Abstract
Three-dimensional structures of NagZ of Bacillus subtilis, the first structures of a two-domain β-N-acetylglucosaminidase of family 3 of glycosidases, were determined with and without the transition state mimicking inhibitor PUGNAc bound to the active site, at 1.84- and 1.40-Å resolution, respectively. The structures together with kinetic analyses of mutants revealed an Asp-His dyad involved in catalysis: His234 of BsNagZ acts as general acid/base catalyst and is hydrogen bonded by Asp232 for proper function. Replacement of both His234 and Asp232 with glycine reduced the rate of hydrolysis of the fluorogenic substrate 4′-methylumbelliferyl N-acetyl-β-d-glucosaminide 1900- and 4500-fold, respectively, and rendered activity pH-independent in the alkaline range consistent with a role of these residues in acid/base catalysis. N-Acetylglucosaminyl enzyme intermediate accumulated in the H234G mutant and β-azide product was formed in the presence of sodium azide in both mutants. The Asp-His dyad is conserved within β-N-acetylglucosaminidases but otherwise absent in β-glycosidases of family 3, which instead carry a “classical” glutamate acid/base catalyst. The acid/base glutamate of Hordeum vulgare exoglucanase (Exo1) superimposes with His234 of the dyad of BsNagZ and, in contrast to the latter, protrudes from a second domain of the enzyme into the active site. This is the first report of an Asp-His catalytic dyad involved in hydrolysis of glycosides resembling in function the Asp-His-Ser triad of serine proteases. Our findings will facilitate the development of mechanism-based inhibitors that selectively target family 3 β-N-acetylglucosaminidases, which are involved in bacterial cell wall turnover, spore germination, and induction of β-lactamase.
Keywords: Enzyme Catalysis, Enzyme Mechanisms, Enzyme Structure, Hydrolases, Kinetics, Site-directed Mutagenesis, X-ray Crystallography, Glycosidases, Cell Wall Recycling, Peptidoglycan
Introduction
Glycosidases that catalyze the hydrolysis of glycosidic linkages under retention of anomeric configuration of substrate and product (retaining glycosidases) proceed via a two-step double displacement mechanism (1, 2). In most cases, the catalytic machinery of these enzymes involves two carboxylic acids, which are located ∼5.5 Å apart in the active site (Fig. 1A) and function as catalytic nucleophile and catalytic acid/base (also general acid/base catalyst). In the first step (glycosylation), a carboxylic acid that hydrogen bonds to the glycosidic oxygen acts as a general acid facilitating the leaving group departure simultaneously with a nucleophilic attack by the second carboxylate that forms a covalent glycosyl enzyme intermediate. In the second step (deglycosylation), the first residue then functions as a general base to activate an incoming water molecule for nucleophilic attack that hydrolyzes the glycosyl enzyme to form a sugar hemiacetal product with overall retention of stereochemistry (1, 2).
FIGURE 1.
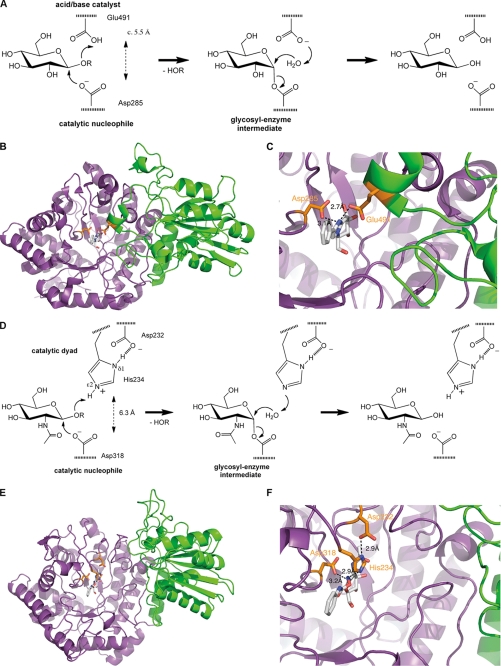
Mechanism and structure of β-exo-glucanase I of H. vulgare (HvExoI) and β-N-acetylglucosaminidase of B. subtilis (BsNagZ) of family 3 of glycosidases. A, schematic of the general two-step double displacement mechanism of retaining β-glycosidases as proposed also for HvExoI. Two catalytic carboxyl groups, which are located ∼5.5 Å apart in the active site, act as the catalytic nucleophile and general acid/base catalyst, respectively. B, ribbon model of HvExoI (PDB identifier 1X38). View of the top of the catalytic N-terminal (β/α)8 (TIM-) barrel domain (magenta) to which the inhibitor glucophenylimidazole is bound in the active site (gray sticks; for chemical structures see supplemental Fig. S1), which carries the catalytic nucleophile (orange stick, left). The C-terminal domain, shown in green, is in close contact with the TIM-barrel domain and also contributes to the active site (15). C, a short helix of the C-terminal domain approaches the bound inhibitor and carries a glutamate residue (Glu491) that acts as the catalytic acid/base residue (orange stick, right). The amino acid sequence of ExoI shows 22% identity with BsNagZ. D, schematic of the modified double displacement mechanism as proposed for BsNagZ. Similar to HvExoI, an asparate residue acts as the catalytic nucleophile (Asp318), however, instead of an acid/base glutamate residue an Asp-His catalytic dyad was now identified as the general acid/base catalyst of β-N-acetylglucosaminidases of family 3. The catalytic groups are located ∼6.3 Å apart in the active site. E, ribbon model of BsNagZ (PDB identifier 3NVD). View on the top of the catalytic N-terminal (β/α)8 (TIM-barrel domain (magenta)) to which the inhibitor PUGNAc is bound (gray sticks; for chemical structures see supplemental Fig. S1) in the active site that carries the catalytic nucleophile (orange stick, left). In contrast to HvExoI, the C-terminal domain of BsNagZ, shown in green, is further apart from the TIM-barrel and does not contribute to the active site. F, in BsNagZ an Asp-His dyad mediates the acid/base function that superimposes with the acid/base glutamate of HvExoI (cf. C). Residues Asp232 and His234 (shown in orange) are in H-bond distance to each other and His234, acid/base catalyst, as well as Asp318, the catalytic nucleophile (shown in orange), are H-bonding the PUGNAc inhibitor.
This mechanism, for instance, is performed by the well studied lysozyme that hydrolyzes the β-glycosidic linkage between N-acetylmuramic acid (MurNAc)4 and N-acetylglucosamine (GlcNAc) of the backbone polysaccharide of the bacterial cell wall compound peptidoglycan (murein). It was shown only recently that lysozyme proceeds though a covalent α-glycosyl enzyme (3) and not a long-lived oxocarbenium-ion intermediate as was proposed earlier (4). We are studying a group of bacterial β-N-acetylglucosaminidases, which hydrolyze the other glycosidic linkage in peptidoglycan, between GlcNAc and MurNAc, and are involved in turnover and recycling of the bacterial cell wall (5–10). These enzymes are classified on the basis of amino acid sequence and secondary structure to family 3 of glycosidases (according to the carbohydrate active enzymes (CAZY) data base), which comprises a heterogeneous group of exo-acting, retaining β-glycosidases that besides β-N-acetylglucosaminidases (EC 3.2.1.52), include β-glucosidases (EC 3.2.1.21), xylan 1,4-β-xylosidases (EC 3.2.1.37), glucan 1,3–1,4-β-glucosidases (EC 3.2.1.58 and 3.2.1.74), α-l-arabinofuranosidases (EC 3.2.1.55), and exo-1,3/1,4-glucanases (EC 3.2.1.-).
Family 3 β-N-acetylglucosaminidase from Vibrio furnisii (VfExoII) as well as related β-glucosidases were shown to proceed through a glycosyl enzyme intermediate. A conserved aspartate residue was identified in all cases as the catalytic nucleophile by trapping the glycosyl enzyme intermediate using slow substrates, proteolytic digestion, and subsequent mass spectrometry of the labeled peptide (11–13). Moreover, for some β-glucosidases of family 3, good evidence is provided by structural or kinetic analyses for a glutamate acting as the acid/base catalyst, e.g. exo-β-glucanase of Hordeum vulgare (14–16), β-glucosidase of Flavobacterium meningosepticum (17, 18), and the β-glucosylceramidase of Paenibacillus sp. TS12 (13). Intriguingly, the structure of the exo-β-glucanase of H. vulgare (HvExoI) reveals that the glutamate acid/base catalyst resides on a short helix on the less conserved C-terminal domain of the protein that comes into close contact to the active site region of the N-terminal domain (Fig. 1, B and C). A conserved glutamate, which may act as the acid/base catalyst, however, was never identified in the subset of β-N-acetylglucosaminidases contained in family 3 (Fig. 2). Moreover, some β-N-acetylglucosaminidases of this family even completely lack a C-terminal domain, and therefore need to provide a different residue for acid/base catalysis. It was shown earlier that family 3 β-N-acetylglucosaminidases are characterized by the highly conserved sequence pattern KH(F/I)PG(H/L)GX(4)D(S/T)H, which lays on the N-terminal domain (Fig. 2) and an involvement in acetamido group binding of the substrate was proposed (11).
FIGURE 2.
Partial multiple sequence alignment of selected glycosidases of family 3, including one domain and two domain β-N-acetylglucosaminidases as well as two domain β-glucosidases. The subfamilies can be distinguished by differences in the sequence pattern next to the conserved KH(F/Y) motif (boxed in black) on the N-terminal domain adjacent to β-strand 5 (=β5=>) of the N-terminal (β/α)8-barrel domain that extends into a loop region. The D(S/T)H motif (boxed in light gray) of β-N-acetylglucosaminidases, which contains the Asp-His catalytic dyad is missing in β-glucosidases of family 3. The C-terminal domain region (extending from β-strand 4 of the C-terminal (a/β/α)8-sandwich domain; =βIV=>) that carries the glutamate general acid/base in β-glucosidases (boxed in dark gray) is not conserved in β-N-acetylglucosaminidases or even missing entirely. The SwissProt data base identifier and protein names from which the partial sequences were obtained are indicated. Structural motifs were from the β-N-acetylglucosaminidase of V. cholerae (36), β-N-acetylglucosaminidase of B. subtilis (this work), and the β-exo-glucanase of H. vulgare (15).
Here we show that the Asp and His within this pattern (bold letters in the sequence shown above) that reside on the N-terminal domain of BsNagZ are directly involved in the mechanism of the β-N-acetylglucosaminidases subfamily of family 3 glycosidases. We present the structure of NagZ of Bacillus subtilis (BsNagZ), the first structure of a two-domain β-N-acetylglucosaminidase, along with kinetic analyses, which provide evidence for participation of the side chains of the conserved Asp and His residues during catalysis. Our results indicate that the histidine, instead of a glutamate, acts as acid/base catalyst, which undergoes hydrogen bonding with the aspartate residue, thereby forming a catalytic dyad that protonates the glycosidic oxygen in the first (glycosylation) step and deprotonates and activates water for nucleophilic attack of the glycosyl enzyme in the second (deglycosylation) step of the reaction (Fig. 1D). The function of this unique Asp-His dyad in glycoside hydrolysis resembles that of the Asp-His-Ser triad of serine proteases (19) as well as the Asp-His dyad of ribonucleases (20).
EXPERIMENTAL PROCEDURES
Bacterial Strains and Plasmids
B. subtilis 168 was obtained from the Bacillus Genetic Stock Center. Escherichia coli BL21(DE3) and E. coli expression vectors pET16b (PT7, Ampr, oripBR322, lacI, N-terminal His10 tag) and pET29b (PT7, Kanr, oripBR322, lacI, C-terminal His6 tag) were from Novagen. The reagents 4′-methylumbelliferyl N-acetyl-β-d-glucosaminide (4-Mu-β-GlcNAc) and 4′-nitrophenyl N-acetyl-β-d-glucosaminide (pNP β-GlcNAc) were obtained from Glycosynth. N-Acetyl-β-d-glucosaminyl azide (β-GlcNAc azide) was from Carbosynth. All other reagents were from Sigma unless otherwise stated.
Cloning and Site-directed Mutagenesis
BsnagZ was cloned as an cytoplasmic construct in which the N-terminal signal sequence was removed and exchanged by a His tag. DNA preparation, restriction enzyme digest, ligation, and transformation were performed according to standard techniques using 5 units of PWO DNA polymerase (Genaxxon Biosciences, Biberach, Germany) and 30 ng of chromosomal DNA. BsnagZ from B. subtilis 168 was cloned into pET16b without signal sequence (pET16b-BsNagZ) using the following primers: BsNagZ/FP, 5′-AAA ACC ATG GGC CAT ATG TTT TTC GGG GCC AGA CAG AC-3′; and BsNagZ/RP, 5′-T TTT CTC GAG TTA AAG CGG TCT TCC CGT TTT G-3′ (underlined are the recognition sites for the endonucleases NdeI and XhoI, respectively). Thirty cycles (15 s at 94 °C, 30 s at 55 °C, and 120 s at 72 °C) were performed in a thermal cycler and revealed a single 1.9-kb fragment (BsnagZ). BsnagZ′ missing the N-terminal signal sequence and the C-terminal domain was amplified using primers with recognition sites for restriction endonucleases NdeI and XhoI, respectively (underlined): BsNagZ′/FP, 5′-AAA ACC ATG GGC CAT ATG TTT TTC GGG GCC AGA CAG-3′ and BsNagZ′/RP 5′-GGG CTC GAG TGC TTT TTC AGC TAA TTT TTT CTC TGC-3′ (Thermo Fisher). Thirty cycles (30 s at 94 °C, 30 s at 50 °C, and 60 s at 72 °C) were performed and the 1.3-kb fragment was cloned into vector pET29b (pET29b-BsNagZ′). The vector pET16b-BsnagZ was used as template for site-directed mutagenesis of H234G. The following degenerated primers (Thermo Fisher) were used: H234G/FP as well as D232G/FP, 5′-GG AGA TAT ACC ATG GGC CAT C-3′; H234G/RP, 5′-C TTG GCC ATG GGA AAC GAG CGG CAG TCC ATA AYM GCT GTC AAC GTC CGT GTC TCC-3′; and D232G/RP, 5′-C TTG GCC ATG GGA AAC GAG CGG CAG TCC ATA ATG GCT CBC AAC GTC CGT GTC TCC ATG TC-3′ (the mutated codon is shown in bold with Y = C or T, M = A or C, and B = C, G, or T; the underlined residue is the restriction site for NcoI). Thirty cycles (30 s at 94 °C, 30 s at 50 °C, and 60 s at 72 °C) were performed. The amplified 758-bp fragments carrying the mutation were cleaved with NcoI and exchanged with the wild type fragment of pET16b-BsNagZ. Two mutants were further processed that give rise to H234G (AYM = ACC) and D232G (CBC = CCC) protein variants.
Overproduction and Purification of Proteins
E. coli strain BL21(DE3) (F− ompT hsdSB(rB−mB−) gal dcm) harboring pET16b-BsNagZ, pET16b-H234G, pET16b-D232G, or pET29b-BsNagZ′ were grown at 37 °C in Luria-Bertani (LB) medium supplemented with ampicillin (100 μg/ml) or kanamycin (50 μg/ml), respectively, under vigorous shaking to log-phase (A600 0.5–0.6). Production of the enzyme was induced by the addition of isopropyl β-d-thiogalactopyranoside to a final concentration of 1 mm. Incubation was continued for a further 3 h and the cells were harvested by centrifugation (4000 × g for 30 min at 4 °C), resuspended in ice-cold phosphate buffer (20 mm Na2HPO4 × 2H2O, 500 mm NaCl, pH 7.5), and lysed by passing the cells three times through a French pressure cell. The lysates were clarified by centrifugation at 100,000 × g for 1 h at 4 °C. The His-tagged proteins in the supernatant were purified by Ni2+-affinity chromatography on a 5-ml HisTrap column (Amersham Biosciences) pre-equilibrated with phosphate buffer and eluted from the column with imidazole (phosphate buffer supplemented with 500 mm imidazole, pH 7.5). To avoid cross-contamination of the proteins, new columns were used for each protein. The purity of the enzymes was assessed by SDS-PAGE. Fractions containing homogeneous protein were pooled, concentrated, and desalted by dialysis against 10 mm sodium acetate, pH 4.5, for crystallization or against 20 mm phosphate buffer, pH 7.5, for enzymatic assay studies at 4 °C. The protein concentrations were measured according to the method of Bradford with bovine serum albumin as a standard. 3 to 5 mg of pure protein was obtained from a 1-liter culture.
Crystallization and Data Collection
Purified BsNagZ was concentrated to 14 mg/ml using an Amicon Ultracentricon (Millipore) with a 10-kDa cut off. Complexes of BsNagZ with the inhibitor for co-crystallization were prepared by mixing PUGNAc with BsNagZ solution at a 10:1 molar ratio and incubating for 30 min at room temperature. Crystals of unliganded BsNagZ and its co-crystals with inhibitor PUGNAc were grown in 0.1 m sodium acetate, pH 4.9, using 24-well hanging drop plates. Presaturated protein drops were prepared by mixing protein and reservoir solution at ratios 1:1, 1:2, and 2:1, yielding final drop volumes of 2–3 μl. The reservoir contained 0.1 m sodium acetate, pH 4.9, and varying concentrations of polyethylene glycol 1000. Crystals were shock-frozen in liquid nitrogen. Data sets were collected at the Swiss Light Source beamline X06SA of the Paul Scherrer Institut, Villigen, Switzerland. Data processing was done with XDS (21) (see Table 1).
TABLE 1.
Crystallographic data collection and refinement statistics
| Parameters | BsNagZ | BsNagZ-PUGNAc |
|---|---|---|
| Crystal information | ||
| Space group | P1 | P1 |
| Solvent content (%) | 48.1 | 47.2 |
| Data collectiona | ||
| Unit cell dimensions (Å) | a = 58.4, b = 73.1, c = 83.6 | a = 58.5, b = 73.2, c = 83.8 |
| Unit cell dimensions | α = 79.8°, β = 69.6°, γ = 88.3° | α = 79.8°, β = 69.4°, γ = 88.2° |
| Wavelength (Å) | 0.9999 | 0.9792 |
| Resolution range (Å) | 49.18–1.40 (1.50–1.40) | 48.55–1.84 (1.95–1.84) |
| Total observations | 1,601,297 (66995) | 207,544 (32774) |
| Unique reflections | 228,046 (25215) | 104,555 (16575) |
| I/σ | 13.3 (2.7) | 7.6 (2.0) |
| Completeness (%) | 90.8 (53.7) | 93.5 (91.5) |
| Rmeasured (%) | 9.0 (57.2) | 10.9 (56.6) |
| Rmerged-F (%) | 8.0 (60.5) | 17.3 (71.1) |
| Refinement | ||
| Resolution range (Å) | 42.84–1.40 (1.42–1.40) | 48.55–1.84 (1.87–1.84) |
| Rworkb | 12.7 (31.5) | 18.7 (27.5) |
| Rfreeb | 16.6 (38.5) | 24.6 (31.5) |
| Model composition | ||
| Protein residues | 1,234 | 1,234 |
| Ligand atoms | 103 | |
| Water molecules | 1,388 | 1,527 |
| Average B (Å2) | 20.9 | 23.5 |
| Root mean square deviations | ||
| Bond length (Å) | 0.007 | 0.017 |
| Bond angles | 0.967° | 1.611° |
| Ramachandran plotc | ||
| Favored regions (%) | 97.8 | 96.8 |
| Allowed regions (%) | 99.8 | 99.8 |
| Twin law | –d | h, -k, h-l |
| Twin fraction | –d | 0.137 |
a Values in parentheses refer to the high resolution shell.
b Rwork and Rfree = Σh‖F(h)obs|−|F(h)c‖/|F(h)o| for reflections in the working and test sets (5% of all data), respectively.
c Regions defined by Molprobity (53).
d The twinning fraction was insignificant and therefore not taken into account during refinement.
Structure Determination
The structure of BsNagZ without ligand was solved first. An ensemble of PDB files (1EX1, 1IEX, 1J8V, 1LQ2, and 1X38 (14–16)) was defined in PHASER (22) as the starting point for molecular replacement. Overall amino acid sequence identities between the target enzyme and the related search models was low (23% and less) (23). Buccaneer (24) was used to build parts of two molecules of BsNagZ (996 out of 1284 residues as poly-Ala model). ARP/wARP (25) and Resolve (26) were used for completing the model. By using Resolve, 2-fold non-crystallographic symmetry (NCS) could be determined and used for phase improvement, leading finally to an almost complete model with 1139 amino acids of the BsNagZ sequence. The resulting electron density map was used for further manual model building in COOT (27) and refinement with Refmac5 and Phenix (28). After modeling water molecules and adding hydrogen atoms, the last anisotropic refinement resulted in a final R-factor of 12.7% (Rfree, 16.6%). The structure of BsNagZ in complex with its inhibitor PUGNAc was determined by using the first structure of BsNagZ as the search model in Molrep (29). The pseudo-merohedral twinning law was identified with the help of the CCP4 (30) program SFCHECK (31). Model refinement was done in Refmac5 and in Phenix, which allows refinement of the twinned structures. Manual modifications were done in COOT. After adding waters and modeling the inhibitor PUGNAc, the last isotropic refinement resulted in a final R-factor of 18.7% (Rfree, 24.6%). Refinement statistics are presented in Table 1. Atomic coordinates and structure factors for unliganded and liganded N-acetylglucosaminidases of B. subtilis have been deposited in the Protein Data Bank under accession code 3BMX and 3NVD, respectively.
Kinetic Studies
All Michaelis-Menten kinetics were carried out at least in triplicates using a discontinuous assay measuring the release of 4-methylumbelliferone from 4-Mu β-GlcNAc. The fluorescence of the released 4-methylumbelliferone was measured using a 96-well plate (Greiner bio-one) in a Spectramax M2 Microplate Reader (Molecular Devices) (excitation, 362 nm; emission, 448 nm) at 37 °C. Enzyme activity was measured using various concentrations of 4-Mu β-GlcNAc in Clark and Lubs solution (0.1 m KH2PO4, 0.1 m NaOH) at pH 5.8. Reactions (300 μl) were initiated by addition of enzyme (0.0158 mg of His, 0.01465 mg of Asp, and 1.5 × 10−5 mg of BsNagZ) and incubated over a period of 5 to 30 min at 37 °C.
The pH activity profiles of wild type BsNagZ and the mutants were determined in 0.1 m citric acid, 0.2 m disodium phosphate buffer (McIlvaine) ranging from 4.0 to 8.0, in 0.2 m sodium acetate/acetic acid buffer ranging from 4.0 to 5.6 and in Clark and Lubs solution in the range of 5.8–8.0. Reactions (300 μl) were initiated by addition of 1.5 mm 4-Mu β-GlcNAc and incubated over a period of 5–30 min at 37 °C. Protein amounts used were 4.5 × 10−5, 0.0114, and 0.01465 mg for BsNagZ, H234G, and D232G, respectively. All reactions were stopped by addition of 270 μl of 0.2 m Na2CO3, pH 10.0, to 30 μl of the reaction mixture. Kinetic parameters were obtained by direct fit of the rate versus substrate concentration data to the Michaelis-Menten equation using Prism 4 software (Graph Pad Software, Inc., La Jolla, CA). The extinction coefficient was determined by calibration using 4′-methylumbelliferone. One unit is defined as the amount of enzyme that hydrolyzes 1 μmol of substrate per min at pH 5.8 at 37 °C.
Mass Spectrometry
H234G was incubated with pNP β-GlcNAc over a period of 10 min until the enzyme/substrate solution exhibited a light yellow color indicating the release of pNP. The mass spectrometry analysis of the trapped intermediate (glycosyl-enzyme) was performed by electrospray ionization time of flight mass spectrometry (ESI-TOF-MS) (at the ZMBH, University of Heidelberg).
HPLC Analysis
High performance liquid chromatography (HPLC) was used for analysis of the β-GlcNAc azide generated by reaction of BsNagZ-H234G and BsNagZ-D232G with pNP β-GlcNAc (6 mm) in 100 mm sodium phosphate, pH 7.5, and 500 mm sodium azide, pH 7.5. In brief a reversed-phase column (Gemini RP-C18, 150 × 4.6 mm, 5 μm particle size, Phenomenex) was used at a flow rate of 0.5 ml/min, isocratic elution with 0.05% trifluoroacetic acid for 5 min, followed by a gradient from 0.05% trifluoroacetic acid to 10% acetonitrile containing 0.035% trifluoroacetic acid over a period of 50 min according to Ref. 32.
RESULTS
Purification, Crystallization, and Structure Determination
BsNagZ was overproduced in E. coli cytoplasm and purified by Ni2+-affinity chromatography to apparent homogeneity (23). The protein retained activity for several months at 4 °C and in the pH range between pH 4.0 and 8.0. BsNagZ tends to precipitate at a pH above 6 and low ionic strength but can be kept soluble at 12–13 mg/ml and high ionic strength (e.g. 20 mm sodium phosphate, pH 7.5, 500 mm sodium chloride). At acidic pH solubility behavior is reversed but the protein shows only low enzymatic activity; BsNagZ is soluble at low ionic strength (e.g. 20 mm sodium acetate, pH 4.5) but precipitates upon addition of 100 mm sodium chloride. Crystals of unliganded BsNagZ and its complex with the transition state-like inhibitor PUGNAc, O-(2-acetamido-2-deoxy-d-glucopyranosylidene)amino-N-phenylcarbamate (chemical structures of inhibitors and substrates of BsNagZ are shown in supplemental Fig. S1) were grown at pH 4.9 and 18 °C by vapor-phase equilibration as described under “Experimental Procedures.” The best results were obtained at polyethylene glycol 1000 reservoir concentrations between 27 and 33% (w/v). Within 2–3 weeks crystals reached a size of approximately 50 μm3 and exhibited triangular, rhombic, or cuboid habits. Data were collected at the Swiss Light Source synchrotron to resolutions of 1.4-Å and 1.84-Å for the native (unliganded) and inhibitor complex (liganded) protein crystals, respectively. All crystals belonged to space group P1 and possessed very similar unit cells with two BsNagZ molecules per cell (cf. Table 1). The inhibitor complex crystals displayed pseudo-merohedral twinning with a twinning fraction of 13.7%. The structures of native BsNagZ and its inhibitor complex were solved by molecular replacement as described under “Experimental Procedures.”
BsNagZ Has a Unique Two-domain Structure
The final atomic model comprises residues 26–642 (native enzyme numbering, corresponding to residues 31–647 of the engineered BsNagZ, lacking the N-terminal His tag) from both monomers of the asymmetric unit (structural parameters and refinement statistics, see Table 1). BsNagZ structure reveals two separate domains (Fig. 1E). The N-terminal domain (residues 26–420) adopts a (β/α)8-barrel fold (TIM-barrel), which is typical for catalytic domains of glycosidases and contain the conserved aspartate, the catalytic nucleophile of family 3 glycosidases. The C-terminal domain (residues 421–642) displays an αβα-sandwich fold. The surface buried between the domains (1407.5 Å2) indicates that both domains are intimately associated (33), however, no part of the C-terminal domain comes into close contact with the substrate/inhibitor binding site on top of the (β/α)8-barrel domain (Fig. 1, E and F). This distinguishes BsNagZ from HvExoI although both display weak sequence similarity (22% overall amino acid sequence identity) (15). As mentioned above, in HvExoI a short helix of the C-terminal domain, which carries the general acid/base catalyst (Glu491), protrudes into the active site (Fig. 1, B and C). This helix is missing in BsNagZ (Fig. 1, E and F).
An Asp-His Dyad in the Inhibitor Binding Site
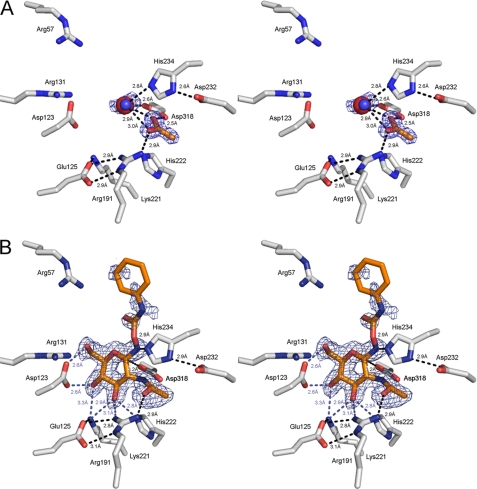
The overall structures of BsNagZ with and without the inhibitor (liganded and unliganded structures) are basically identical, including the inhibitor binding site arrangement. An acetate molecule in the unliganded BsNagZ structure (Fig. 3A) superimposes with the N-acetamido group of PUGNAc of the liganded structure (Fig. 3B). In the unliganded structure, a sodium ion lies within H-bond distance (2.6 Å) to the catalytic nucleophile Asp318 and an active site water localizes at 2.8 Å distance to His234 (Fig. 3A). In the liganded structure, the inhibitor sits in a cavity formed by the N-terminal (β/α)8-barrel and the GlcNAc moiety of PUGNAc makes multiple H-bond contacts with amino acid site chains constituting the active site of BsNagZ. The average B-factor of 12.9 for the pyranose ring of PUGNAc including the imine nitrogen is consistent with the observed set of H-bonding contacts to amino acids within the binding pocket. The high average B-factor of 56.4 and lack of continuous electron density of the aglycon of PUGNAc is likely caused by high flexibility due to the lack of H-bonding contacts. Intriguingly, Nϵ2 of His234 comes within hydrogen bonding distance (2.9 Å) to the imine nitrogen of PUGNAc (Fig. 3B). The histidine superimposes with the glutamate residue of HvExoI (cf. Fig. 1) and is a likely candidate for the general acid/base catalyst. Nδ1 of His234, furthermore, forms an H-bond to Asp232 (distance of 2.9 Å) (Fig. 3B).
FIGURE 3.
Stereo view of the native and liganded BsNagZ (PDB entries 3BMX and 3NVD, respectively). The Fo − Fc omit maps of the ligands within the active sites are shown, as calculated from the models and contoured at the 2.5-σ level. The orientation of catalytic residues Asp318 and Asp232/His234 as well as further residues in the binding pocket (Asp123, Glu125, Arg131, Arg191, Lys221, and His222) that are highly conserved among β-N-acetyl-glucosaminidases of family 3 (54), are almost identical to the native, unliganded (A) and liganded structure (B). Putative hydrogen bonds between protein and ligands, identified by using the criteria of proper geometry and a distance cut off of 3.2 Å are indicated. The blue mesh shows the maximum likelihood electron density map for acetate, sodium, and active water (A) and PUGNAc (B) contoured at 0.2 e−/Å3. Nϵ2 of His234 forms a hydrogen bond (2.8 Å) to a water molecule (blue sphere) that is situated in proximity of a bound acetate molecule in A or to the imine nitrogen (2.9 Å) of the transition state inhibitor PUGNAc (gray sticks) in B. The acetate molecule in the binding site in the native structure (A) is located at a position that superimposes with the acetamido group of the PUGNAc in the liganded structure (B). Arg57, which is highly conserved among members of the glucosidase, might be involved in binding of the natural substrate (muropeptides). The catalytic nucleophile (Oδ1 of Asp318) is in 2.6 or 3.2 Å distance to an enzyme bound sodium ion (red sphere) or C1 of the GlcNAc part of the inhibitor, respectively. His234 is in hydrogen bond distance to the nitrogen atom between GlcNAc and the aglycon ring of the inhibitor and Nδ1 of His234 is H-bonded to Oδ1 of Asp232.
Kinetic Properties
Besides natural peptidoglycan substrates (cf. Ref. 23, depicted in supplemental Fig. S1), BsNagZ readily hydrolyzes the chromogenic and fluorogenic substrates pNP β-GlcNAc and 4-Mu β-GlcNAc), respectively, which are convenient substrates for continuous assays and kinetic studies. Kinetics were determined with 4-Mu β-GlcNAc, because this substrate generates a fluorogenic product that can be measured with much higher sensitivity than chromogenic products. Hydrolysis of 4-Mu β-GlcNAc by wild type BsNagZ and mutants obey Michaelis-Menten kinetics and the kinetic constants are given in Table 2. To investigate the role of the Asp-His dyad in family 3 β-N-acetylglucosaminidases, His234 and Asp232 of BsNagZ were exchanged by a glycine. Both mutants showed severely reduced activity. As shown in previous studies with β-glycosidases, glycosylation (the first irreversible step that is reflected through kcat/Km) requires major assistance in protonation of the glycosidic oxygen by the general acid/base catalyst for cleavage of the substrates that have poor leaving groups. By contrast, the second step (deglycosylation, reflected through kcat) depends on the general acid/base catalyst functioning as base at this stage independent of the leaving group of the substrate (34). Exchanging His234 with glycine resulted in a 1900-fold reduction in kcat but only a 80-fold reduction in kcat/Km compared with wild type BsNagZ and with 4-Mu β-GlcNAc as substrate (a fairly good substrate; pKa of methylumbelliferone = 7.79 compared with the natural muropeptide substrates with an estimated pKa of about 14). The apparent second-order rate constant (kcat/Km) for substrate hydrolysis of 4-Mu β-GlcNAc by H234G thus was much less affected than the apparent first-order rate constant (kcat), indicating less impairment of the glycosylation step (reflected in kcat/Km) than the deglycosylation step (reflected in kcat), indicating a particularly important role of His234 in base catalysis. This leads to a significant accumulation of the covalent glycosyl-enzyme intermediate (cf. Fig. 4), which is reflected in a 24-fold reduced Michaelis-Menten constant (Km) for H234G compared with the wild type enzyme.
TABLE 2.
Kinetic parameters for substrate hydrolysis by BsNagZ and mutants
| Enzyme | Substrate | Km | kcat | kcat/Km | Km (wt)/Km | kcat (wt)/kcat | (kcat/Km (wt))/(kcat/Km) |
|---|---|---|---|---|---|---|---|
| μm | s−1 | s−1/mm | |||||
| WT | 4-Mu β-GlcNAc | 109.6 ± 4.3 | 6.42 ± 0.07 | 58.58 | 1 | 1 | 1 |
| H234G | 4-Mu β-GlcNAc | 4.57 ± 0.39 | 3.37 × 10−3 ± 4.8 × 10−5 | 0.74 | 24 | 1905 | 79.2 |
| D232G | 4-Mu β-GlcNAc | 56.24 ± 3.56 | 1.40 × 10−3 ± 2.6 × 10−5 | 0.025 | 1.95 | 4586 | 2343 |
FIGURE 4.
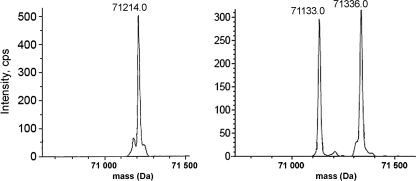
Transform of the electrospray mass spectrum of BsNagZ (left) and H234G (right) after incubation with pNP β-GlcNAc for 10 min, respectively. The peak with a molecular mass of 71,214 amu corresponds to BsNagZ (the calculated value for [BsNagZ]+ lacking methionine is 71214 amu). The peak with a molecular mass of 71,133 amu is the H234G mutant (the calculated value for [H234G]+ with His6 tag and lacking methionine is 71133.6 amu), the peak with a molecular weight of 71,336 amu is the H234G with covalently bound GlcNAc (the calculated value for [H234G]+ with GlcNAc and lacking methionine is 71336 amu). The mass shift of 203 Da corresponds to the GlcNAc residue covalently bound to H234G.
The rate of 4-Mu β-GlcNAc hydrolysis by the D232G mutant was 4500-fold reduced compared with wild type, which is an even larger reduction compared with the effect of the H234G mutation (Table 2). However, the Km was only a little affected by the D232G mutation (Table 2). This indicates a shift in the rate-determining step in the D232G mutant compared with H234G from deglycosylation in the latter to glycosylation in the former. This might be explained by a larger impairment of the protonation of the leaving group of the substrate (glycosylation step) in D232G compared with H234G, presumably because Asp232 mostly is required for His234 to function as a proton donor. Protonation of the glycosidic oxygen in catalysis by the H234G mutant might be substituted by small organic acids of the buffer (phosphate, acetate), which, however, cannot substitute general base catalysis (deglycosylation step). Congruently, the activity of the H234G mutant of BsNagZ in the Tris buffer was lower than in phosphate or acetate buffer but to low to determine kinetic parameters.
pH Dependence of Catalysis
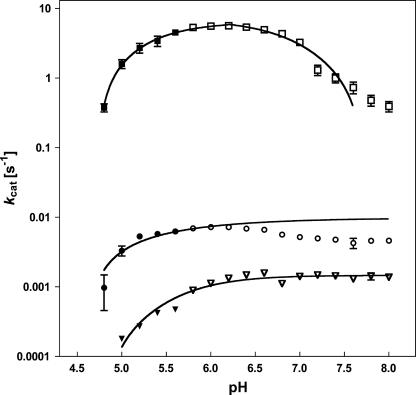
The pH activity profile of wild type BsNagZ was compared with H234G and D232G mutants. The plots of log kcat as a function of pH are shown in Fig. 5 and supplemental Fig. S3. The pH activity profile for the BsNagZ-catalyzed hydrolysis of 4-Mu β-GlcNAc resembles a bell-shaped curve as expected due to involvement of two ionizable groups in catalysis. The maximal catalytic activity ranges from 5.8 to 6.2 in Clark and Lubs solution (pH 5.8–8.0) as well as in McIlvaine buffer (pH 4.0–8.0). The pH activity profile of BsNagZ indicates acid ionization constants of the nucleophile and the general acid/base catalytic residue (pKa1 and pKa2) around 5.0 and 7.0, respectively. Hydrolysis of 4-Mu β-GlcNAc by H234G and D232G mutants was extremely slow and the pH profiles for both protein variants retained activity at alkaline pH, suggesting the elimination of a catalytic acid/base residue with a pKa value of ∼7.0 in the protein variants.
FIGURE 5.
pH activity profiles of BsNagZ and the mutants. kcat values at different pH were determined for wild type BsNagZ (■, □) and mutants H234G (●, ○) and D232G (▾, ▿). The buffers were: 0.1 m NaAc, pH 4.0–5.6, solid symbols; and 0.1 m KHPO4, 0.1 m NaOH, pH 5.8–8.0, open symbols. Data were from a representative experiment measured in triplicate (cf. supplemental Fig. S3). Solid lines are fits of the data supposing that kcat values depend on the enzyme being in an acid form (H234G, D232G) or depending on two residues being in an acidic and basis form (BsNagZ).
Accumulation of the Glycosyl-Enzyme Intermediate
Removing the general acid/base catalysts in family 3 glycosidases leads to accumulation of the glycosyl-enzyme intermediate. This was also the case in the H234G mutant as reflected by a small Km value. Furthermore, direct evidence that His234 functions as the acid/base catalyst was performed by ESI-MS measurement. After reaction of the H234G mutant with the substrate pNP β-GlcNAc two protein species were observed (Fig. 4). The mass of 71.133 kDa corresponds to the unmodified protein, whereas the mass of 71.336 kDa corresponds to the glycosyl-enzyme intermediate. The mass difference of 203 Da is in accordance with the mass of bound N-acetylglucosaminyl.
Chemical Rescue
As shown above, deglycosylation is the rate-limiting step in hydrolysis of 4-Mu β-GlcNAc by BsNagZ H234G, which leads to accumulation of the glycosyl-enzyme intermediate. It has been shown for many glycosidases that the addition of external small anionic nucleophiles like azide can compensate for the loss of the base residue, resulting in rescue of activity (13, 18). However, activity of the H234G mutant of BsNagZ could not be restored by the addition of azide, although replacement of His234 by the small glycine should allow accommodation of small compounds in the active site (see supplemental Table S1). Possibly, binding of the small anionic azide to the active site is disfavored by its negative charge. However, β-azide product formation was identified upon pNP β-GlcNAc cleavage by D232G (and to some extent also by H234G) in the presence of azide (supplemental Fig. S4). With D232G a 2-fold rate enhancement was observed in the presence of azide (supplemental Table S1), possibly due to accelerating the rate-determining glycosylation step in these mutant.
DISCUSSION
N-Acetylglucosaminidases of family 3 of glycosidases are involved in peptidoglycan turnover and cell wall recycling of bacteria (23, 32). In this process they liberate, for instance, inducers of chromosomal β-lactamases in Gram-negative bacteria (35, 36) as well as spore germinants in the Gram-positive bacterium B. subtilis (37). A detailed understanding of the mechanism of these enzymes is a prerequisite for the design of potent selective inhibitors that may serve as novel therapeutic agents. The identification of evolutionary and structural relationships within glycosidases, which led to the classification into families (CAZY), has greatly facilitated the efforts toward the rational design of such inhibitors. Given that structure is more conserved than sequence, it is assumed that the mechanism of glycoside hydrolysis is identical for all members of a family, which makes this classification particularly valuable. Catalytic residues identified in one member of a family, in general, allowed the prediction of catalytic residues in others. In almost all retaining glycosidases, in which the general acid/base catalyst has been reliably identified to date, it was an enzymic glutamic or aspartic acid residue (cf. CAZY). This, however, does not hold for all members of family 1 glycosidases. Myrosinases of this family cleave the highly reactive S-glycoside sinigrin without requiring protonic assistance for the intermediate glycosylation step in catalysis, hence lacking a general acid/base residue. The subsequent deglycosylation step, however, depends on the unusual coenzyme ascorbic acid that acts as a base catalyst in the hydrolysis of the glycosyl-enzyme intermediate (38). We recognized that a glutamate residue that acts as acid/base residue is also missing in the β-N-acetylglucosaminidase subfamily of family 3 glycosidases (cf. Fig. 2). It would have been possible that substrate participation in the hydrolysis of natural substrates (see supplemental Fig. S1) by these enzymes may not require assistance by a general acid/base catalyst.
This study now showed that family 3 β-N-acetylglucosaminidases, instead of a glutamate general acid/base, involve an Asp-His catalytic dyad, which is unique for glycosidases. The crystal structure of BsNagZ, the first structure of a two-domain β-N-acetylglucosaminidase of family 3 glycosidases, revealed that the Asp-His dyad superimposes with a glutamate residue that had been identified as the general acid/base catalyst in other members of family 3 of glycosidases. Most intriguing evidence for the Asp232–His234 dyad functioning as acid/base catalyst in family 3 β-N-acetylglucosaminidases was obtained from kinetic studies with protein variants. Removal of His234 or Asp232 of the dyad by site-directed mutagenesis resulted in disappearance of the characteristic bell-shaped pH profile of glycosidases and rendered activity pH-independent at higher pH ranges. It therefore suggests that a group was deleted that is responsible for pH dependence in the basic range. Moreover, the first-order rate constants were decreased compared with wild type enzyme by 1900- and 4500-fold for 4-Mu β-GlcNAc on exchange of His234 or Asp232 with glycine, respectively, which are reductions in kcat values that are typically determined for general acid/base mutant glycosidases (13, 18). The second-order rate constants kcat/Km, which reflects the first irreversible step (34), is relatively little affected in the H234G mutant but severely affected in the D232G mutant. This indicates a shift in the rate-limiting step in the His mutant toward deglycosylation, which is also indicated by accumulation of the N-acetylglucosaminyl-enzyme intermediate by the H234G mutant. The glycosylation step, which involves acid catalysis, however, is the rate-limiting step of wild type BsNagZ as well as BsNagZ-D232G. Together, all these results, structural as well as kinetic data, provide evidence that His234 of the Asp-His dyad of family 3 β-N-acetylglucosaminidases acts as the general acid/base that is assisted by Asp232. This contrasts with a study on Clostridium paraputrificum M-21 β-N-acetylglucosaminidase (Nag3A). A conserved aspartate residue (Asp175) on the N-terminal domain was proposed as the acid/base catalyst (39) and replacement of Asp175 with Ala abolished the activity of Nag3A. However, clear kinetic evidence for a role as acid/base catalyst of the above mentioned residue is lacking.
The amino acid histidine is perfectly qualified as a general acid/base because it has a pKa near neutrality. Nevertheless, involvement of a histidine residue, respectively, in an Asp-His dyad, is unique in glycosidases. It is, however, commonly found in enzymes cleaving phosphodiester bonds, e.g. ribonucleases (20, 40). The major role of the aspartate of the dyad in ribonucleases may be to orient the proper tautomer of the histidine for catalysis. Notably, in bovine ribonucleases Nδ1 rather that Nϵ2 faces the phosphodiester (20). The His-Asp-Ser catalytic triad is renowned for serine proteases (19) and lipases (41) and the Asp-His hydrogen bond in the catalytic triad is known to contribute greatly to catalysis, potentially via forming of a short, strong (low-barrier) hydrogen bond (42). There is no evidence for the formation of a low-barrier hydrogen bond between His234 and Asp232 for the family 3 β-N-acetylglucosaminidases. Possibly the major role of Asp232 in BsNagZ is its influence on proton dissociation of Nϵ2 of His234 for catalysis. Frank and Wen (43) suggested a cooperative behavior in chains of hydrogen-bonded molecules that may sharpen the acid/base behavior of the His for catalysis (reviewed in Ref. 44). The catalytic triad His57-Asp102-Ser195 of chymotrypsin (bovine chymotrypsin numbering) operates in this way: Ser195 functions as a nucleophilic catalyst, assisted by Nϵ2 of His57 that serves as acid/base catalyst, and residue Asp102 assists in acid/base catalysis by hydrogen bonding to Nδ1 of His57 increasing the pKa of His57. In the native BsNagZ structure (Fig. 3A), Asp232 is in short H-bond distance (2.6 Å) to His234 and electron density that can be attributed to a water molecule in the H-bond distance to Nϵ2 of His234. Although, at 1.4-Å resolution this has to be taken with caution, the situation clearly resembles that of chymotrypsin, in which the serine of the triad forms a short H-bond with the His57 (19). In the liganded structure His234 H-bonds to the glycosidic oxygen-mimicking nitrogen of PUGNAc and the Asp232–His234 distance is significantly larger (2.9 Å). According to the suggested mechanism (Fig. 1D), in BsNagZ the catalytic His234 forms an ion pair with Asp232 thereby allowing protonation of the extracyclic oxygen of the glycosidic bond and facilitating removal of the leaving group upon nucleophilic attack of Asp318. In a second step, the His removes a proton of an incoming water molecule thereby hydrolyzing the glycosyl-enzyme intermediate.
The catalytic nucleophile and the Asp-His dyad are located ∼6.3 Å apart, which is in the range of catalytic residues involved in bond cleavage via a two-step double displacement mechanism in glycosidases (1). They reside on the N-terminal domain of BsNagZ and are positioned at the carboxyl-terminal ends of β-strands 5 and 7, respectively. The Asp-His dyad lays on an extended loop that occupies a position after strand 4 in the BsNagZ molecule due to the shortened strand 4 loop, thereby resembling the situation of the 4/7-superfamily of glycoside hydrolases (clan GH-A glycoside hydrolases) in which the nucleophile is positioned at the C terminus of β-strand 7 as in family 3 glycosidases but the acid/base catalyst lays in a loop extending β-strand 4 (45, 46).
Recently, the crystal structure of a single domain β-N-acetylglucosaminidases of family 3 of glycosidases from Vibrio cholerae, VcNagZ, in the presence of the competitive transition state-like inhibitor PUGNAc has been reported (36). This structure, however, provided no information regarding the identity of a putative acid/base catalyst. Apparently in this structure, a flexible loop carrying the Asp-His dyad is flipped outward. Moreover, the aspartate nucleophile within this structure is distorted, indicating a non-physiological conformation of the active site in the crystal or an unproductive binding of the inhibitor (cf. Fig. 1E). We recently solved a further BsNagZ structure, in which a short loop of the protein that carries the Asp-His dyad is moved outwards away from the active site.5 It can be speculated that the proper orientation of the Asp-His dyad in this enzymes is induced upon substrate binding, providing a high degree of substrate specificity.
The obvious question is why the sub-family 3 β-N-acetylglucosaminidases apparently are the only glycosidases that act by a catalytic mechanism involving an Asp-His dyad. One rational for the replacement of an acid/base glutamate for a dyad might be the negative charge of the natural substrates of these enzymes, which are MurNAc or 1,6-anhydroMurNAc containing cell wall fragments as mentioned above. The negative charge of the carboxylic acid of these molecules might interfere with the use of a negative charged acid/base catalyst in the active site. A similar situation holds for sialidases, which have been shown to utilize tyrosine as a catalytic nucleophile rather than a carboxylate nucleophile (47, 48). It was argued that the anomeric center of the sialic acid sugars bears an anionic carboxylate residue and the nucleophile attack by an anionic nucleophile is therefore disfavored. Sialytransferases of family 42 glycosyltransferases have been reported to utilize a histidine as the base catalyst that abstracts the proton from the nucleophilic hydroxyl group of the sugar acceptor, thereby facilitating attack on the CMP-Neu5Ac donor nucleotide (49, 50). Further precedence for the role of histidine residues as base catalysts are reported (51, 52).
The residues His234 and Asp232 are completely conserved in the subfamily of β-N-acetylglucosaminidases of family 3 glycosidases located in the conserved sequence pattern KH(F/I)PG(H/L)GX(4)D(S/T)H, which is used as an identifier for members of the subfamily. The Asp-His dyad is suitably positioned to act as the acid/base catalyst (cf. supplemental Fig. S2) and may substitute the “normal” carboxylic acid acid/base catalyst to act on substrates bearing a negative charged residue. Our findings will facilitate the development of mechanism-based inhibitors that selectively target family 3 β-N-acetylglucosaminidases, which are involved in cell wall turnover, release of spore germinants, and induction of β-lactamase in bacteria.
Supplementary Material
Acknowledgments
We appreciate the help of the Swiss Light Source beamline staff of the Paul Scherrer Institute, Villingen, Switzerland, with data collection. We thank David Vocadlo, Simon-Fraser University, Burnaby, Canada, for providing PUGNAc inhibitor and Jocelyne Fiaux, ZMBH, University of Heidelberg, for measuring ESI-MS.
This work was supported in part by Grant MA2436/4 and a Heisenberg stipend (MA2436/3) from the Deutsche Forschungsgemeinschaft (to C. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S4.
The atomic coordinates and structure factors (codes 3BMX and 3NVD) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
S. Litzinger, M. Krug, S. Fischer, K. Diederichs, W. Welte, and C. Mayer, unpublished results.
- MurNAc
- N-acetylmuramic acid (2-acetamido-2-deoxy-3-O-[(R)-1-carboxyethyl]-d-glucopyranose)
- PUGNAc
- O-(2-acetamido-2-deoxy-d-glucopyranosylidene)amino N-phenylcarbamate
- 4-Mu β-GlcNAc
- 4′-methylumbelliferyl N-acetyl-β-d-glucosaminide
- pNP β-GlcNAc
- 4′-nitrophenyl N-acetyl-β-d-glucosaminide
- N-acetyl-β-d-glucosaminyl azide
- (β-GlcNAc azide)
- BsNagZ
- NagZ of Bacillus subtilis.
REFERENCES
- 1.Rye C. S., Withers S. G. (2000) Curr. Opin. Chem. Biol. 4, 573–580 [DOI] [PubMed] [Google Scholar]
- 2.Zechel D. L., Withers S. G. (2000) Acc. Chem. Res. 33, 11–18 [DOI] [PubMed] [Google Scholar]
- 3.Vocadlo D. J., Davies G. J., Laine R., Withers S. G. (2001) Nature 412, 835–838 [DOI] [PubMed] [Google Scholar]
- 4.Phillips D. C. (1967) Proc. Natl. Acad. Sci. U.S.A. 57, 416–436 [Google Scholar]
- 5.Mayer C., Vocadlo D. J., Mah M., Rupitz K., Stoll D., Warren R. A., Withers S. G. (2006) FEBS J. 273, 2929–2941 [DOI] [PubMed] [Google Scholar]
- 6.Dahl U., Jaeger T., Nguyen B. T., Sattler J. M., Mayer C. (2004) J. Bacteriol. 186, 2385–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaeger T., Arsic M., Mayer C. (2005) J. Biol. Chem. 280, 30100–30106 [DOI] [PubMed] [Google Scholar]
- 8.Uehara T., Suefuji K., Jaeger T., Mayer C., Park J. T. (2006) J. Bacteriol. 188, 1660–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaeger T., Mayer C. (2008) Cell Mol. Life Sci. 65, 928–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaeger T., Mayer C. (2008) J. Bacteriol. 190, 6598–6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vocadlo D. J., Mayer C., He S., Withers S. G. (2000) Biochemistry 39, 117–126 [DOI] [PubMed] [Google Scholar]
- 12.Dan S., Marton I., Dekel M., Bravdo B. A., He S., Withers S. G., Shoseyov O. (2000) J. Biol. Chem. 275, 4973–4980 [DOI] [PubMed] [Google Scholar]
- 13.Paal K., Ito M., Withers S. G. (2004) Biochem. J. 378, 141–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hrmova M., Varghese J. N., De Gori R., Smith B. J., Driguez H., Fincher G. B. (2001) Structure 9, 1005–1016 [DOI] [PubMed] [Google Scholar]
- 15.Varghese J. N., Hrmova M., Fincher G. B. (1999) Structure Fold Des. 7, 179–190 [DOI] [PubMed] [Google Scholar]
- 16.Hrmova M., De Gori R., Smith B. J., Vasella A., Varghese J. N., Fincher G. B. (2004) J. Biol. Chem. 279, 4970–4980 [DOI] [PubMed] [Google Scholar]
- 17.Chir J., Withers S., Wan C. F., Li Y. K. (2002) Biochem. J. 365, 857–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y. K., Chir J., Tanaka S., Chen F. Y. (2002) Biochemistry 41, 2751–2759 [DOI] [PubMed] [Google Scholar]
- 19.Hedstrom L. (2002) Chem. Rev. 102, 4501–4524 [DOI] [PubMed] [Google Scholar]
- 20.Schultz L. W., Quirk D. J., Raines R. T. (1998) Biochemistry 37, 8886–8898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 22.Storoni L. C., McCoy A. J., Read R. J. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 432–438 [DOI] [PubMed] [Google Scholar]
- 23.Litzinger S., Duckworth A., Nitzsche K., Risinger C., Wittmann V., Mayer C. (2010) J. Bacteriol. 192, 3132–3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cowtan K. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 25.Morris R. J., Perrakis A., Lamzin V. S. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 968–975 [DOI] [PubMed] [Google Scholar]
- 26.Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1937–1940 [DOI] [PubMed] [Google Scholar]
- 27.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 28.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 29.Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 30.Collaborative Computational Project. N. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 31.Vaguine A. A., Richelle J., Wodak S. J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 191–205 [DOI] [PubMed] [Google Scholar]
- 32.Cheng Q., Li H., Merdek K., Park J. T. (2000) J. Bacteriol. 182, 4836–4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bahadur R. P., Chakrabarti P., Rodier F., Janin J. (2004) J. Mol. Biol. 336, 943–955 [DOI] [PubMed] [Google Scholar]
- 34.Wolfenden R. (1976) Annu. Rev. Biophys. Bioeng. 5, 271–306 [DOI] [PubMed] [Google Scholar]
- 35.Jacobs C., Frère J. M., Normark S. (1997) Cell 88, 823–832 [DOI] [PubMed] [Google Scholar]
- 36.Stubbs K. A., Balcewich M., Mark B. L., Vocadlo D. J. (2007) J. Biol. Chem. 282, 21382–21391 [DOI] [PubMed] [Google Scholar]
- 37.Shah I. M., Laaberki M. H., Popham D. L., Dworkin J. (2008) Cell 135, 486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burmeister W. P., Cottaz S., Rollin P., Vasella A., Henrissat B. (2000) J. Biol. Chem. 275, 39385–39393 [DOI] [PubMed] [Google Scholar]
- 39.Li H., Zhao G., Miyake H., Umekawa H., Kimura T., Ohimiya K., Sakka K. (2006) Biosci. Biotechnol. Biochem. 70, 1127–1133 [DOI] [PubMed] [Google Scholar]
- 40.Quirk D. J., Raines R. T. (1999) Biophys. J. 76, 1571–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brady L., Brzozowski A. M., Derewenda Z. S., Dodson E., Dodson G., Tolley S., Turkenburg J. P., Christiansen L., Huge-Jensen B., Norskov L., et al. (1990) Nature 343, 767–770 [DOI] [PubMed] [Google Scholar]
- 42.Ash E. L., Sudmeier J. L., De Fabo E. C., Bachovchin W. W. (1997) Science 278, 1128–1132 [DOI] [PubMed] [Google Scholar]
- 43.Frank H. S., Wen W. Y. (1957) Discussions Faraday Soc. 24, 133–140 [Google Scholar]
- 44.Ludwig R. (2001) Angew. Chem. Int. Ed. Engl. 40, 1808–1827 [PubMed] [Google Scholar]
- 45.Henrissat B., Davies G. (1997) Curr. Opin. Struct. Biol. 7, 637–644 [DOI] [PubMed] [Google Scholar]
- 46.Jenkins J., Lo Leggio L., Harris G., Pickersgill R. (1995) FEBS Lett. 362, 281–285 [DOI] [PubMed] [Google Scholar]
- 47.Watts A. G., Damager I., Amaya M. L., Buschiazzo A., Alzari P., Frasch A. C., Withers S. G. (2003) J. Am. Chem. Soc. 125, 7532–7533 [DOI] [PubMed] [Google Scholar]
- 48.Watts A. G., Oppezzo P., Withers S. G., Alzari P. M., Buschiazzo A. (2006) J. Biol. Chem. 281, 4149–4155 [DOI] [PubMed] [Google Scholar]
- 49.Chan P. H., Lairson L. L., Lee H. J., Wakarchuk W. W., Strynadka N. C., Withers S. G., McIntosh L. P. (2009) Biochemistry 48, 11220–11230 [DOI] [PubMed] [Google Scholar]
- 50.Ni L., Chokhawala H. A., Cao H., Henning R., Ng L., Huang S., Yu H., Chen X., Fisher A. J. (2007) Biochemistry 46, 6288–6298 [DOI] [PubMed] [Google Scholar]
- 51.Whiteson K. L., Chen Y., Chopra N., Raymond A. C., Rice P. A. (2007) Chem. Biol. 14, 121–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Legler P. M., Massiah M. A., Mildvan A. S. (2002) Biochemistry 41, 10834–10848 [DOI] [PubMed] [Google Scholar]
- 53.Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 54.Cournoyer B., Faure D. (2003) J. Mol. Microbiol. Biotechnol. 5, 190–198 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.