies revealed that the N/C interaction contributes to this stabilization of the AR by hormone binding (41). These findings suggest that inhibition of the N/C interaction would lead to increased turnover of the mutant AR, likely explaining the effect of high-dose bicalutamide on decreasing AR steady-state protein levels. In addition, we observed that, although the F23A mutation did not alter the half-life of unliganded AR, it modestly decreased the half-life of DHT-bound AR (Fig. 7), supporting the idea that stabilization of the AR with the N/C interaction may contribute to its role in disease pathogenesis.
The mechanism by which the N/C interaction leads to AR stabilization is unknown. However, others have shown effects of FXXLF mutation on AR stabilization by hormone (41), and a more recent study suggests that the FXXLF motif may function as a degron (42). Thus, the degron function of the FXXLF motif may be masked by its binding to the carboxyl terminus within the N/C interaction. This finding raises questions about the effect of FXXLF mutation on AR degradation in our studies. However, although substantial increases in AR protein were observed upon “triple” mutation of the FXXLF motif (23AXXAA27) (42), more modest effects were found upon mutation of only one or two of the conserved amino acids within this sequence, albeit within the context of a truncated huntingtin protein. Moreover, these observations suggest that FXXLF mutation would reduce the efficiency of expanded (polyQ) AR degradation by the 26S proteasome, resulting in further accumulation of the mutant AR and enhancement of its toxicity and aggregation. Our studies of FXXLF mutant AR indicated a modest effect of the F23A mutation on DHT-bound AR turnover, but by decreasing (Fig. 7) rather than increasing its half-life, consistent with a negligible effect on degron function and a major effect on the N/C interaction. In contrast, the L26A/F27A mutation led to the modest stabilization of both unliganded and liganded AR (Fig. 7, data not shown). Nonetheless, the finding that 23AXXLF27 and 23FXXAA27 similarly inhibit AR aggregation and toxicity strongly suggests that the main effect of these mutations in modulating disease pathogenesis is through their effects on the N/C interaction.
Interdomain interactions have been shown to be an important determinant for regulating the neurotoxicity of another neurodegenerative disease-causing protein, α-synuclein. In the native state, this interaction masks the exposure of the central, hydrophobic NAC domain, thus preventing α-synuclein aggregation (27, 28). In contrast to findings in α-synuclein, we found that the N/C interaction of the AR contributes to, and substantially impacts, aggregation and toxicity. How this interaction contributes to toxicity is a matter of speculation. It may be that the amino-terminal region of the AR contains structural motifs that may contribute to its role in disease. The amino terminus of the AR, which includes the polyglutamine tract, has been shown to contain significant regions of intrinsically disordered structure (48), and polyglutamine expansion increases the α-helical content of the region (21). The FXXLF motif itself has been shown to form an amphipathic α-helix that binds to the AF-2 domain via a primary charge clamp (19). Our demonstration of a role for this interaction in expanded AR aggregation and toxicity implicates a distinct conformation of the AR amino-terminal region in its abnormal metabolism. Working together, an expanded polyQ repeat and the N/C interaction may promote a highly stable AR structure refractory to its normal metabolism and/or degradation and render it prone to aggregation.
Our data reveal that, in addition to having a major impact on AR aggregation and toxicity, the N/C interaction of expanded polyQ AR is required for the phosphorylation of the AR on Ser-81 and Ser-308. Both genetic mutation of the FXXLF motif and the use of AR ligands that prevent the N/C interaction confirmed the necessity of the N/C interaction for phosphorylation of these two sites. Moreover, we noted that low levels of bicalutamide produced a detectable N/C interaction, Ser-81/Ser-308 phosphorylation and expanded AR aggregation; all were decreased with increasing bicalutamide levels, consistent with a direct relationship between these events. The mechanism by which low bicalutamide levels lead to these events is under investigation. Thus, phosphorylation of Ser-81 and Ser-308 represents a useful tool in reporting on the N/C interaction. This finding also suggests that disruption of the N/C interaction affects downstream phosphorylation events that require the interaction with the AR of nuclear kinases or phosphatases.
Another distinct set of protein interactions with the AR includes p160 coactivators and chromatin remodeling complexes, which govern the ability of the AR to promote transcription. GRIP1, as well as F-src-1 and CBP (CREB-binding protein), have been shown to enhance the N/C interaction (15, 50), indicating that this conformation of the AR is not only an important first step in the metabolism of the AR but may itself be further stabilized by coactivator binding, as suggested previously (51). Moreover, the finding that the melanoma antigen gene protein, MAGE-11, plays a role in modulating both AR metabolism and coregulator binding in response to the N/C interaction (49, 52, 53) suggests another candidate protein interaction through which the N/C interaction might play a role in disease. Additional studies are needed to understand whether a role exists for MAGE-11 and/or other coregulators in the pathogenesis of SBMA.
The AR plays an important role in both neural and muscle functions; thus, maintaining AR function represents an important component of an ideal therapeutic strategy for SBMA. The recent identification of alternative AR ligands that promote AR transcriptional activity while preventing the N/C interaction, combined with our results revealing the essential role of the N/C interaction in the altered metabolism of the expanded AR, suggested to us that these ligands could represent a powerful new avenue to explore for therapeutic tools to treat SBMA. Our finding that at least two of these ligands inhibit AR aggregation and toxicity in PC12 cells and primary mouse SBMA motor neurons supports this idea and suggests an exciting new avenue for therapeutic development. Further studies are currently under way to identify optimal N/C-inhibiting ligands for the treatment of SBMA.
Acknowledgments
We thank Dr. Karen Knudsen (Kimmel Cancer Center, Thomas Jefferson University) and members of the Merry laboratory for helpful suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 NS32214 (to D. E. M.), DK07705 (to H. L. M.), CA139818 (to D. P. M.), and HD16910 (to E. M. W.).
E. Heine and D. E. Merry, unpublished results.
- SBMA
- spinal and bulbar muscular atrophy
- polyQ
- polyglutamine
- AR
- androgen receptor
- DHT
- 5α-dihydrotestosterone
- N/C
- amino/carboxyl
- NAC
- non-Aβ component of Alzheimer disease amyloid
- SARMs
- selective androgen receptor modulators
- ANOVA
- analysis of variance
- MMTV
- mouse mammary tumor virus.
REFERENCES
- 1.Kennedy W. R., Alter M., Sung J. H. (1968) Neurology 18, 671–680 [DOI] [PubMed] [Google Scholar]
- 2.La Spada A. R., Wilson E. M., Lubahn D. B., Harding A. E., Fischbeck K. H. (1991) Nature 353, 77–79 [DOI] [PubMed] [Google Scholar]
- 3.Pinsky L., Trifiro M., Kaufman M., Beitel L. K., Mhatre A., Kazemi-Esfarjani P., Sabbaghian N., Lumbroso R., Alvarado C., Vasiliou M., et al. (1992) Clin. Invest. Med. 15, 456–472 [PubMed] [Google Scholar]
- 4.Merry D. E., Kobayashi Y., Bailey C. K., Taye A. A., Fischbeck K. H. (1998) Hum. Mol. Genet. 7, 693–701 [DOI] [PubMed] [Google Scholar]
- 5.Li M., Miwa S., Kobayashi Y., Merry D. E., Yamamoto M., Tanaka F., Doyu M., Hashizume Y., Fischbeck K. H., Sobue G. (1998) Ann. Neurol. 44, 249–254 [DOI] [PubMed] [Google Scholar]
- 6.Orr H. T., Zoghbi H. Y. (2007) Annu. Rev. Neurosci. 30, 575–621 [DOI] [PubMed] [Google Scholar]
- 7.Sobue G., Hashizume Y., Mukai E., Hirayama M., Mitsuma T., Takahashi A. (1989) Brain 112, 209–232 [DOI] [PubMed] [Google Scholar]
- 8.Chevalier-Larsen E. S., O'Brien C. J., Wang H., Jenkins S. C., Holder L., Lieberman A. P., Merry D. E. (2004) J. Neurosci. 24, 4778–4786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katsuno M., Adachi H., Kume A., Li M., Nakagomi Y., Niwa H., Sang C., Kobayashi Y., Doyu M., Sobue G. (2002) Neuron 35, 843–854 [DOI] [PubMed] [Google Scholar]
- 10.Takeyama K., Ito S., Yamamoto A., Tanimoto H., Furutani T., Kanuka H., Miura M., Tabata T., Kato S. (2002) Neuron 35, 855–864 [DOI] [PubMed] [Google Scholar]
- 11.Walcott J. L., Merry D. E. (2002) J. Biol. Chem. 277, 50855–50859 [DOI] [PubMed] [Google Scholar]
- 12.Pratt W. B., Welsh M. J. (1994) Semin. Cell Biol. 5, 83–93 [DOI] [PubMed] [Google Scholar]
- 13.He B., Kemppainen J. A., Voegel J. J., Gronemeyer H., Wilson E. M. (1999) J. Biol. Chem. 274, 37219–37225 [DOI] [PubMed] [Google Scholar]
- 14.He B., Kemppainen J. A., Wilson E. M. (2000) J. Biol. Chem. 275, 22986–22994 [DOI] [PubMed] [Google Scholar]
- 15.Ikonen T., Palvimo J. J., Jänne O. A. (1997) J. Biol. Chem. 272, 29821–29828 [DOI] [PubMed] [Google Scholar]
- 16.Schaufele F., Carbonell X., Guerbadot M., Borngraeber S., Chapman M. S., Ma A. A., Miner J. N., Diamond M. I. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9802–9807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langley E., Kemppainen J. A., Wilson E. M. (1998) J. Biol. Chem. 273, 92–101 [DOI] [PubMed] [Google Scholar]
- 18.Langley E., Zhou Z. X., Wilson E. M. (1995) J. Biol. Chem. 270, 29983–29990 [DOI] [PubMed] [Google Scholar]
- 19.He B., Gampe R. T., Jr., Kole A. J., Hnat A. T., Stanley T. B., An G., Stewart E. L., Kalman R. I., Minges J. T., Wilson E. M. (2004) Mol. Cell 16, 425–438 [DOI] [PubMed] [Google Scholar]
- 20.Chen S., Ferrone F. A., Wetzel R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11884–11889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davies P., Watt K., Kelly S. M., Clark C., Price N. C., McEwan I. J. (2008) J. Mol. Endo. 41, 301–314 [DOI] [PubMed] [Google Scholar]
- 22.Perutz M. F. (1996) Curr. Opin. Struct. Biol. 6, 848–858 [DOI] [PubMed] [Google Scholar]
- 23.Sánchez I., Mahlke C., Yuan J. (2003) Nature 421, 373–379 [DOI] [PubMed] [Google Scholar]
- 24.Klement I. A., Skinner P. J., Kaytor M. D., Yi H., Hersch S. M., Clark H. B., Zoghbi H. Y., Orr H. T. (1998) Cell 95, 41–53 [DOI] [PubMed] [Google Scholar]
- 25.Thakur A. K., Jayaraman M., Mishra R., Thakur M., Chellgren V. M., Byeon I. J., Anjum D. H., Kodali R., Creamer T. P., Conway J. F., Gronenborn A. M., Wetzel R. (2009) Nat. Struct. Mol. Biol. 16, 380–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu X., Greiner E. R., Mishra R., Kodali R., Osmand A., Finkbeiner S., Steffan J. S., Thompson L. M., Wetzel R., Yang X. W. (2009) Neuron 64, 828–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertoncini C. W., Fernandez C. O., Griesinger C., Jovin T. M., Zweckstetter M. (2005) J. Biol. Chem. 280, 30649–30652 [DOI] [PubMed] [Google Scholar]
- 28.Ehrnhoefer D. E., Bieschke J., Boeddrich A., Herbst M., Masino L., Lurz R., Engemann S., Pastore A., Wanker E. E. (2008) Nat. Struct. Mol. Biol. 15, 558–566 [DOI] [PubMed] [Google Scholar]
- 29.Roy J., Minotti S., Dong L., Figlewicz D. A., Durham H. D. (1998) J. Neurosci. 18, 9673–9684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bailey C. K., Andriola I. F., Kampinga H. H., Merry D. E. (2002) Hum. Mol. Genet. 11, 515–523 [DOI] [PubMed] [Google Scholar]
- 31.Montie H. L., Cho M. S., Holder L., Liu Y., Tsvetkov A. S., Finkbeiner S., Merry D. E. (2009) Hum. Mol. Genet. 18, 1937–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li M., Chevalier-Larsen E. S., Merry D. E., Diamond M. I. (2007) J. Biol. Chem. 5, 3157–3164 [DOI] [PubMed] [Google Scholar]
- 33.Mojsilovic-Petrovic J., Nedelsky N., Boccitto M., Mano I., Georgiades S. N., Zhou W., Liu Y., Neve R. L., Taylor J. P., Driscoll M., Clardy J., Merry D., Kalb R. G. (2009) J. Neurosci. 29, 8236–8247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gioeli D., Ficarro S. B., Kwiek J. J., Aaronson D., Hancock M., Catling A. D., White F. M., Christian R. E., Settlage R. E., Shabanowitz J., Hunt D. F., Weber M. J. (2002) J. Biol. Chem. 277, 29304–29314 [DOI] [PubMed] [Google Scholar]
- 35.Guo Z., Dai B., Jiang T., Xu K., Xie Y., Kim O., Nesheiwat I., Kong X., Melamed J., Handratta V. D., Njar V. C., Brodie A. M., Yu L. R., Veenstra T. D., Chen H., Qiu Y. (2006) Cancer Cell 10, 309–319 [DOI] [PubMed] [Google Scholar]
- 36.Zhou Z. X., Kemppainen J. A., Wilson E. M. (1995) Mol. Endocrinol. 9, 605–615 [DOI] [PubMed] [Google Scholar]
- 37.Zhu Z., Becklin R. R., Desiderio D. M., Dalton J. T. (2001) Biochem. Biophys. Res. Commun. 284, 836–844 [DOI] [PubMed] [Google Scholar]
- 38.Chen S., Xu Y., Yuan X., Bubley G. J., Balk S. P. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 15969–15974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masiello D., Cheng S., Bubley G. J., Lu M. L., Balk S. P. (2002) J. Biol. Chem. 277, 26321–26326 [DOI] [PubMed] [Google Scholar]
- 40.Sathya G., Chang C. Y., Kazmin D., Cook C. E., McDonnell D. P. (2003) Cancer Res. 63, 8029–8036 [PubMed] [Google Scholar]
- 41.He B., Bowen N. T., Minges J. T., Wilson E. M. (2001) J. Biol. Chem. 276, 42293–42301 [DOI] [PubMed] [Google Scholar]
- 42.Chandra S., Shao J., Li J. X., Li M., Longo F. M., Diamond M. I. (2008) J. Biol. Chem. 283, 23950–23955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He B., Lee L. W., Minges J. T., Wilson E. M. (2002) J. Biol. Chem. 277, 25631–25639 [DOI] [PubMed] [Google Scholar]
- 44.Norris J. D., Joseph J. D., Sherk A. B., Juzumiene D., Turnbull P. S., Rafferty S. W., Cui H., Anderson E., Fan D., Dye D. A., Deng X., Kazmin D., Chang C. Y., Willson T. M., McDonnell D. P. (2009) Chem. Biol. 16, 452–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Furutani T., Takeyama K., Tanabe M., Koutoku H., Ito S., Taniguchi N., Suzuki E., Kudoh M., Shibasaki M., Shikama H., Kato S. (2005) J. Pharmacol. Exp. Ther. 315, 545–552 [DOI] [PubMed] [Google Scholar]
- 46.Katsuno M., Adachi H., Doyu M., Minamiyama M., Sang C., Kobayashi Y., Inukai A., Sobue G. (2003) Nat. Med. 9, 768–773 [DOI] [PubMed] [Google Scholar]
- 47.Kemppainen J. A., Lane M. V., Sar M., Wilson E. M. (1992) J. Biol. Chem. 267, 968–974 [PubMed] [Google Scholar]
- 48.Lavery D. N., McEwan I. J. (2008) Biochemistry 47, 3360–3369 [DOI] [PubMed] [Google Scholar]
- 49.Askew E. B., Bai S., Blackwelder A. J., Wilson E. M. (2010) J. Biol. Chem. 285, 21824–21836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen H. C., Buchanan G., Butler L. M., Prescott J., Henderson M., Tilley W. D., Coetzee G. A. (2005) Biol. Chem. 386, 69–74 [DOI] [PubMed] [Google Scholar]
- 51.He B., Gampe R. T., Jr., Hnat A. T., Faggart J. L., Minges J. T., French F. S., Wilson E. M. (2006) J. Biol. Chem. 281, 6648–6663 [DOI] [PubMed] [Google Scholar]
- 52.Bai S., He B., Wilson E. M. (2005) Mol. Cell. Biol. 25, 1238–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bai S., Wilson E. M. (2008) Mol. Cell. Biol. 28, 1947–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]