Abstract
Transplantation of mesenchymal stem cells (MSCs) is a promising therapy for ischemic injury; however, inadequate survival of implanted cells in host tissue is a substantial impediment in the progress of cellular therapy. Secreted Frizzled-related protein 2 (sFRP2) has recently been highlighted as a key mediator of MSC-driven myocardial and wound repair. Notably, sFRP2 mediates significant enhancement of MSC engraftment in vivo. We hypothesized that sFRP2 improves MSC engraftment by modulating self-renewal through increasing stem cell survival and by inhibiting differentiation. In previous studies we demonstrated that sFRP2-expressing MSCs exhibited an increased proliferation rate. In the current study, we show that sFRP2 also decreased MSC apoptosis and inhibited both osteogenic and chondrogenic lineage commitment. sFRP2 activity occurred through the inhibition of both Wnt and bone morphogenic protein (BMP) signaling pathways. sFRP2-mediated inhibition of BMP signaling, as assessed by levels of pSMAD 1/5/8, was independent of its effects on the Wnt pathway. We further hypothesized that sFRP2 inhibition of MSC lineage commitment may reduce heterotopic osteogenic differentiation within the injured myocardium, a reported adverse side effect. Indeed, we found that sFRP2-MSC-treated hearts and wound tissue had less ectopic calcification. This work provides important new insight into the mechanisms by which sFRP2 increases MSC self-renewal leading to superior tissue engraftment and enhanced wound healing.
Keywords: Apoptosis, Bone Morphogenetic Protein (BMP), Cell Differentiation, Hypoxia, Stem Cell, Wnt Pathway, Self-renewal, Wound Repair
Introduction
Bone marrow-derived mesenchymal stem cells (MSCs)2 are an attractive candidate for cell-mediated wound repair. Because of their plasticity, MSCs have been utilized in several preclinical and clinical trials of tissue regeneration (1).3 MSCs have been able to repair infarcted myocardium, bone, and soft tissue, albeit with varying degrees of success (3). These promising results are inconsistent, partly due to the low levels of engraftment of MSCs within the injured tissues (4). Hence, strategies to increase survival and engraftment within the wound may enhance MSC therapy.
Self-renewal is an intrinsic property of stem cells that allows them to give rise to non-differentiated daughter cells by proliferating, preventing apoptosis, and avoid lineage commitment (5, 6). This process is important for the maintenance of a stem cell pool that, in the case of MSCs, can exert a more robust effect within the context of a wound. Although several cytokines, growth factors, adhesion molecules, and extracellular matrix components have been identified as cues that signal MSCs to differentiate, the molecular signals that modulate MSC self-renewal remain unknown (5). Data from the hematopoietic stem cell (HSC) field have documented the involvement of Wnt, Notch, and BMP signaling cascades in self-renewal; these pathways are implicated in the expansion of undifferentiated HSCs that upon transplantation into lethally irradiated mice successfully reconstitute the cleared bone marrow (7–9). Although no data are available to demonstrate the role of these pathways in MSC self-renewal, the Wnt and BMP cascades are involved in MSC lineage commitment. Canonical Wnt signaling directs osteogenic differentiation of MSCs by stimulating the expression of osteocalcin (10), this pathway is also involved in early chondrogenesis (11). The BMP pathway also modulates osteogenic differentiation of MSCs by controlling osteocalcin (12), and BMP2 induces chondrocyte fate determination (13). The Wnt cascade is involved in other cellular processes besides differentiation; canonical Wnt signaling inhibition, through the activity of Dkk-1, increases human MSC proliferation without overt differentiation (14). The mechanisms by which MSCs modulate these signaling events during growth and/or lineage commitment remain unknown. The data herein, describe the ability of sFRP2 to promote MSC self-renewal by inhibition of both the Wnt and BMP pathways.
Secreted frizzled-related protein-2 (sFRP2) has recently been implicated by our group and others as a mediator of MSC-driven myocardial and wound repair; however, the mechanisms are unclear. sFRP2-mediated regeneration can be attributed, at least in part, to its paracrine role in mediating myocardial survival by inhibiting apoptosis (15). However, there are compelling data that sFRP2 has direct effects on MSCs themselves. Overexpression of sFRP2 by MSCs results in increased MSC proliferation and long-term engraftment in vivo (16).
The members of the secreted frizzled-related protein (sFRPs) family contain a region with high homology to the cysteine-rich domain (CRD) of the Wnt-pathway frizzled receptors (17). sFRPs bind Wnt glycoproteins through the CRD, preventing them from reaching their cognate receptors (18). The five mammalian members, sFRP1–5, have been associated with several developmental and disease processes (19, 20). Their documented effect thus far has been that of Wnt inhibition (18).
Here, we demonstrate that sFRP2 plays dual and important roles in increasing MSC self-renewal by inhibiting both the Wnt and BMP pathways. We show that sFRP2 confers apoptotic resistance to MSCs in parallel to Wnt pathway inhibition. sFRP2 affects the rate of MSC multilineage differentiation; processes directed by Wnt and BMP signaling. This article is the first to demonstrate that sFRP2 directly inhibits BMP signaling in MSCs. The data contained within describe the mechanisms by which sFRP2 enhances MSC self-renewal to promote MSC-mediated wound healing.
EXPERIMENTAL PROCEDURES
Antibodies
β-Catenin (BD Transduction Labs, 610153), Active Caspase 3 (Promega, G748A), Caspase 3 (Cell Signaling, 9665), Smad 1/5 (Santa Cruz Biotechnology, sc-6201), phospho-Smad1/5/8 (Cell Signaling, 9511).
Recombinant Proteins
From R&D Systems: BMP2 (355-BEC), sFRP2 (1169-FR), sFRP3 (592-FR), Wnt3a (1324-WN).
Small Molecule Inhibitors
The Wnt inhibitor, pyrvinium, was a gift from Dr. Ethan Lee, Vanderbilt University.4 The BMP inhibitor, dorsomorphin, was a gift from Dr. Charles Hong, Vanderbilt University (21).
Animals/Surgical Interventions
All procedures were carried out in accordance with Vanderbilt Institutional Animal Care and Use Committee. NOD/SCID and NOD/SCID/β-glucuronidase-deficient (β-gluc−/−) maintained by PPY. Surgical interventions were performed as previously described (16).
Cells
Primary MSCs were generated from pooled BM from n = 3 mice as previously described (22). For sFRP2 overexpression, MSCs were infected with retrovirus constructs LZRS-sFRP2-GFP or LZRS-GFP as control as previously described (16). GFP-positive cells were sorted using BDFACS Aria 2 days after transduction. At least four independent stable lines were tested; sorted cells were used for a maximum of five passages.
Immunofluorescence
MSCs on coverslips were fixed with cold acetone, blocked with 10% goat serum, and incubated with primary antibody. Secondary antibody conjugated to Cy5 was incubated in the dark. Coverslips were affixed onto slides using Vectashield Hard Set mounting medium with Dapi (Vector H-1500).
β-Catenin and Caspase 3 Immunoblotting
400,000 MSCs were plated onto 10-cm dishes in 2% FBS-containing medium. The cells were grown in hypoxic conditions (5% O2) for 24 h. Cytoplasmic extracts were obtained by lysing the cells in 20 mm Tris-HCl, pH 7.5 and removing the membrane fraction via centrifugation at 100,000 × g for 2 h. After BCA Protein Assay (Pierce) proteins were resolved by SDS/PAGE and transferred to nitrocellulose. Membranes were incubated with primary antibody diluted in 5% milk-TBST at 4 °C overnight (β-catenin 1:1000 and caspase 3 1:1000). Species-specific secondary antibodies conjugated to HRP were used and chemiluminescence (PerkinElmer, NEL104) was detected by film. ImageJ version 1.38× (National Institutes of Health) software was used for densitometry analysis of the appropriate lanes; values are normalized to β-actin loading control.
pSMAD 1/5/8 (SMAD 1/5) Immunoblotting
30 h serum-starved MSCs plated onto 10-cm dishes were treated and, at the appropriate times, cellular proteins were extracted with RIPA buffer containing NaF and Na3VO4. After the BCA Protein assay, proteins were resolved by SDS/PAGE and transferred to nitrocellulose. Membranes were incubated with primary antibody diluted in 5% milk-TBST at 4 °C overnight (1.5:1000). Species-specific secondary antibodies conjugated to HRP were used and chemiluminescence (PerkinElmer, NEL104) was detected by film. ImageJ version 1.38× (National Institutes of Health) software was used for densitometry analysis of the appropriate lanes; values are normalized to β-actin loading control.
Annexin V Flow Cytometry Analysis
Annexin V-PE apoptosis detection kit from BD Biosciences (559763) was used. MSCs were grown in 2% serum, serum-free conditions or in hypoxic conditions for 48–72 h prior to analysis. To stain, 1 × 105 cells in 100 μl of binding buffer were incubated with 5 μl of Annexin V-PE and 5 μl of 7-AAD for 15 min at room temperature in the dark. Flow cytometry analysis was performed in the LSMII flow cytometry and subsequently analyzed using FACSDiva v5.02 software (Becton Dickinson).
In Vitro Differentiation and RNA Isolation
MSCs were plated at minimal cell density in differentiation media with or without Wnt3a (50 ng/ml) or BMP2 (100 ng/ml) as previously described (16). Following differentiation, Trizol (Invitrogen) was used to obtain RNA. cDNA was synthesized (iScript, Bio-Rad) and real-time PCR for osteocalcin, Collagen XI, PPARγ and Runx2 was performed in triplicate for each sample (iCycler; Bio-Rad) (FastStart SYBR Green; Roche). Reactions were normalized to 18S.
BMP Signaling/ID-1 Luciferase
MSCs were transiently transfected using Lipofectamine 2000 (Invitrogen) with a reporter construct containing ID-1 promoter with SMAD binding elements upstream of luciferase (23). A CMV-driven β-galactosidase (β-gal) reporter was used to control for transfection efficiency. Cells were collected, and luciferase activity measured with the Dual Luciferase assay (Promega).
Histology and Morphometry
PVA sponges were cut in half and embedded with cut surface down for histology. Von Kossa staining was performed on sponge sections. Five random images from each section were photographed with a CoolSNAP Hq CCD camera (Photometrics). The area of differentiated tissue for each field was quantified using MetaMorph (Molecular Devices Corp., Sunnyvale, CA) by outlining tissue and calculating total area per field. MSC-treated heart sections were stained with Von Kossa. Two, blinded pathologist scored the amount and quality of the positive areas within the myocardial scar. The scores ranged from zero to three, where zero meant no Von Kossa-positive areas observed and three meant extensive and brightly stained areas. The average score between the two pathologists is presented.
Statistical Analysis
The statistical significance between experimental groups and control were determined by Student's t test or ANOVA followed by Newman-Keuls multiple comparison test, unless otherwise noted, using GraphPad Prism (San Diego, CA). Graphs show mean ± S.D., and p ≤ 0.05 is statistically significant.
RESULTS
sFRP2 Increases MSC Engraftment and Inhibits Canonical Wnt Signaling to Protect MSCs from Undergoing Apoptosis
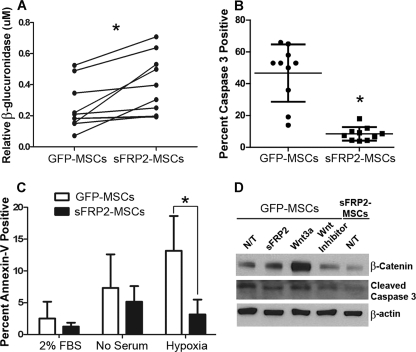
sFRP2 has been identified as a key factor responsible for the biogenesis of a superior MSC phenotype (15, 16). We traced the ability of MSCs to remain within PVA sponges 2 weeks following implantation in the ventral subcutaneous space of β-gluc−/− mice. The activity of β-glucuronidase within the extracted sponges served as a surrogate marker for the MSCs. As seen in Fig. 1A, sFRP2-overexpressing MSCs have statistically significant increased engraftment within granulation tissue. In this figure, each mouse corresponds to one line, a clear trend in the increase of enzyme activity in the sFRP2-MSC-loaded sponges is observed. Overall, sFRP2 overexpression resulted in an approximate 2-fold increase in engraftment. In a previous study, we showed a similar (2–3-fold) increase in engraftment in an in vivo model of myocardial infarction and wound repair (16). To understand the cellular basis of the enhanced engraftment, we assessed if this effect was due, in part, to decreased apoptosis. We determined basal levels of activated caspase 3 by indirect immunofluorescence (IF) of MSCs stably overexpressing sFRP2 (sFRP2-MSCs) versus those stably transduced with empty vector (GFP-MSCs). Approximately 47 ± 18% of GFP-MSCs were undergoing apoptosis under high-confluency in vitro conditions, whereas only 8.5 ± 4.3 (p = 0.002) percent of sFRP2-MSCs were positive for this effector caspase (Fig. 1B). To detect apoptosis by an independent method, we assessed Annexin V levels on MSCs grown under 2% serum, 0% serum, or low oxygen conditions, by flow cytometry. The level of apoptosis was increased to ∼7% when MSCs were grown in no serum as compared with low serum (∼2.5%). Hypoxia (5% O2) further doubled the level of apoptosis. An overall 2-fold decrease in Annexin V-positive cells was observed in sFRP2-MSCs regardless of the in vitro conditions. Statistical significance was only achieved when the cells were exposed to hypoxia (Fig. 1C).
FIGURE 1.
sFRP2 enhances engraftment and protects MSCs from undergoing apoptosis by inhibiting canonical Wnt signaling. A, levels of β-glucuronidase enzyme activity, normalized to DNA content, are higher within sFRP2-MSC-loaded PVA sponges. Data show the enzymatic activity within the different sponges implanted in the same mouse. *, p = 0.041, Mann Whitney U-Test, n = 11. B, basal levels of activated caspase 3 positivity in sFRP2-MSCs are reduced as compared with GFP-MSCs, as determined by IF. *, p = 0.002, Mann Whitney U-Test, n = 10. C, quantification of flow cytometry analysis for annexin V in GFP-MSCs and sFRP2-MSCs grown in 2% serum, serum-starved, or hypoxic conditions. n = 9; one-way ANOVA with Bonferroni's multiple comparison test. D, representative Western blot analysis of cytoplasmic lysates of GFP-MSCs treated with sFRP2 (100 ng/ml), Wnt3a (50 ng/ml), or pyrvinium (100 nm). N/T, no treatment, n = 3. sFRP2 inhibits canonical Wnt signaling and concomitantly reduces apoptosis.
To determine if the ability of sFRP2 to protect MSCs from undergoing apoptosis was, in part, due to its Wnt inhibitory activity, we grew GFP-MSCs and sFRP2-MSCs under hypoxic conditions. GFP-MSCs were treated with recombinant sFRP2, Wnt3a, or the small molecule Wnt inhibitor pyrvinium. Pyrvinium causes inhibition of axin degradation and promotes degradation of both β-catenin and pygopus thereby leading to functional inhibition of the canonical Wnt cascade.4 Western blot analysis of the cytoplasmic extracts of these cells revealed that sFRP2-MSCs had decreased Wnt activity, as observed by the levels of β-catenin, when compared with GFP-MSCs. As seen in Fig. 1D, a decrease in Wnt signaling correlated with a decrease in activated caspase 3 levels; sFRP2-MSCs had ∼44% lower levels of the cleaved caspase 3 fragment.
sFRP2 Inhibits Chondrogenic and Osteogenic Differentiation of MSCs in Vitro
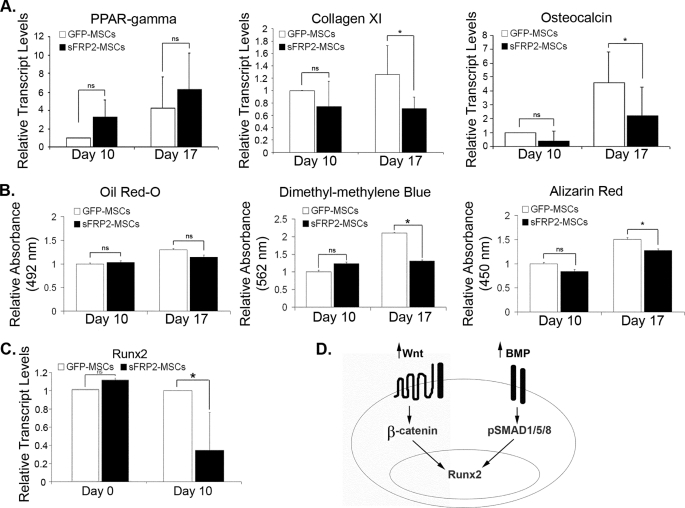
Control of the balance between stem cell self-renewal and subsequent differentiation is crucial to the therapeutic efficacy of MSCs. To enhance their engraftment within wounds, MSCs must promote their survival and delay differentiation. We assessed the effect of sFRP2 overexpression on the in vitro multilineage commitment by two different methods. First, we performed quantitative real-time PCR (qRT-PCR) analysis of peroxisome proliferator-activated receptor γ (PPAR-γ), collagen XI, and osteocalcin as specific markers of the adipogenic, chondrogenic, and osteogenic lineages, respectively. The transcript levels were normalized to 18S content. Second, quantification of biochemical markers for adipogenic, chondrogenic, and osteogenic differentiation was also carried out: Oil Red-O, dimethyl methylene blue (sulfated glycosamino glycans) and Alizarin Red, respectively. Fig. 2A depicts the changes in the transcript levels of the different lineage markers, and the quantification of the stains is seen in Fig. 2B. sFRP2-MSCs had no difference in the levels of PPAR-γ or Oil Red-O stain compared with GFP-MSCs. On the other hand, sFRP2 had an inhibitory effect on the chondrogenic and osteogenic differentiation. Decreased chondrogenesis was seen in the lower levels of collagen XI and sulfated glycosamino glycans observed in sFRP2-MSCs. The decreased osteogenic differentiation of sFRP2-MSCs was observed in the statistical reduction of osteocalcin transcript levels and the amount of calcified matrix (as determined by Alizarin Red staining) compared with GFP-MSCs. We confirmed the involvement of Wnt and BMP signaling in MSC lineage commitment in vitro and in vivo (supplemental Fig. S1, A—C). The addition of recombinant Wnt3a or BMP2 was not sufficient to increase the in vitro osteogenic or chondrogenic differentiation of sFRP2-MSCs (supplemental Fig. S1D), thus sFRP2 could decrease differentiation by inhibiting either pathway.
FIGURE 2.
sFRP2 decreases the chondrogenic and osteogenic differentiation of MSCs, not on adipogenesis. A, relative transcript levels of PPAR-γ (adipogenic marker, n = 3), collagen XI (chondrogenic marker, n = 4), and osteocalcin (osteogenic marker, n = 5) increase in relation to time in GFP-MSCs and sFRP2-MSCs under differentiating conditions. There is no effect of sFRP2 in the adipogenic potential of the MSCs. There is decreased chondrogenic and osteogenic commitment of sFRP2-MSCs compared with GFP-MSCs as observed by the lower levels of collagen XI and osteocalcin. ns, not statistically significant, *, p ≤ 0.05. B, quantification of Oil Red-O stain demonstrates no difference in the oil droplet formation between GFP-MSCs and sFRP2-MSCs. The amount of sulfated glycosaminoglycans quantified by the use of dimethylmethylene blue dye in sFRP2-MSCs is lower compared with GFP-MSCs. There is decreased extracellular calcification (as quantified by the Alizarin Red stain) in sFRP2-MSCs, compared with GFP-MSCs. *, p ≤ 0.05, n = 4. C, relative transcript levels of Runx2, a transcription factor that controls osteocalcin expression, is not changed in non-differentiated MSCs. During osteogenic differentiation, Runx2 levels are greater in GFP-MSCs. *, p ≤ 0.05, n = 4. D, model figure depicting that canonical Wnt and BMP signaling cascades are involved in the regulation of Runx2 in MSCs.
To further determine the effect of sFRP2 on the decreased osteogenic differentiation of MSCs, we quantified the levels of Runx2 transcription factor by qRT-PCR. This transcription factor is essential for bone formation; its deletion completely ablates ossification (24). Bone-specific activation of the osteocalcin promoter is regulated by Runx2 (25). Wnt signaling, as well as BMP signaling, are involved in osteogenic lineage commitment, in part by regulating the expression of Runx2 (10, 26). Although there was no difference in the baseline levels of Runx2 transcripts, Runx2 levels were decreased in sFRP2-MSCs undergoing osteogenic differentiation (Fig. 2C). We came up with a working model shown in Fig. 2D where we hypothesize that sFRP2 could be regulating Runx2 expression by inhibiting either the Wnt or the BMP pathways. This model prompted us to determine whether sFRP2 plays a role in BMP signaling.
sFRP2 Inhibits Phosphorylation of Nuclear SMAD 1/5/8 in a Wnt-independent Manner
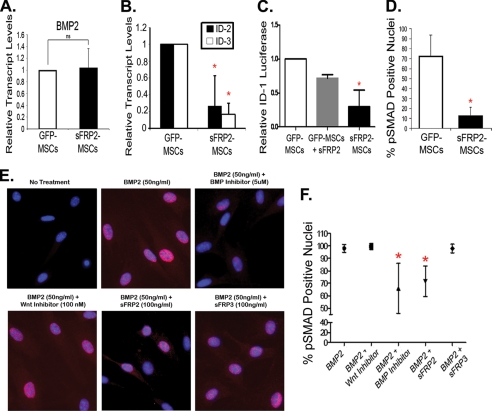
Data from the chick, Xenopus, and zebrafish homologues of sFRP2 indicate that this protein could inhibit the BMP pathway (27). BMPs mediate their signals through phosphorylation of specific receptor-associated Smads, which then complex with other Smads, translocate to the nucleus, and affect gene transcription. We explored whether, under basal conditions, sFRP2 overexpression altered the transcript levels of BMP2, known to play a role in both osteogenic and chondrogenic commitment (12, 13). Baseline relative transcript levels of BMP2 were similar in both groups (Fig. 3A). We explored whether BMP downstream targets were differentially regulated in sFRP2-MSCs compared with GFP-MSCs and found a significant decrease in the transcript levels of both ID-2 and ID-3 transcript levels (Fig. 3B). To further explore if sFRP2 altered basal BMP signaling we utilized a BMP-responsive luciferase reporter construct driven by the ID-1 gene (23). GFP-MSCs and sFRP2-MSCs were co-transfected with the ID-1 reporter construct as well as a β-gal expression vector employed to ensure equal transfection efficiency. The addition of recombinant sFRP2 to GFP-MSCs led to a decrease in luciferase production. Moreover, sFRP2-MSCs had significantly reduced levels of luciferase activity (Fig. 3C), suggesting a role for sFRP2 in BMP signaling inhibition, an effect that has not been previously documented for this mammalian protein. Recombinant BMP2 treatment increases the luciferase signal in both cell types (data not shown). To assess BMP signaling by an independent method, we quantified the number of nuclei positive for phosphorylated SMAD 1, 5, and 8 (pSmad1/5/8) by IF. sFRP2-MSCs had a statistically significant, 7-fold decrease in the percentage pSMAD-positive nuclei under basal conditions (Fig. 3D). Fig. 3E shows representative images of a series of IF studies where GFP-MSCs were serum-starved prior to treatment with recombinant BMP2 for 1 h and subsequent treatment for 30 min with either recombinant sFRP2, sFRP3, pyrvinium, or the BMP inhibitor dorsomorphin. Dorsomorphin has been shown to selectively inhibit BMP type 1 receptors, and subsequent pSMAD1/5/8 signaling, presumably by blocking the ALK2, ALK3, and ALK6 receptor kinase function (28). Only dorsomorphin and sFRP2 statistically decreased the percentage of pSMAD-positive nuclei. Wnt inhibition through the activity of either pyrvinium or sFRP3 did not affect the nuclear localization of pSMAD1/5/8. The quantification of these experiments is seen in Fig. 3F. The capacity of sFRP2 to inhibit nuclear pSMAD1/5/8 was concentration dependent (data not shown). Our data showed that while BMP2 transcript levels were similar in sFRP2-MSCs, BMP signaling was significantly down-regulated relative to GFP-MSCs.
FIGURE 3.
sFRP2 inhibits phosphorylation of nuclear SMAD 1/5/8 in a Wnt-independent manner and causes functional inhibition of BMP signaling. A, no changes in the relative transcript levels of BMP2 in serum-starved GFP-MSCs and sFRP2-MSCs. n = 4. B, significant decrease in the relative transcript levels of BMP downstream target genes ID-2 and ID-3 in serum-starved sFRP2-MSCs compared with GFP-MSCs. n = 3, *, p < 0.01. C, involvement of sFRP2 in BMP signaling demonstrated through transcriptional inhibition of BMP-driven luciferase reporter. sFRP2-MSCs had lower basal BMP activity. Addition of recombinant sFRP2 reduced luciferase activity in GFP-MSCs. n = 4, *, p < 0.0001. D, sFRP2-MSCs exhibit decreased basal phosphorylation of nuclear SMAD1/5/8 as determined by indirect immunofluorescent analysis. Average percentage of positive nuclei per high power field is shown, at least 4 fields counted in three independent experiments. *, p = 0.006. E, representative photomicrographs demonstrating modulation of BMP pathway in MSCs in the presence of BMP2 (50 ng/ml), alone or with combination treatments: pyrvinium (100 nm), dorsomorphin (5 μm), sFRP2 (100 ng/ml), or sFRP3 (100 ng/ml). pSMAD 1/5/8 in red, DAPI in blue. F, data (n = 5 independent experiments) quantifying the average percentage of MSCs containing nuclear pSMAD 1/5/8 were graphed and show that only dorsomorphin and recombinant sFRP2 reduced BMP2-induced activation of nuclear pSMAD in MSCs. Mean ± 95%, confidence interval; *, p < 0.0001.
Inhibition of Phosphorylated SMAD 1/5/8 Accumulation Is Not a BMP-Wnt Cross-talk Event
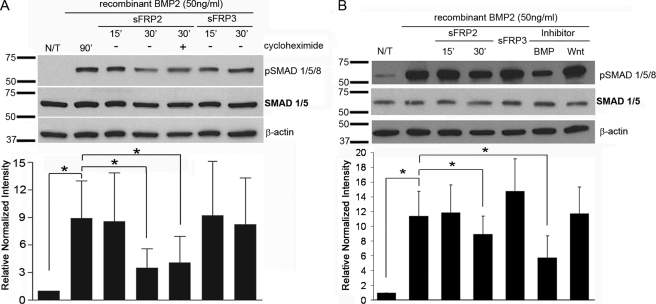
Although a novel BMP-inhibitory role of sFRP2 was suggested by the above studies, it was important to determine if it was due to a translational cross-talk between Wnt and BMP. We performed immunoblot analysis to determine the time course of phosphorylation of Smads1/5/8. Serum-starved MSCs were treated with recombinant BMP2 for 1 h prior to treatment with either sFRP2 or sFRP3. The addition of sFRP2, but not sFRP3, decreased pSMAD1/5/8 levels by 43 ± 17% within 30 min of treatment (Fig. 4A). sFRP2-mediated BMP pathway inhibition did not require new protein synthesis as addition of protein synthesis inhibitor, cycloheximide, did not abrogate sFRP2 effect. We examined whether pyrvinium could affect the levels of pSMAD1/5/8; whereas dorsomorphin significantly reduced BMP2-induced phosphorylation of Smads1/5/8 by 62 ± 14%, there was no significant diminution by pyrvinium (Fig. 4B). The ability of sFRP2 to inhibit pSMAD accumulation was not affected in the presence of pyrvinium (supplemental Fig. S2). Together, these data support a concept that sFRP2 directly modulates the BMP signaling pathway independent of its effect on the Wnt pathway.
FIGURE 4.
Wnt-independent inhibition of phosphorylated SMAD1/5/8 accumulation. A, time course of sFRP2-induced SMAD1/5/8 phosphorylation in mouse MSCs. MSCs were serum-starved for 24 h and stimulated with recombinant BMP2 (50 ng/ml) for 1 h prior to addition of recombinant sFRP2 (100 ng/ml) or sFRP3 (100 ng/ml) for 15 or 30 min. sFRP2-mediated BMP-signaling inhibition was unaltered by the addition of protein synthesis inhibitor, cycloheximide (10 μm). Representative Western blot analysis and the quantification of the relative pSMAD1/5/8 protein abundance in cell lysates normalized to β-actin; n = 3. B, serum-starved MSCs were pretreated with BMP2 (50 ng/ml) for 1 h before addition of designated treatments: sFRP2 (100 ng/ml), sFRP3 (100 ng/ml), pyrvinium (100 nm), or dorsomorphin (5 μm). Only sFRP2 and dorsomorphin decreased BMP-induced pSMAD levels. sFRP3 and a Wnt inhibitor did not alter BMP signaling. Representative Western blot analysis and the quantification of the relative pSMAD1/5/8 protein abundance in total cell lysates normalized to β-actin; n = 3.
sFRP2-MSCs Demonstrate Decreased Osteogenic Differentiation in Vivo
We have demonstrated that sFRP2-MSCs have a significant (2–3-fold) increase in engraftment compared with GFP-MSCs in two separate in vivo models of wound repair (16). Using the PVA sponge model of granulation tissue formation, we examined whether the increase in engraftment correlated with a decrease in differentiated progeny by visualizing matrix calcification within MSC-loaded sponges. Fig. 5A shows representative images of sections of GFP- and sFRP2-MSC-loaded PVA sponges stained for Von Kossa (as an indicator of calcified matrix) and clearly demonstrate the decreased osteogenic differentiation of sFRP2-MSCs. Von-Kossa-positive areas were quantified with morphometric analysis, and the data are shown in Fig. 5B. The inhibition of osteogenic differentiation observed in vitro translates to an in vivo setting. These data indicate that increased engraftment was due to the maintenance of un-differentiated MSCs, further demonstrating the effects of sFRP2 on self-renewal.
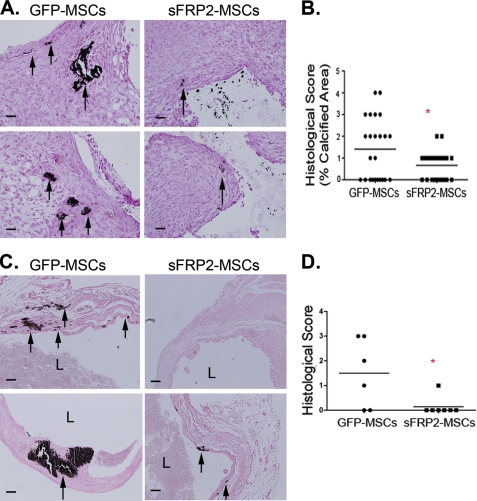
FIGURE 5.
Overexpression of sFRP2 causes decreased ectopic calcification within infarcted myocardium in vivo. A, sFRP2-MSCs, which have higher levels of engraftment within PVA-sponges, have decreased osteogenic differentiation in vivo. Representative images of calcified matrix within granulation tissue of MSC-loaded PVA sponges. Black arrows point to Von Kossa-positive areas (40×). B, quantification of the total percentage Von Kossa-positive areas within the granulation tissue in PVA sponges shows a decrease in the amount of calcified matrix within the sFRP2-loaded sponges. Wilcoxon sum ranks test; *, p = 0.0012, GFP-MSC n = 6, sFRP2-MSC n = 7. C, representative images of GFP- and sFRP2-MSC-treated hearts of NOD/SCID mice stained for Von Kossa to visualize ectopic calcification. Black arrows point to calcified matrix. L, lumen (10×). D, histological score of the amount and the quality of Von Kossa-positive staining within MSC-treated hearts. sFRP2-MSC-treated hearts, which demonstrated statistically improved cardiac function, have statistically decreased ectopic calcification within the infarcted myocardium. Mann-Whitney U-Test, GFP-MSC n = 6, sFRP2-MSC n = 7.
sFRP2 Expression Reduces Ectopic Calcification of MSC-treated Hearts
A serious reported complication of MSC-mediated myocardial therapy following injury is ectopic calcification and/or ossification (29). Based on our data that showed that sFRP2 decreases osteogenic differentiation in vitro as well as in vivo within PVA sponges, we predicted that sFRP2 may ameliorate this complication of MSC therapy. In a previous study we demonstrated that in the setting of myocardial infarcts induced through coronary artery ligation, sFRP2-MSCs reduced infarct size, prevented adverse remodeling and improved cardiovascular function. sFRP2-MSC-treated hearts contained a higher density of blood vessels and greater numbers of MSCs at 30 days post infarct and cell therapy (16). We examined histologic sections stained with Von Kossa to detect calcifications within both GFP- and sFRP2-MSC treated hearts (Fig. 5C). Von Kossa-positive calcified matrix within the left-ventricular scar tissue was found in almost 70% of GFP-MSC-treated (4/6) hearts but only in 14% of sFRP2-MSC-treated hearts (1/7) (Fig. 5D). These data suggest that sFRP2 not only drives more effective MSC-mediated repair but may reduce the frequency by which MSC-mediated therapy may induce ectopic myocardial calcifications.
DISCUSSION
MSCs are utilized in a variety of preclinical models to ameliorate wound healing. Enhanced cardiac tissue repair mediated by MSCs, in concert with clinical studies suggesting that their clinical use is feasible and safe, have spurred a number of clinical trials using bone marrow-derived MSCs as regenerative cell therapy. At least 14 trials are currently ongoing using MSCs to treat myocardial disease (clinicaltrials.gov).3 However, our understanding on how MSCs mediate cardiac and soft tissue repair, including aspects that regulate MSC self renewal, remains incomplete.
sFRP2 has been identified as a factor mediating myocardial repair and wound granulation tissue formation by MSCs (15, 16). Although sFRP2 has been postulated to function as a paracrine modulator of cardiomyocyte apoptosis (30), we have provided compelling data that sFRP2 is an important autocrine factor for MSCs themselves. sFRP2 overexpression directly up-regulated MSC proliferation in vitro and enhanced their long-term engraftment in mouse models of myocardial injury and wound granulation tissue. In these studies we sought to better clarify how sFRP2 was coordinating repair by MSCs at the cellular level. Our studies showed that sFRP2 is an important modulator of MSC self-renewal. In addition to its published role to enhance proliferation (16), we report that it also inhibits both MSC apoptosis and their differentiation along osteogenic and chondrogenic lineages.
Our previous study showed that sFRP2 resulted in a dose-dependent increase in human and murine MSC proliferation in vitro. Conversely, addition of recombinant Wnt3a (or its overexpression in MSCs) resulted in a decreased proliferation rate. Dkk1, an inhibitor of canonical Wnt signaling, has been demonstrated to increase human MSC proliferation and entry into the cell cycle in vitro (14). Taken together with our findings that sFRP2 was able to decrease functional canonical Wnt signaling in MSCs (16), these data suggested that sFRP2 increased proliferation through inhibition of canonical Wnt signaling.
The sFRP family has also been termed the secreted apoptosis-related proteins, or SARPs, and as their name suggests has been documented to play important roles in the cytoprotection of distinct cells (31). Herein we demonstrated that sFRP2 served to protect MSCs from apoptosis under basal and hypoxic conditions. This pro-survival effect was likely to be mediated by sFRP2 inhibition of canonical Wnt signaling as sFRP2 decreased both β-catenin and cleaved caspase 3 levels. The anti-apoptotic action of sFRP2 in MSCs closely mirrors its recently reported paracrine anti-apoptotic effects on cardiomyocytes through direct binding of Wnt3a and inhibition of Wnt3a-induced activation of caspase activities (30).
MSCs are capable of differentiating into multiple connective tissue lineages, in particular bone, cartilage and fat. Differentiation of MSCs along these lineages occurs at very low levels spontaneously but is accelerated by specific differentiation-inducing culture conditions. By testing for biochemical and genetic markers characteristic of adipose, cartilage, and osteogenic lineages, we showed that sFRP2 inhibited differentiation of MSCs along both cartilage and osteogenic lineages in vitro. We further demonstrated a sFRP2-dependent decrease in transcript levels of Runx2, a transcription factor necessary for osteogenic commitment. Wnt and BMP pathways regulate Runx2 transcript levels, and BMP is a known critical regulator of both osteogenic and chondrogenic differentiation of MSCs, we evaluated the effect of sFRP2 on the BMP pathway. Our data provided evidence that sFRP2 inhibited BMP signaling: sFRP2 inhibited nuclear levels of BMP effectors, pSMAD1/5/8, as well as BMP signaling-dependent Id-1 driven reporter gene. Inhibition of pSMAD1/5/8 occurred within 30 min, independently of protein synthesis, and was not affected by the addition of the Wnt inhibitor, pyrvinium. These data suggested that sFRP2 inhibition of BMP signaling did not result from cross-talk between the Wnt/BMP pathways and occurred independently of its effect on Wnt inhibition. Moreover, BMP signaling inhibition occurred both with the addition of recombinant sFRP2 as well as in cells overexpressing sFRP2, suggesting that sFRP2 inhibited BMP in a non-self-autonomous manner.
The mechanism by which sFRP2 directly inhibits BMP signaling in mammalian cells is not known and, until this report, only clues as to its involvement in BMP signaling were available. For example, the non-mammalian homologue of sFRPs, Sizzled (Szl), establishes dorsal-ventral patterning in the Drosophila melanogaster, Xenopus, and zebrafish by regulating gradients of BMPs in the developing embryos (27). The vertebrate dorsal center secretes BMP antagonists, among which chordin is known to sequester BMP ligands to prevent receptor binding (32). Szl inhibits BMP signaling by inhibiting the activity of Xolloid-related (Xlr), the metalloproteinase that degrades chordin. In this developmental process the effect of Szl on Xlr allows sequestration of BMPs by chordin (33) and ultimately BMP pathway inhibition. In a cell-free in vitro system, mouse sFRP2 interacted with Xenopus Xlr to prevent the degradation of Xenopus chordin (34). Another recent report suggests sFRP2 may play a role in BMP inhibition in the developing embryo; unilateral electroporation of murine BMP and sFRP2 into the chick neural tube blocked the induction of BMP downstream targets (35). This study did not address whether the observed BMP inhibition was an indirect or a direct effect of sFRP2.
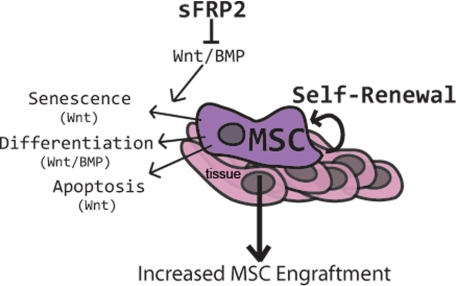
In this report, we show direct, Wnt-independent inhibition of BMP signaling molecules, accomplished not only by overexpression of sFRP2 by MSCs, but by the addition of recombinant mouse sFRP2 to the extracellular space. This biochemical inhibition led to functional inhibition of BMP signaling observed by a decrease in both chondrogenic and osteogenic lineage commitment of MSCs. Compiling our data with the information available about sFRP2 and its non-mammalian homologues, we speculate that the effects of sFRP2 as a BMP inhibitor are carried out in the extracellular space. Our results support the following model (Fig. 6): Increased expression of sFRP2 in MSCs inhibits both canonical Wnt and BMP signaling. The resulting cellular effects of sFRP2 on proliferation, apoptosis, and differentiation impact MSC self-renewal and ultimately engraftment within the wound.
FIGURE 6.
Model of the proposed mechanism of action of sFRP2 in MSC biology. In the context of a wound, sFRP2 expression by MSCs inhibits Wnt and BMP signaling leading to a decreased senescence (increased proliferation), differentiation, and apoptosis. Inhibition of Wnt and BMP signaling by sFRP2 thereby increases MSC self-renewal and increases their tissue engraftment.
Recent reports have highlighted the potential complication of spontaneous in situ osteogenic differentiation of MSCs following cardiac cell therapy. We assessed heterotopic calcification in two, independent in vivo models following MSC cell therapy. Our data showed that in the setting of myocardial infarction and within experimental granulation tissue generated in PVA sponges, sFRP2-expressing MSCs demonstrated significantly fewer foci of ectopic calcification than GFP-MSCs. These data suggest that sFRP2-mediated inhibition of differentiation may reduce the risk of heterotopic cartilage and/or bone tissues observed with MSC therapy.
Despite direct injection of large numbers of cells into tissues, preclinical studies show poor engraftment with only small numbers or no MSCs remaining after 30 days. Hence, a better understanding of the molecular players, such as sFRP2, that favorably modulate MSC self-propagation, engraftment, and differentiation may alter the overall efficacy of MSC cardiac and wound therapy.
Supplementary Material
Acknowledgments
MI surgeries were performed in the Cardiovascular Pathophysiology and Complications Core of the Vanderbilt Mouse Metabolic Phenotyping Center, supported by NIDDK, National Institutes of Health Grant U24 DK-59637.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-HL088424, HL08842402S1, and R01-GM081635; Veterans Affairs merit award (to P. P. Y.); National Cancer Institute GI SPORE P50CA95103 (to E. L.); and American Heart Association Grant 09PRE2010035 (to M. P. A.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
U.S. National Library of Medicine, U.S. Department of Health & Human Services, Clinicaltrials.gov, Lister Hill National Center for Biomedical Communications.
Thorne, C. A., Hanson, A. J., Schneider, J., Tahinci, E., Orton, D., Cselenyi, C. S., Jernigan, K. K., Meyers, K. C., Hang, B. I., Waterson, A. G., Kim, K., Melancon, B., Ghidu, V. P., Sulikowski, G. A., LaFleur, B., Salic, A., Lee, L. A., Miller, 3rd, D. M., and Lee, E. (October 3, 2010) Nat. Chem. Biol. 10.1038/nchembio.453.
- MSC
- mesenchymal stem cell
- sFRP2
- secreted Frizzled-related protein 2
- BMP
- bone morphogenic protein
- HSC
- hematopoietic stem cell
- CRD
- cysteine-rich domain.
REFERENCES
- 1.Jorgensen C., Gordeladze J., Noel D. (2004) Curr. Opin. Biotechnol. 15, 406–410 [DOI] [PubMed] [Google Scholar]
- 2.Deleted in proof
- 3.Orlic D., Kajstura J., Chimenti S., Jakoniuk I., Anderson S. M., Li B., Pickel J., McKay R., Nadal-Ginard B., Bodine D. M., Leri A., Anversa P. (2001) Nature 410, 701–705 [DOI] [PubMed] [Google Scholar]
- 4.Iso Y., Spees J. L., Serrano C., Bakondi B., Pochampally R., Song Y. H., Sobel B. E., Delafontaine P., Prockop D. J. (2007) Biochem. Biophys. Res. Commun. 354, 700–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Satija N. K., Gurudutta G. U., Sharma S., Afrin F., Gupta P., Verma Y. K., Singh V. K., Tripathi R. P. (2007) Stem Cells Dev. 16, 7–23 [DOI] [PubMed] [Google Scholar]
- 6.Schofield R. (1983) Biomed. Pharmacother. 37, 375–380 [PubMed] [Google Scholar]
- 7.Reya T., Clevers H. (2005) Nature 434, 843–850 [DOI] [PubMed] [Google Scholar]
- 8.McReynolds L. J., Gupta S., Figueroa M. E., Mullins M. C., Evans T. (2007) Blood 110, 3881–3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki T., Chiba S. (2005) Int. J. Hematol. 82, 285–294 [DOI] [PubMed] [Google Scholar]
- 10.Gaur T., Rich L., Lengner C. J., Hussain S., Trevant B., Ayers D., Stein J. L., Bodine P. V., Komm B. S., Stein G. S., Lian J. B. (2006) J. Cell Physiol. 208, 87–96 [DOI] [PubMed] [Google Scholar]
- 11.Goldring M. B., Tsuchimochi K., Ijiri K. (2006) J. Cell Biochem. 97, 33–44 [DOI] [PubMed] [Google Scholar]
- 12.Manton K. J., Leong D. F., Cool S. M., Nurcombe V. (2007) Stem Cells 25, 2845–2854 [DOI] [PubMed] [Google Scholar]
- 13.Denker A. E., Haas A. R., Nicoll S. B., Tuan R. S. (1999) Differentiation 64, 67–76 [DOI] [PubMed] [Google Scholar]
- 14.Gregory K. E., Ono R. N., Charbonneau N. L., Kuo C. L., Keene D. R., Bächinger H. P., Sakai L. Y. (2005) J. Biol. Chem. 280, 27970–27980 [DOI] [PubMed] [Google Scholar]
- 15.Mirotsou M., Zhang Z., Deb A., Zhang L., Gnecchi M., Noiseux N., Mu H., Pachori A., Dzau V. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1643–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alfaro M. P., Pagni M., Vincent A., Atkinson J., Hill M. F., Cates J., Davidson J. M., Rottman J., Lee E., Young P. P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 18366–18371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rattner A., Hsieh J. C., Smallwood P. M., Gilbert D. J., Copeland N. G., Jenkins N. A., Nathans J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 2859–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finch P. W., He X., Kelley M. J., Uren A., Schaudies R. P., Popescu N. C., Rudikoff S., Aaronson S. A., Varmus H. E., Rubin J. S. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 6770–6775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ezan J., Leroux L., Barandon L., Dufourcq P., Jaspard B., Moreau C., Allières C., Daret D., Couffinhal T., Duplàa C. (2004) Cardiovasc. Res. 63, 731–738 [DOI] [PubMed] [Google Scholar]
- 20.Hoang B., Moos M., Jr., Vukicevic S., Luyten F. P. (1996) J. Biol. Chem. 271, 26131–26137 [DOI] [PubMed] [Google Scholar]
- 21.Hao J., Daleo M. A., Murphy C. K., Yu P. B., Ho J. N., Hu J., Peterson R. T., Hatzopoulos A. K., Hong C. C. (2008) PLoS ONE 3, e2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tropel P., Noël D., Platet N., Legrand P., Benabid A. L., Berger F. (2004) Exp. Cell Res. 295, 395–406 [DOI] [PubMed] [Google Scholar]
- 23.Zilberberg L., ten Dijke P., Sakai L. Y., Rifkin D. B. (2007) BMC Cell Biol. 8, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R. T., Gao Y. H., Inada M., Sato M., Okamoto R., Kitamura Y., Yoshiki S., Kishimoto T. (1997) Cell 89, 755–764 [DOI] [PubMed] [Google Scholar]
- 25.Ducy P. (2000) Dev. Dyn. 219, 461–471 [DOI] [PubMed] [Google Scholar]
- 26.Lee K. S., Hong S. H., Bae S. C. (2002) Oncogene 21, 7156–7163 [DOI] [PubMed] [Google Scholar]
- 27.Collavin L., Kirschner M. W. (2003) Development 130, 805–816 [DOI] [PubMed] [Google Scholar]
- 28.Yu P. B., Hong C. C., Sachidanandan C., Babitt J. L., Deng D. Y., Hoyng S. A., Lin H. Y., Bloch K. D., Peterson R. T. (2008) Nat. Chem. Biol. 4, 33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breitbach M., Bostani T., Roell W., Xia Y., Dewald O., Nygren J. M., Fries J. W., Tiemann K., Bohlen H., Hescheler J., Welz A., Bloch W., Jacobsen S. E., Fleischmann B. K. (2007) Blood 110, 1362–1369 [DOI] [PubMed] [Google Scholar]
- 30.Zhang Z., Deb A., Zhang Z., Pachori A., He W., Guo J., Pratt R., Dzau V. J. (2009) J. Mol. Cell Cardiol. 46, 370–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melkonyan H. S., Chang W. C., Shapiro J. P., Mahadevappa M., Fitzpatrick P. A., Kiefer M. C., Tomei L. D., Umansky S. R. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 13636–13641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasai Y., Lu B., Steinbeisser H., Geissert D., Gont L. K., De Robertis E. M. (1994) Cell 79, 779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Robertis E. M., Kuroda H. (2004) Annu. Rev. Cell Dev. Biol. 20, 285–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee H. X., Ambrosio A. L., Reversade B., De Robertis E. M. (2006) Cell 124, 147–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Misra K., Matise M. P. (2010) Dev. Biol. 337, 74–83 [DOI] [PubMed] [Google Scholar]
- 36.Deleted in proof
- 37.Deleted in proof
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.