Abstract
Defective or imbalanced expression of spliceosomal factors has been linked to human disease; however, how a defective spliceosome affects intron-containing gene transcripts in human cells is largely unknown. DEAH-box protein DHX16 is a human orthologue of Saccharomyces cerevisiae spliceosomal protein Prp2, an RNA-dependent ATPase that activates the spliceosome before the first catalytic step of splicing. Yeast prp2 mutants accumulate unspliced RNAs from the vast majority of intron-containing genes. Here we used a genomic tiling microarray to screen transcripts from four chromosomes in human cells expressing a dominant negative DHX16 mutant and identified a number of gene transcripts that retained their introns. The mutant protein also affected gene transcripts that are sensitive to pladienolide, an SF3b inhibitor. The unspliced RNAs were retained in the nucleus, and block of nonsense-mediated decay did not affect their accumulation. Thus, a perturbation of human PRP2/DHX16 results in accumulation of unspliced transcripts, similar to the outcome in yeast prp2 mutants. The results further suggest that mutant DHX16/hPRP2 causes a defective spliceosome to retain unspliced gene transcripts in the nuclei of human cells.
Keywords: ATPases, Nuclear RNA, RNA Helicase, RNA Splicing, Spliceosome, DExD/H-box Protein, Genomic Tiling Microarray, Nonsense-mediated Decay, Unspliced RNA
Introduction
Split genes are common features of eukaryotic genomes, and their transcripts are only functional after the introns are removed by RNA splicing (1, 2). Intron-containing pre-mRNA and other mRNA-like RNA polymerase II transcripts are spliced by the spliceosome, which is composed of five small nuclear RNAs and ∼140 proteins (3–5). Although some splicing factors are dispensable, mutations in genes encoding spliceosomal proteins or small nuclear RNAs abolish splicing and lead to growth defects or cell death in yeasts (6–8). Defective or imbalanced expression of spliceosomal factors has also been linked to human disease (9). However, how a defective spliceosome affects the expression of intron-containing genes in human cells is largely unknown.
DExD/H-box RNA helicases play an essential role in pre-mRNA splicing and function to ensure that the correct conformational rearrangements occur during the spliceosome cycle (10–12). Human DHX16 (DEAH-box protein 16) was initially identified as DBP2 (13) and resides on chromosome 6p21.3 in the major histocompatibility complex region, which is linked to a number of malignant and autoimmune diseases. DHX16 is homologous to Schizosaccharomyces pombe Cdc28/Prp8, a protein involved in cell cycle progression and pre-mRNA splicing (14), and DHX16 partially rescues a prp8 mutant (13). The DHX16 protein is found in human spliceosomes assembled in nuclear extracts (15–18). We have recently shown that DHX16 contributes to human pre-mRNA splicing in vitro (19) and shares characteristics with Saccharomyces cerevisiae Prp2 (pre-mRNA processing factor 2), an RNA-dependent ATPase required for the onset of splicing in the assembled spliceosome (20).
Here, we investigated how intron-containing gene transcripts are affected by mutant DHX16 in human cells. By using a human genomic tiling microarray that covers four chromosomes, we identified gene transcripts that showed intron retention in cells expressing DHX16-G724N, which has a dominant negative mutation in the conserved motif VI of the helicase domain of DHX16 (19). We also detected fully unspliced pre-mRNA as well as intron-containing gene transcripts that have been shown to be accumulated by pladienolide, an SF3b inhibitor (21). The intron-containing RNAs were enriched in the nucleus, and their level of accumulation was not altered by inhibiting nonsense-mediated decay. The results suggest that mutant DHX16/hPRP2 causes a defective spliceosome to retain unspliced gene transcripts in the nuclei of human cells.
EXPERIMENTAL PROCEDURES
Plasmids, Cell Lines, and Transfection
Plasmids to express wild type DHX16 and mutant DHX16-G724N have been described (19). HSPi15, HSPi16, PRPi27, PRPi28, and SFRSi2 minigenes were constructed by amplifying the respective portions of HSPH1, PRPF8, or SFRS1 and inserting them into the SalI/SmaI sites of pEGFP-N1. The primers used are listed in supplemental Table 1. All constructs were confirmed by sequencing. A plasmid carrying an shRNA construct targeting UPF1 was a kind gift from Douglas Black (UCLA, Los Angeles, CA). The plasmid carrying an shRNA construct targeting GFP at the ACGACTTCTTCAAGTCCGC sequence was constructed by inserting a double-stranded synthetic DNA carrying the hairpin sequence into pBlsH1. The pBlsH1 plasmid has a modified H1 promoter (GAAGACTAAAAA), which can be recognized by BbsI to facilitate cloning. Cells were cultured in DMEM (Irvine Scientific) and transfected in Lipofectamine 2000 (Invitrogen) as described previously (19). Cells were lysed 72 h after transfection unless otherwise specified.
RNA Isolation, RT-PCR, and Northern Hybridization
Total RNA was isolated using the RNeasy minikit and with DNase treatment (Qiagen), and reverse transcription was carried out with Superscript II/III reverse transcriptase (Invitrogen) and random hexamers (New England Biolabs) as described previously (19). To enrich for poly(A) RNA, total RNA (120 μg) was incubated with conditioned oligo(dT)-cellulose beads (8 mg; Amersham Biosciences) in 240 μl of binding buffer (1 m NaCl, 20 mm Tris-HCl, pH 8.0, 1 mm EDTA, 0.2% Triton X-100) at 4 °C for 30 min. The supernatant (the unbound fraction) was collected after a brief centrifugation, and the beads were washed in 0.3 m NaCl, 20 mm Tris-HCl, pH 8.0. The bound RNA was eluted with 300 μl of water and precipitated with ethanol. The precipitate was resuspended to obtain poly(A)-enriched RNA (the bound fraction). Primers for PCR are listed in supplemental Table 1. PCR products were resolved on agarose or in acrylamide gels and visualized by staining with ethidium bromide or SYBR Green (Invitrogen). For Northern hybridization, total RNA extracted from cells using an RNeasy kit or TRIzol (Invitrogen) or poly(A)-enriched RNA was separated on a 1% agarose gel containing 1× MOPS and 2% formaldehyde. RNA Millennium markers (Ambion) were used as size markers. After electrophoresis, RNA was transferred to Hybond-N+ membrane (GE Healthcare) by capillary overnight in 10× SSC buffer and immobilized onto the membrane by irradiation in a Stratalinker 1800 UV cross-linker (Stratagene). The position of the RNA marker was marked under UV light. Oligonucleotide probes were labeled with [γ-32P]ATP with T4 polynucleotide kinase and purified using a G25 column (GE Healthcare). The membrane was soaked in PerfectHyb Plus hybridization buffer (Sigma) for 30 min and incubated with a 32P-labeled probe overnight at 40–42 °C. The membrane was washed in 2× SSC, 0.1% SDS and then with 0.5× SSC, 0.1% SDS at 40–42 °C before exposure to an x-ray film or PhosphorImager.
Nuclear and Cytoplasmic Fractionation, Protein Isolation, and Western Blotting
Nuclear and cytoplasmic fractionation was performed by incubating 2 × 106 HEK293 cells for 5 min in 100 μl of lysis buffer (10 mm Tris-HCl, pH 7.8, 150 mm NaCl, 0.4% Nonidet P-40) on ice. The mixture was then centrifuged at 1,000 × g for 3 min to separate the nuclei from the cytoplasm. The nuclei were washed once in lysis buffer and recentrifuged before RNA or protein isolation. Total protein lysates were isolated in RIPA buffer, and typically 20–30 μg of protein from the lysate was used in Western immunoblot analysis as described previously (19). Antibodies used were affinity-purified anti-DHX16 antibody (19), anti-GAPDH (Ambion), and anti-UPF1 (Bethyl Laboratory).
Microarray Hybridization and Data Analysis
The quality of the RNA was checked on an Agilent Bioanalyzer. For the GeneChip Human Tiling 2.0R G Array (Affymetrix), total RNA (7 μg) was used, and hybridization and data collection were performed according to Affymetrix protocols at the Functional Genomics Core of City of Hope. Two independent experiments were performed for both DHX16-WT-transfected cells and DHX16-G724N-transfected cells. Analysis of the data were done with Tiling Analysis Software, version 1.1.02 (Affymetrix). Files were normalized together; signal intensity values were converted to log 2 scale, and sliding window analysis was performed with the following parameters: bandwidth = 50, MaxGap = 100, and MinRun = 100. Results were visualized by the Integrated Genome Browser (Affymetrix). Intervals that were enriched in DHX16-G724N at a threshold of 2.3-fold were determined. Two approaches were used to estimate the false discovery rate of the identified intervals, a randomization-based approach and a parametric approach.2 Briefly, the first approach was to randomize the log 2 ratios between dominant negative and wild type along the genomic locations and rerun the analysis using the same set of parameters. Because the data were randomized, any identified intervals will be false positives. The randomization was repeated 20 times. Using the same set of parameters, there was no interval identified in randomized data sets, indicating that the intervals that we identified could not have been identified by chance. The parametric approach was developed based on binomial distribution. The number of false intervals was calculated by the summation of the probability of identifying intervals with longer length and smaller gaps times the total number of probes on the array. The estimated number of false intervals using this parametric approach was less than 1 using our interval selection parameters. We also analyzed the two wild type/G724N pairs separately using the same parameters and threshold. The number of the common genes between the two replicates is 86, and the p value was less than 2.2 × 10−16 using Fisher's exact test. Nearly 80% of the genes identified using the mean values were found in this list.
RESULTS
Detection of Intron-containing Transcripts from Endogenous Genes Using Genomic Tiling Microarray
We have reported that expression of several DHX16 helicase domain mutants, including the G724N mutant, blocks splicing of minigene transcripts (19). To investigate whether transcripts from endogenous genes would be affected by DHX16 mutants, we used an Affymetrix whole genome tiling array to detect spliced and unspliced transcripts from 3,561 RefSeq genes on human chromosomes 10, 13, 14, and 17 (NCBI build 36.2). This microarray had 25-nt probes tiled at a 35-bp distance on the average (i.e. a 10-bp gap between two adjacent oligonucleotide probes), intended to cover all non-repetitive genomic sequences.
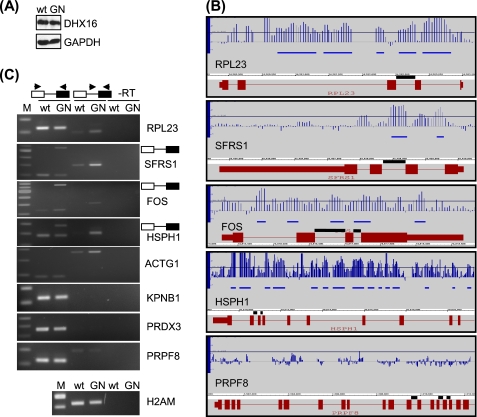
We transfected HEK293 cells with plasmid carrying either wild type DHX16 or mutant DHX16-G724N. An immunoblot showed that cells transfected with the wild type or the mutant G724N plasmid expressed the corresponding DHX16 at a similar level (Fig. 1A). We isolated RNA from transfected cells and analyzed it using the genomic tiling microarray. The array data from two biological replicas were analyzed by a sliding window approach using the tiling analysis software. The parameters were set to detect genomic regions or intervals longer than 100 bp for which signal was increased in the mutant as compared with the wild type by 2.3-fold. At this chosen threshold, the computed false discovery rate was <1 (see “Experimental Procedures”), thus giving a conservative but reliable estimate of the differences between mutant and wild type DHX16 samples.
FIGURE 1.
Intron-containing transcripts are enriched in cells expressing the DHX16-G724N mutant. HEK293 cells were transfected with plasmid carrying wild type DHX16 (wt) or mutant G724N (GN); protein and RNA were isolated and analyzed by immunoblot (A), genomic tiling microarray (B), or RT-PCR (C). A, protein lysates were probed with anti-DHX16 and anti-GAPDH antibodies. B, RNA was analyzed by genomic tiling array. Hybridization signals of mutant over wild type were calculated and displayed on the Integrated Genome Browser. Five genes are depicted: RPL23, SFRS1, FOS, HSPH1, and PRPF8. The gene structure is shown in red; boxes represent exons, and lines represent introns. FOS is transcribed from left to right, whereas the other four genes are transcribed from right to left. For HSPH1 and PRPF8, only the 3′ portion of the gene is shown. The vertical blue lines in each panel represent the ratios of signal intensity in log 2 scale between DHX16-G724N and wild type DHX16; intervals up-regulated in cells expressing DHX16-G724N over the threshold are marked with horizontal blue bars below the vertical ratio lines. The threshold is set at 2.3-fold (1.2 in log 2 scale) and is represented by a line across the graph. The introns that were assayed by RT-PCR are indicated with black lines directly above the gene structure. C, RNA was analyzed by RT-PCR. Exonic and intronic primers used in the PCRs are depicted above the gels, with arrowheads denoting primers and boxes for exons and lines for introns. Reactions performed without reverse transcriptase (−RT) using intronic/exonic primer pairs are shown in the last two lanes of each panel. PCR bands corresponding to the intron-containing RNA produced with the flanking exonic primers are indicated to the right of the panels. H2AM contains no intron. DNA size markers are shown in the first lane (M) of each panel.
The detection of RNA by tiling array could be visualized by the Integrated Genome Browser. In Fig. 1B, we plotted the signal intensity ratio between mutant- and wild type-expressing cells for individual oligonucleotide probes (blue vertical lines) as well as the sequence regions or intervals that exhibited a difference in signal above the threshold (blue horizontal lines) for five genes. Integrated Genome Browser presentation of 16 additional genes is shown in supplemental Fig. 1. In RPL23 (transcription is from right to left), five up-regulated intervals were detected in the first three introns, and oligonucleotide probes in the last intron also showed greater intensity in RNA samples from DHX16-G742N-expressing cells; on the contrary, signals in the exons showed little difference between the mutant and wild type, suggesting that the total amounts of RPL23 transcripts remained relatively unchanged (Fig. 1B, top panel). Introns in SFRS1, FOS, and HSPH1 also showed greater intensity in samples from DHX16-G724N-expressing cells (Fig. 1B, middle panels). No significant changes in expression were detected for exons of SFRS1 or HSPH1, whereas significant increases were detected in exons of FOS. It appeared that the total amount of transcript for FOS was greater in mutant DHX16-expressing cells (see below). We did not detect up-regulated intervals or a consistent increase in signal intensity within the introns of PRPF8 in DHX16-G724N-expressing cells (Fig. 1B, bottom panel). Thus, the genomic tiling array detected an increase of intron-containing transcripts from some endogenous genes in mutant DHX16-expressing cells.
Of the four chromosomes covered by the microarray, we detected 193 up-regulated intervals in the DHX16-G724N-expressing cells; 179 mapped to introns of known or predicted genes, 11 mapped to exons of known genes, and three were intergenic (supplemental Table 2). The detected 179 intron intervals mapped to 56 genes, and 21 of these genes showed increased signals in the majority of their introns in DHX16-G724N-expressing cells (Table 1). The 11 exons mapped to five genes, CSTF2T, PEO1, FOS, FLJ10587, and SMCR8 (supplemental Table 2), which probably resulted from an increase in transcription or decrease in RNA turnover of these genes in the DHX16-G724N-expressing cells. The three intergenic intervals were not investigated. The observation that nearly 93% of the intervals with elevated signals reside in introns is consistent with the notion that the major defect brought about by mutant DHX16 is in pre-mRNA splicing.
TABLE 1.
List of genes on chromosomes 10, 13, 14, and 17 that show intron intervals up-regulated at a threshold of 2.3-fold in DHX16-G724N-transfected HEK293 cells
| Gene | Gene description | No. of enriched intervals | Introns (total/detected) |
|---|---|---|---|
| ACTG1 | γ-Actin | 1 | 5/1 |
| AHSA1 | Activator of 90-kDa heat shock protein ATPase homolog 1 | 2 | 8/1 |
| APEX1 | AP endonuclease 1 | 2 | 4/2 |
| BAG3 | BCL2-associated athanogene 3 | 14 | 3/3 |
| BAG5 | BCL2-associated athanogene 5 | 1 | 2/1 |
| BC100293 | snoRNA-containing non-coding gene | 5 | 3/3 |
| BMS1L | BMS1-like, ribosome assembly protein | 1 | 22/1 |
| C1QBP | Complement component 1 Q subcomponent-binding protein | 5 | 5/3 |
| CCNB1IP1 | Cyclin-B1-interacting protein 1, E3 ubiquitin-protein ligase | 4 | 8/3 |
| CLTC | Clathrin heavy chain 1 | 1 | 31/1 |
| DDX21 | DEAD box protein 21, nucleolar RNA helicase 2 | 1 | 14/1 |
| DDX5 | RNA helicase p68 | 3 | 12/2 |
| DIP2C | Disco-interacting protein 2 homolog C | 1 | 36/1 |
| DLC2 | Dynein light chain 2 | 1 | 2/1 |
| EIF3S10 | Eukaryotic translation initiation factor 3 subunit 10 | 1 | 21/1 |
| EIF4A1 | Eukaryotic translation initiation factor 4A | 6 | 10/6 |
| EIF5 | Eukaryotic translation initiation factor 5 | 3 | 12/3 |
| EIF5A | Eukaryotic translation initiation factor 5A | 5 | 5/3 |
| FOS | Proto-oncogene protein c-Fos | 5 | 3/3 |
| GABARAP | GABA(A) receptor-associated protein | 2 | 3/2 |
| GEMIN4 | Gemin 4 | 1 | 1 |
| GTPBP4 | G protein-binding protein CRFG | 1 | 16/1 |
| HNRPF | Heterogeneous nuclear ribonucleoprotein F | 3 | 3/1 |
| HSPA14 | Heat shock protein hsp70-related protein | 1 | 4/1 |
| HSPCA | Heat shock protein HSP 90-α | 7 | 10/7 |
| HSPH1 | Heat-shock protein 105 kDa | 26 | 17/12 |
| MIS12 | MIS12 homologue | 1 | 2/1 |
| IMP-1 | Insulin-like growth factor 2 mRNA-binding protein | 1 | 14/1 |
| NET1 | Neuroepithelial cell-transforming gene 1 protein | 1 | 9/1 |
| PEO1 | Progressive external ophthalmoplegia 1 protein | 6 | 4/4 |
| POLR1D | DNA-directed RNA polymerase I subunit D | 1 | 1 |
| PRKAR1A | cAMP-dependent protein kinase type I-α regulatory subunit | 1 | 11/1 |
| PRO1855 | Leucine-rich repeat-containing protein 59 | 1 | 6/1 |
| PSMD3 | Proteasome 26 S non-ATPase subunit 3 | 2 | 11/1 |
| PTDSR | Jumonji domain-containing 6 | 2 | 5/2 |
| RAD51C | RAD51 homolog C isoform 2 | 1 | 1 |
| RPL19 | Ribosomal protein L19 | 5 | 5/5 |
| RPL21 | Ribosomal protein L21 | 4 | 5/2 |
| RPL23 | Ribosomal protein L23 | 5 | 4/3 |
| RPL23A | Ribosomal protein L23A | 7 | 4/4 |
| RPL26 | Ribosomal protein L26 | 6 | 3/3 |
| RPL27 | Ribosomal protein L27 | 1 | 4/1 |
| RPL38 | Ribosomal protein L38 | 6 | 4/3 |
| RPS24 | Ribosomal protein S24 | 3 | 5/2 |
| SAP18 | Sin3-associated polypeptide, 18 kDa | 1 | 3/1 |
| SFRS1 | Pre-mRNA-splicing factor SF2/ASF | 2 | 3/2 |
| SMCR8 | Smith-Magenis syndrome chromosome region | 3 | 1 |
| SMNDC1 | Survival of motor neuron-related splicing factor 30 | 1 | 5/1 |
| SUPV3L1 | Suppressor of Var1, 3-like 1 | 1 | 14/1 |
| TEX14 | Testis expressed sequence 14 | 1 | 32/1 |
| THOC4 | THO complex subunit 4; transcriptional coactivator Aly/REF | 3 | 5/2 |
| TPT1 | Tumor protein, translationally controlled 1 | 2 | 5/2 |
| TSR1 | Homologue of yeast TSR1 | 8 | 14/7 |
| UBB | Ubiquitin B precursor | 1 | 1 |
| YWHAE | 14-3-3 protein ϵ isoform transcript variant 1 | 2 | 5/2 |
| ZNF22 | Zinc finger protein 22 | 2 | 1 |
Because there were only two replicates, tiling analysis software analysis using the p value was not possible. To further examine the significance of the results, we used Fisher's exact test, which is a statistical significance test used in the analysis of contingency tables where sample sizes are small. We used tiling analysis software to compare signals in each pair of mutant versus wild type samples and obtained two lists of genes having up-regulated intervals (543 intervals in 255 genes and 861 intervals in 263 genes, respectively, and 272 intervals in 86 genes were overlapped). Fisher's exact test yielded a p value of <2.2 × 10−16, indicating that the results from these two experiments overlapped significantly. The number of overlapping genes in this analysis is 86 (supplemental Table 3), which is more than the 56 genes identified when using the average values for the comparison (Table 1) (45 genes were common on both lists). The difference is probably caused by the fact that one analysis used the average value to compare the mutant and the wild type, and the other analysis compared the mutant and the wild type in each pair and then looked for a denominator.
To verify the microarray results, we performed RT-PCR to detect spliced and unspliced RNA for five genes that showed increases in intron signals (RPL23, SFRS1, FOS, HSPH1, and ACTG1) as well as three genes that did not show an increase (KPNB1, PRDX3, and PRPF8) in the DHX16-G724N-expressing cells (Fig. 1C). We also included H2AM, an intronless histone 2A gene, in the analysis. We designed primers for two adjacent exons in the respective transcripts to detect the spliced RNAs; these primers could also detect unspliced RNAs when the intron was relatively short. We also performed separate RT-PCRs with intronic and exonic primer pairs to detect specifically the transcripts that contained introns. Fig. 1C showed that RNA isolated from DXH16-G724N-transfected cells exhibited higher levels of unspliced RPL23, SFRS1, FOS, HSPH1, and ACTG1 transcripts, as detected by the intron/exon primer pair (Fig. 1C, middle lanes). For SFRS1, FOS, and HSPH1, the respective unspliced RNA was also detected by the exonic primer pair (Fig. 1C, left lanes). Intron-containing transcripts were hardly detected for KPNB1 and PRDX3, whereas unspliced PRPF8 RNA was detected at similar levels in both wild type- and DHX16-G724N-expressing cells (Fig. 1C, middle lanes). Thus, the RT-PCR result was in agreement with the microarray data, supporting the notion that unspliced transcripts of endogenous genes were accumulated in the mutant-expressing cells.
To investigate when the unspliced RNA can be detected after the expression of the mutant protein, we isolated total RNA after 24, 48, and 72 h post-transfection (supplemental Fig. 2). RT-PCR analysis showed that the unspliced RNA from HSPH1 was detected as early as 24 h, and the level of accumulation was consistent through 72 h, whereas the amount of unspliced RNA from PRPF8 was similar between cells expressing wild type or mutant DHX16 and remained unchanged throughout the course of the experiment (supplemental Fig. 2). Thus, the splicing inhibition brought about by the mutant was quite rapid and sustainable.
DHX16-G724N Inhibited Splicing of Endogenous Transcripts from Pladienolide-sensitive Genes as Well as Minigene Transcripts Containing PRPF8 Introns
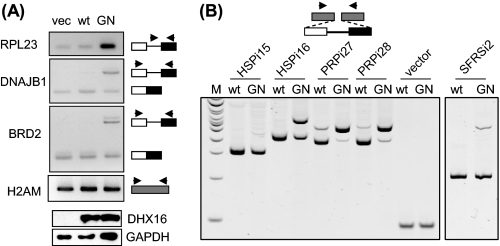
Unspliced or partially spliced transcripts from DNAJB1 (on chromosome 19), BRD2 (on chromosome 6), and RIOK3 (on chromosome 18) have been shown to accumulate in cells treated with pladienolide, a small molecule that inhibits another spliceosomal protein, SF3b (21). To test whether DHX16-G724N would also affect pladienolide-sensitive transcripts, we assayed for intron retention in transcripts from DNAJB1 and BRD2 in RNA samples isolated from wild type- and mutant-transfected cells (Fig. 2A). Increases in intron-containing RNA from both DNAJB1 and BRD2 were observed in mutant samples, similar to the increase in RPL23. Thus, the accumulation of unspliced or intron-containing RNA in G724N-expressing cells is likely to reflect a general inhibition of the spliceosome by the mutant protein.
FIGURE 2.
DHX16-G724N causes intron retention in transcripts from pladienolide-sensitive endogenous genes and from PRPF8 minigenes. A, HEK293T cells were transfected with vector, wild type DHX16, and G724N, respectively. RNA isolated from cells 48 h post-transfection was analyzed by RT-PCR using primers (depicted by arrowheads) specific for RPL23 (intronic and exonic), DNAJB1 (exonic), and BRD2 (exonic). H2AM was analyzed as a control. The bottom two panels show Western blots of protein lysates probed with anti-DHX16 and anti-GAPDH antibodies. B, HEK293 cells were transfected with a minigene along with wild type DHX16 (wt) or DHX16-G724N (GN). The minigene constructs contain inserts carrying HSPH1 intron 15 (HSPi15), HSPH1 intron 16 (HSPi16), PRPF8 intron 27 (PRPi27), PRPF8 intron 28 (PRPi28), or SFRS1 intron 2 (SFRSi2) with flanking exon sequences (white and black boxes for exons and line for intron). Vector is represented by the gray boxes and has no insert. RNA was isolated from cells 48 h post-transfection and analyzed by RT-PCR using a common pair of primers for the flanking vector sequences. The major band detected in RNA from wild type-transfected cells corresponded to the spliced RNA; in addition, a band with a bigger size was detected in RNA from mutant-transfected cells, which corresponded to the unspliced, intron-containing RNA.
We noticed that many genes showing intron retention in our assay encoded ubiquitous proteins, such as ribosomal proteins, translation factors, and heat shock proteins (Table 1). Thus, it appeared that gene expression levels might contribute to the accumulation of intron-containing RNA. To examine genome-wide expression levels, we used the Affymetrix Gene 1.0ST array to estimate the relative transcript level of each gene in wild type DHX16-transfected HEK293 cells (supplemental Fig. 3). The result indicated that most gene transcripts that exhibited intron retention were indeed abundantly expressed in those cells.
It was curious that unspliced transcripts from certain highly expressed genes (e.g. HSPH1) were accumulated, whereas unspliced transcripts from other genes (e.g. PRPF8) were not (Fig. 1 and supplemental Fig. 2). In the cell at steady state, the amount of an unspliced RNA is the function of transcription, splicing, and degradation (see “Discussion”). We suspected that the disparity might be due to difference in their transcription and/or degradation rather than in their intron sequences. To test this, we inserted introns of similar size with their flanking exon sequences from HSPH1 and PRPF8 into a common vector and tested for intron retention in cells expressing the mutant DHX16. The resulting minigenes, HSPi15, HSPi16, PRPi27, and PRPi28, were transfected into HEK293 cells together with either wild type DHX16 or mutant DHX16-G724N. RNA was isolated and subjected to RT-PCR analysis using vector-specific primers that detected both spliced and unspliced minigene transcripts. As shown in Fig. 2B, the amount of spliced transcripts decreased, and unspliced transcripts accumulated in all four minigenes when comparing mutant DHX16 with wild type DHX16-transfected cells. Sequencing of the PCR products indicated that introns were present in the unspliced forms and were correctly excised in the spliced forms. A minigene construct containing an intron from SFRS1 (SFRSi2) showed a similar result (Fig. 2B, right), although to a different extent, these five minigene transcripts all accumulated unspliced transcripts with a lower amount of spliced RNA in mutant-expressing cells (Fig. 2B, lanes labeled GN) regardless of the source of the intron. The change of the pre-mRNA/mRNA ratio is a hallmark of splicing inhibition (22, 23). Thus, this result suggests that the differential accumulation of unspliced gene transcripts is not likely to be determined by the intron sequences but rather by transcription and/or degradation of the unspliced transcripts (see “Discussion”).
The Presence of Fully Unspliced Pre-mRNA in DHX16-G724N-expressing Cells
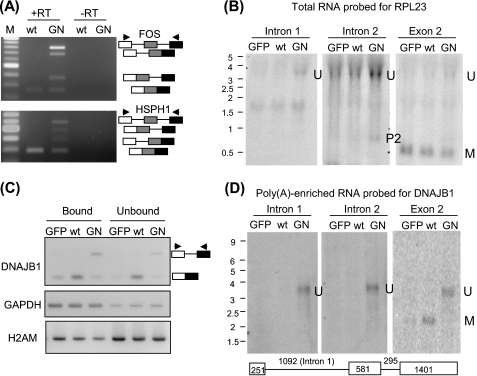
We noticed that gene transcripts showing intron retention had increased signals in most if not all of their introns (Fig. 1B and supplemental Fig. 1). To investigate whether multiple introns detected by microarray could reside in a single unspliced transcript, we performed RT-PCR using primers to exons flanking two introns in two genes, FOS and HSPH1 (Fig. 3A). In both cases, four PCR products were produced, corresponding to RNAs that contained both introns, either intron, and no intron in the RNA sample isolated from DHX16-G724N-expressing cells. Significantly, for both genes, the transcripts containing two introns were enriched compared with the transcripts containing either intron, indicating that these two pairs of adjacent introns were likely to be simultaneously retained in the same transcript.
FIGURE 3.
Transcripts containing multiple introns are present in DHX16-G724N mutant expressing cells. RNA isolated from DHX16-transfected (wt), DHX16-G724N-transfected (GN), or pEGFP-C1-transfected (GFP) cells were analyzed for the presence of transcripts containing multiple introns. Total RNA was used for A and B. Total RNA was incubated with oligo(dT)-cellulose, and the bound and unbound fractions were collected. The oligo(dT)-bound fraction was enriched with poly(A) RNA and was used for D. A, RNA was assayed by RT-PCR using exonic primers flanking two introns and an exon. The two adjacent introns that were assayed in FOS and HSPH1 are marked with black bars in Fig. 1B. PCR products corresponding to RNA containing two introns, one intron, or no intron are depicted on the right. The arrowheads denote primer positions. B, RNA was analyzed by Northern hybridization using oligonucleotides complementary to intron 1, intron 2, or exon 2 of RPL23 (see Fig. 1B). Total RNA (15 μg) was electrophoresed on a 1% agarose formaldehyde gel, transferred, and hybridized with 32P-labeled oligonucleotides. RNA size markers in kilobases are marked on the left. U, fully unspliced RNA; M, fully spliced RNA; P2, a partially spliced RNA containing intron 2. C, RNA that bound to oligo(dT) (Bound) or remained in the supernatant (Unbound) was assayed by RT-PCR for DNAJB1, GAPDH, and H2AM RNA. D, RNA that bound to the oligo(dT) that was enriched for poly(A) RNA (∼0.8 μg) was probed with oligonucleotides complementary to intron 1, intron 2, or exon 2 of DNAJB1. A diagram of the gene structure is depicted below the Northern blot; the lengths of the introns and exons are numbered in bases. U, fully unspliced RNA; M, fully spliced RNA.
We used Northern hybridization to further investigate whether an unspliced pre-mRNA could be retained in mutant cells. We transfected HEK293T cells with wild type DHX16, mutant G724N, or a GFP plasmid and isolated RNA after 48 h of transfection. The RNA was probed with oligonucleotides complementary to intron 1 or intron 2 of RPL23 (Fig. 3B). A ∼3.5-kb RNA with a size similar to the unspliced RPL23 pre-mRNA (labeled U) was detected with both intron probes in samples from cells transfected with mutant DHX16. The intron 2 probe also detected in the mutant sample a ∼0.8-kb RNA with a size similar to that of a partially spliced, intron 2-containing RPL23 RNA (labeled P2). The blot was reprobed with an exon 2 oligonucleotide probe in which spliced SPL23 mRNA was readily detected in all three samples (Fig. 3B, right). Together with the tiling array data, these results indicated that unspliced pre-mRNA was accumulated in DHX16-G724N-expressing cells.
To test whether the unspliced RNA is polyadenylated, we incubated the total RNA with oligo(dT)-cellulose and separated the RNA into the bound fraction and the unbound fraction. RNA from both fractions was assayed by RT-PCR (Fig. 3C). The GAPDH mRNA was enriched in the bound fraction, and the H2AM mRNA lacking poly(A) was enriched in the unbound fraction. The spliced and the intron 2-containing DNAJB1 RNAs were found in the bound fraction. We then analyzed the RNA from the bound fractions by Northern hybridization, probing for DNAJB1 using intron 1 or intron 2 probe (Fig. 3D). A ∼3.6-kb RNA with a size similar to that of the fully unspliced DNAJB1 (labeled U) was detected with both probes in the sample from cells transfected with the mutant DHX16. The blots were reprobed with an exon 2 probe, and the fully spliced DNAJB1 mRNA (labeled M) was detected in samples from cells transfected with GFP or wild type DHX16. Thus, the result indicated that unspliced, polyadenylated pre-mRNAs were accumulated in mutant DHX16-G724N-expressing cells.
Unspliced Transcripts in DHX16-G724N-expressing Cells Are Retained in the Nucleus and Not Affected by Cytoplasmic Nonsense-mediated Decay
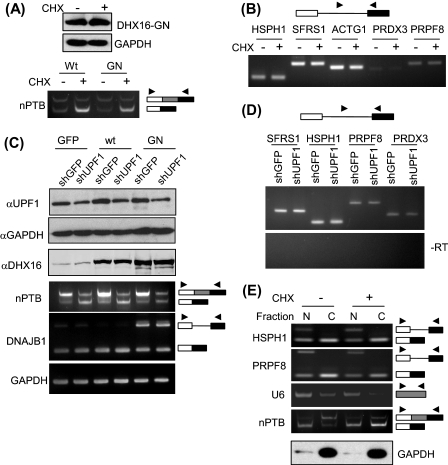
Because unspliced or incompletely processed RNAs are detrimental to the cell, we further investigated the fate of the intron-containing transcripts that accumulated in cells expressing mutant DHX16 by testing whether the unspliced RNA was subjected to nonsense-mediated decay (NMD).3 We treated the transfected cells with cycloheximide to inhibit protein synthesis and NMD (24, 25) and tested whether that treatment would affect the level of unspliced RNA accumulation. To verify that NMD remained active in the transfected cells, we assayed a transcript known to be degraded via NMD, the exon 10-skipping isoform of nPTB. Skipping of nPTB exon 10 results in a transcript containing a premature stop codon, and the amount of this skipped isoform is elevated by blocking NMD (26, 27). First, we showed that the amount of DHX16-G724N in the transfected cells was not affected by 6 h of cycloheximide treatment (Fig. 4A). A pronounced increase in the skipped nPTB isoform upon cycloheximide treatment confirmed that NMD was active in both transfected cells (Fig. 4A). Under these conditions, cycloheximide treatment of DHX16-G724N-transfected cells did not alter the amount of intron-containing RNA from the DHX16-sensitive genes (HSPH1, SFRS1, and ACTG1) or the DHX16-insensitive genes (PRDX3 and PRPF8) (Fig. 4B).
FIGURE 4.
Intron-containing transcripts in cells expressing DHX16-G724N are not subject to NMD and are retained in the nucleus. To inhibit NMD with cycloheximide, HEK293 cells were transfected with wild type DHX16 (Wt) or DHX16-G724N (GN) and then treated with 20 μm of cycloheximide (+CHX) or solvent (−CHX) for 6 h. To inhibit NMD by knocking down UPF1, HEK293T cells were co-transfected for 48 h with a plasmid carrying an shRNA gene (shGFP or shUPF1, at 3 μg for 8 × 105 cells) and a plasmid carrying a DHX16 construct (wild type, mutant G724N, or pEGFP-C1 control (GFP), at 0.3 μg for 8 × 105 cells). A, top, Western blots of protein lysates from G724N-transfected HEK293 cells with or without cycloheximide treatment were probed with anti-DHX16 or anti-GAPDH antibodies. Bottom, RNA from the transfected cells was analyzed by RT-PCR for nPTB exon 10-skipping and exon 10-containing isoforms. B, RT-PCR for the indicated genes was performed with RNA from HEK293 cells transfected with DHX16-G724N and either treated with cycloheximide or not. Intronic/exonic primer pairs were used in all cases, as depicted in the diagram above the gel. C, top three panels, Western blots of protein lysates from HEK293T cells co-transfected with an shRNA-containing plasmid and a DHX16-expressing plasmid were probed with antibodies against UPF1, GAPDH, or DHX16. Bottom three panels, RNA from the co-transfected cells was analyzed by RT-PCR for nPTB exon 10 isoforms, for unspliced DNAJB1 RNA, and for GAPDH mRNA. D, RT-PCR for the indicated genes was performed with RNA from HEK29T cells co-transfected with DHX16-G724N and an shRNA-containing plasmid. Intronic/exonic primer pairs were used in all cases. E, the intron-containing transcripts in cells expressing DHX16-G724N were retained in the nucleus. HEK293 cells transfected with DHX16-G724N and treated with or without cycloheximide were lysed. The lysate was fractionated into two fractions: nuclear (N) and cytoplasmic (C). RNA and protein were isolated from each fraction. Top four panels, RNA was analyzed by RT-PCR for HSPH1, PRPF8, U6, and nPTB RNAs using exonic primer pairs shown as arrowheads; the unspliced and spliced RNAs are indicated (box, exon; line, intron). Bottom panel, Western blots of nuclear and cytoplasmic protein fractions were probed with anti-GAPDH antibody.
We also used another strategy to inhibit NMD by knocking down the expression of UPF1, a key factor in NMD (25). We transfected HEK293T cells with two plasmids: one carrying an shRNA construct targeting UPF1 (26) and one carrying the mutant DHX16 cDNA. We also used a plasmid carrying a GFP shRNA construct and a plasmid expressing wild type DHX16 or EGFP as controls (Fig. 4C). By probing the protein lysates from the two-plasmid-transfected cells using antibodies, we showed that the level of UPF1 protein decreased in the shUPF1 (small nuclear RNA targeting UPF1)-transfected cells (Fig. 4C, top panel). RT-PCR analysis of the RNA from the co-transfected cells showed an increase of the nPTB exon 10-excluded isoform, indicating NMD was inhibited in shUPF1-transfected cells (Fig. 4C, fourth panel) (26). The expression of the plasmid-borne DHX16 was confirmed by the immunoblotting analysis (Fig. 4C, third panel). RT-PCR analysis showed an accumulation of unspliced DNAJB1 RNA in cells expressing the mutant DHX16-G724N, indicating that splicing inhibition brought about by DHX16-G724N was not affected by inhibiting UPF1 and NMD (Fig. 4C, fifth panel). We then analyzed whether inhibiting NMD by knocking down UPF1 would affect the level of intron-containing RNAs by RT-PCR and found virtually no changes in DHX16-G724N-sensitive transcripts (SFRS1 and HSPH1) or insensitive transcripts (PRPF8 and PRDX3) (Fig. 4D). Finally, we also analyzed RNA isolated from the transfected, cycloheximide-treated cells by genomic tiling microarray and found a similar set of genes accumulating unspliced transcripts (data not shown). Thus, these results suggested that blocking cytoplasmic NMD did not affect the accumulation of intron-containing RNA by the DHX16 mutant.
Because DHX16/hPRP2 is associated with the spliceosome (18, 19) and that mutant DHX16 arrests a precatalytic spliceosome in vitro (19), we suspected that the unspliced RNA present in DHX16-G724N-expressing cells escaped cytoplasmic NMD by being retained in the nucleus. To test this, we isolated nuclear and cytoplasmic RNA from HEK293 cells transfected with DHX16-G724N with or without cycloheximide treatment. U6 small nuclear RNA was found enriched in the nuclear fraction, and the GAPDH protein was found predominantly in the cytoplasmic fraction, verifying the fractionation (Fig. 4E). We also analyzed nuclear and cytoplasmic fractions for the nPTB RNA by RT-PCR. The amounts of both nPTB isoforms were not altered in the nuclear fraction, but the amount of the exon 10-skipping, premature stop codon-containing isoform increased in the cytoplasmic fraction upon cycloheximide treatment, verifying NMD inhibition as well as the nucleus-cytoplasm fractionation (Fig. 4E). Under these conditions, the HSPH1 intron-containing transcript that was accumulated in DHX16-G724N-expressing cells (as shown in Fig. 1C) was in the nuclear fraction (Fig. 4E, lanes labeled N) and was not detected in the cytoplasmic fraction, even upon treating the cells with cycloheximide (lanes labeled C). The PRPF8 intron-containing transcript that was present in wild type- and DHX16-G724N-expressing cells (as shown in Fig. 1C) was also in the nuclear fraction (Fig. 4E, lanes N) and was not detected in the cytoplasmic fraction, regardless of cycloheximide treatment (lanes labeled C). Considering there was no increase in intron-containing RNA of HSPH1 in the cytoplasmic fraction (in contrast to the increase in nPTB premature stop codon-containing RNA) when NMD was inhibited by cycloheximide treatment, this result indicated that the intron-containing transcripts in cells expressing mutant DHX16 were retained in the nucleus.
DISCUSSION
We showed in this work that hindering the function of DHX16, the human orthologue of yeast Prp2 spliceosomal ATPase, led to accumulation of unspliced RNA from intron-containing genes. The genomic tiling microarray was effective at detecting intronic signals and thus intron-containing RNA. The unspliced RNA remained in the nucleus and the level of accumulation was not affected by perturbing cytoplasmic nonsense-mediated decay. Because introns from genes showing more or little intron retention were all retained in minigene transcripts, DHX16/hPRP2 is more likely to be a general splicing factor. The disparity in intron detection perhaps was influenced by additional nuclear events, such as transcription and RNA degradation (28, 29) (see below).
Accumulation of intron-containing transcripts is a common feature of splicing mutants of unicellular organisms like yeast (30). In fact, screening for conditional mutants that accumulate unspliced RNA is very effective in identifying genes required for RNA splicing (31, 32). S. cerevisiae prp2 conditional mutants and mutants overexpressing dominant negative prp2 accumulate unspliced pre-mRNA (33, 34). Genome-wide analysis using an oligonucleotide microarray designed to detect individual introns and spliced exons revealed that transcripts from most of the intron-containing genes retained their introns in prp2 mutants (35, 36). Genomic tiling arrays have been used to detect and discover introns in S. cerevisiae (37–39) and in Arabidopsis thaliana (40). Thus, it might be somewhat unexpected that our genomic tiling microarray detected a relatively small number of intron-containing transcripts that were accumulated in cells expressing mutant DHX16/hPRP2. We believe that the intensity threshold and the sliding window setting we used were too stringent, and our data represent a conservative estimate that may miss many introns with less robust accumulation in mutant DHX16-expressing cells.
The majority of genes showing retention of introns in our study were ubiquitously expressed, including ribosomal protein genes, heat-shock factor genes, and genes involved in protein folding. Korol and colleagues (41) have reported that three groups of highly expressed genes (ribosomal proteins, heat shock proteins, and amino-acyl tRNA synthetases) have different purine-pyrimidine composition when compared with randomly selected genes in 12 eukaryotic species, including humans. It is not clear whether these nucleotide sequence characteristics contribute to the intron retention observed in DHX16 mutant cells. The fact that some introns from an insensitive gene (PRPF8) were well retained when expressed from a minigene suggests that intron retention by DHX16 mutants was probably not determined by the intron sequence.
In the cell at steady state, the amount of the unspliced pre-mRNA (P) is determined by three rate constants, transcription (kT), splicing (kS), and degradation (kD), and can be expressed in the following equation.
This relationship was described by Pikielny and Rosbash (22) and Fouser and Friesen (23) when they investigated the in vivo efficiency of pre-mRNA splicing in yeast; it has been further discussed by David Horowitz.4 According to this model, when splicing is impeded (kS becomes smaller), the amount of unspliced RNA may not change significantly if the unspliced RNA is quickly degraded (a large kD) (42). Moreover, unspliced transcripts from a highly expressed gene (a large kT) will have a greater chance of being detected. The transcription rate constant is a characteristic of a gene, whereas the splicing rate constant could be a characteristic of an intron. However, it is not clear whether the degradation constant is a characteristic of a gene or of an intron, and it can be quite complicated for genes that contain multiple introns, like most human genes. If the degradation constant were a gene characteristic, it could explain why some genes for which RNA splicing was inhibited accumulated most of their introns (Fig. 1B).
It is formally possible that DHX16/hPRP2 may differentially affect the splicing of different introns or different genes. A transcript-specific effect on splicing by core spliceosome components has been documented in Drosophila (43) and in S. cerevisiae (36). The S. cerevisiae study also indicated that ribosomal protein genes may be regulated in specific ways by constitutive splicing factors and suggested that their transcripts fundamentally differ from other transcripts in their susceptibility to splicing mutations. Furthermore, the splicing of ribosomal protein gene transcripts is regulated in response to environmental stress (44). By switching promoters, Christine Guthrie and her colleagues show that splicing of ribosomal protein RNAs is regulated by a promoter-dependent and intron-independent mechanism in budding yeast.5 It is interesting to note that ribosomal protein genes and genes related to ribosome biogenesis (either protein-coding or small nucleolar RNA-containing) were enriched in the group of genes for which we detected intron retention, raising the possibility that regulation of splicing of these gene transcripts may be conserved between yeasts and humans. This is rather remarkable considering that virtually all intron-containing genes in S. cerevisiae have only one intron, whereas the vast majority of human genes have multiple introns. Furthermore, our results support the idea that inhibition of a general spliceosomal factor could result in differential accumulation of unspliced gene transcripts (45).
In our assay, the unspliced transcripts produced in mutant DHX16-expressing cells were not subject to nonsense-mediated decay, probably because they were retained in the nucleus. It is possible that this nuclear retention is directly related to the fact that DHX16/hPRP2 functions after spliceosome formation (19). Transcripts produced from stably transfected β-globin genes carrying splicing or 3′-end processing mutations are retained at the transcription site (46). Primary transcripts are attached to RNA polymerase II even after their 3′-ends are cleaved; the cleaved transcripts are released after polyadenylation, but the release requires the completion of splicing (47). However, splicing deficiency does not always retain all of the unspliced RNA. For example, when splicing factor SF3b is impaired by gene knockdown or by spliceostatin A, some unspliced or partially spliced transcripts are exported and translated to produce truncated proteins (48). In yeast, mutations at the 5′-splice site or at the branch point (49) and alterations in the early splicing factor BBP/SF1 (50) lead to leakage of unspliced RNA to the cytoplasm. Furthermore, a recent study using yeast genomic tiling arrays has revealed that many unspliced pre-mRNAs accumulate in the cytoplasm when NMD is defective and has further indicated that NMD plays a previously unappreciated role in discarding precursors of regulated or suboptimally spliced transcripts (39). Thus, it remains to be investigated how intron-containing RNAs accumulated in mutant DHX16/hPRP2-producing cells are handled by the gene expression machineries in the nucleus.
Supplementary Material
Acknowledgments
We thank Douglas Black (UCLA, Los Angeles, CA) and Joachim Lingner (Swiss Institute for Experimental Cancer Research) for gifts of shRNA constructs. We are grateful to David Horowitz (Uniformed Services University of the Health Science) for sharing an unpublished manuscript and to Peter Stoilov and Douglas Black for help with data analysis and insightful discussions. We thank Ning Ye Zhou, Zheng Liu, and Xuejun Li (City of Hope Microarray and Bioinformatics Core) for help with the microarray assay and analysis and Keely Walker for editing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM40639. This work was also supported by a City of Hope Cancer Center grant (to R.-J. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–3 and Figs. 1–3.
X. Wu, unpublished observations.
D. Horowitz, personal communication.
C. Guthrie, personal communication.
- NMD
- nonsense-mediated decay.
REFERENCES
- 1.Sharp P. A. (2005) Trends Biochem. Sci. 30, 279–281 [DOI] [PubMed] [Google Scholar]
- 2.Brent M. R. (2008) Nat. Rev. Genet 9, 62–73 [DOI] [PubMed] [Google Scholar]
- 3.Black D. L. (2003) Annu. Rev. Biochem. 72, 291–336 [DOI] [PubMed] [Google Scholar]
- 4.Jurica M. S., Moore M. J. (2003) Mol. Cell 12, 5–14 [DOI] [PubMed] [Google Scholar]
- 5.Wahl M. C., Will C. L., Lührmann R. (2009) Cell 136, 701–718 [DOI] [PubMed] [Google Scholar]
- 6.Ruby S. W., Abelson J. (1991) Trends Genet. 7, 79–85 [DOI] [PubMed] [Google Scholar]
- 7.Guthrie C. (1991) Science 253, 157–163 [DOI] [PubMed] [Google Scholar]
- 8.Käufer N. F., Potashkin J. (2000) Nucleic Acids Res. 28, 3003–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper T. A., Wan L., Dreyfuss G. (2009) Cell 136, 777–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Staley J. P., Guthrie C. (1998) Cell 92, 315–326 [DOI] [PubMed] [Google Scholar]
- 11.Silverman E., Edwalds-Gilbert G., Lin R. J. (2003) Gene 312, 1–16 [DOI] [PubMed] [Google Scholar]
- 12.Cordin O., Banroques J., Tanner N. K., Linder P. (2006) Gene 367, 17–37 [DOI] [PubMed] [Google Scholar]
- 13.Imamura O., Saiki K., Tani T., Ohshima Y., Sugawara M., Furuichi Y. (1998) Nucleic Acids Res. 26, 2063–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lundgren K., Allan S., Urushiyama S., Tani T., Ohshima Y., Frendewey D., Beach D. (1996) Mol. Biol. Cell 7, 1083–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Z., Licklider L. J., Gygi S. P., Reed R. (2002) Nature 419, 182–185 [DOI] [PubMed] [Google Scholar]
- 16.Rappsilber J., Ryder U., Lamond A. I., Mann M. (2002) Genome Res. 12, 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deckert J., Hartmuth K., Boehringer D., Behzadnia N., Will C. L., Kastner B., Stark H., Urlaub H., Lührmann R. (2006) Mol. Cell. Biol. 26, 5528–5543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bessonov S., Anokhina M., Will C. L., Urlaub H., Lührmann R. (2008) Nature 452, 846–850 [DOI] [PubMed] [Google Scholar]
- 19.Gencheva M., Kato M., Newo A. N., Lin R. J. (2010) Biochem. J. 429, 25–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S. H., Lin R. J. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 888–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kotake Y., Sagane K., Owa T., Mimori-Kiyosue Y., Shimizu H., Uesugi M., Ishihama Y., Iwata M., Mizui Y. (2007) Nat. Chem. Biol. 3, 570–575 [DOI] [PubMed] [Google Scholar]
- 22.Pikielny C. W., Rosbash M. (1985) Cell 41, 119–126 [DOI] [PubMed] [Google Scholar]
- 23.Fouser L. A., Friesen J. D. (1986) Cell 45, 81–93 [DOI] [PubMed] [Google Scholar]
- 24.Carter M. S., Doskow J., Morris P., Li S., Nhim R. P., Sandstedt S., Wilkinson M. F. (1995) J. Biol. Chem. 270, 28995–29003 [DOI] [PubMed] [Google Scholar]
- 25.Chang Y. F., Imam J. S., Wilkinson M. F. (2007) Annu. Rev. Biochem. 76, 51–74 [DOI] [PubMed] [Google Scholar]
- 26.Boutz P. L., Stoilov P., Li Q., Lin C. H., Chawla G., Ostrow K., Shiue L., Ares M., Jr., Black D. L. (2007) Genes Dev. 21, 1636–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ni J. Z., Grate L., Donohue J. P., Preston C., Nobida N., O'Brien G., Shiue L., Clark T. A., Blume J. E., Ares M., Jr. (2007) Genes Dev. 21, 708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore M. J., Proudfoot N. J. (2009) Cell 136, 688–700 [DOI] [PubMed] [Google Scholar]
- 29.Houseley J., Tollervey D. (2009) Cell 136, 763–776 [DOI] [PubMed] [Google Scholar]
- 30.Abelson J. (1989) Harvey Lect. 85, 1–42 [PubMed] [Google Scholar]
- 31.Vijayraghavan U., Company M., Abelson J. (1989) Genes Dev. 3, 1206–1216 [DOI] [PubMed] [Google Scholar]
- 32.Potashkin J., Li R., Frendewey D. (1989) EMBO J. 8, 551–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosbash M., Harris P. K., Woolford J. L., Jr., Teem J. L. (1981) Cell 24, 679–686 [DOI] [PubMed] [Google Scholar]
- 34.Plumpton M., McGarvey M., Beggs J. D. (1994) EMBO J. 13, 879–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burckin T., Nagel R., Mandel-Gutfreund Y., Shiue L., Clark T. A., Chong J. L., Chang T. H., Squazzo S., Hartzog G., Ares M., Jr. (2005) Nat. Struct. Mol. Biol. 12, 175–182 [DOI] [PubMed] [Google Scholar]
- 36.Pleiss J. A., Whitworth G. B., Bergkessel M., Guthrie C. (2007) PLoS Biol. 5, e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Juneau K., Palm C., Miranda M., Davis R. W. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1522–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z., Hesselberth J. R., Fields S. (2007) Genome Res. 17, 503–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sayani S., Janis M., Lee C. Y., Toesca I., Chanfreau G. F. (2008) Mol. Cell 31, 360–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ner-Gaon H., Fluhr R. (2006) DNA Res. 13, 111–121 [DOI] [PubMed] [Google Scholar]
- 41.Paz A., Mester D., Nevo E., Korol A. (2007) J. Mol. Evol. 64, 248–260 [DOI] [PubMed] [Google Scholar]
- 42.Bousquet-Antonelli C., Presutti C., Tollervey D. (2000) Cell 102, 765–775 [DOI] [PubMed] [Google Scholar]
- 43.Park J. W., Parisky K., Celotto A. M., Reenan R. A., Graveley B. R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 15974–15979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pleiss J. A., Whitworth G. B., Bergkessel M., Guthrie C. (2007) Mol. Cell 27, 928–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nilsen T. W. (2007) Mol. Cell 28, 715–720 [DOI] [PubMed] [Google Scholar]
- 46.Custódio N., Carmo-Fonseca M., Geraghty F., Pereira H. S., Grosveld F., Antoniou M. (1999) EMBO J. 18, 2855–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rigo F., Martinson H. G. (2009) RNA 15, 823–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaida D., Motoyoshi H., Tashiro E., Nojima T., Hagiwara M., Ishigami K., Watanabe H., Kitahara T., Yoshida T., Nakajima H., Tani T., Horinouchi S., Yoshida M. (2007) Nat. Chem. Biol. 3, 576–583 [DOI] [PubMed] [Google Scholar]
- 49.Legrain P., Rosbash M. (1989) Cell 57, 573–583 [DOI] [PubMed] [Google Scholar]
- 50.Rutz B., Séraphin B. (2000) EMBO J. 19, 1873–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.