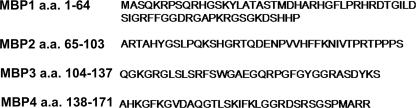Abstract
Accumulation of amyloid β-protein (Aβ) into brain parenchymal plaques and the cerebral vasculature is a pathological feature of Alzheimer disease and related disorders. Aβ peptides readily form β-sheet-containing oligomers and fibrils. Previously, we reported a strong interaction between myelin basic protein (MBP) and Aβ peptides that resulted in potent inhibition of fibril assembly (Hoos, M. D., Ahmed, M., Smith, S. O., and Van Nostrand, W. E. (2007) J. Biol. Chem. 282, 9952–9961; Hoos, M. D., Ahmed, M., Smith, S. O., and Van Nostrand, W. E. (2009) Biochemistry 48, 4720–4727). MBP is recognized as a highly post-translationally modified protein. In the present study, we demonstrate that human MBP purified from either brain or a bacterial recombinant expression system comparably bound to Aβ and inhibited Aβ fibril assembly indicating that post-translational modifications are not required for this activity. We also show that purified mouse brain MBP and recombinantly expressed mouse MBP similarly inhibited Aβ fibril formation. Through a combination of biochemical and ultrastructural techniques, we demonstrate that the binding site for Aβ is located in the N-terminal 64 amino acids of MBP and that a stable peptide (MBP1) comprising these residues was sufficient to inhibit Aβ fibrillogenesis. Under conditions comparable with those used for Aβ, the fibrillar assembly of amylin, another amyloidogenic peptide, was not inhibited by MBP1, although MBP1 still bound to it. This observation suggests that the potent inhibitory effect of MBP on fibril formation is not general to amyloidogenic peptides. Finally, MBP1 could prevent the cytotoxic effects of Aβ in primary cortical neurons. Our findings suggest that inhibition of Aβ fibril assembly by MBP, mediated through its N-terminal domain, could play a role in influencing amyloid formation in Alzheimer disease brain and corresponding mouse models.
Keywords: Alzheimer Disease, Amyloid, Electron Microscopy (EM), Neuron, Peptide Conformation, Myelin Basic Protein
Introduction
The histopathology of Alzheimer disease (AD)2 is characterized by the prominent presence of plaques of amyloid β-protein (Aβ) in the brain parenchyma (1). Dystrophic neurons and neuronal loss are often associated with Aβ accumulation leading to the clinical manifestation of cognitive decline and memory loss. Aβ is derived from the amyloid β-protein precursor (AβPP) through sequential proteolysis by β- and γ-secretases yielding peptides of between 39 and 43 residues (2–5). These peptides exhibit a high propensity to self-assemble into β-sheet-containing oligomers and fibrils of which the oligomeric forms are widely believed to be the most toxic species of the peptide, responsible for the majority of neuronal loss (6). A condition known as cerebral amyloid angiopathy (CAA) is prevalently found in AD and is characterized by fibrillar deposition of Aβ along arteries and arterioles of the cerebral cortex, leptomeninges, and cerebral microvasculature (6–8). Apart from the more typical CAA, which commonly occurs in AD, familial forms of CAA exist that are the result of specific point mutations within the Aβ sequence of the AβPP gene (9–15). Two well studied examples of familial CAA are Dutch type, resulting from an E22Q substitution in Aβ (9, 10), and Iowa type, resulting from a D23N substitution in Aβ (15).
Elucidating the molecules and processes in brain that regulate Aβ assembly and accumulation as parenchymal plaques and cerebral vascular deposits is important for understanding the pathogenesis of AD and related disorders as well as for providing opportunities for development of potential therapeutic interventions. Previously, we found that myelin basic protein (MBP) strongly bound to a highly fibrillogenic Aβ40 peptide that contains both the Dutch and Iowa mutations (Aβ40DI) compared with the less fibrillogenic wild-type Aβ40 peptide (Aβ40WT) (16). The presence of both of these CAA mutations together in the same Aβ peptide is known to enhance its fibrillogenic and pathogenic properties in vitro (17). It was shown that the strong interaction of MBP with Aβ40DI potently inhibited its fibril formation (16). We also demonstrated that MBP strongly binds the longer wild-type Aβ42 peptide (Aβ42WT) and similarly blocks its fibril formation (18). Because the Aβ42WT peptide contains no substitutions, we postulated that MBP was likely not interacting with the mutated residues of Aβ40DI but rather some oligomeric conformer that is commonly formed by both Aβ42WT and Aβ40DI.
The gene encoding MBP belongs to a large family of developmentally regulated genes called the Golli complex (genes of the oligodendrocyte lineage) (19–21). Members of this family are primarily produced by oligodendrocytes in the central nervous system and Schwann cells in the peripheral nervous system and are most often associated with the formation and maintenance of myelin sheaths. The Golli locus contains two distinct start sites under independent regulation and consists of 11 exons that can be alternatively spliced to form the various Golli/MBP proteins. Included in the Golli locus are seven exons that encode the MBP proteins (20). The major species of MBP have molecular masses of 21.5, 20.2, 18.5, and 17.2 kDa (22). These variations can result from deamidation, phosphorylation, C-terminal arginine loss, and deimination of arginyl residues. The 21.5-, 20.2-, and 17.2-kDa proteins are found in fetal and developing brain. In adults, the 18.5- and 17.2-kDa species are predominant (23). In addition to the different molecular mass species, at least eight charge isoforms have been shown to exist for the prominent adult 18.5-kDa protein. These numerous post-translational modifications in many cases are thought to regulate myelin assembly, MBP-ligand interactions, and signaling functions (24). It is not known whether post-translational modifications are required for the ability of MBP to bind Aβ and inhibit its fibrillar assembly or what region of the MBP protein is responsible for this activity.
In the present study, we show that unmodified human MBP purified from bacterial recombinant protein expression also bound to Aβ and exhibited the same fibril assembly inhibiting properties as MBP purified from human brain indicating that post-translational modifications are not necessary for these activities. Similar results were observed with purified mouse brain MBP and recombinantly expressed mouse MBP protein demonstrating that this function is shared across these two species. Furthermore, we demonstrate through a combination of biochemical and ultrastructural techniques that the binding site for Aβ is located in the N-terminal 64 amino acids of MBP and that a stable peptide (MBP1) comprising these residues is responsible for Aβ binding and sufficient to inhibit the fibrillogenesis of Aβ40DI and Aβ42WT. The active MBP1 fragment did not inhibit fibril formation of a different amyloidogenic peptide, islet amyloid polypeptide (IAPP), although MBP1 could bind to IAPP, suggesting that fibril inhibition is somewhat specific for Aβ peptides. Lastly, we show that MBP1 can prevent the cytotoxic effects of Aβ in primary cortical neurons. These findings further our understanding of this novel function of MBP and warrant its further investigation as to what role it may play in the pathogenesis of AD and related disorders involving Aβ assembly and deposition.
EXPERIMENTAL PROCEDURES
Reagents and Chemicals
Aβ peptides were synthesized on an ABI 430A solid phase peptide synthesizer (Applied Biosystems, Foster City, CA) using t-butoxycarbonyl chemistry, purified, and structurally characterized as described previously (25). Aβ peptides were initially prepared in 1,1,1,3,3,3-hexafluoro-2-propanol, flash frozen, lyophilized to remove solvent, and resuspended in dimethyl sulfoxide (DMSO) (26). Thioflavin T was purchased from Sigma-Aldrich. IAPP was a kind gift from the laboratory of Dr. Dan Raleigh (Stony Brook University).
Isolation and Purification of MBP from Normal Human Brain or Mouse Brain
MBP was purified from normal human white matter or pooled mouse brains following procedures described previously (27). The predominant human 18.5-kDa MBP or mouse 14.3-kDa MBP exists as a family of charge isoforms that differ in net charge and result from various post-translational modifications. To isolate the individual charge isoforms of MBP, the human or mouse brain homogenates were loaded onto a CM52 cation exchange column, and the components were eluted with a 0–0.2 m NaCl gradient. Component 8 was found in the void volume, whereas the more cationic components (C5, C4, C3, C2, and C1) eluted with an increasing salt gradient. The components were dialyzed against water, lyophilized, and stored at −80 °C. The most abundant MBP C1 component was used in all subsequent studies. Yields were ≈2.5 mg of MBP from 20 g of human brain white matter and ≈500 μg of MBP from 20 g of pooled mouse brains.
Recombinant Human and Mouse MBP Expression and Purification
The cDNA for human 18.5-kDa or mouse 14-kDa MBP was cloned into the vector pPROEXHT (Invitrogen). This vector allowed production of recombinant MBP protein with an N-terminal His6 tag (His-MBP). Plasmid was transformed into competent Escherichia coli BL21 DE3 cells by heat shock. Cells were grown at 37 °C in 1-liter cultures of LB broth containing 0.1 mg/ml ampicillin until an optical density of 0.600 absorbance unit at 600 nm was reached. Expression of the fusion protein was induced with 0.5 mm isopropyl β-d-1-thiogalactopyranoside, and then growth was allowed to proceed for an additional 3 h at 37 °C. Cells were lysed by dissolving in 20 mm Tris-HCl, pH 7.9, 6 m urea, 5 mm imidazole, 500 mm NaCl by stirring for 16 h at 4 °C. This material was centrifuged at 5,000 × g for 30 min at 4 °C and then passed over a column of HisBind resin (Invitrogen). The column was washed with 10 column volumes of lysis buffer and lysis buffer containing 20 mm imidazole sequentially. Bound recombinant His-MBP was eluted with 1 m imidazole in lysis buffer. The His-MBP-containing fractions were then dialyzed against 6 m urea, 80 mm glycine, pH 10.5; passed over a CM52 column; washed; and eluted with 80 mm glycine buffer, pH 10.5 containing 200 mm NaCl. Protein refolding was achieved by removal of denaturant and salt by slow dialysis into distilled water. Yields were typically 300–500 μg of purified MBP from 1-liter bacterial cultures.
Recombinant MBP Peptide Expression
MBP-derived peptide gene sequences were cloned into a pTYB11 plasmid vector (New England Biolabs, Ipswich, MA) and transformed into competent E. coli BL21 DE3 cells by heat shock. Cells were grown at 37 °C in 1-liter cultures of LB broth containing 0.1 mg/ml ampicillin until an optical density of 0.600 absorbance unit at 600 nm was reached. Expression of the fusion protein was induced with either 0.3 or 0.5 mm isopropyl β-d-1-thiogalactopyranoside at 26 °C for either 3 or 18 h. Cells were harvested by centrifugation at 5,000 × g for 30 min at 4 °C and cracked in a French press in 20 mm Tris-HCl, pH 9.0, 0.5 m NaCl, 1 mm EDTA containing Complete protease inhibitor (Roche Applied Science). Cell lysate was clarified by centrifugation and passed over chitin beads (New England Biolabs) equilibrated with 20 mm Tris-HCl, pH 9.0, 0.5 m NaCl, 1 mm EDTA (EQ buffer). The column was washed with EQ buffer containing 0.05% Triton X-100, and the peptide was cleaved and eluted from the intein fusion protein by incubation of the column in 40 mm dithiothreitol (DTT) according to the manufacturer's instruction. The eluate was diluted 10-fold into 50 mm glycine, pH 9.0 and passed over a CM52 column equilibrated with 50 mm glycine, pH 9.0, 50 mm NaCl (CM EQ). The column was washed in CM EQ and eluted with high salt. Fractions were analyzed by SDS-PAGE, pooled, dialyzed against water, lyophilized, and stored at −70 °C.
Solid Phase Binding Assay
12.5 μm Aβ or IAPP in 100 μl of PBS was coated on flat bottom 96-well plates (Fisher Scientific) by incubation at 37 °C overnight. Each well was blocked in 1% BSA, PBS for 1 h at RT. Then 1.56 μm purified MBP or MBP1 peptide was added and incubated at 37 °C overnight. After washing 3 × 5 min with 1% BSA, PBS, 0.05% Tween 20 (PBS-T), rabbit polyclonal antibody to MBP in PBS-T was added for 1 h at RT. Wells were washed 3 × 5 min with PBS-T. Secondary horseradish peroxidase-conjugated sheep anti-rabbit IgG was then added to each well (1:5,000; Amersham Biosciences); wells were then washed 3 × 5 min with 1% BSA, PBS-T. SureBlue tetramethylbenzidine microwell peroxidase substrate (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, MD) was added and developed, and the reaction was terminated by adding 1 n HCl. Absorbance of the samples was measured at a wavelength of 450 nm in a SpectraMax spectrofluorometer (Molecular Devices, Sunnyvale, CA) using SoftMax Pro control software.
Thioflavin T Fluorescence Assay
Lyophilized Aβ40DI peptide was first resuspended with DMSO to 2.5 mm, diluted to 12.5 μm in PBS, and then incubated at 37 °C with rocking either alone or with 1.56 μm MBP peptides. Control samples containing 0.5% DMSO and 1.56 μm MBP peptides in PBS were also included. At each time point, 100-μl samples of each reaction were placed in a 96-well microplate in triplicate, and 5 μl of 100 μm thioflavin T was added. The plate was mixed and incubated at 25 °C in the dark for 10 min. Fluorescence was measured at 490 nm using an excitation wavelength of 446 nm in a SpectraMax spectrofluorometer (Molecular Devices) using SoftMax Pro control software. For studies with IAPP, the peptide was resuspended to 2.5 mm in DMSO and then diluted to 3.13 μm in PBS containing 5 μm thioflavin T either alone or with 0.391 μm MBP1. Aliquots of 100 μl were placed in triplicate into a 96-well microplate. Fluorescence was measured at 25 °C at 5-min intervals at 490 nm using an excitation wavelength of 446 nm in a SpectraMax spectrofluorometer. Plates were mixed for 5 s before each reading.
Surface Plasmon Resonance
All runs were performed on a Biacore 2000 instrument (Uppsala, Sweden) with 10 mm HEPES, pH 7.4, 150 mm NaCl, 3 mm EDTA, 0.005% (v/v) Tween 20 as running buffer and diluent. N-terminally biotinylated Aβ peptides were resuspended in DMSO to 2.5 mm and serially diluted to 100 nm immediately before application. Biotinylated Aβ was immobilized to a Biacore CM4 sensor chip coated with streptavidin at 10 μl/min to achieve an average Rmax of ∼400 resonance units for each analyte leaving flow cell 1 as a reference. This chip preparation procedure was found to result in a surface that minimized mass transfer effects for kinetic interaction studies. Purified analyte was passed over flow cells at 5, 10, 25, 50, and 100 nm in triplicate at flow rates of 30 μl/min. Faster flow rates did not significantly improve the quality of data. Surfaces were regenerated with 0.2 m glycine, pH 2.0, 150 mm NaCl between runs. The resulting sensorgrams were analyzed by BiaCore Analysis software.
Transmission Electron Microscopy
Sample mixtures were deposited onto carbon-coated copper mesh grids (EM Sciences, Hatfield, PA) and negatively stained with 2% (w/v) uranyl acetate. The samples were viewed with an FEI Tecnai 12 BioTwin transmission electron microscope, and digital images were taken with an Advanced Microscopy Techniques camera.
Atomic Force Microscopy
AFM was carried out using a LifeScan controller developed by LifeAFM (Port Jefferson, NY) interfaced with a Digital Instruments (Santa Barbara, CA) MultiMode microscope fitted with an E scanner. AFM samples were first titrated to pH 4 using dilute HCl and then adsorbed onto freshly cleaved ruby mica (S & J Trading, Glen Oaks, NY). The lower pH allows for better adsorption of Aβ peptides to the negatively charged mica surface. Samples were imaged under hydrated conditions using supersharp silicon probes (SSS-Cont, Nanosensors, Neuchatel, Switzerland) that were modified for magnetic retraction by attaching samarium cobalt particles (LifeAFM). We estimate the effective diameter of the supersharp silicon probes to be 4 ± 1 nm at a height of 2 nm. Data analysis and graphics were performed using Interactive Display Language 5.0 (Research Systems Inc., Boulder, CO). In the Z scale bars, numbers in each color square indicate the Z-value at the middle of the range for that color.
Neuronal Toxicity Assay
Embryonic rat cortical neuronal cultures were preparing using six to eight E18 pups essentially as described previously (28). Isolated neurons were plated on poly-d-lysine-coated 48-well plates (BD Biosciences) at a density of 3 × 104 cells/ml in Neurobasal medium (Invitrogen) with 1× B27 (Invitrogen), 10 μm Ara C (Roche Applied Science), 1× Glutamax (Invitrogen), 100 μg/ml gentamicin (Sigma) for 5 days.
Prior to neuronal treatments, Aβ42WT peptide (700 μm) was incubated in the absence or presence of purified MBP1 (85 μm) for 2 h at 37 °C. Then the neuronal cultures were treated with the preincubated Aβ42WT alone, MBP1 alone, or the Aβ42WT + MBP1 at 10 μm and 1.25 μm, respectively, for 18 h at 37 °C. To measure neuronal viability the cells were analyzed for mitochondrial activity using a standard 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay kit (CGD-1 kit, Sigma-Aldrich). MTT was added at a final concentration of 0.5 mg/ml, and the cells were incubated for 4 h. To determine the cellular reduction of MTT or MTT formazan, the reaction was terminated by addition of 200 μl of a cell lysis solution, 0.1 n HCl isopropanol solution, and the plate was shaken at 25 °C to allow the MTT formazan precipitates to dissolve. The assay was then quantified by measuring the absorbance at 570 nm using a SpectraMax spectrofluorometer (Molecular Devices).
RESULTS
Human MBP Purified from Brain or Bacterial Recombinant Expression Similarly Binds to and Inhibits Aβ Fibril Formation
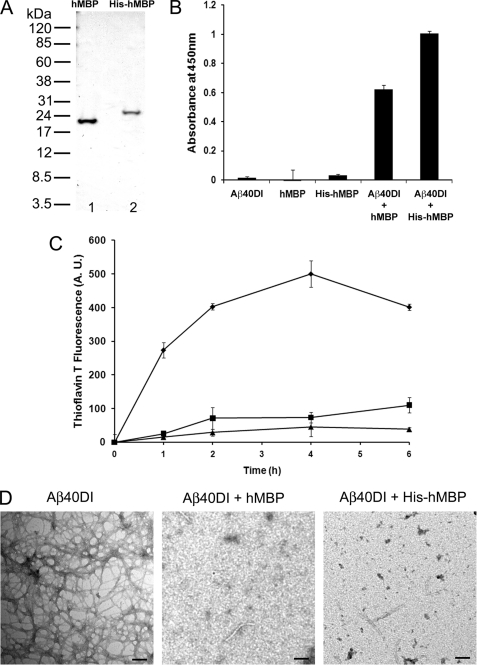
We first compared the ability of purified MBP proteins from human brain white matter and bacterial recombinant expression to bind to Aβ and inhibit its fibrillar assembly. Isolation from human brain white matter or bacterial recombinant expression yielded highly purified MBP proteins (Fig. 1A). In Fig. 1B, a solid phase binding assay was performed showing that purified human brain MBP and His-tagged human MBP similarly bound to the fibrillogenic Aβ40DI peptide. In addition, the binding affinities of brain purified and His-tagged recombinant human MBP proteins for fibrillogenic Aβ40DI and Aβ42WT peptides were found to be similar as measured by SPR spectroscopy (1.56 and 0.89 nm for human brain MBP and 1.39 and 2.73 nm for His-tagged recombinant human MBP, respectively) (data not shown). Next, using a thioflavin T fibrillogenesis assay, the inhibitory activities of purified brain and recombinant forms of human MBP were assessed (Fig. 1C). For these studies we used the highly fibrillogenic Aβ40DI peptide as described previously (16). Both MBP proteins showed a similar ability to inhibit the fibril formation of the Aβ40DI peptide over 6 h at the same substoichiometric ratio of 1:8 (MBP:Aβ). Lastly, inhibition of fibril assembly was confirmed by transmission electron microscopy (TEM) analysis of Aβ40DI fibrils alone and in the presence of either brain purified or recombinant human MBP demonstrating no difference in this activity between the different sources of human MBP (Fig. 1D). These findings suggest that there is no discernible difference between human MBP purified from brain white matter and bacterial recombinant expression lacking post-translational modifications with regard to binding to Aβ and inhibiting fibril assembly.
FIGURE 1.
Inhibition of Aβ fibril formation by purified human brain MBP and recombinant His-tagged human MBP. A, MBP was either purified from human white matter (lane 1) or recombinantly expressed in bacteria as a His-tagged protein (lane 2) and assessed by SDS-PAGE. B, interaction of Aβ40DI with human brain MBP (hMBP) or His-tagged human MBP was analyzed by solid phase binding assay. The data shown are the mean ± S.D. of triplicate determinations. C, inhibition of Aβ40DI (12.5 μm) fibrillogenesis by either purified human brain MBP (1.56 μm) or purified His-tagged human MBP (1.56 μm) as assessed by thioflavin T binding and fluorescence. Aβ40DI alone, ♦; Aβ40DI + human brain MBP, ■; Aβ40DI + His-tagged human MBP, ▴. The data shown are the mean ± S.D. of triplicate determinations. D, TEM analysis of Aβ40DI with and without human brain MBP or His-tagged human MBP. Both forms of human MBP inhibit Aβ40DI fibril formation. Scale bars, 100 nm. A.U., absorbance units.
Mouse MBP Purified from Brain or Bacterial Recombinant Expression Binds to and Inhibits Aβ Fibril Formation Similarly to Human MBP
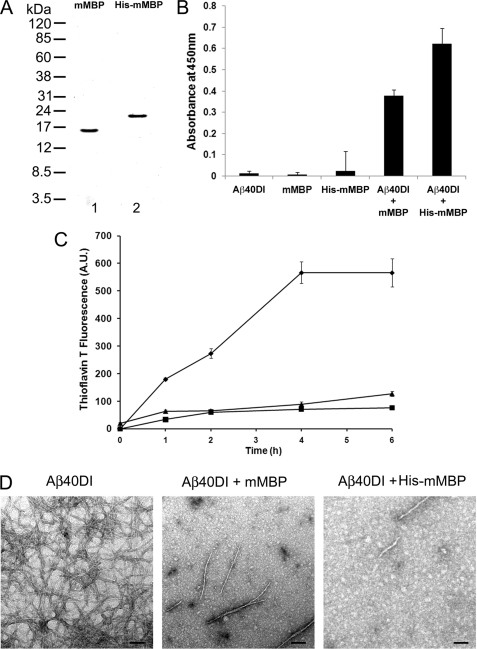
Mouse MBP was purified from brain to determine whether it binds to Aβ and inhibits its fibrillar assembly in a manner similar to human MBP. Because a mouse brain is small and contains relatively little white matter compared with human brain, we pooled ≈45 mouse brains to obtain 20 g of starting material for the purification. This amount was sufficient to yield ≈500 μg of highly purified mouse MBP for analysis. In addition, we also purified His-tagged mouse MBP from a bacterial recombinant expression system (Fig. 2A). As above with the purified human MBP proteins, we found that mouse MBP purified from brain or His-tagged mouse MBP also bound to Aβ40DI as shown in Fig. 2B. Both mouse MBP proteins exhibited similar binding to fibrillogenic Aβ40DI and Aβ42WT peptides as assessed by SPR analysis (2.2 and 7.2 nm, respectively) (data not shown). Similarly, mouse brain MBP or His-tagged mouse MBP inhibited the fibril formation of Aβ40DI at the same substoichiometric ratio of 1:8 as measured by the thioflavin T fluorescence assay (Fig. 2C). Again, amyloid fibril inhibition was confirmed by TEM images (Fig. 2D). These data demonstrate that mouse brain MBP or His-tagged mouse MBP exhibits binding and inhibition of Aβ fibril assembly comparable with that observed with the human MBP proteins.
FIGURE 2.
Inhibition of Aβ fibril formation by purified mouse brain and recombinant His-tagged mouse MBP. A, MBP was either purified from mouse brain (lane 1) or recombinantly expressed in bacteria as a His-tagged protein (lane 2) and assessed by SDS-PAGE. B, interaction of Aβ40DI with mouse brain MBP (mMBP) or His-tagged mouse MBP was analyzed by solid phase binding assay. The data shown are the mean ± S.D. of triplicate determinations. C, inhibition of Aβ40DI (12.5 μm) by purified mouse brain MBP (1.56 μm) or purified His-tagged mouse MBP (1.56 μm) as assessed by thioflavin T binding and fluorescence. Aβ40DI alone, ♦; Aβ40DI + mouse MBP, ■; Aβ40DI + His-tagged mouse MBP, ▴. The data shown are the mean ± S.D. of triplicate determinations. D, TEM analysis of Aβ40DI with and without mouse brain MBP or His-tagged mouse MBP. Both forms of mouse MBP inhibit Aβ40DI fibril formation. Scale bars, 100 nm. A.U., absorbance units.
Localization of Aβ Binding to N Terminus of MBP
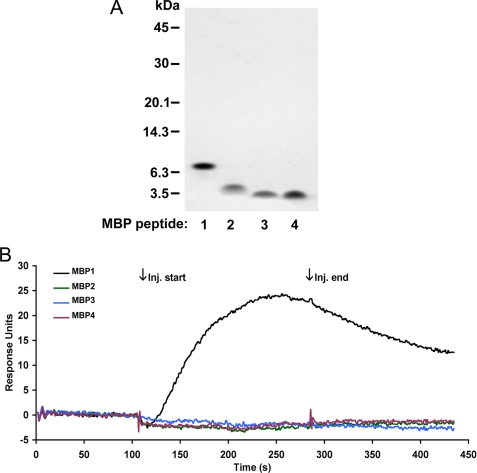
To determine the location of the binding region for Aβ in MBP, we created shorter recombinant peptides from the larger human MBP protein. Gene sequences of four different peptides representing different non-overlapping sequences (MBP1, amino acids 1–64; MBP2, amino acids 65–103; MBP3, amino acids 104–137; and MBP4, amino acids 138–171) (Fig. 3) were recombinantly expressed in bacteria and purified. The purity of each recombinant MBP fragment was assessed by SDS-PAGE (Fig. 4A). We next used SPR analysis to qualitatively determine the relative binding affinity of the purified MBP fragments to immobilized Aβ40DI. Fig. 4B shows representative sensorgrams after the injection of each of the MBP fragments over the Aβ40DI ligand surface. Only the MBP1 fragment composed of amino acids 1–64 displayed any detectable affinity for Aβ40DI. We concluded from these results that the binding domain for Aβ is located in the N-terminal 64 residues of MBP.
FIGURE 3.
Sequence of recombinant MBP peptides. MBP was divided into four peptides for recombinant bacterial expression and purification. a.a., amino acids.
FIGURE 4.
Binding of MBP peptides to Aβ as assessed by SPR binding analysis. A, each of the MBP peptides was expressed in bacteria, purified, and assessed by SDS-PAGE. B, each of the purified MBP peptides (50 nm) was passed over immobilized Aβ40DI ligand. Representative sensorgrams were base line-corrected and plotted as overlays. Binding is identified by an increase in response during injection (Inj.) (association) followed by a gradual decrease in response (dissociation). MBP1, black line; MBP2, green line; MBP3, blue line; MBP4, red line). Only MBP1 demonstrated binding to the Aβ40DI ligand.
MBP1 Inhibits Fibril Formation of Aβ42 or Aβ40DI
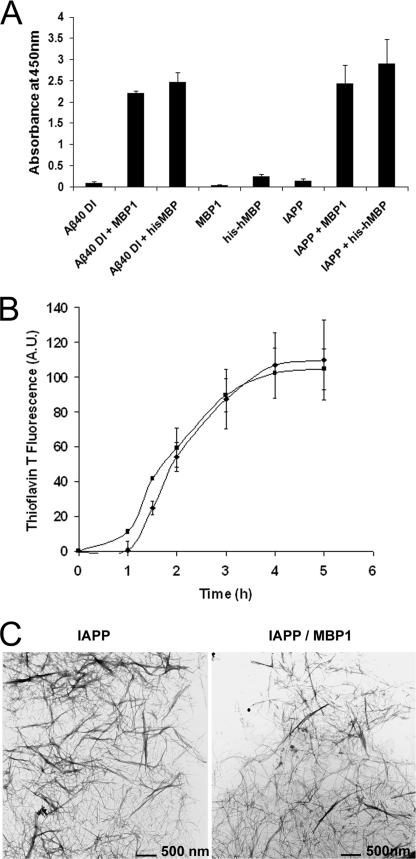
After demonstrating that MBP1 exhibited affinity for Aβ40DI through SPR analysis, we next determined whether this interaction could similarly inhibit Aβ40DI fibrillogenesis. Aβ40DI was incubated in the presence of each of the four purified MBP fragments to assess their effects on fibril formation as measured by thioflavin T fluorescence. Fig. 5 shows that only MBP1, the fragment that exhibited Aβ binding (Fig. 4), effectively blocked fibril formation. We previously showed that MBP binds to and inhibits the fibrillar assembly of Aβ42WT (18). The solid phase binding assay showed that MBP1 bound to Aβ42WT as well as Aβ40DI (Fig. 6A). Ultrastructural confirmation of Aβ fibril inhibition by MBP and MBP1 was obtained by directly visualizing fibrils using TEM (Fig. 6, B–G). To better visualize the existing structures of Aβ40DI after treatment with MBP1 or MBP, we additionally performed sensitive AFM analysis. As shown in Fig. 6, H–J, MBP1 and MBP comparably blocked amyloid fibril assembly preserving oligomeric assemblies as we reported previously (16). Together, these data indicate that MBP1 contains the necessary domain to bind to and inhibit Aβ42WT or Aβ40DI fibril formation.
FIGURE 5.
Thioflavin T analysis of inhibition of Aβ fibril formation by purified MBP peptides. Aβ40DI was treated with hexafluoroisopropanol, resuspended to a concentration of 2.5 mm in DMSO, and then diluted to a concentration of 12.5 μm in PBS in the absence (□) or presence of 1.56 μm MBP peptides. Aβ40DI + MBP1, ●; Aβ40DI + MBP2, ♦; Aβ40DI + MBP3, ■; Aβ40DI + MBP4, ▴). At specific time points aliquots were collected from each sample and assayed by thioflavin T binding and fluorescence to determine fibrillar assembly. MBP1 is as capable as intact MBP at inhibiting Aβ40DI fibrillogenesis. The data shown are the mean ± S.D. of triplicate samples. A.U., absorbance units.
FIGURE 6.
MBP1 binds to and inhibits Aβ peptide fibril formation. A, interaction of Aβ42WT and Aβ40DI with MBP or MBP1 was analyzed by solid phase binding assay. The data shown are the mean ± S.D. of triplicate determinations. B–D, 100 μm Aβ42WT was incubated for 24 h in the absence or presence of 12.5 μm MBP1 or MBP. Samples were imaged by TEM (scale bars, 100 nm). E–J, 12.5 μm Aβ40DI was incubated for 6 h in the absence or presence of 1.56 μm MBP1 or MBP. Samples were imaged by TEM (scale bars in E–G, 100 nm) and by AFM (scale bars in H–J, 100 nm). Both MBP1 and MBP are capable of binding and inhibiting Aβ42WT or Aβ40DI fibrillogenesis.
Selective Inhibition of Aβ Fibril Formation by MBP1
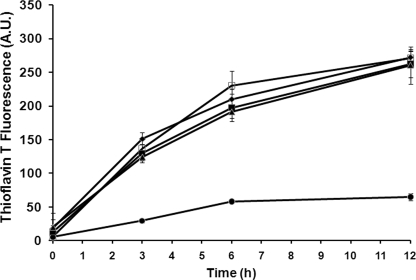
To determine whether the fibril assembly inhibiting properties of MBP extend to amyloidogenic peptides other than Aβ, we examined the effects of MBP1 on another well studied amyloidogenic peptide, IAPP (29, 30). Fig. 7A shows that MBP1 bound to IAPP similarly to its binding to Aβ40DI. However, when IAPP and MBP1 were incubated under similar stoichiometric ratios and conditions as were used with Aβ40DI and MBP1, fibril formation was not inhibited as assessed by thioflavin T fluorescence binding (Fig. 7B). The inability of MBP1 to inhibit IAPP fibril formation was further confirmed by TEM analysis (Fig. 7C). These results suggest that MBP1 is not a general amyloid formation inhibitor but exhibits some specificity for Aβ peptides.
FIGURE 7.
MBP binds to amyloidogenic IAPP but does not inhibit its fibril formation. A, interaction of Aβ40DI and IAPP with MBP and MBP1 was analyzed by solid phase binding assay. The data shown are the mean ± S.D. of triplicate determinations. B, IAPP peptide was treated with hexafluoroisopropanol, resuspended to a concentration of 2.5 mm in DMSO, and then diluted to a concentration of 3.13 μm in PBS in the absence (■) or presence (♦) of 0.391 μm MBP1. At specific time points, aliquots were collected from each sample and assayed by thioflavin T binding and fluorescence to determine fibrillar assembly. The data shown are the mean ± S.D. of triplicate samples. C, at the conclusion of the thioflavin T assay, samples were imaged by TEM. Scale bars, 500 nm. MBP1 does not inhibit the fibrillogenesis of IAPP. hMBP, human MBP.
Protective Effect of MBP1 on Aβ Neuronal Toxicity
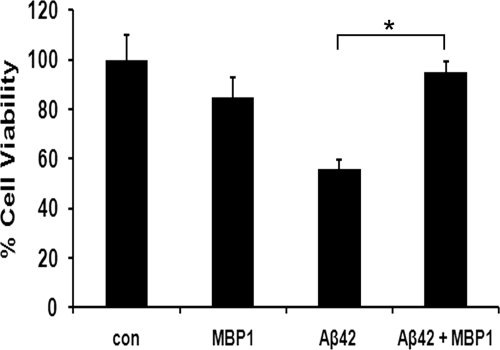
Assembled forms of Aβ are considered to account for the neurotoxic effects of the peptide (31). Therefore, the inhibition of Aβ assembly may prevent its toxicity. To examine this, we treated primary rat cortical neurons with Aβ42WT in the presence and absence of purified MBP1. Although Aβ42WT treatment alone caused a robust 53% loss in viability, co-incubation with MBP1 effectively abrogated this cytotoxic response (Fig. 8).
FIGURE 8.
MBP1 protects neurons from Aβ42WT toxicity. In the cell viability assay, primary rat cortical neurons were treated with 10 μm Aβ42WT in the absence of presence of 1.25 μm MBP1 for 18 h. Cell viability was determined using the MTT assay. The data shown are the mean ± S.D. of six independent samples. *, p < 0.001. con, control.
DISCUSSION
Accumulation of Aβ into fibrillar deposits in the brain is a hallmark of AD, CAA, and other related disorders (1, 6). These deposits are often associated with neuronal loss, cerebrovascular pathology, and cellular dysfunction leading to cognitive decline. Blocking or reversing Aβ assembly has been a central theme in the development of treatments for AD and CAA. Our earlier studies showed that MBP exhibits high affinity binding toward fibrillogenic Aβ peptides and is capable of potently inhibiting Aβ fibrillogenesis (16, 18). The precise mechanism of this inhibition remains unknown but fundamental to understanding the potential role MBP may play in AD and related disorders.
To begin with, because MBP in brain is a highly post-translationally modified protein, we wanted to determine whether these modifications of MBP are required to mediate Aβ binding and fibril inhibiting activity. Therefore, we expressed and purified a recombinant His-tagged human MBP protein, which lacked any post-translational modifications, and compared its activity with that of a purified human brain MBP. We expressed the 18.5-kDa isoform of MBP because this is the predominant species found in adult human brain (22, 32). With regard to Aβ binding and inhibition of fibrillogenesis, the recombinant human MBP protein was comparable with MBP purified from human brain white matter. Because the bacterial expression system used is unable to introduce post-translational modifications to MBP that are commonly found in mammalian brain, this comparison indicates that the Aβ interacting activities do not require these modifications.
Although the human and mouse MBP proteins are highly homologous, differences do exist in the sequence as well as in the size of the MBP proteins that are commonly expressed in adults of each species (33, 34). Therefore, we purified MBP from adult mouse brain tissue and investigated its interactions with human Aβ peptides. With regard to Aβ binding and inhibition of amyloid assembly, purified mouse brain MBP was found to be comparable with purified human brain MBP in these activities. These findings are significant in that the study of AD and CAA involves the extensive use of transgenic mouse models that express human AβPP and produce human Aβ peptides (35–37). The present results suggest that mouse MBP could play a role in influencing the quantitative and spatial accumulation of assembled Aβ deposits in these human AβPP transgenic mouse models. This idea is consistent with the paucity of fibrillar Aβ in white matter of human AβPP transgenic mouse brain and warrants future investigation into altering MBP expression in these models to determine whether this modulates the characteristic amyloid pathology.
We next sought to determine the region on MBP that was responsible for its interaction with Aβ peptides by recombinantly expressing smaller sections of the human MBP protein, which was divided into four peptides named MBP1, MBP2, MBP3, and MBP4, which were 64-, 39-, 34-, and 34-residue sequential segments, respectively (Fig. 3). These were purified, and their ability to bind Aβ was determined by using SPR spectroscopy. Fig. 4 clearly demonstrates that MBP1, the first 64 amino acids of MBP, bound to Aβ. This finding was consistent with the subsequent biochemical and ultrastructural experiments showing that only the MBP1 fragment could inhibit Aβ fibrillogenesis as shown in Fig. 5. MBP1 and intact MBP were capable of inhibiting fibril formation indicating that this N-terminal region of MBP is sufficient for this activity. This is also consistent with mouse brain MBP having activity comparable with that of human brain MBP. In adult mouse brain, the 14.3-kDa MBP predominates, whereas in adult human brain, the predominant form is the 18.5-kDa MBP (32–34, 38). The major difference between these particular human and mouse MBP proteins resides in their C-terminal regions. The first 64 amino acids of human and mouse MBP are 95% homologous. Therefore, it appears this highly conserved N-terminal region of human and mouse MBP is responsible for its interactions with Aβ.
In previous studies, we demonstrated that MBP interacts with both wild-type and CAA mutant forms of Aβ (16, 18). However, it is unknown whether this interaction is specific to Aβ peptides or whether MBP can inhibit assembly of other known amyloidogenic proteins. Therefore, we determined whether MBP1 could inhibit assembly of IAPP, another well studied amyloidogenic peptide (39, 40). Fig. 7 shows that although MBP1 could bind to IAPP it was not capable of inhibiting the fibrillar assembly of IAPP under conditions in which Aβ assembly was inhibited. These data suggest that the interaction between MBP1and Aβ requires more than the presence of β-sheet amyloid and that the interaction is more likely mediated at the level of the primary sequence. Aβ toxicity has been hypothesized to initiate the pathogenesis of AD. Assembled forms of Aβ have long been considered to account for the neurotoxicity (31). However, more recent research found that the most potent neurotoxic assemblies appear to be oligomeric rather than fibrillar (28, 41–43). It was reported that MBP could counteract the surface structure of Aβ fibril-mediated cytotoxicity (44). In our study, we show that the MBP1 fragment was biologically active on Aβ42WT-induced cell death in primary rat cortical neurons (Fig. 8). Because MBP1 exhibited binding and inhibition of Aβ fibril assembly comparable with that of MBP, this region of the protein is likely involved in this cytoprotective activity toward Aβ. However, investigating the role of MBP on amyloid assembly and deposition in vivo awaits future studies using mouse models of Aβ deposition and pathology.
In summary, we demonstrate that the N-terminal region of purified MBP contains the domain responsible for binding Aβ and inhibiting its assembly into fibrillar amyloid. Moreover, this region does not require post-translational modification for its activity and is shared with MBP expressed in adult mouse brain. Importantly, this MBP1 fragment was capable of blocking the neurotoxic effects of Aβ in vitro. The identification of a functionally equivalent shorter segment of full-length MBP provides a smaller molecule with which to use in further investigations of the structural mechanisms involved in this interaction and will facilitate the future study of MBP on amyloid assembly and deposition in in vivo models.
This work was supported, in whole or in part, by National Institutes of Health Grants NS35781 (to W. E. V. N.) and AG27317 (to S. O. S.). This work was also supported by Alzheimer's Association Grant IIRG-06-26805, a Collaborative MS Research Award, and a grant from the Cure Alzheimer's Disease Fund.
- AD
- Alzheimer disease
- Aβ
- amyloid β-protein
- Aβ42WT
- wild-type Aβ42 peptide
- Aβ40DI
- Dutch/Iowa double CAA mutant Aβ40 peptide
- AβPP
- amyloid β-protein precursor
- CAA
- cerebral amyloid angiopathy
- MBP
- myelin basic protein
- DMSO
- dimethyl sulfoxide
- SPR
- surface plasmin resonance
- TEM
- transmission electron microscopy
- AFM
- atomic force microscopy
- IAPP
- islet amyloid polypeptide
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
REFERENCES
- 1.Selkoe D. J. (2001) Physiol. Rev. 81, 741–766 [DOI] [PubMed] [Google Scholar]
- 2.Kang J., Lemaire H. G., Unterbeck A., Salbaum J. M., Masters C. L., Grzeschik K. H., Multhaup G., Beyreuther K., Müller-Hill B. (1987) Nature 325, 733–736 [DOI] [PubMed] [Google Scholar]
- 3.Goldgaber D., Lerman M. I., McBride O. W., Saffiotti U., Gajdusek D. C. (1987) Science 235, 877–880 [DOI] [PubMed] [Google Scholar]
- 4.Tanzi R. E., Gusella J. F., Watkins P. C., Bruns G. A., St George-Hyslop P., Van Keuren M. L., Patterson D., Pagan S., Kurnit D. M., Neve R. L. (1987) Science 235, 880–884 [DOI] [PubMed] [Google Scholar]
- 5.Robakis N. K., Ramakrishna N., Wolfe G., Wisniewski H. M. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 4190–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masters C. L., Simms G., Weinman N. A., Multhaup G., McDonald B. L., Beyreuther K. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 4245–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vinters H. V. (1987) Stroke 18, 311–324 [DOI] [PubMed] [Google Scholar]
- 8.Jellinger K. A. (2002) J. Neural Transm. 109, 813–836 [DOI] [PubMed] [Google Scholar]
- 9.Thal D. R., Ghebremedhin E., Orantes M., Wiestler O. D. (2003) J. Neuropathol. Exp. Neurol. 62, 1287–1301 [DOI] [PubMed] [Google Scholar]
- 10.Levy E., Carman M. D., Fernandez-Madrid I. J., Power M. D., Lieberburg I., van Duinen S. G., Bots G. T., Luyendijk W., Frangione B. (1990) Science 248, 1124–1126 [DOI] [PubMed] [Google Scholar]
- 11.Van Broeckhoven C., Haan J., Bakker E., Hardy J. A., Van Hul W., Wehnert A., Vegter-Van der Vlis M., Roos R. A. (1990) Science 248, 1120–1122 [DOI] [PubMed] [Google Scholar]
- 12.Hendriks L., van Duijn C. M., Cras P., Cruts M., Van Hul W., van Harskamp F., Warren A., McInnis M. G., Antonarakis S. E., Martin J. J., Hofman A., Van Broeckhoven C. (1992) Nat. Genet. 1, 218–221 [DOI] [PubMed] [Google Scholar]
- 13.Kamino K., Orr H. T., Payami H., Wijsman E. M., Alonso M. E., Pulst S. M., Anderson L., O'dahl S., Nemens E., White J. A., Sadovnick A. D., Ball M. J., Kaye J., Warren A., McInnis M., Antonarakis S. E., Korenberg J. R., Sharma V., Kukull W., Larson E., Heston L. L., Martin G. M., Bird T. D., Schellenberg G. D. (1992) Am. J. Hum. Genet. 51, 998–1014 [PMC free article] [PubMed] [Google Scholar]
- 14.Miravalle L., Tokuda T., Chiarle R., Giaccone G., Bugiani O., Tagliavini F., Frangione B., Ghiso J. (2000) J. Biol.Chem. 275, 27110–27116 [DOI] [PubMed] [Google Scholar]
- 15.Grabowski T. J., Cho H. S., Vonsattel J. P., Rebeck G. W., Greenberg S. M. (2001) Ann. Neurol. 49, 697–705 [DOI] [PubMed] [Google Scholar]
- 16.Hoos M. D., Ahmed M., Smith S. O., Van Nostrand W. E. (2007) J. Biol. Chem. 282, 9952–9961 [DOI] [PubMed] [Google Scholar]
- 17.Van Nostrand W. E., Melchor J. P., Cho H. S., Greenberg S. M., Rebeck G. W. (2001) J. Biol. Chem. 276, 32860–32866 [DOI] [PubMed] [Google Scholar]
- 18.Hoos M. D., Ahmed M., Smith S. O., Van Nostrand W. E. (2009) Biochemistry 48, 4720–4727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campagnoni A. T., Pribyl T. M., Campagnoni C. W., Kampf K., Amur-Umarjee S., Landry C. F., Handley V. W., Newman S. L., Garbay B., Kitamura K. (1993) J. Biol. Chem. 268, 4930–4938 [PubMed] [Google Scholar]
- 20.Pribyl T. M., Campagnoni C. W., Kampf K., Kashima T., Handley V. W., McMahon J., Campagnoni A. T. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 10695–10699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Givogri M. I., Bongarzone E. R., Campagnoni A. T. (2000) J. Neurosci. Res. 59, 153–159 [PubMed] [Google Scholar]
- 22.Roth H. J., Kronquist K. E., Kerlero de Rosbo N., Crandall B. F., Campagnoni A. T. (1987) J. Neurosci. Res. 17, 321–328 [DOI] [PubMed] [Google Scholar]
- 23.Baumann N., Pham-Dinh D. (2001) Physiol. Rev. 81, 871–927 [DOI] [PubMed] [Google Scholar]
- 24.Zand R., Li M. X., Jin X., Lubman D. (1998) Biochemistry 37, 2441–2449 [DOI] [PubMed] [Google Scholar]
- 25.Burdick D., Soreghan B., Kwon M., Kosmoski J., Knauer M., Henschen A., Yates J., Cotman C., Glabe C. (1992) J. Biol. Chem. 267, 546–554 [PubMed] [Google Scholar]
- 26.Stine W. B., Jr., Dahlgren K. N., Krafft G. A., LaDu M. J. (2003) J. Biol. Chem. 278, 11612–11622 [DOI] [PubMed] [Google Scholar]
- 27.Wood D. D., Moscarello M. A. (1989) J. Biol. Chem. 264, 5121–5127 [PubMed] [Google Scholar]
- 28.Ahmed M., Davis J., Aucoin D., Sato T., Ahuja S., Aimoto S., Elliott J. I., Van Nostrand W. E., Smith S. O. (2010) Nat. Struct. Mol. Biol. 17, 561–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Höppener J. W., Oosterwijk C., van Hulst K. L., Verbeek J. S., Capel P. J., de Koning E. J., Clark A., Jansz H. S., Lips C. J. (1994) J. Cell. Biochem. 55, (suppl.) 39–53 [DOI] [PubMed] [Google Scholar]
- 30.Abedini A., Raleigh D. P. (2005) Biochemistry 44, 16284–16291 [DOI] [PubMed] [Google Scholar]
- 31.Pike C. J., Burdick D., Walencewicz A. J., Glabe C. G., Cotman C. W. (1993) J. Neurosci. 13, 1676–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harauz G., Ishiyama N., Hill C. M., Bates I. R., Libich D. S., Farès C. (2004) Micron 35, 503–542 [DOI] [PubMed] [Google Scholar]
- 33.Barbarese E., Carson J. H., Braun P. E. (1978) J. Neurochem. 31, 779–782 [DOI] [PubMed] [Google Scholar]
- 34.Kamholz J., Toffenetti J., Lazzarini R. A. (1988) J. Neurosci. Res. 21, 62–70 [DOI] [PubMed] [Google Scholar]
- 35.Davis J., Xu F., Deane R., Romanov G., Previti M. L., Zeigler K., Zlokovic B. V., Van Nostrand W. E. (2004) J. Biol. Chem. 279, 20296–20306 [DOI] [PubMed] [Google Scholar]
- 36.Hsiao K., Chapman P., Nilsen S., Eckman C., Harigaya Y., Younkin S., Yang F., Cole G. (1996) Science 274, 99–102 [DOI] [PubMed] [Google Scholar]
- 37.Wilcock D. M., Lewis M. R., Van Nostrand W. E., Davis J., Previti M. L., Gharkholonarehe N., Vitek M. P., Colton C. A. (2008) J. Neurosci. 28, 1537–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Ferra F., Engh H., Hudson L., Kamholz J., Puckett C., Molineaux S., Lazzarini R. A. (1985) Cell 43, 721–727 [DOI] [PubMed] [Google Scholar]
- 39.Betsholtz C., Svensson V., Rorsman F., Engström U., Westermark G. T., Wilander E., Johnson K., Westermark P. (1989) Exp. Cell Res. 183, 484–493 [DOI] [PubMed] [Google Scholar]
- 40.Brender J. R., Lee E. L., Cavitt M. A., Gafni A., Steel D. G., Ramamoorthy A. (2008) J. Am. Chem. Soc. 130, 6424–6429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirkitadze M. D., Bitan G., Teplow D. B. (2002) J. Neurosci. Res. 69, 567–577 [DOI] [PubMed] [Google Scholar]
- 42.Walsh D. M., Klyubin I., Fadeeva J. V., Cullen W. K., Anwyl R., Wolfe M. S., Rowan M. J., Selkoe D. J. (2002) Nature 416, 535–539 [DOI] [PubMed] [Google Scholar]
- 43.Walsh D. M., Klyubin I., Fadeeva J. V., Rowan M. J., Selkoe D. J. (2002) Biochem. Soc. Trans. 30, 552–557 [DOI] [PubMed] [Google Scholar]
- 44.Yoshiike Y., Akagi T., Takashima A. (2007) Biochemistry 46, 9805–9812 [DOI] [PubMed] [Google Scholar]