Abstract
Angiotensin-I-converting enzyme (ACE) plays a critical role in the regulation of blood pressure through its central role in the renin-angiotensin and kallikrein-kinin systems. ACE contains two domains, the N and C domains, both of which are heavily glycosylated. Structural studies of ACE have been fraught with severe difficulties because of surface glycosylation of the protein. In order to investigate the role of glycosylation in the N domain and to create suitable forms for crystallization, we have investigated the importance of the 10 potential N-linked glycan sites using enzymatic deglycosylation, limited proteolysis, and mass spectrometry. A number of glycosylation mutants were generated via site-directed mutagenesis, expressed in CHO cells, and analyzed for enzymatic activity and thermal stability. At least eight of 10 of the potential glycan sites are glycosylated; three C-terminal sites were sufficient for expression of active N domain, whereas two N-terminal sites are important for its thermal stability. The minimally glycosylated Ndom389 construct was highly suitable for crystallization studies. The structure in the presence of an N domain-selective phosphinic inhibitor RXP407 was determined to 2.0 Å resolution. The Ndom389 structure revealed a hinge region that may contribute to the breathing motion proposed for substrate binding.
Keywords: Glycosylation, Metalloprotease, Mutagenesis Mechanisms, Protease Inhibitor, Protein Domains, Protein Structure, Angiotensin-converting Enzyme, Phosphinic Peptide, Thermal Stability
Introduction
Angiotensin-I-converting enzyme (ACE4; EC 3.4.15.1) consists of two domains, the N and C domains, both of which are able to cleave the tensor peptides angiotensin I and bradykinin in vitro, although recent findings indicate that the C domain is the major site of angiotensin I cleavage in vivo (1, 2). ACE is also known to cleave a number of other substrates, including substance P, angiotensin 1–7, gonadotropin-releasing hormone, and acetyl-seryl-aspartyl-lysyl-proline (Ac-SDKP), of which the last three are cleaved predominantly by the N domain (3, 4). Ac-SDKP is a regulatory peptide that displays a number of beneficial functions, including the prevention of or reversal of cardiac, vascular, and renal inflammation and fibrosis (5). Increased levels of circulating Ac-SDKP, following ACE inhibition, are thought to contribute to the beneficial effects of ACE inhibitors by a novel mechanism whereby Ac-SDKP inhibits collagen deposition in the left ventricle of the heart following vascular injury (6). These findings highlight a role for the development of inhibitors selective for the N domain of ACE.
A highly specific phosphinic inhibitor, Ac-Asp-l-PheΨ(PO2CH2)-l-Ala-Ala-NH2 (RXP407), displaying ∼200-fold selectivity for the N domain, has been developed (7). Although this inhibitor is not a good drug candidate due to its large size and poor bioavailability, it was able to increase plasma Ac-SDKP levels without affecting blood pressure in a rat model, thus illustrating proof of concept for selective N domain inhibition (1). Recent work that made use of N domain active site mutants has shown that two S2 pocket residues, Tyr369 and Arg381, are likely to play an important role in conferring the N domain selectivity of RXP407 (8).
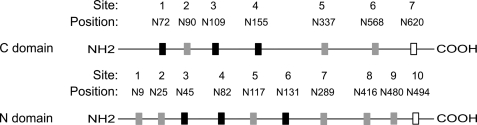
Both the N and C domains of ACE are heavily glycosylated, with the N domain containing 10 and the C domain containing seven potential N-glycosylation sites, although the most C-terminal site is not glycosylated in the C domain and is predicted to be unglycosylated in the N domain (9, 10). It should be noted that the locations of these potential N-glycosylation sites are unique to each domain, with the exception of sites 1, 3, and 4 in the C domain, which map to equivalent positions for sites 3, 4, and 6 in the N domain (Fig. 1). Glycosylation has been shown to have a prominent role in the folding, localization, and stability of glycoproteins as well as conveying resistance to proteolysis (11, 12). In this regard, when ACE is expressed in bacterial cells that lack complex eukaryotic glycosylation machinery, or when expressed in the presence of the glycosylation inhibitor tunicamycin, the expressed protein is inactive and rapidly degraded (13).
FIGURE 1.
Schematic representation of the potential N-linked glycosylation sites on the N and C domains of ACE. Black boxes, conserved glycosylation sequons; gray boxes, sequons unique to each domain; open boxes, unglycosylated sequons.
A significant difference between the two domains is their thermal stability. The N domain has been shown to have a Tm of 70 °C, 15 °C higher than that of the C domain (Tm = 55 °C), rendering it more thermostable (10, 14). It has previously been suggested that this difference in thermal stability is attributable to the fact that the N domain has a greater number of α-helices, a greater degree of glycosylation, and an increased proline content (14). Recent work has shown that the N-linked glycans of the C domain contribute significantly to its thermal stability, whereas in contrast, the presence of O-linked glycans had no effect (10).
Given the importance of the N domain in the cleavage of Ac-SDKP and the potential for N domain-selective inhibitors, it is desirable to carry out high throughput enzyme-inhibitor crystallization studies for structure-based drug design. Although a crystal structure of the N domain has been determined (15), the crystallization process was not readily reproducible. Because glycosylation interferes with protein crystallization and subsequent diffraction study, we have determined the minimal glycosylation requirements and the role of glycosylation in the N domain in order to generate a variant of the N domain suitable for high throughput enzyme-inhibitor crystallization. From one of these mutants, we have elucidated the structure of the N domain in complex with the domain-specific phosphinic peptide inhibitor RXP407. Furthermore, thermal denaturation studies have revealed that the N-terminal glycans are important for the stability of the ACE N domain.
EXPERIMENTAL PROCEDURES
Materials
Peptide:N-glycosidase F (PNGase F; proteomics grade), trypsin (modified, sequencing grade), and endoproteinase Glu-C (proteomics grade) were purchased from Sigma. Synthesis of RXP407 was as described previously (7).
Expression and Purification of Recombinant N Domain Proteins
Constructs were transfected into Chinese hamster ovary-K1 (CHO) cells as described previously (16). Soluble recombinant N domain proteins were purified from conditioned medium by lisinopril affinity chromatography, as described previously (16), with the exception that the harvested medium and wash buffer were adjusted to 800 mm NaCl (17).
Deglycosylation of Purified ACE N Domain Protein
Purified N domain (1 mg/ml) in 50 mm KH2PO4, pH 7.5 (0.2% SDS, 1% β-mercaptoethanol) was denatured by boiling for 5 min and then cooled on ice. Digestion with PNGase F (100 units/ml) was carried out at 37 °C overnight.
In-gel Protease Digestion for Mass Spectrometry Analysis
Deglycosylated ACE N domain was fractionated using 10% SDS-PAGE, and protein was visualized by Coomassie staining. The N domain band was excised, cut into 1-mm3 gel slices, and destained in 200 mm NH4HCO3, 50% acetonitrile. Gel slices were dehydrated before reduction with 5 mm triscarboxyethyl phosphine in 100 mm NH4HCO3 for 30 min at 56 °C. Excess triscarboxyethyl phosphine was removed, and the gel slices were dehydrated. Cysteines were protected with the addition of 100 mm iodoacetamide in 100 mm NH4HCO3 and incubated at room temperature in the dark for 30 min. Gel slices were dehydrated and washed with 50 mm NH4HCO3 and dehydrated again. Gel slices were rehydrated by the addition of 20 ng/μl protease (trypsin or endoproteinase Glu-C in 50 mm NH4HCO3) and digested at 37 °C overnight. Peptides were solubilized with 50 μl of 0.1% trifluoroacetic acid (TFA). The peptides were lyophilized, rehydrated in 50 μl of water, and concentrated to less than 20 μl to remove residual NH4HCO3.
Mass Spectrometry
Mass spectra were recorded on an ABI 4800 MALDI-TOF/TOF mass spectrometer (Applied Biosystems Foster City, CA), using positive reflector mode. Spectra were generated with 400 laser shots/spectrum, at an intensity of 3800 (arbitrary units), with a grid voltage of 16 kV. All peptide-containing spots were internally calibrated using trypsin autolytic fragments. The matrix used was α-cyano-4-hydroxycinnamic acid, at a final concentration of 5 mg/ml matrix in 40% acetonitrile, 0.1% TFA, 10 mm NH4H2PO4 (Fluka, Buchs, Switzerland).
Construction of N Domain Glycosylation Mutants
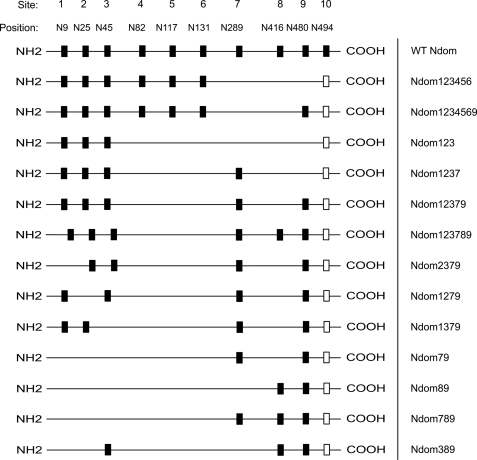
Site-directed mutagenesis of the glycosylation sequons was carried out in pBSKII-D629 (15, 18, 19), by converting the Asn residues in the various Asn-Xaa-Ser/Thr glycosylation sequons to Gln (see Fig. 4). Oligonucleotide primers containing the mutations of interest were synthesized by Inqaba Biotech (Pretoria, South Africa), and reagents used for site-directed mutagenesis were purchased from Promega (Madison, WI) and Kapa Biotech (Cape Town, South Africa). Successful mutagenesis was confirmed by automated DNA sequencing (Macrogen Inc., Seoul, Korea), after which constructs were cloned into pcDNA3.1+ (Invitrogen), using the EcoRI and XbaI restriction sites. The final sequence comprised residues 1–629 of the mature somatic ACE sequence and contained two mismatch mutations, P576L and Q545R.
FIGURE 4.
Schematic representation of the N domain glycosylation sites and the different glycoforms generated. Black boxes, intact N-glycosylation sites; open boxes, the unglycosylated sequon 10 (Asn494), which was not modified during mutagenesis.
RT-PCR
Total RNA was isolated from CHO cells using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. RNA concentration was determined by spectrophotometry at 260 nm. Reverse transcription of total mRNA was carried out using the Advantage RT-for-PCR kit (Clontech), in a Hybaid PCR Sprint thermocycler (Mandel Scientific Company Inc., Guelph, Canada), according to the manufacturer's instructions. Specific amplification of ACE N domain cDNA was achieved using the forward and reverse primers 5′-TTTGCCTGGGAGGGCTGGCA-3′ and 5′-TGGTCCACCAAGTAGCCAAA-3′, which correspond to nucleotides 581–600 and 1421–1440, respectively, of ACE N domain (GenBankTM accession number NM_000789.2). Ten microliters of total cDNA product was used as a template in each 50-μl PCR. The cycling parameters consisted of one cycle of 95 °C for 2 min and then 20 cycles of 95 °C for 20 s, 58 °C for 0.5 min, 68 °C for 1.5 min, followed by a final extension cycle of 3 min at 68 °C. PCR products were electrophoresed on a 1% agarose gel using O'GeneRuler DNA ladder mix (Fermentas, Burlington, Canada) as a molecular size standard.
Enzymatic Activity Assay and Western Blot
ACE activity was assayed using the substrate benzyloxycarbonyl-phenylalanyl-histidyl-leucine (Z-FHL) (Bachem, Budendorf, Switzerland) as described previously (20, 21), and data were plotted using GraphPad Prism 4.01 (GraphPad Software, La Jolla, CA). Immunoblotting of purified recombinant ACE N domain proteins was carried out as described previously (22). The membrane was probed with an N domain monoclonal antibody, 4G6 (kind donation from S. Danilov) and developed with the Immun-StarTM WesternCTM chemiluminescence kit (Bio-Rad). Detection was carried out on a G:Box iChemiTM chemiluminescence imager (Syngene, Cambridge, UK) and analyzed using the GeneSnapTM software package (Syngene).
Thermal Denaturation Assay
Purified recombinant ACE proteins were incubated at 55 °C for up to 90 min in 50 mm HEPES, pH 7.4. Residual activity was determined using the aforementioned ACE assay.
Crystallization of Ndom389 Glycosylation Mutant-RXP407 Complex
Initial conditions for the N domain ACE with RXP407 inhibitor were found from screening using a Phoenix protein crystallization robot (Art Robbins Instruments, Sunnyvale, CA) in sitting drop/vapor diffusion at 16 °C. Crystals were observed at 1, 2, and 4 mg/ml in five conditions of the Morpheus screen (MDL Ltd., Suffolk, UK). Condition A9 (0.06 m divalent cations, 0.1 m Tris/Bicine, pH 8.5, 30% PEG 550MME/PEG 20000) was refined using the Hampton Research (Aliso Viejo, CA) silver bullets additive screen. This yielded crystals in many drops of which several diffracted. The best diffracting crystals were obtained with 300 nl of 4 mg/ml ACE N domain in complex with 0.5 mm RXP407 mixed with 150 nl of condition A9 and 150 nl of additive G10 (0.2% (w/v) 1,4-cyclohexanedicarboxylic acid, 0.2% (w/v) 2,5-pyridinedicarboxylic acid, 0.2% (w/v) glutaric acid, 0.2% (w/v) trans-1,2-cyclohexanedicarboxylic acid, 0.2% (w/v) trans-aconitic acid, 0.02 m sodium HEPES, pH 6.8).
X-ray Diffraction Study
Diffraction data up to 2.0 Å were collected using a Quantum-4 CCD detector (ADSC, Poway, CA) on station IO2 at the Diamond Light Source (Oxon, UK). No cryoprotectant was used, although the crystal was cooled to 100 K by liquid nitrogen vapor during data collection. Raw data images were indexed and scaled with the HKL2000 software package (23). Data reduction was carried out by using the CCP4 program TRUNCATE (24) (Table 1). Initial phases for structure solution were obtained using the molecular replacement routines of the PHASER program (25). The native ACE N domain (Protein Data Bank code 2C6F) (15) structure was used as the search model. The resultant model was refined using REFMAC5 (26). One percent of reflections were separated as the Rfree set and used for cross-validation (27). Manual adjustments of the model were made using COOT (28) and its validation tools (29). Based on electron density interpretation, one RXP407 molecule per monomer was added to the structure, and further refinement was carried out. The coordinate and restraint parameter files for the inhibitor were generated using the PRODRG server (30). TLS refinement was also included at a later stage. TLS restraints were calculated from the TLSMD Web site (31), and five TLS groups were used. On the basis of Fo − Fc electron density, side chain atoms were omitted at some positions, and some double conformations were included. Water molecules were added at positions where Fo − Fc electron density peaks exceeded 3σ. A few peaks that did not fit the stereochemical requirements for water molecule were left unmodeled. Validation of the structure showed that there were no residues in the disallowed region of the Ramachandran plot. Data collection and refinement statistics are given in Table 1. Figures were drawn with PyMOL (DeLano Scientific, San Carlos, CA). Hydrogen bonds were verified with the program HBPLUS (32).
TABLE 1.
X-ray data collection and refinement statistics
| ACE N domain-RXP407 inhibitor complex | |
|---|---|
| Resolution (Å) | 1.99 |
| Space group (2 molecules) | P1 |
| Cell dimensions | a = 72.89, b = 76.69, c = 82.65 Å; α = 88.6, β = 64.2, γ = 75.7° |
| Total no. of observations | 211,317 |
| No. of unique reflections | 106,429 |
| Completeness (outer shell)a (%) | 96.0 (96.1) |
| I/σ(I) (outer shell) | 12.7 (1.7) |
| Rsymmb (outer shell) | 5.4 (39.7) |
| Rcrystc/Rfreed | 19.4/23.7 |
| Multiplicity | 2.0 |
| Deviation from ideality | |
| Bond lengths (Å) | 0.007 |
| Bond angles (degrees) | 1.1 |
| B-factor analysis (Å2) | |
| Wilson B-factor | 28.1 |
| Protein all atoms (Mol A/Mol B) | 19.5/21.6 |
| Inhibitor RXP407 atoms (Mol A/Mol B) | 27.1/25.8 |
| Zn2+ ion (Mol A/Mol B) | 24.0/21.7 |
| Solvent atoms (563 in total) | 26.0 |
| Glycosylated sugars (Mol A/Mol B) | 52.4/56.3 |
| Ramachandran plot statistics | Allowed 98%, generously allowed 1.83%, outlier 0.17%. The two slight outliers are Asn45 in Mol A and B with a carbohydrate moiety (with clear density) attached to this residue |
a Outer shell, 2.04-1.99 Å.
b Rsymm = ΣhΣi|Ii(h) − 〈I(h)〉|/ΣhΣiIi(h), where Ii is the ith measurement and 〈I(h)〉 is the weighted mean of all of the measurements of I(h).
c Rcryst = Σh|Fo − Fc|/ΣhFo, where Fo and Fc are observed and calculated structure factor amplitudes of reflection h, respectively.
d Rfree is equal to Rcryst for a randomly selected 1% subset of reflections.
RESULTS
Mapping Glycans to Specific N-Linked Glycosylation Sites in the ACE N Domain Protein
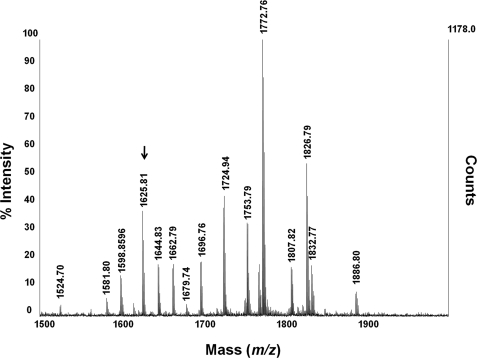
There are 10 potential N-linked glycosylation sites on the N domain, and a neural network-based N-glycosylation prediction tool, NetNGlyc 1.0 (available on the World Wide Web), indicates that nine of these sites are likely to be glycosylated. Mapping of the N-glycosylation sites was achieved by deglycosylating purified recombinant N domain with PNGase F. To maximize coverage of potential glycopeptide sequences, the deglycosylated protein was digested with either trypsin or endoproteinase Glu-C, and the resulting peptides were analyzed by MALDI-TOF/TOF (Fig. 2). Analysis of peptides resulting from the Glu-C digest revealed the presence of glycosylation at sites 1 (Asn9), 2 (Asn25), and 3 (Asn45) (Table 2), whereas the tryptic digests identified glycosylation at sites 4 (Asn82), 5 (Asn117), 6 (Asn131), 8 (Asn416), and 9 (Asn480). Additionally, site 10 (Asn494) was detected in its non-glycosylated form in a number of tryptic digests.
FIGURE 2.
Representative MALDI-TOF/TOF spectrum of tryptic peptides from PNGase F-digested ACE N domain. Masses indicate singly charged ions. The glycopeptide containing site 8 (Asn416), m/z 1625.81, is indicated.
TABLE 2.
Observed molecular ions for peptides from tryptic and endoproteinase Glu-C digests of PNGase F-treated ACE N domain
Singly charged ions are indicated. PNGase F deamidates the Asn (114 Da) residue of the glycosylation sequon to an Asp (115 Da). This increase of 1 Da can be used to identify the presence of N-glycosylation at potential glycosylation sites.
| Glycan site | Glycan site residue | Peptide residues | Calculated mass [MH]+a | Observed mass (m/z) [MH]+ | Peptide identification (determined by MS/MS) | Digest type |
|---|---|---|---|---|---|---|
| Da | Da | |||||
| 1 | Asn9 | 1–14 | 1459.54 | 1460.56 | LDPGLQPGDFSADE | Glu-C |
| 2 | Asn25 | 15–29 | 1543.62 | 1544.60 | AGAQLFAQSYDSSAE | Glu-C |
| 3 | Asn45 | 30–49 | 2188.06 | 2188.92 | QVLFQSVAASWAHDTDITAE | Glu-C |
| 4 | Asn82 | 74–89 | 2049.01 | 2050.01 | ELYEPIWQDFTDPQLR | Trypsin |
| 5 | Asn117 | 109–120 | 1424.70 | 1425.70 | QQYNALLSDMSR | Trypsin |
| 6 | Asn131 | 121–132 | 1393.51 | 1394.62 | NDb | Trypsin |
| 8 | Asn416 | 414–427 | 1624.81 | 1625.81 | DDTESDINYLLK | Trypsin |
| 9 | Asn480 | 480–489 | 1089.50 | 1090.70 | DETHFDAGAK | Trypsin |
| 10 | Asn494 | 490–500 | 1342.73 | 1342.76 | FHVPNVTPYIR | Trypsin |
a Refers to calculated mass if the peptide was not glycosylated and therefore not deamidated by PNGase F.
b ND, not determined.
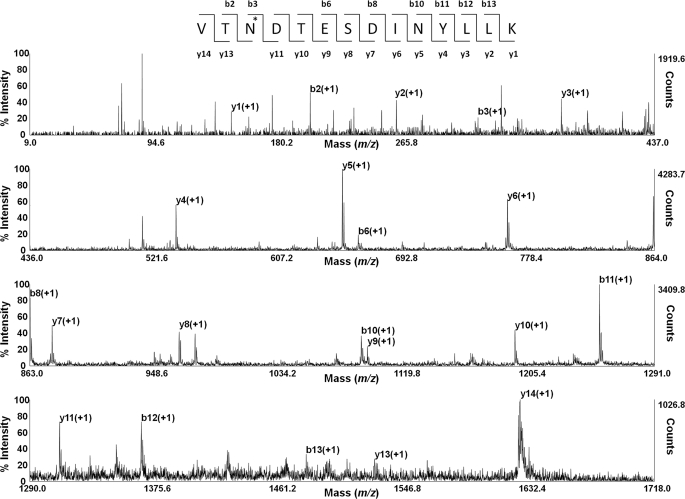
These data show that eight of the 10 potential N-linked sites are glycosylated. These results were confirmed by subjecting the relevant peptides to MS/MS to fragmentation (Fig. 3), where the identification of the peptides confirmed an Asn to Asp conversion in each case, with the exception of the peptide containing the unglycosylated site 10 (Asn494), where the conversion was not seen.
FIGURE 3.
Representative spectrum of MALDI-TOF/TOF-MS/MS sequencing of the fragmented glycopeptide, m/z 1625.81. The y and b ion series are shown. The asterisk indicates a deamidated Asn (Asn416), causing a mass increase of 1 Da.
Effect of Different Glycosylation Profiles on Enzymatic Activity
Glycosylation sites on the N domain were sequentially disrupted to determine which sites were important for the expression of active protein as well as to determine the minimum glycosylation requirements for the folding of active N domain. The effect of the removal of specific glycosylation sequons on protein expression was assessed using the ACE substrate Z-FHL, as described in Ref. 21, and by Western blot analysis.
It had been previously found that only one or two of the N-terminal sugars were necessary and sufficient for the expression of a functional C domain construct (22). Considering the high level of sequence and structural identity between two domains, we decided to begin by removing sites from the C terminus of the N domain (Fig. 4).
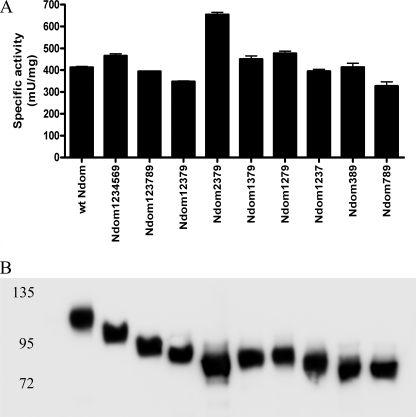
Removal of the three C-terminal sites (Ndom123456), resulted in a complete loss of enzymatic activity, in contrast to what was seen with the C domain (data not shown). However, Ndom1234569 was active (Fig. 5A), indicating that C-terminal sites are generally more important for the production of active protein. This is contrary to the C domain, where C-terminal sites were not required for the expression of correctly folded, active protein (22).
FIGURE 5.
Expression of N domain glycosylation variants. A, specific activity of ACE N domain and various glycosylation mutants are shown. Error bars, S.E. B, Western blot of purified recombinant N domain proteins, probed with an anti-N domain antibody (4G6).
Therefore, we decided to reintroduce the C-terminal sites 7, 8, and 9 into a hypoglycosylated N domain variant, Ndom123. Ndom123 was not active, but the newly generated variants Ndom1237, Ndom12379, and Ndom123789 (Fig. 4) were all active and had similar specific enzyme activities compared with that of the wild-type N domain (Fig. 5A). In addition, Western blot analysis of the glycosylation mutants showed increases in electrophoretic mobility that correlated with the decreases in the number of intact glycosylation sequons, confirming the reduced N-glycan content of the proteins (Fig. 5B). These findings suggest that C-terminal glycosylation sites are required for expression of a functional N domain.
Removal of individual N-terminal glycosylation sites of the hypoglycosylated mutant Ndom12379 (Ndom2379, Ndom1379, and Ndom1279) had no significant effect on enzymatic activity (Fig. 5A), indicating that individually these sites were not critical for the production of active protein. Further mutagenesis was carried out to produce Ndom79 and Ndom89, both of which were inactive. The reintroduction of either site 3 or site 7 into Ndom89 (Ndom389 and Ndom789) was able to rescue the enzymatic activity and produced functional protein in both cases (Fig. 5, A and B). Inactive proteins Ndom123456, Ndom123, Ndom79, and Ndom89 could not be detected by Western blot, although mRNA expression was confirmed by RT-PCR (data not shown), indicating that protein detection may have been hampered by rapid intracellular degradation.
Effect of Glycosylation on the Thermal Stability of the N Domain
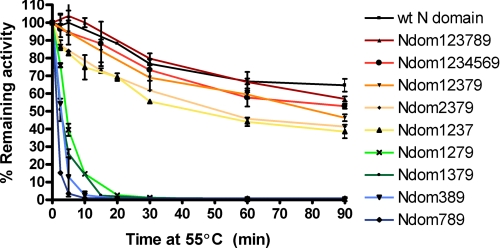
The thermal stability of the N domain and its hypoglycosylated mutants was assessed by incubation at 55 °C, followed by determination of residual activity using the ACE substrate Z-FHL. Wild-type N domain showed remarkable thermal stability, retaining almost 80% of its activity after 30 min of incubation at 55 °C; even after 90 min at 55 °C, there was little change in enzyme activity (Fig. 6). The removal of sites 7 and 8 (Ndom1234569) and sites 4, 5, and 6 (Ndom123789) did not have a notable affect on thermal stability, with Ndom123789 retaining about 80% of its activity after 30 min. The further removal of site 8 (Ndom12379) and sites 8 and 9 (Ndom1237) reduced the stability to about 70 and 55%, respectively, after 30 min. The removal of either site 2 or site 3 from Ndom12379 caused a marked reduction in thermal stability, with only 1.2% of their activity remaining after a 30-min thermal inactivation. The removal of site 1 (Ndom2379), however, did not greatly alter the thermal stability from that of Ndom12379, with the variant retaining 55% of its activity after 30 min at 55 °C. The hypoglycosylated mutants Ndom789 and Ndom389 were the most unstable of all of the glycosylation variants and effectively had no residual activity after a 10-min thermal inactivation.
FIGURE 6.
Thermal denaturation curve for ACE N domain and various glycosylation mutants. The percentage of remaining activity is plotted against time incubated at 55 °C. Error bars, S.E. of at least two experiments carried out in triplicate.
Crystal Structure of Ndom389 in Complex with RXP407
The crystallization of a complex of the N domain with RXP407 has been under investigation for some time. No progress could be achieved using the previously crystallized N domain construct expressed in the presence of glycosylation inhibitors (15) with either previous crystallization conditions or new screens. Small crystals could be grown, but all attempts to produce large or well diffracting crystals failed. By contrast, the first screen set up for the Ndom389 produced hits in several wells. Although this variant could not be reproduced manually, further optimization with an additive screen produced crystals in many of the additive conditions. Several of these were tested and found to vary in diffraction strength (although of the same space group), and the results presented here are from the best diffracting crystal. Interestingly, not only did the crystals form at a high pH (pH 8.5) when previous human ACE structures have been crystallized at pH 4–5, but the current space group, P1, is different from that of the previously described N domain structure in C2221 space group (15).
In the present structure, there are two N domain molecules in the asymmetric unit, both with the compact ellipsoid form as observed previously in the native N domain structure (15). Residues 1–610 are visible in both molecules, although residues 131 and 132, part of a highly disordered loop, are not modeled in molecule A. Density for glycosyl chains was visible at the three remaining glycosylation sites in both molecules, although the quality of the density varied, and no more than four sugar residues were observed at any one site.
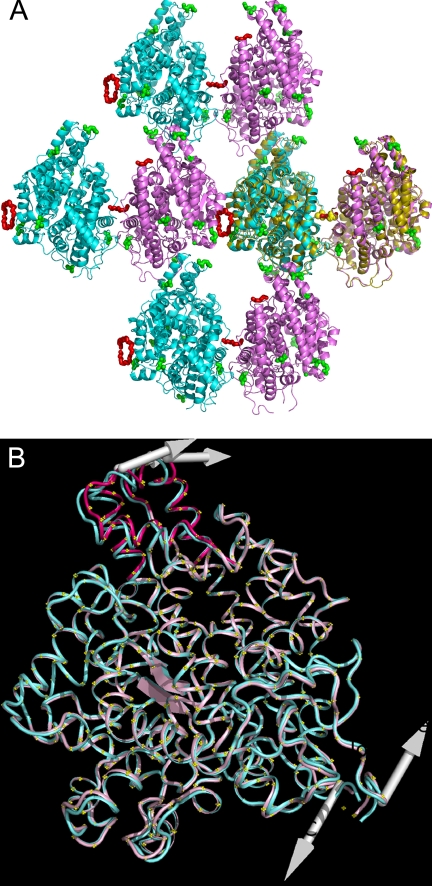
Crystal Packing and Glycosylation
The protein interfaces, surfaces, and assemblies service (PISA) at the European Bioinformatics Institute (available on the World Wide Web) (33) identified six interfaces within the crystal, although the symmetry allows the two molecules to approach each other from eight directions. In one plane where the molecules form configurations, head (i.e. the N-terminal area)-to-tail and side-to-side (Fig. 7A), the symmetrical side-to-side interactions both incorporate a non-protein element. Large peaks in the difference map at these positions were eventually modeled as fragments of PEG because these best fitted the shape of the density. They also have reasonable crystallographic thermal parameters (B-factors) (40–50 Å2), and most make direct or water-mediated interactions with the protein. The interface with the largest fragments of PEG does not involve any direct protein-protein contact and is therefore not recognized by PISA. The other of the two interfaces (protein-protein interface of 1868 Å2), which also incorporates a small fragment of PEG, has a glycosyl chain (Asn480, site 9) from each molecule contributing to the edge of the interface. Interestingly, the dimeric structure generated by this interface is also observed in the symmetry of the previously determined N domain native structure in the orthorhombic space group C2221 (Fig. 7A) (15). In the C2221 form, partial glycosyl chains (site 9) are also observed contributing to this interface, and instead of PEG, two peaks in the difference map were modeled as symmetry-related acetate ions at this position. Additionally, two glycerol molecules were also modeled within that interface.
FIGURE 7.
A, arrangement of protein molecules in the crystal with molecule A in pink and molecule B in blue. The residues that are glycosylated in the native N domain are shown as green spheres, and the PEG fragments are modeled as red spheres. An overlay of two symmetry-related molecules from the previously determined crystal structure in C2221 (Protein Data Bank Code 2C6F) (15) is shown in olive, with the two acetate ions superposing the central PEG as yellow spheres. B, superposition of molecules A (pink) and B (blue). The shift in the N terminus of molecule A is shown in dark pink. The arrows indicate the direction of movement predicted by the ANM server (35).
Other involvements of glycosylation in crystal packing in the current structure include the glycosyl chain at site 8 in molecule A that contributes to a smaller “head-to-tail” interface and the glycosyl chain at site 3 that is part of a small interface near the head regions in molecule A but not in B.
Observation of the N Domain Hinge
It has previously been proposed (34) that ACE domains may have a breathing motion that allows the substrate to enter the buried active site, but so far no structure of human ACE has displayed this. However, in the present structure, despite both molecules being in the “closed” state, a hinge in the N domain was observed. Normal mode analysis of the N domain by the Anisotropic Network Model Web server (35) indicates that the molecule can undergo a large twisting motion between the N terminus and the C terminus. The observed difference between molecules A and B in the present structure indicates a slight twist in molecule A, of which the most obvious feature is a small shift of the N-terminal helices away from the opening for the active site at this end of the molecule. This movement is a partial version of that predicted by the normal mode analysis server (Fig. 7B). The N-terminal shift was explored further using DynDom (36), which revealed the shifted domain to comprise residues 9–25 and 65–99, rotated by a modest 4.1° away from the active site opening. It is interesting to note that the top part of the hinged region is bracketed by glycosylation at sites 1 and 2 (mutated to glutamine in the present structure), of which we have shown site 2 to be important for thermal stability.
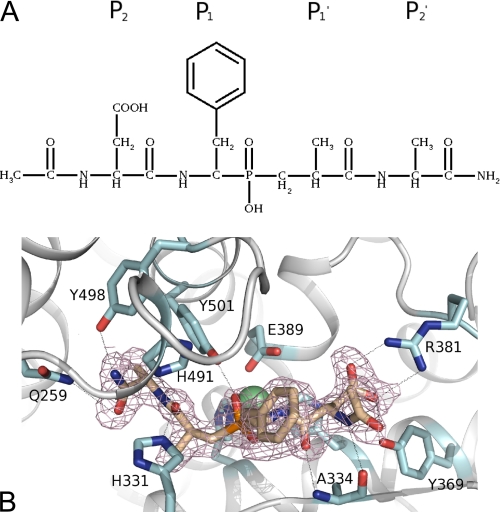
Binding of RXP407 Inhibitor
RXP407 is one of several inhibitors (Fig. 8A) designed to be specific to the N domain by means of using a weaker zinc binding moiety, phosphate, combined with extension out into the S2 and S2′ subsites, where the N and C domains differ in amino acid composition. This inhibitor has been shown to have orders of magnitude higher affinity for the N domain than for the C domain in vitro (7). The conformation of the inhibitor (Fig. 8B) was identical for both molecules A and B, with the exception of the “N-terminal” aldehyde, for which there is evidence for two conformations. The water structure in the S2 subsite differed between A and B, suggesting that the aldehyde oxygen can be on either side because it has no direct interaction with the protein. Hydrogen bond interactions calculated with HBPLUS (32) indicates 12 direct hydrogen bonds (Table 3) between the inhibitor and the protein (in addition to the interaction with the zinc), which is five more than observed for its complex with the Drosophila melanogaster angiotensin-converting enzyme homologue despite the same configuration (37). These bonds have arisen from nine residues: Arg381, Tyr369, and Ala334 in the S2 subsite; Tyr501 with the phosphate; His331 and His491 in the S1′ subsite; and Tyr498, His491, and Gln259 in the S2′ subsite (Fig. 8, A and B). Of these residues, Arg381 and Tyr369 are not conserved in the C domain, being replaced by Glu403 and Phe391, respectively, indicating that the selectivity of RXP407 for the N domain lies in the S2 subsite as proposed previously (8).
FIGURE 8.
A, chemical structure of RXP407, showing residue positions in the context of active site binding regions. B, close up of ACE N domain active site showing RXP407 and residues interacting with it. RXP407 is shown in wheat; the zinc and chloride ions are shown as green and red spheres, respectively; and the omit map for the RXP407 at 1σ is shown in dirty pink. Hydrogen bonds identified by HBPLUS (32) are shown as lines.
TABLE 3.
Hydrogen bonds between RXP407 inhibitor and the protein atoms in molecules A and B as calculated by HBPLUS (32)
| Residue | Atom | RXP407 atom | Distance Mol A | Distance Mol B | |
|---|---|---|---|---|---|
| Å | Å | ||||
| P2 | |||||
| Arg381 | NH1 | OAG | 3.2 | 3.3 | |
| Arg381 | NH2 | OAK | 2.5 | 2.7 | |
| Arg381 | NH2 | OAG | 3.2 | 3.3 | |
| Tyr369 | OH | OAG | 2.5 | 2.5 | |
| Ala334 | N | OAI | 2.8 | 2.8 | |
| Phosphate | |||||
| Tyr501 | OH | OAJ | 2.6 | 2.5 | |
| P1′ | |||||
| His331 | NE2 | OAH | 2.7 | 2.7 | |
| His491 | NE2 | OAH | 3.0 | 3.0 | |
| P2′ | |||||
| Tyr498 | OH | NAD | 2.7 | 2.5 | |
| His491 | NE2 | NAD | 3.3 | 3.3 | |
| Gln259 | NE2 | O | 3.2 | 3.2 | |
| Ala334 | O | NAU | 3.0 | 3.1 | |
In comparison with the native N domain structure, both active site waters, including the one bound to the zinc, are displaced by the RXP407. However, it is possible to observe many more waters in the active site of this structure because the resolution is much higher. The peak in the difference map modeled as acetate in the native structure has the same position as, although different orientation from, the C-terminal amide group of the RXP407. Therefore, the hydrogen bond with Tyr498 is conserved between them, but the interaction of the acetate ion with Lys498 is not replicated in the RXP407 structure. Instead, the oxygen of the amide group makes water-mediated interactions with Asp255 and Thr358.
DISCUSSION
We have determined that at least eight of the 10 N-glycan sites on the N domain are glycosylated, namely sites 1 (Asn9), 2 (Asn25), 3 (Asn45), 4 (Asn82), 5 (Asn117), 6 (Asn131), 8 (Asn416), and 9 (Asn480), with site 10 (Asn494) being found in the unglycosylated form. These findings are consistent with work carried out by Yu et al. (9) on somatic ACE purified from human kidney cells, where N domain glycosylation sites 1, 2, 4, 5, and 9 were detected in the glycosylated form. Thus, the N domain has a greater degree of glycosylation site occupancy than does the C domain, which has three sites that are always glycosylated and three that are partially glycosylated (9).
The glycosylation pattern of the C domain shows a distinct preference for N-terminal sugars (9), whereas the N domain has a strong dependence on C-terminal glycans because only three C-terminal sugars (Ndom789) were required for the folding and processing of active protein. Interestingly, Ndom389 was active, whereas Ndom79 and Ndom89 were not. Thus, it would seem the presence of glycosylation at the C terminus is vital, although some redundancy exists with respect to the precise combination of glycosylation sites required for the production of active N domain.
In terms of thermal stability, the different N domain glycosylation variants can be broadly placed into two groups. The first group consists of glycosylation mutants that showed only a marginal reduction in thermal stability, whereas the second group comprises those constructs that have greatly reduced thermal stability. The first group includes the glycosylation variants Ndom1234569, Ndom123789, Ndom12379, Ndom1237, and Ndom2379. When taken together, these variants indicate that the removal of glycosylation at the respective sites results in only a slight decrease in thermal stability. The second group includes Ndom1279, Ndom1379, Ndom389, and Ndom789. The major characteristic distinguishing these variants from their more stable counterparts is that they lack glycosylation at either site 2 or site 3 or both, showing that N-terminal glycosylation is required for maintaining thermal stability in the N domain. Interestingly, the identification of sites 2 and 3 as contributing to thermal stability is consistent with the observation of site 2 forming part of the hinge identified in the crystal structure because this site may help protect the lid region from thermal denaturation.
Both the observed change in the space group of the crystallized N domain and the increase in resolution that have resulted in the elucidation of the N domain RXP407 structure can be attributed to the difference in the glycosylation of the Ndom389 construct. However, the difference in the space group is also interesting for the evaluation of interactions between N domain molecules within the crystal. The observation of a large common interface in two structures of differing space group indicates that this interface may be significant. Analysis of this interface using the PISA web server (33), despite indicating nine hydrogen bonds and a negative ΔG value, does not identify it or any other interface as significant. However, the PISA analysis is incomplete because it does not recognize the contribution to the interface of the glycosylation at site 9 nor that of PEG. This is important because the evidence (18) that the N domain may dimerize, or form aggregates, indicates that the interaction may be mediated via glycosylation. One problem with analyzing the interfaces in our structure, however, is that we have two PEG-mediated interfaces, whereas the natural ACE would never encounter PEG. Furthermore, the glycosylation of our protein is more limited than that which occurs naturally. The N domain used for the crystal structures is recombinantly expressed in CHO cells, and the glycosylation has been modified to aid crystallization either by glycosidase inhibitors (in the case of the C2221 structure) or by mutation of glycosylation sites. On the other hand, the fact that the conserved interface is contributed to by glycosyl chains in our minimally glycosylated form allows for the possibility that it could be further stabilized by more extensive glycosylation in vivo. This is also consistent with our mutational studies, which highlight site 9 as important for correct folding and processing.
The proposal of this interface as a region of dimerization in vivo would be most convincing if it agreed with the previous monoclonal antibody data regarding this process. The proposed carbohydrate binding site, based on antibody binding studies, was indicated to be in the region of residues 521–572 (18) and is therefore distal to the conserved interface described here. The PEG-mediated interface on the other side of the molecule, however, is proximal to the carbohydrate binding site and incorporates the mutated glycosylation site 7. Because we have shown that both of these glycosylation sites (sites 9 and 7) might be important in stability during folding and processing of the active protein, and both of these sites are unique to the N domain (the C domain was not observed to form these dimers (18)), it is likely that either may contribute to any glycosyl-mediated dimerization.
CONCLUSIONS
The presence of C-terminal glycosylation in the N domain is vital for its folding and processing, whereas the presence of N-terminal glycans is crucial for maintaining its high level of thermal stability. The minimally glycosylated Ndom389 structure of the variant was determined at 2.0 Å resolution and revealed a hinge in the N domain for the first time as well as a conserved protein-protein interface. This mutant protein is also amenable to crystallization with inhibitors, as demonstrated by this complex with RXP407.
Acknowledgments
We thank the scientists at station IO2 of Diamond Light Source, Didcot, Oxon (United Kingdom), Kenneth Holbourn, and Nethaji Thiyagarajan for support during x-ray diffraction data collection and Daniel Holloway for constructive criticisms of the manuscript. We also thank M. Vlok and B. Kekana at the Centre for Proteomic and Genomic Research for assistance with the mass spectrometry.
This work was supported by the Wellcome Trust (United Kingdom) through Senior International Research Fellowship 070060, the National Research Foundation of South Africa, the Ernst and Ethel Erikson Trust, the Deutscher Austauschdienst, and the University of Cape Town (to E. D. S.). This work was also supported by Medical Research Council (United Kingdom) Grant 81272 and a Royal Society (United Kingdom) Industry Fellowship (to K. R. A.).
The atomic coordinates and structure factors (code 3NXQ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- ACE
- angiotensin-I-converting enzyme
- Z-
- benzyloxycarbonyl-
- RXP407
- Ac-Asp-l-Phe(PO2CH2)-l-Ala-Ala-NH2
- Bicine
- N,N-bis(2-hydroxyethyl)glycine
- TLS
- translation-libration-screw.
REFERENCES
- 1.Junot C., Gonzales M. F., Ezan E., Cotton J., Vazeux G., Michaud A., Azizi M., Vassiliou S., Yiotakis A., Corvol P., Dive V. (2001) J. Pharmacol. Exp. Ther. 297, 606–611 [PubMed] [Google Scholar]
- 2.Fuchs S., Xiao H. D., Hubert C., Michaud A., Campbell D. J., Adams J. W., Capecchi M. R., Corvol P., Bernstein K. E. (2008) Hypertension 51, 267–274 [DOI] [PubMed] [Google Scholar]
- 3.Jaspard E., Wei L., Alhenc-Gelas F. (1993) J. Biol. Chem. 268, 9496–9503 [PubMed] [Google Scholar]
- 4.Rousseau A., Michaud A., Chauvet M. T., Lenfant M., Corvol P. (1995) J. Biol. Chem. 270, 3656–3661 [DOI] [PubMed] [Google Scholar]
- 5.Sharma U., Rhaleb N. E., Pokharel S., Harding P., Rasoul S., Peng H., Carretero O. A. (2008) Am. J. Physiol. Heart Circ. Physiol. 294, H1226–H1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng H., Carretero O. A., Liao T. D., Peterson E. L., Rhaleb N. E. (2007) Hypertension 49, 695–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dive V., Cotton J., Yiotakis A., Michaud A., Vassiliou S., Jiracek J., Vazeux G., Chauvet M. T., Cuniasse P., Corvol P. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 4330–4335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kröger W. L., Douglas R. G., O'Neill H. G., Dive V., Sturrock E. D. (2009) Biochemistry 48, 8405–8412 [DOI] [PubMed] [Google Scholar]
- 9.Yu X. C., Sturrock E. D., Wu Z., Biemann K., Ehlers M. R., Riordan J. F. (1997) J. Biol. Chem. 272, 3511–3519 [DOI] [PubMed] [Google Scholar]
- 10.O'Neill H. G., Redelinghuys P., Schwager S. L., Sturrock E. D. (2008) Biol. Chem. 389, 1153–1161 [DOI] [PubMed] [Google Scholar]
- 11.Hammond C., Braakman I., Helenius A. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 913–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helenius A., Aebi M. (2004) Annu. Rev. Biochem. 73, 1019–1049 [DOI] [PubMed] [Google Scholar]
- 13.Sadhukhan R., Sen I. (1996) J. Biol. Chem. 271, 6429–6434 [DOI] [PubMed] [Google Scholar]
- 14.Voronov S., Zueva N., Orlov V., Arutyunyan A., Kost O. (2002) FEBS Lett. 522, 77–82 [DOI] [PubMed] [Google Scholar]
- 15.Corradi H. R., Schwager S. L., Nchinda A. T., Sturrock E. D., Acharya K. R. (2006) J. Mol. Biol. 357, 964–974 [DOI] [PubMed] [Google Scholar]
- 16.Ehlers M. R., Chen Y. N., Riordan J. F. (1991) Protein Expr. Purif. 2, 1–9 [DOI] [PubMed] [Google Scholar]
- 17.Deddish P. A., Wang J., Michel B., Morris P. W., Davidson N. O., Skidgel R. A., Erdös E. G. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 7807–7811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kost O. A., Balyasnikova I. V., Chemodanova E. E., Nikolskaya I. I., Albrecht R. F., 2nd, Danilov S. M. (2003) Biochemistry 42, 6965–6976 [DOI] [PubMed] [Google Scholar]
- 19.Papworth C., Bauer J. C., Braman J., Wright D. A. (1996) Strategies 9, 3–4 [Google Scholar]
- 20.Friedland J., Silverstein E. (1976) Am. J. Clin. Pathol. 66, 416–424 [DOI] [PubMed] [Google Scholar]
- 21.Schwager S. L., Carmona A. K., Sturrock E. D. (2006) Nat. Protoc. 1, 1961–1964 [DOI] [PubMed] [Google Scholar]
- 22.Gordon K., Redelinghuys P., Schwager S. L., Ehlers M. R., Papageorgiou A. C., Natesh R., Acharya K. R., Sturrock E. D. (2003) Biochem. J. 371, 437–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 24.Collaborative Computational Project 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 25.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 27.Brünger A. T. (1992) Nature 355, 472–475 [DOI] [PubMed] [Google Scholar]
- 28.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 29.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schüttelkopf A. W., van Aalten D. M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 31.Painter J., Merritt E. A. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 [DOI] [PubMed] [Google Scholar]
- 32.McDonald I. K., Thornton J. M. (1994) J. Mol. Biol. 238, 777–793 [DOI] [PubMed] [Google Scholar]
- 33.Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 34.Watermeyer J. M., Sewell B. T., Schwager S. L., Natesh R., Corradi H. R., Acharya K. R., Sturrock E. D. (2006) Biochemistry 45, 12654–12663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eyal E., Yang L. W., Bahar I. (2006) Bioinformatics 22, 2619–2627 [DOI] [PubMed] [Google Scholar]
- 36.Hayward S., Kitao A., Berendsen H. J. (1997) Proteins 27, 425–437 [DOI] [PubMed] [Google Scholar]
- 37.Akif M., Georgiadis D., Mahajan A., Dive V., Sturrock E. D., Isaac R. E., Acharya K. R. (2010) J. Mol. Biol. 400, 502–517 [DOI] [PubMed] [Google Scholar]