Abstract
Streptococcus pneumoniae are commensals of the human nasopharynx with the capacity to invade mucosal respiratory cells. PspC, a pneumococcal surface protein, interacts with the human polymeric immunoglobulin receptor (pIgR) to promote bacterial adherence to and invasion into epithelial cells. Internalization of pneumococci requires the coordinated action of actin cytoskeleton rearrangements and the retrograde machinery of pIgR. Here, we demonstrate the involvement of Src protein-tyrosine kinases (PTKs), focal adhesion kinase (FAK), extracellular signal-regulated kinase (ERK), and c-Jun N-terminal kinase (JNK) but not p38 mitogen-activated protein kinases (MAPK) in pneumococcal invasion via pIgR. Pharmacological inhibitors of PTKs and MAPKs and genetic interference with Src PTK and FAK functions caused a significant reduction of pIgR-mediated pneumococcal invasion but did not influence bacterial adhesion to host cells. Furthermore, pneumococcal ingestion by host cells induces activation of ERK1/2 and JNK. In agreement with activated JNK, its target molecule and DNA-binding protein c-Jun was phosphorylated. We also show that functionally active Src PTK is essential for activation of ERK1/2 upon pneumococcal infections. In conclusion, these data illustrate the importance of a coordinated signaling between Src PTKs, ERK1/2, and JNK during PspC-pIgR-mediated uptake of pneumococci by host epithelial cells.
Keywords: Bacterial Signal Transduction, ERK, MAP Kinases (MAPKs), Src, Tyrosine-protein Kinase (Tyrosine Kinase), JNK, Focal Adhesion Kinase, Pneumococci
Introduction
Streptococcus pneumoniae (pneumococci) are encapsulated Gram-positive human pathogens colonizing the nasopharynx. Pneumococci can remain asymptomatic without causing any clinical symptoms but may also, depending on the strain and host susceptibility, spread from the nasopharynx into the middle ear or lower respiratory tract and cause severe middle ear or lung infections. Moreover, pneumococci can also gain access to the bloodstream after breaching the epithelial barrier and cause life-threatening invasive pneumococcal diseases such as septicemia and bacterial meningitis (1). Pneumococci produce a wide variety of virulence factors that are thought to contribute to its pathogenesis (2, 3). These bacterial factors, which are adapted successfully to different host niches, are involved either predominantly in nasopharyngeal colonization or transmigration of host tissue barriers and immune evasion (4, 5).
Pneumococcal adherence to host cells is facilitated by serum or extracellular matrix proteins such as complement factor H, human thrombospondin-1, and vitronectin, which were shown to act as molecular bridges linking the pathogen with its host cell (6–8). In addition, pneumococci produce adhesins, which interact directly with cellular receptors and consequently, these interactions promote bacterial adherence to and invasion into host cells (4, 9). The major adhesin of S. pneumoniae, the pneumococcal surface protein C (PspC)4 (also designated as CbpA or SpsA) represents a multifunctional surface-exposed choline-binding protein and plays an important role in invasion and pathogenesis of this versatile pathogen. The functions attributed to PspC include a direct and human-specific interaction with the free secretory component (SC), SC as part of the apically transported human polymeric immunoglobulin receptor (pIgR), or SC as part of secretory IgA (SIgA) (10). One or two hexapeptide motifs in the amino-terminal part of the PspC protein are critical for binding to human-specific amino acids in the immunoglobulin-like ectodomains D3 and D4 of pIgR (10–12). Within the host, the physiological role of pIgR is the transport of immunoglobulins (IgA and IgM) across the mucosal epithelial barriers from the basolateral to apical surface (13–15). The pIgR-dIgA or polymeric IgA (pIgA) complex, which is endocytosed and translocated via vesicular transport, is proteolytically cleaved off at the apical surface of epithelial cells. This results in release of free SC or SIgA. Alternatively, unloaded pIgR is recycled and can be transported on a retrograde pathway from apical to basal (13–18). Strikingly, pneumococci can adopt this retrograde transport machinery of pIgR and after PspC-mediated binding to pIgR, pneumococci are ingested and transcytosed through mucosal epithelial cells (11, 19).
Recently we demonstrated that pneumococcal invasion of host epithelial cells via PspC-hpIgR interaction requires the small GTPase member Cdc42, phosphatidylinositol 3-kinase (PI3K) and Akt activity (20). Under physiological conditions and bacteria-free settings binding of polymeric IgA to pIgR at the basolateral surface of the cells was shown to cause transient activation of protein-tyrosine kinase (PTK) activity. Moreover, inhibitors of the Src family of PTKs inhibited pIgA-mediated stimulation of pIgR transcytosis (21). However, it is currently unknown whether PTKs are involved in pIgR-mediated endocytosis of pneumococci.
Here, we have assessed the role of PTKs and downstream signaling molecules during PspC-hpIgR-mediated pneumococcal invasion into host cells. We demonstrate that the Src family PTKs and FAK are important for pIgR-initiated pneumococcal internalization by host cells. In addition, we show that ERK1/2 activation depends on functionally active Src PTKs. Taken together, our data indicate that PspC-pIgR-mediated invasion of pneumococci requires the coordinated signaling of Src PTKs, FAK, ERK1/2, and JNK.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture Conditions
S. pneumoniae (NCTC10319; serotype 35A) or NCTC10319 transformed with plasmid pMV158 (22) and expressing GFP were cultured in Todd-Hewitt broth (Oxoid, Basingstoke, UK) supplemented with 0.5% yeast extract (THY) to mid-log phase or grown on blood agar plates (Oxoid).
Cell Culture, Infection Experiments, and Inhibitor Studies
Madin-Darby canine kidney (ATCC CCL-34) epithelial cells stably transfected with the hpIgR cDNA in pCB6 (MDCK-hpIgR) (23) and the pIgR-expressing human lung epithelial cell line Calu-3 (ATCC HTB-55) were cultured in Eagle's minimum essential medium supplemented with 10% fetal bovine serum (FBS), 2 mm glutamine, penicillin G (100 IU ml−1), and streptomycin (100 μg ml−1) (all from PAA) at 37 °C under 5% CO2. The medium for Calu-3 cells was further supplemented with 1 mm sodium pyruvate and 0.1 mm non-essential amino acids (PAA).
Epithelial cells were seeded on glass coverslips (diameter 12 mm) or directly in wells of a 24-well plate (Cellstar, Greiner, Germany). 5 × 104 cells per well were cultivated to cell monolayers with ∼2 × 105 cells per well. Prior to infection with pneumococci the cells were washed three times with Dulbecco's modified Eagle's medium containing HEPES (DMEM-HEPES, PAA Laboratories, Coelbe, Germany) supplemented with 1.0% FBS. Host cells were infected as described using a multiplicity of infection of 50 bacteria per host cell (20). To enhance the numbers of adherent bacteria in our signaling studies a centrifugation step at 120 × g was conducted for 3 min. Infections were carried out for 1 h at 37 °C and 5.0% CO2. To remove unbound bacteria from the supernatant of the infection experiment, host cells were thoroughly washed with DMEM-HEPES. The infection dose (colony forming units) per infection and cell culture well was controlled by serial plating of pneumococci on blood agar plates. Pharmacological inhibitors used here to study the impact of PTKs on pneumococcal invasion were solved in DMSO and host cells were preincubated for 1 h at 37 °C with PD98059 or for 30 min with one of the other inhibitors prior to host cell infection. The infection assays were performed in the presence of the inhibitors. To evaluate the effect of DMSO, control experiments were performed in which the host cells were incubated with DMSO alone and infected as indicated. Our results revealed no effect of DMSO alone on bacterial and host cell viability, and pneumococcal adherence as determined by immunofluorescence microscopy (data not shown).
Reagents and Antibodies
Genistein, PP2, PD98059, LY294002, RGD peptide, RGE peptide, and JNK inhibitor II were purchased from Calbiochem (Calbiochem). Cytochalasin D was purchased from MP Biomedicals. The inhibitors were reconstituted in DMSO and stored according to the manufacturer's instructions. Secramine A, a specific inhibitor of Cdc42, was kindly provided by Tom Kirchhausen, Immune Disease Institute, Harvard Medical School, Boston, MA (24), and used as described (20). The following antibodies were used: mouse-anti Csk, anti-FAK (both from BD Biosciences), goat anti-GAPDH (Abnova via Biozol, Eching, Germany), rabbit anti-phospho-ERK1/2 (Thr202/Tyr204), rabbit anti-phospho-JNK, rabbit anti-phospho-c-Jun (all from Cell Signaling Technology), rabbit anti-ERK1/2, anti-JNK, anti-Src (all from Santa Cruz Biotechnology), HRP-conjugated goat anti-rabbit (Dako, Hamburg, Germany), and HRP-conjugated anti-mouse IgG (Jackson Laboratories).
Transfection Experiments
Expression constructs encoding kinase-inactive c-Src (Src K297M), Csk WT, and kinase-inactive Csk (Csk K222M) were employed (25, 26). MDCK-hpIgR cells were transfected using Lipofectamine LTX reagent (Invitrogen) according to the manufacturer's instructions. Uptake of pneumococci by transiently transfected MDCK-hpIgR was evaluated by infecting the host cells for 1 h with S. pneumoniae serotype 35A.
shRNA Construction and Cloning
Recombinant lentiviral particles were generated using the systems developed by the groups of Trono (27) and Van Parijs (28). The plasmids used (pLL3.7, pMD2.G, and psPAX2) were obtained from Addgene and maintained in Escherichia coli STBL4 (Invitrogen). Using the algorithm AAGN18TT we identified sequences that could silence expression of FAK. According to this prediction, two complementary primers were synthesized: sh-hFAK sense, 5′-TGCTGATTGGAGTCATCACAGATTCAAGAGATCTGTGATGACTCCAATCAGCTTTTTTC-3′ and sh-hFAK anti, 5′-TCGAGAAAAAAGCTGATTGGAGTCATCACAGATCTCTTGAATCTGTGATGACTCCAATCAGCA-3′. Furthermore, a scrambled control shRNA was generated with sh-scrambled sense, 5′-TGTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGACTTTTTTC-3′and sh-scrambled anti, 5′-TCGAGAAAAAAGTCTCCGAACGTGTCACGTTCTCTTGAAACGTGACACGTTCGGAGACA-3′. The primers were annealed and cloned into the XhoI and HpaI sites of pLL3.7 generating pLL3.7-shFAK and pLL3.7-sh scrambled. The correct insertion of the shRNA cassette was verified by sequencing.
Lentiviral Production, Titration, and in Vitro Application
Lentiviral particles were produced, concentrated, and titrated as described previously (29). For in vitro transduction of epithelial cells a multiplicity of infection of 50 infectious particles per cell was used and the GFP-positive cells were sorted using a BD FACSAria (BD Biosciences).
Antibiotic Protection Assays
To quantify the total number of ingested pneumococci after infecting the host cells, antibiotic protection assays were performed as described (20). Briefly, epithelial cells were infected with pneumococci (multiplicity of infection of 50) and thereafter, the infected and washed host cells were incubated for 1 h with DMEM-HEPES containing 100 μg ml−1 of gentamicin and 100 units ml−1 of penicillin G at 37 °C and 5.0% CO2 to kill extracellular and non-adherent pneumococci. Invasive and viable pneumococci were recovered from the intracellular compartments of the host cells by a saponin-mediated host cell lysis (1.0% w/v) and the total number of invasive pneumococci was monitored after plating sample aliquots on blood agar plates, followed by colony formation and enumeration. Each experiment was repeated at least three times and results were expressed as mean ± S.D.
Fluorescence Staining and Microscopy
To determine the number of adherent bacteria per host cell, infected host cells were fixed on glass coverslips (diameter, 12 mm) with 3.0% paraformaldehyde. The immunofluorescence staining of pneumococci attached to the host cells and in some case also the differentiation between extracellular and intracellular bacteria was carried out as described recently (8). Briefly, extra- and intracellular pneumococci were stained using a polyclonal anti-pneumococcal antiserum and secondary goat anti-rabbit IgG coupled to Alexa 488 (green) or Alexa 568 (red) (Invitrogen). To visualize phosphorylated ERK, pIgR-expressing epithelial cells were infected with GFP-expressing pneumococci (NCTC 10319 transformed with plasmid pMV158) (22), and stained post-infection with rabbit anti-phospho-ERK1/2 (Thr202/Tyr204) and secondary goat anti-rabbit IgG coupled to Alexa 568 (red). The nucleus was stained blue with Hoechst 33258 dye. The samples were viewed with a confocal laser scanning microscope (Zeiss LSM-510Meta) and the appropriate software (LSM-510) was used for image acquisition. Each bar in the images represents 10 μm.
Cell Lysis and Western Blotting
Host cells were washed at the indicated time points post-infection with ice-cold PBS and lysed with lysis buffer (10 mm Tris, 100 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1 mm NaF, 20 mm Na4P2O7, 2 mm, Na3VO4, 0.1% SDS, 1.0% Triton X-100, 10% glycerol, 0.5% deoxycholate) containing a complete protease inhibitor mixture tablet (Roche). The amount of protein in the samples was determined using the Bradford protein quantification method (Sigma) and equal amounts of protein lysates were separated by SDS-PAGE. After protein transfer using a semi-dry blotting apparatus (Bio-Rad) membranes were blocked with 5% skim milk (Roth) prior to incubations with specific antibodies at 4 °C overnight. Membranes were washed and incubated with HRP-conjugated secondary antibodies for 1 h at room temperature. Following washing, antibody binding was detected using enhanced chemiluminescence (ECL, Amersham Biosciences). To confirm equal protein amounts in each sample, blots were stripped and reprobed or aliquots of lysates from different time points were analyzed with antibodies specific for the non-phosphorylated form of the protein.
Statistical Evaluation
The infection experiments were performed at least three times, each in duplicate and the data were expressed as mean ± S.D. Differences in adherence and internalization of pneumococci were analyzed by the two-tailed unpaired Student's t test. In all analyzes, p values of <0.05 were considered statistically significant.
RESULTS
PspC-hpIgR-mediated Pneumococcal Invasion Relies on Host Protein-tyrosine Kinase Activities
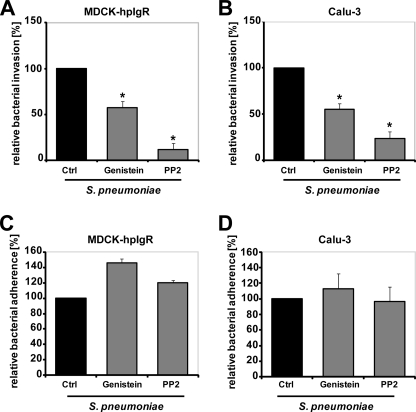
PTKs and especially members of the Src family of PTKs are implicated in the uptake of pathogenic bacteria by host cells. A role of Src PTKs for invasion of Staphylococcus aureus, Listeria monocytogenes, Helicobacter pylori, or Neisseria meningitidis and the pathogenic fungus Paracoccidioides brasiliensis has been reported (25, 30–33). To assess the role of PTKs and Src family PTKs for pIgR-mediated ingestion of pneumococci by human pIgR-expressing MDCK (MDCK-hpIgR) or Calu-3 cells, we determined pneumococcal internalization in the presence of the general PTK inhibitor genistein. The results revealed a significant reduction of intracellular pneumococci recovered from the infected host cells (Fig. 1, A and B). Similar, treatment of host cells with PP2, a specific Src PTK inhibitor, significantly reduced PspC-pIgR-mediated pneumococcal internalization by epithelial cells (Fig. 1, A and B). However, no significant differences were observed for pneumococcal adherence to inhibitor-treated eukaryotic cells, when the number of attached pneumococci was compared with those attached to untreated host cells (Fig. 1, C and D). These result suggested that PTKs, especially Src family kinases, are involved in pneumococcal uptake by epithelial cells.
FIGURE 1.
Host protein-tyrosine kinase activity is required for PspC-hpIgR-mediated invasion of pneumococci into MDCK-hpIgR and Calu-3 cells. A and B, invasion and intracellular survival of the bacteria in MDCK-hpIgR and Calu-3 cells was monitored in the absence (Ctrl) or presence of the broad spectrum protein-tyrosine kinase inhibitor genistein (50 μm) or Src family of tyrosine kinase inhibitor PP2 (5 μm) by a antibiotic protection assay. Pneumococcal invasion in the absence of the inhibitors was set to 100%. *, p < 0.001 relative to infections carried out in the absence of inhibitor. C and D, pneumococcal adherence to MDCK-hpIgR and Calu-3 cells was determined in the absence (Ctrl) or presence of genistein or PP2. Adherence of S. pneumoniae in the absence of an inhibitor was set to 100%.
Functionally Active Src Kinase Is Critical for pIgR-mediated Pneumococcal Internalization
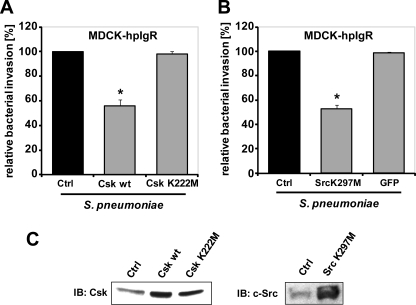
To investigate whether Src PTK activity is involved in pIgR-mediated internalization of pneumococci, a genetic interference approach was exploited. Plasmids encoding wild-type C-terminal Src kinase (Csk WT), which is a negative regulator of Src protein-tyrosine kinase, and a kinase-inactive form of Csk (Csk K222M) were transiently transfected into host epithelial cells (25, 26). Expression of wild-type Csk inhibits Src protein-tyrosine kinase activity and hence Src kinase-dependent cellular signaling events are impeded (34). Antibiotic protection assays 1 h post-infection demonstrated a significant reduction of pneumococcal ingestion by host cells over-expressing Csk WT compared with control transfected cells (Fig. 2, A and C). In contrast, transient expression of kinase-inactive Csk K222M in pIgR-expressing host cells was not associated with changes in bacterial invasion (Fig. 2, A and C). Hence, these results are in agreement with the data obtained with pharmacological inhibitors and support the hypothesis that Src kinase activity is required during pneumococcal uptake by mucosal epithelial cells. To corroborate these data, a kinase-inactive form of Src (Src K297M) was transiently overexpressed in human pIgR-expressing MDCK cells. Again, a significant reduction of pneumococcal internalization was observed, confirming the critical role of Src kinase activity for pneumococcal host cell invasion via the PspC-hpIgR interaction at the host cellular surface (Fig. 2, B and C).
FIGURE 2.
Src family kinase function promotes PspC-hpIgR-mediated invasion by pneumococci. A, MDCK-hpIgR cells were transfected with constructs encoding wild-type C-terminal Src kinase (Csk wt) or a kinase-inactive form of Csk (Csk K222M). Transiently transfected host cells were infected with S. pneumoniae and the number of invasive bacteria was determined by the antibiotic protection assay. Pneumococcal invasion of non-transfected cells (Ctrl) was set to 100%. *, p < 0.05 relative to infections carried out in non-transfected cells. B, MDCK-hpIgR cells were transfected with constructs encoding a kinase-inactive form of c-Src (Src K297M). Transiently transfected cells infected with pneumococci and bacterial invasion was monitored after performing the antibiotic protection assay. Pneumococcal invasion of non-transfected cells (Ctrl) was set to 100%. *, p < 0.05 relative to infections carried out in non-transfected cells. C, immunoblot (IB) analysis of Csk wt, Csk K222M, or Src K297M expression is shown in transiently transfected MDCK-hpIgR cells.
Focal Adhesion Kinase (FAK) Is Involved in pIgR-mediated Uptake of Pneumococci
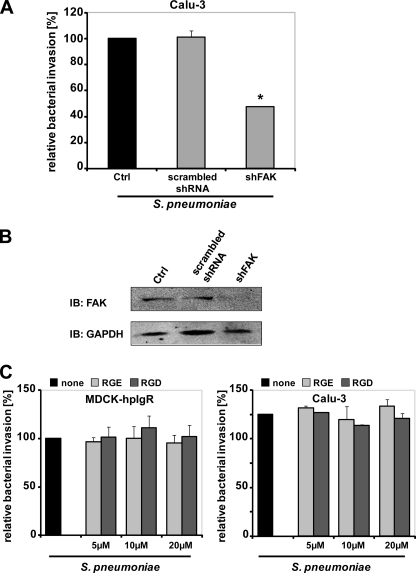
Focal adhesion kinase plays an important role in the regulation of various cellular processes and moreover, FAK activity is required for invasion of host cells by extracellular and intracellular pathogens such as S. aureus, Group A streptococci, E. coli, Shigella flexneri, and Chlamydia pneumoniae (35–40). To investigate whether FAK is also involved in pIgR-mediated uptake of pneumococci, FAK expression was knocked down in hpIgR-expressing Calu-3 cells by RNA interference using short hairpin RNA (shRNA) directed against FAK (shFAK). Antibiotic protection assays indicated a significant decrease (45%) in the number of internalized pneumococci as compared with scrambled shRNA-transfected or non-transfected Calu-3 cells (Fig. 3A), demonstrating the contribution of FAK to pneumococcal uptake via hpIgR. Reduction of FAK expression in shFAK-treated cells was verified by immunoblot analysis (Fig. 3B). Similar to the other infection assays, pneumococcal adherence was not affected (data not shown). As FAK is often, but not exclusively, connected to integrin engagement, we explored whether integrins facilitate pneumococcal uptake in hpIgR-expressing cells. However, blocking of ligand binding by integrins with RGD peptides did not affect bacterial uptake as compared with cells infected in the presence of the control peptide RGE or in the absence of a peptide (Fig. 3C). Immunofluorescence microscopy showed that pneumococcal attachment also remained unaffected by this treatment (Fig. 4D). Taken together, the results suggest that FAK is involved in pneumococcal uptake by pIgR-expressing cells and that the role of FAK in this context is independent of integrin engagement.
FIGURE 3.
FAK is required for efficient PspC-hpIgR-mediated invasion by pneumococci. A, Calu-3 epithelial cells expressing hpIgR were transduced with shFAK or scrambled shRNA and infected with S. pneumoniae. Uptake of bacteria was determined by the antibiotic protection assays. Pneumococcal invasion of non-transfected cells was set to 100%. *, p < 0.05 relative to infections carried out in non-transfected cells. B, immunoblot analysis of FAK expression in control (Ctrl), scrambled shRNA, or FAK shRNA-transduced Calu-3 epithelial cells. C, invasion and intracellular survival of the pneumococci in MDCK-hpIgR and Calu-3 cells was monitored in the absence (none) or presence of RGE or RGD peptides (5, 10, and 20 μm) by the antibiotic protection assay. Pneumococcal invasion in the absence of the peptides was set to 100%.
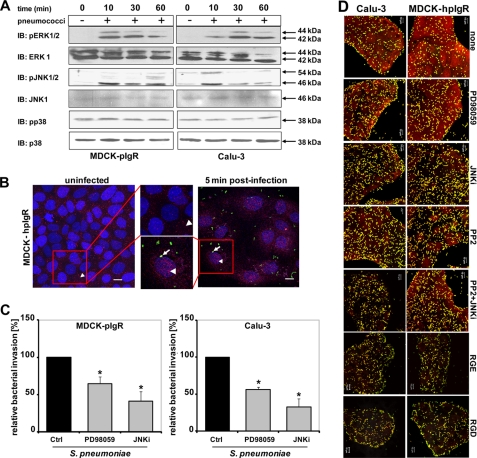
FIGURE 4.
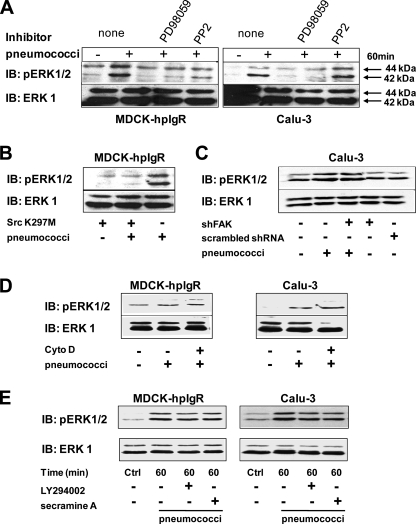
Activity of MAP kinases is essential for pIgR-initiated uptake of pneumococci. A, host cell lysates of infected MDCK-hpIgR and Calu-3 cells separated by 10% SDS-PAGE and analyzed using antibodies against the phosphorylated forms of ERK1/2, JNK isoforms p54 and p46, and p38, respectively. Total ERK1/2, JNK1, or p38 served as loading controls. B, translocation and concentration of activated ERK1/2 (arrowheads) in the nucleus of pIgR-expressing epithelial cells in response to the infection with GFP-pneumococci (arrows). Bar, 10 μm. C, invasion and intracellular survival of pneumococci into MDCK-hpIgR and Calu-3 cells was monitored in the absence (Ctrl) or presence of MAP kinase kinase (MEK1) inhibitor (PD98059, 100 μm) or c-Jun N-terminal kinase inhibitor (JNKi, 5 μm) by the antibiotic protection assay. Pneumococcal invasion in the absence of an inhibitor was set to 100%. *, p < 0.005 relative to infections carried out in the absence of inhibitor. D, immunofluorescence microscopy of pneumococcal adherence to MDCK-hpIgR and Calu-3 cells in the absence (none) or presence of Src kinase, MEK1, JNK, Src/JNK inhibitors, or RGD/RGE peptides. Bar, 10 μm in all panels.
PspC-hpIgR-mediated Internalization of Pneumococci Requires Activity of MAPKs
MAPK family members are involved in host cell invasion by several pathogenic bacteria such as Staphylococcus aureus, C. pneumoniae, L. monocytogenes, enteropathogenic E. coli, and Salmonella enterica serovar Typhimurium (41–45). Moreover, JNK activation has been associated with the invasion Porphyromonas gingivalis in gingival cells, Neisseria gonorrhoeae in epithelial cells, and N. meningitidis infection of human brain-derived microvascular endothelial cells (46–48). To assess the role of MAPKs during pneumococcal cell infections, serum-starved hpIgR-expressing host cells were infected with pneumococci for 10 min up to 1 h. At the indicated time points, the phosphorylation levels of ERK1/2, JNK isoforms p54 and p46, and p38 MAPK were analyzed by immunoblotting using antibodies specifically recognizing activated ERK1/2, JNK, or p38 MAPK. The results demonstrated a time-dependent phosphorylation and hence activation of both ERK1/2 and JNK after infecting host cells with pneumococci, whereas p38 was not activated (Fig. 4A). In addition, immunofluorescence microscopy demonstrated that pERK is translocated and concentrated in the nucleus of pneumococci-stimulated pIgR-expressing cells as visualized 5 min post-infection using GFP-pneumococci (Fig. 4B). To confirm the role of MAPKs during pneumococcal uptake by host cells, bacterial invasion by human pIgR-expressing cells was monitored in the presence of PD98059, a specific inhibitor of MEK1/2, a MAPK kinase required to activate ERK1/2, or JNK inhibitor II, a selective and reversible inhibitor of the JNK signal transduction pathway (49). Treatment of hpIgR expressing cells with MEK1 inhibitor PD98059 or JNK inhibitor significantly reduced PspC-hpIgR-mediated pneumococcal uptake by host epithelial cells (Fig. 4C). Importantly, the presence of these inhibitors did not alter pneumococcal adherence to host cells expressing human pIgR as determined by immunofluorescence staining (Fig. 4D). Taken together these results demonstrated the essential role of MAPKs for pneumococcal internalization into host epithelial cells expressing human pIgR.
Src Kinase Activity Is Cross-linked with Pneumococcal-induced ERK1/2 Activation
Various studies have suggested that ERK1/2 can be a downstream effector of Src kinases (50, 51). Therefore, we investigated the relationship between Src kinase activity and ERK1/2 activation during pneumococcal infections of human pIgR-expressing epithelial cells. Activation of ERK1/2 was monitored after 60 min of infection of pIgR-expressing cells pretreated with the Src family PTK specific inhibitor PP2. In the presence of this inhibitor phosphorylation levels of ERK1/2 did not increase upon infection in both MDCK-hpIgR as well as Calu-3 cells (Fig. 5A). A similar level of ERK inhibition was seen upon treatment with PD98059 suggesting that Src family PTKs are the critical upstream regulators of MEK-dependent activation of ERK1/2 (Fig. 5A). Importantly, pretreatment of cells with either PP2 or PD98059 had no effect on the total endogenous ERK levels of host cells (Fig. 5A). To further verify a role of Src kinase in this process, ERK1/2 phosphorylation was investigated in cells transiently transfected with Src K297M, a kinase-inactive mutant of Src. Notably, Src K297M expressing host cells infected with pneumococci showed no activation of ERK1/2 60 min post-infection (Fig. 5B). In contrast, knockdown of FAK expression by shFAK suggested that FAK is not directly involved in pneumococci-induced phosphorylation of ERK (Fig. 5C). Together, these results demonstrated that the PspC-hpIgR-triggered activation of ERK1/2 depends on Src PTK-initiated activation of MEK.
FIGURE 5.
ERK1/2 activation during PspC-h-pIgR-mediated pneumococcal adherence and invasion relies on Src kinase activity. A, phosphorylation of ERK1/2 (upper panel) was analyzed in the absence (none) or presence of PD98059 (100 μm) and PP2 (5 μm), respectively, after 60 min of infection with S. pneumoniae. The membrane was stripped and reprobed with total anti-ERK antibodies that served as a loading control (lower panel). B, phosphorylation of ERK1/2 (upper panel) was analyzed in MDCK-hpIgR cells transiently transfected with a plasmid encoding the kinase-inactive c-Src (Src K297M), after 60 min of infection with S. pneumoniae. As a loading control total ERK was detected (lower panel). C, Calu-3 cells were transduced with shFAK or scrambled shRNA, infected with S. pneumoniae, and phosphorylation of ERK1/2 (upper panel) was analyzed. As a loading control total ERK was detected (lower panel). D, pneumococcal adherence via PspC is sufficient to induce ERK1/2 phosphorylation. Invasion of pneumococci was blocked by inhibiting actin cytoskeleton polymerization with cytochalasin D (Cyto D, 125 nm) as described (20), and ERK1/2 activation was analyzed. E, activation of ERK1/2 during pneumococcal infections is independent on Cdc42 and PI3K. Phosphorylation of ERK1/2 was analyzed in the presence of Cdc42 inhibitor secramine A (10 μm) or the PI3K inhibitor LY294002 (50 μm), respectively, during pneumococcal host cell infection. Total ERK served as loading control (lower panel). IB, immunoblot.
Similar to our previous observation that pneumococcal adherence is sufficient to activate Akt (20), inhibition of the actin cytoskeleton dynamics by cytochalasin D reduced significantly pneumococcal ingestion by host cells (data not shown), whereas this treatment had no effect on adherence and ERK1/2 activation (Fig. 5D). Finally, we were interested whether there is a cross-link between the PI3K kinase-Akt induced pathway (20) and Src-initiated signaling transduction pathways during PspC-mediated pneumococcal invasion of host cells. Inhibition of PI3K by its specific inhibitor LY294002 did not significantly reduce phosphorylation of ERK1/2 (Fig. 5E). In addition, inhibition of Cdc42, which was recently shown to be essential for pneumococcal uptake in hpIgR-expressing cells (20), did not significantly change phosphorylation of ERK1/2 in infected pIgR-expressing host cells (Fig. 5E). Taken together, these results demonstrate that activation of ERK1/2 is mediated via Src kinase and MEK1, whereas this pathway is not interconnected with the PI3K-Akt pathway or Cdc42 activity. Furthermore, these inhibition experiments confirm that pneumococcal adherence to pIgR is sufficient to trigger independent signal transduction cascades (20).
Activation of JNK in Response to Pneumococcal Infections Requires Src Kinase Activity
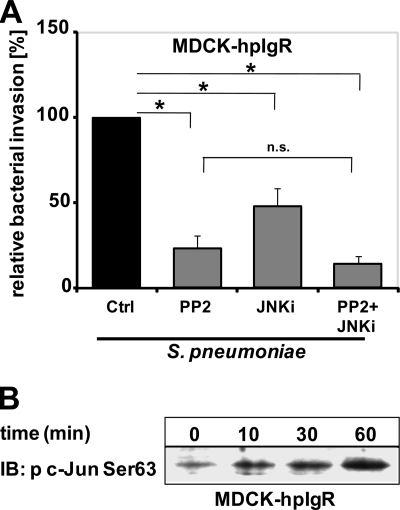
Src kinase and JNK activities are required for pneumococcal invasion into host cells. Therefore, we were also interested in the interplay between these two signaling molecules. The cross-talk between Src kinase and JNK pathways was analyzed by determining the invasion of pneumococci in hpIgR-expressing epithelial cells in the presence of a combination of Src kinase and JNK inhibitors. In addition, each inhibitor was employed separately and their individual effects on pneumococcal invasion were assessed. In general, simultaneous inhibitions of two independent signaling pathways are thought to cause additive effects on bacterial ingestion by host cells. Consequently, inhibition of signaling molecules belonging to the same signal transduction cascade will not show any additive effect on bacterial invasion. The results confirmed that inhibition of Src protein-tyrosine kinase or JNK activities decreased significantly PspC-hpIgR-initiated pneumococcal invasion of epithelial cells (Fig. 6A), whereas adherence is not affected (Fig. 4D). However, simultaneous inhibition of Src kinase and JNK showed no additive effect on pneumococcal internalization when compared with those experiments in which only one kinase activity, either Src kinase or JNK, was abrogated (Fig. 6A). Consequently, these data point to a direct link between these two kinases and moreover, the inhibition data suggest that in this PspC-hpIgR initiated signal transduction pathway activation of Src kinase occurs upstream of JNK activation.
FIGURE 6.
Sequential activation of Src and JNK MAPK during pneumococcal invasion of pIgR-expressing host cells. A, pneumococcal invasion of MDCK-hpIgR cells was monitored in the absence (Ctrl) or presence of Src kinase inhibitor (PP2, 5 μm), c-Jun N-terminal kinase inhibitor (JNKi, 5 μm), or a combination of both inhibitors. Bacterial invasion was determined by the antibiotic protection assay. Invasion of pneumococci in the absence of inhibitor was set to 100%. *, p < 0.005 relative to infections carried out in the absence of inhibitors. B, activation of transcription factor c-Jun during pneumococcal infections. Host cell lysates were prepared at the indicated time points post-infection of MDCK-hpIgR cells with pneumococci, and proteins were separated by 10% SDS-PAGE. Phosphorylation studies of c-Jun were performed by immunoblotting (IB) using specific p-c-Jun antibodies recognizing phosphorylated Ser63.
Pneumococcal Host Cell Infections via pIgR Induce c-Jun
One of the major target molecules of JNK is the DNA-binding protein c-Jun. Phosphorylation of serine 63 and 73 of c-Jun through JNK activity results in an enhanced transcriptional activity of activator protein-1 (52). Activation of c-Jun in pIgR-expressing host epithelial cells was monitored in infection experiments. Immunoblot analyzes demonstrated a time-dependent increase in c-Jun phosphorylation in response to pneumococcal host cell infections (Fig. 6B). Taken together, these data demonstrate a specific host cell receptor-mediated pneumococcal invasion mechanism that requires the coordinated signaling of kinases such as ERK and JNK downstream of Src kinase.
DISCUSSION
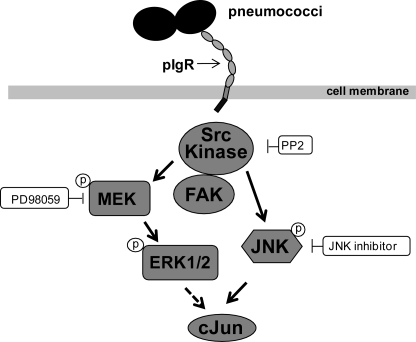
Pathogens employ a variety of strategies to subvert and control normal host cellular functions. The ability to influence key host intracellular-signaling molecules is a major advantage for many bacterial pathogens with respect to invasion and intracellular fate (53). Pneumococcal PspC was identified as a major adhesin promoting adherence to and invasion into host cells via its human-specific interaction with the pIgR of respiratory epithelial cells (4, 19). However, the endocytotic mechanism(s) of pneumococcal uptake per se and the induced host signal transduction cascades as well as the consequences for both the host cell and pathogen have not been completely explored. Recently we were able to demonstrate that PspC-hpIgR-mediated pneumococcal uptake by respiratory epithelial cells requires the activities of the small GTPase Cdc42, PI3K, and Akt (20). Here in this study we demonstrate that non-receptor-associated protein-tyrosine kinases are also involved in pneumococcal internalization by host cells expressing human pIgR. In particular, FAK and Src PTKs and downstream MAP kinases including ERK1/2 and JNK, which were phosphorylated during pneumococcal adherence, are shown to be pivotal for pneumococcal invasion into host cells (Fig. 7).
FIGURE 7.
Schematic model of pneumococci-induced signaling cascades. Solid arrows depict activated signaling molecules due to invasion of respiratory epithelial cells by pneumococci via the PspC-pIgR interaction. The dotted arrow depicts the known downstream effect of ERK (60, 62, 63).
Src PTKs are critical signal transducers modulating a wide variety of cellular functions. Activities of Src family PTKs were also shown to play a critical role in various bacterial and viral infections. For example, it has been demonstrated that activation of Src PTKs is important for host cell infections with extracellular pathogens such as Streptococcus pyogenes, S. aureus, and N. meningitidis but also for intracellular pathogens such as L. monocytogenes (25, 30, 33, 54). In addition, Src family kinases Lck and Lyn contribute to HIV type 1 and Epstein-Barr virus pathogenesis, respectively (55, 56). Although the Src PTK p62yes has been shown to be involved in regulation of the rodent-pIgR-dimeric IgA transcytosis in vivo (16), no information is currently available concerning the role of Src PTKs in human pIgR-mediated pneumococcal infections of host epithelial cells. Here, we showed that Src kinases are key non-receptor PTKs regulating PspC-hpIgR-mediated pneumococcal invasion of epithelial cells. Pneumococcal invasion into pIgR-expressing epithelial cells was severely diminished after pharmacological inhibition of or genetic ablation of Src family PTKs, suggesting a critical role for these enzymes in pneumococcal uptake via pIgR. Although genetic interference with Src PTK function showed statistically significant effects and was in agreement with our pharmacological inhibitor studies, the level of reduction in uptake was not as drastic as measured when specific inhibitors were used. This difference is most likely due to transfection efficiencies below 50%, suggesting that only a portion of the transfected cell population could be affected by treatment. Strikingly, S. pyogenes serotype M3 invasion also requires Src kinase activity and is in this respect similar to pneumococcal invasion via pIgR. Nevertheless, S. pyogenes enters cells independent of PI3K activity (54), whereas pIgR-initiated pneumococcal invasion requires both, Src kinase (this study) and PI3K activity as shown recently (20).
In addition to Src kinase activity, our data clearly demonstrates that FAK activity is involved in PspC-hpIgR dependent uptake of pneumococci. Upon phosphorylation, FAK tyrosine residue Tyr397 provides a high affinity binding site for the Src PTK SH2 domain and the active FAK-Src complex is responsible for multiple downstream events including ERK and JNK activation (57). However, it appears that in the context of pneumococcal pIgR engagement, FAK is not critical for the Src PTK-dependent stimulation of mitogen-activated protein and stress-activated protein (SAP) kinase cascades.
Clearly, our investigation points to an involvement of multiple kinase cascades in human pIgR-mediated pneumococcal uptake by host epithelial cells. MAPKs include, in addition to others, the extracellular-regulated kinases 1 and 2 (ERK1 (p44 MAPK) and ERK2 (p42 MAPK)), whereas SAP kinases include c-Jun amino-terminal kinases (JNK1/2). MAP and SAP kinases have been demonstrated to be involved in host cell invasion and cytokine release induced by different pathogenic bacteria. For example, activation of several MAPKs were found in response to epithelial cell infection with S. aureus, C. pneumoniae, L. monocytogenes, enteropathogenic E. coli, and S. enterica serovar Typhimurium (41–45). In particular JNK activation was described to be associated with the invasion of Porphyromonas gingivalis in gingival cells, N. gonorrhoeae in epithelial cells, and N. meningitidis infection of human brain-derived microvascular endothelial cells (46–48). S. pneumoniae was demonstrated to induce JNK, MAPK, and activator protein-1-dependent IL-8 release by lung epithelial BEAS-2B cells (58). In addition, nonencapsulated pneumococci R6x induced p38 MAPK and JNK-mediated caspase-dependent apoptosis in human endothelial cells (59). However, these host cell responses were independent of PspC and pIgR, because the used cell lines BEAS-2B, HEK-293, and human endothelial cells line from lung microvasculature and umbilical vein do not express pIgR.
Our inhibition data suggest that MAP kinase kinase (MEK1) and JNK play a significant role in pIgR-mediated pneumococcal uptake by respiratory epithelial cells. A key downstream effector of MEK1 is the serine-threonine kinase ERK1/2, which is phosphorylated in response to MEK activation and in turn regulates the activity of a number of other target molecules including kinases, transcription factors including activator protein-1 component c-Jun (for JNK, p38 MAPK, or ERK pathways depending on the stimulus), and other regulatory molecules (60). Infection experiments demonstrated phosphorylation of ERK1/2, JNK isoforms p54 and p46, and transcription factor c-Jun. Activation of ERK1/2 was dependent on MEK activity as demonstrated in inhibition assays with PD98059 followed by immunoblot analysis. Moreover, phosphorylation of ERK by extracellular stimuli directs ERK into the nucleus, where it activates transiently the expression of specific genes (61). Consistent with these findings, stimulation of epithelial cells by pneumococci also result in translocation of activated ERK in the nucleus. Taken together the results revealed the important role played by ERK1/2 and JNK MAPK during pneumococcal infection of hpIgR expressing cell lines. Signal transduction cascades are highly complex and tightly regulated pathways in which activation of one signaling molecule leads to downstream activation or deactivation of various effector proteins or stimulation of other signaling pathways. Activation of ERK1/2 in pIgR expressing host cells after infections with pneumococci was significantly reduced in the presence of PP2, which inhibits specifically Src family PTKs. Notably, inhibition of ERK1/2 activation during pneumococcal infections was demonstrated when a dominant-negative, kinase-inactive version of Src (Src K297M) was overexpressed in human pIgR expressing cells. The result indicated that Src kinase facilitates ERK1/2 activation during pIgR-mediated pneumococcal infection. The Src kinase-initiated signal transduction and activation of ERK1/2 was independent of PI3K and Cdc42 activity as shown in inhibition experiments. These data and the recently described requirement of PI3K and Cdc42 (20) demonstrate the complexity of induced signal transduction cascades upon pneumococcal adherence and suggest that at least two independent signal transduction cascades are involved in pneumococcal internalization by human pIgR-expressing host cells. In addition, infection experiments performed in the presence of individual inhibitors or with a combination of inhibitors suggested interplay between the Src kinase and JNK pathway. The results revealed that Src kinase is activated upstream of the JNK pathway in the signal transduction cascade induced during hpIgR-mediated pneumococcal infection. In conclusion, our data demonstrate that pneumococcal invasion via PspC-hpIgR is a highly complex process and involves the concerted role of protein-tyrosine kinases as depicted in the schematic model in Fig. 7. We have clearly shown the key role of the Src kinase and MAPK pathways during this specific uptake mechanism employed by pneumococci. However, further investigation is required to elucidate the cross-talk between various host cell signaling pathways and the process of pneumococcal endocytosis via pIgR.
Acknowledgments
We are grateful to Petra Hildebrandt (Interfaculty Institute for Genetics and Functional Genomics, Dept. Functional Genomics, University of Greifswald, Germany) for FACS of transduced epithelial cells and Claudia Weber (Dept. Genetics of Microorganisms, University of Greifswald, Germany) for critical discussions.
This work was supported in part by Deutsche Forschungsgemeinschaft Grants DFG Ha 3125/2-1 and Ha 3125/4-1 and European Union Grant FP7 CAREPNEUMO EU-CP223111.
- PspC
- pneumococcal surface protein C
- pIgR
- polymeric immunoglobulin receptor
- SC
- secretory component
- FAK
- focal adhesion kinase
- JNK
- c-Jun N-terminal kinase
- PTKs
- protein-tyrosine kinases
- DMSO
- dimethyl sulfoxide
- MDCK
- Madin-Darby canine kidney cells
- shFAK
- short hairpin FAK.
REFERENCES
- 1.Cartwright K. (2002) Eur. J. Pediatr. 161, 188–195 [DOI] [PubMed] [Google Scholar]
- 2.Bergmann S., Hammerschmidt S. (2006) Microbiology 152, 295–303 [DOI] [PubMed] [Google Scholar]
- 3.Kadioglu A., Weiser J. N., Paton J. C., Andrew P. W. (2008) Nat. Rev. Microbiol. 6, 288–301 [DOI] [PubMed] [Google Scholar]
- 4.Hammerschmidt S. (2006) Curr. Opin. Microbiol. 9, 12–20 [DOI] [PubMed] [Google Scholar]
- 5.Orihuela C. J., Radin J. N., Sublett J. E., Gao G., Kaushal D., Tuomanen E. I. (2004) Infect. Immun. 72, 5582–5596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergmann S., Lang A., Rohde M., Agarwal V., Rennemeier C., Grashoff C., Preissner K. T., Hammerschmidt S. (2009) J. Cell Sci. 122, 256–267 [DOI] [PubMed] [Google Scholar]
- 7.Hammerschmidt S., Agarwal V., Kunert A., Haelbich S., Skerka C., Zipfel P. F. (2007) J. Immunol. 178, 5848–5858 [DOI] [PubMed] [Google Scholar]
- 8.Rennemeier C., Hammerschmidt S., Niemann S., Inamura S., Zähringer U., Kehrel B. E. (2007) FASEB J. 21, 3118–3132 [DOI] [PubMed] [Google Scholar]
- 9.Bagnoli F., Moschioni M., Donati C., Dimitrovska V., Ferlenghi I., Facciotti C., Muzzi A., Giusti F., Emolo C., Sinisi A., Hilleringmann M., Pansegrau W., Censini S., Rappuoli R., Covacci A., Masignani V., Barocchi M. A. (2008) J. Bacteriol. 190, 5480–5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammerschmidt S., Tillig M. P., Wolff S., Vaerman J. P., Chhatwal G. S. (2000) Mol. Microbiol. 36, 726–736 [DOI] [PubMed] [Google Scholar]
- 11.Elm C., Braathen R., Bergmann S., Frank R., Vaerman J. P., Kaetzel C. S., Chhatwal G. S., Johansen F. E., Hammerschmidt S. (2004) J. Biol. Chem. 279, 6296–6304 [DOI] [PubMed] [Google Scholar]
- 12.Lu L., Lamm M. E., Li H., Corthesy B., Zhang J. R. (2003) J. Biol. Chem. 278, 48178–48187 [DOI] [PubMed] [Google Scholar]
- 13.Johansen F. E., Pekna M., Norderhaug I. N., Haneberg B., Hietala M. A., Krajci P., Betsholtz C., Brandtzaeg P. (1999) J. Exp. Med. 190, 915–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mostov K. E. (1994) Annu. Rev. Immunol. 12, 63–84 [DOI] [PubMed] [Google Scholar]
- 15.Shimada S., Kawaguchi-Miyashita M., Kushiro A., Sato T., Nanno M., Sako T., Matsuoka Y., Sudo K., Tagawa Y., Iwakura Y., Ohwaki M. (1999) J. Immunol. 163, 5367–5373 [PubMed] [Google Scholar]
- 16.Luton F., Mostov K. E. (1999) Mol. Biol. Cell 10, 1409–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mostov K. E., Friedlander M., Blobel G. (1984) Nature 308, 37–43 [DOI] [PubMed] [Google Scholar]
- 18.Piskurich J. F., Blanchard M. H., Youngman K. R., France J. A., Kaetzel C. S. (1995) J. Immunol. 154, 1735–1747 [PubMed] [Google Scholar]
- 19.Zhang J. R., Mostov K. E., Lamm M. E., Nanno M., Shimida S., Ohwaki M., Tuomanen E. (2000) Cell 102, 827–837 [DOI] [PubMed] [Google Scholar]
- 20.Agarwal V., Hammerschmidt S. (2009) J. Biol. Chem. 284, 19427–19436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luton F., Cardone M. H., Zhang M., Mostov K. E. (1998) Mol. Biol. Cell 9, 1787–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nieto C., Espinosa M. (2003) Plasmid 49, 281–285 [DOI] [PubMed] [Google Scholar]
- 23.Tamer C. M., Lamm M. E., Robinson J. K., Piskurich J. F., Kaetzel C. S. (1995) J. Immunol. 155, 707–714 [PubMed] [Google Scholar]
- 24.Pelish H. E., Peterson J. R., Salvarezza S. B., Rodriguez-Boulan E., Chen J. L., Stamnes M., Macia E., Feng Y., Shair M. D., Kirchhausen T. (2006) Nat. Chem. Biol. 2, 39–46 [DOI] [PubMed] [Google Scholar]
- 25.Agerer F., Michel A., Ohlsen K., Hauck C. R. (2003) J. Biol. Chem. 278, 42524–42531 [DOI] [PubMed] [Google Scholar]
- 26.Sieg D. J., Ilić D., Jones K. C., Damsky C. H., Hunter T., Schlaepfer D. D. (1998) EMBO J. 17, 5933–5947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naldini L., Blömer U., Gallay P., Ory D., Mulligan R., Gage F. H., Verma I. M., Trono D. (1996) Science 272, 263–267 [DOI] [PubMed] [Google Scholar]
- 28.Rubinson D. A., Dillon C. P., Kwiatkowski A. V., Sievers C., Yang L., Kopinja J., Rooney D. L., Zhang M., Ihrig M. M., McManus M. T., Gertler F. B., Scott M. L., Van Parijs L. (2003) Nat. Genet. 33, 401–406 [DOI] [PubMed] [Google Scholar]
- 29.Muenzner P., Bachmann V., Hentschel J., Zimmermann W., Hauck C. R. (2010) Science 329, 1197–1201 [DOI] [PubMed] [Google Scholar]
- 30.Hoffmann I., Eugène E., Nassif X., Couraud P. O., Bourdoulous S. (2001) J. Cell Biol. 155, 133–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwok T., Zabler D., Urman S., Rohde M., Hartig R., Wessler S., Misselwitz R., Berger J., Sewald N., König W., Backert S. (2007) Nature 449, 862–866 [DOI] [PubMed] [Google Scholar]
- 32.Maza P. K., Straus A. H., Toledo M. S., Takahashi H. K., Suzuki E. (2008) Microbes Infect. 10, 540–547 [DOI] [PubMed] [Google Scholar]
- 33.Sousa S., Cabanes D., Bougnères L., Lecuit M., Sansonetti P., Tran-Van-Nhieu G., Cossart P. (2007) Cell. Microbiol. 9, 2629–2643 [DOI] [PubMed] [Google Scholar]
- 34.Imamoto A., Soriano P. (1993) Cell 73, 1117–1124 [DOI] [PubMed] [Google Scholar]
- 35.Agerer F., Lux S., Michel A., Rohde M., Ohlsen K., Hauck C. R. (2005) J. Cell Sci. 118, 2189–2200 [DOI] [PubMed] [Google Scholar]
- 36.Humtsoe J. O., Kim J. K., Xu Y., Keene D. R., Höök M., Lukomski S., Wary K. K. (2005) J. Biol. Chem. 280, 13848–13857 [DOI] [PubMed] [Google Scholar]
- 37.Ozeri V., Rosenshine I., Ben-Ze'Ev A., Bokoch G. M., Jou T. S., Hanski E. (2001) Mol. Microbiol. 41, 561–573 [DOI] [PubMed] [Google Scholar]
- 38.Reddy M. A., Prasadarao N. V., Wass C. A., Kim K. S. (2000) J. Biol. Chem. 275, 36769–36774 [DOI] [PubMed] [Google Scholar]
- 39.Reddy M. A., Wass C. A., Kim K. S., Schlaepfer D. D., Prasadarao N. V. (2000) Infect. Immun. 68, 6423–6430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watarai M., Funato S., Sasakawa C. (1996) J. Exp. Med. 183, 991–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coombes B. K., Mahony J. B. (2002) Cell. Microbiol. 4, 447–460 [DOI] [PubMed] [Google Scholar]
- 42.Czerucka D., Dahan S., Mograbi B., Rossi B., Rampal P. (2001) Infect. Immun. 69, 1298–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ellington J. K., Elhofy A., Bost K. L., Hudson M. C. (2001) Infect. Immun. 69, 5235–5242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hobbie S., Chen L. M., Davis R. J., Galán J. E. (1997) J. Immunol. 159, 5550–5559 [PubMed] [Google Scholar]
- 45.Tang P., Sutherland C. L., Gold M. R., Finlay B. B. (1998) Infect. Immun. 66, 1106–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Naumann M., Rudel T., Wieland B., Bartsch C., Meyer T. F. (1998) J. Exp. Med. 188, 1277–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sokolova O., Heppel N., Jägerhuber R., Kim K. S., Frosch M., Eigenthaler M., Schubert-Unkmeir A. (2004) Cell. Microbiol. 6, 1153–1166 [DOI] [PubMed] [Google Scholar]
- 48.Watanabe K., Yilmaz O., Nakhjiri S. F., Belton C. M., Lamont R. J. (2001) Infect. Immun. 69, 6731–6737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dickens M., Rogers J. S., Cavanagh J., Raitano A., Xia Z., Halpern J. R., Greenberg M. E., Sawyers C. L., Davis R. J. (1997) Science 277, 693–696 [DOI] [PubMed] [Google Scholar]
- 50.Schlaepfer D. D., Hauck C. R., Sieg D. J. (1999) Prog. Biophys. Mol. Biol. 71, 435–478 [DOI] [PubMed] [Google Scholar]
- 51.Schlaepfer D. D., Hunter T. (1997) J. Biol. Chem. 272, 13189–13195 [DOI] [PubMed] [Google Scholar]
- 52.Bogoyevitch M. A., Kobe B. (2006) Microbiol. Mol. Biol. Rev. 70, 1061–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bhavsar A. P., Guttman J. A., Finlay B. B. (2007) Nature 449, 827–834 [DOI] [PubMed] [Google Scholar]
- 54.Nerlich A., Rohde M., Talay S. R., Genth H., Just I., Chhatwal G. S. (2009) J. Biol. Chem. 284, 20319–20328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rovedo M., Longnecker R. (2008) J. Virol. 82, 8520–8528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strasner A. B., Natarajan M., Doman T., Key D., August A., Henderson A. J. (2008) J. Immunol. 181, 3706–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mitra S. K., Hanson D. A., Schlaepfer D. D. (2005) Nat. Rev. Mol. Cell Biol. 6, 56–68 [DOI] [PubMed] [Google Scholar]
- 58.Schmeck B., Moog K., Zahlten J., van Laak V., N′Guessan P. D., Opitz B., Rosseau S., Suttorp N., Hippenstiel S. (2006) Respir. Res. 7, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.N′Guessan P. D., Schmeck B., Ayim A., Hocke A. C., Brell B., Hammerschmidt S., Rosseau S., Suttorp N., Hippenstiel S. (2005) Thromb. Haemost. 94, 295–303 [DOI] [PubMed] [Google Scholar]
- 60.Krishna M., Narang H. (2008) Cell. Mol. Life Sci. 65, 3525–3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Costa M., Marchi M., Cardarelli F., Roy A., Beltram F., Maffei L., Ratto G. M. (2006) J. Cell Sci. 119, 4952–4963 [DOI] [PubMed] [Google Scholar]
- 62.Morton S., Davis R. J., McLaren A., Cohen P. (2003) EMBO J. 22, 3876–3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pulverer B. J., Kyriakis J. M., Avruch J., Nikolakaki E., Woodgett J. R. (1991) Nature 353, 670–674 [DOI] [PubMed] [Google Scholar]