Abstract
The BzdR transcriptional regulator that controls the PN promoter responsible for the anaerobic catabolism of benzoate in Azoarcus sp. CIB constitutes the prototype of a new subfamily of transcriptional regulators. Here, we provide some insights about the functional-structural relationships of the BzdR protein. Analytical ultracentrifugation studies revealed that BzdR is homodimeric in solution. An electron microscopy three-dimensional reconstruction of the BzdR dimer has been obtained, and the predicted structures of the respective N- and C-terminal domains of each BzdR monomer could be fitted into such a reconstruction. Gel retardation and ultracentrifugation experiments have shown that the binding of BzdR to its cognate promoter is cooperative. Different biochemical approaches revealed that the effector molecule benzoyl-CoA induces conformational changes in BzdR without affecting its oligomeric state. The BzdR-dependent inhibition of the PN promoter and its activation in the presence of benzoyl-CoA have been established by in vitro transcription assays. The monomeric BzdR4 and BzdR5 mutant regulators revealed that dimerization of BzdR is essential for DNA binding. Remarkably, a BzdRΔL protein lacking the linker region connecting the N- and C-terminal domains of BzdR is also dimeric and behaves as a super-repressor of the PN promoter. These data suggest that the linker region of BzdR is not essential for protein dimerization, but rather it is required to transfer the conformational changes induced by the benzoyl-CoA to the DNA binding domain leading to the release of the repressor. A model of action of the BzdR regulator has been proposed.
Keywords: Bacterial Transcription, Gene Regulation, Gene Transcription, Repressor Protein, Transcription Repressor, Azoarcus, BzdR, Anaerobic, Benzoate
Introduction
The anaerobic catabolism of aromatic compounds by microorganisms is relevant in the global biochemical cycles because many contaminated environments are anoxic (1, 2). Benzoate is a common carbon source in nature that has been used as a model compound for studying the anaerobic catabolism of aromatic compounds. This compound is funneled directly to the widely distributed benzoyl-coenzyme A (benzoyl-CoA) central pathway (3, 4). In the β-proteobacterium Azoarcus sp. CIB, the genes responsible for the anaerobic benzoate degradation are organized in the bzd cluster constituted by a single catabolic operon (bzdNOPQMSTUVWXYZA) under the control of the PN promoter. Upstream from the bzd catabolic operon, and divergently transcribed as an independent transcriptional unit, there is a regulatory gene, bzdR, whose product controls the expression of the catabolic operon at the level of the PN promoter (5–7). The BzdR regulator binds to three different operator regions in the PN promoter, i.e. region I (63 bp), spanning positions −32 to +31 with respect to the transcription start site; region II (21 bp), spanning positions −83 to −63; and region III (21 bp) spanning positions −146 to −126. The three protected regions contain direct repetitions of a sequence, TGCA, that forms part of longer palindromic structures (5). The BzdR binding to the PN promoter inhibits the expression of the bzd catabolic operon, but this specific repression is alleviated in the presence of the first intermediate of the pathway, the inducer molecule benzoyl-CoA (5–7). An oxygen-dependent activator, the AcpR protein, a Fnr ortholog, is also needed to activate the PN promoter under anaerobic conditions (6).
Analysis of the primary structure of the BzdR regulator (298 amino acids) revealed a unique molecular architecture that makes this protein the prototype of a new subfamily of transcriptional regulators (7). Thus, BzdR exhibits two domains, the N-terminal domain (N-BzdR, residues 1–90) and the C-terminal domain (C-BzdR, residues 131–298). N-BzdR contains a DNA binding region similar to that of the xenobiotic response element (XRE)7 transcriptional regulators (8). The XRE family of transcriptional regulators constitute a large family of proteins with a helix-turn-helix DNA-binding motif similar to that of the CI repressor and the Cro proteins of λ bacteriophage (9–11), the Bacillus subtilis SinR protein (12), and the restriction modification system control proteins such as C.PvII and C.AhdI (13, 14). These proteins make specific DNA contacts through their N-terminal domains, but their DNA binding affinity is modulated by their C-terminal domains (15, 16). The members of the helix-turn-helix XRE family are monomers, dimers, or tetramers in solution and bind to the DNA as homodimers or heterodimers (8, 17, 18). Homology modeling of the three-dimensional structure of N-BzdR has been accomplished using the SinR protein as template (7, 19).
On the other hand, C-BzdR is homologous to shikimate kinases, enzymes that catalyze the conversion of shikimate to shikimate 3-phosphate using ATP as co-substrate (20). Remarkably, the protein fold of shikimate kinases is highly versatile because it can accommodate a wide array of sequences without significant structural changes (21). Homology modeling of the three-dimensional structure of C-BzdR using the shikimate kinase I of Escherichia coli as template (7) revealed a consensus nucleotide binding fold, which includes the characteristic phosphate binding loop (P-loop or Walker A motif) (22, 23). Moreover, this model allowed the visualization of the predicted complex between C-BzdR and the inducer molecule benzoyl-CoA (7). Manual docking with benzoyl-CoA was easily performed because the predicted C-BzdR fold exhibits a conserved nucleotide-binding site where the ADP moiety of benzoyl-CoA can be fitted properly. Moreover, the pantothenate and β-mercaptoethylamine units of benzoyl-CoA fit into a predicted deep groove of C-BzdR that ends in a cavity equivalent to the shikimate-binding site described for shikimate kinases (22, 24).
In addition to the bzdR gene from Azoarcus sp. CIB, in silico analysis revealed homologous genes that code for BzdR-like proteins located in the vicinity or integrated into the anaerobic or CoA-dependent aerobic benzoate degradation clusters in α- and β-proteobacteria (7, 25), suggesting that BzdR is a prototype of a new subfamily of prokaryotic transcriptional regulators that control CoA-dependent aromatic degradation pathways. Because there are no reports about the biochemical characterization of any member of the BzdR subfamily, we provide here new insights on the functional-structural relationships of the BzdR protein from Azoarcus sp. CIB.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Growth Conditions
The E. coli strains, as well as the plasmids used in this work, are listed in Table 1. E. coli cells were grown at 37 °C on Luria-Bertani (LB) medium (26). Where appropriate, antibiotics were added at the following concentrations: ampicillin, 100 μg ml−1; kanamycin, 50 μg ml−1.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant phenotype and/or genotypea | Ref. or source |
|---|---|---|
| E. coli strains | ||
| DH5α | endA1 hsdR17 supE44 thi-1 recA1 gyrA(Nalr) | 26 |
| relA1Δ(argF-lac) U169 depR φ80dlacΔ(lacZ)M15 | ||
| M15 | Strain for regulated high level expression with pQE vectors | Qiagen |
| Plasmids | ||
| pQE32 | Apr, oriColE1, T5 promoter lac operator, λ to/E. coli rrnB T1 terminators, N-terminal His6 | Qiagen |
| pQE32-His6BzdR | Apr, pQE32 derivative harboring the His6-bzdR gene under the control of T5 promoter lac operator | 7 |
| pQE32-His6BzdR3 | Apr, pQE32 derivative harboring the His6-bzdR3 gene under the control of T5 promoter lac operator | This work |
| pQE32-His6BzdR4 | Apr, pQE32 derivative harboring the His6-bzdR4 gene under the control of T5 promoter lac operator | This work |
| pQE32-His6BzdR5 | Apr, pQE32 derivative harboring the His6-bzdR5 gene under the control of T5 promoter lac operator | This work |
| pQE32-His6BzdRΔL | Apr, pQE32 derivative harboring the His6-bzdRΔL gene under the control of T5 promoter lac operator | This work |
| pJCD-PN | Apr, pJCD01 derivative harboring a 585-bp EcoRI fragment that includes the PN promoter | 6 |
| pQE60-His6-FNR* | Apr, pQE60 derivative that harbors the His6-fnr* gene under the control of T5 promoter lac operator | 6 |
| pREP4 | Kmr, plasmid that expresses the lacI repressor | Qiagen |
| pECOR7 | Apr, pUC19 harboring a 7.1-kb EcoRI DNA fragment containing the bzdRNO genes | 5 |
| pGEM-Teasy | Apr, oriColE1, lacZα | Promega |
| pGEMT-BzdRΔL | Apr, pGEM-Teasy derivative harboring the BzdRΔL gene | This work |
a Apr, ampicillin-resistant, Kmr, kanamycin-resistant, Nalr, nalidixic acid-resistant.
Molecular Biology Techniques
Recombinant DNA techniques were carried out by published methods (26). Plasmid DNA was prepared with a High Pure plasmid isolation kit (Roche Applied Science). DNA fragments were purified with Gene-Clean Turbo (Qbiogene, Inc.). Oligonucleotides were supplied by Sigma. All cloned inserts and DNA fragments were confirmed by DNA sequencing through an ABI Prism 377 automated DNA sequencer (Applied Biosystems Inc.). Transformation of E. coli cells was carried out by using the RbCl method or by electroporation (Gene Pulser, Bio-Rad) (26). Proteins were analyzed by SDS-PAGE as described previously (27).
Sequence Data Analyses
Protein sequence comparison analyses were performed by using the BLAST algorithm (28) at National Center for Biotechnology Information (www.ncbi.nlm.nih.gov). Protein secondary structure prediction was performed by using JPred3 program (29).
Construction of the bzdR3, bzdR4, bzdR5, and bzdRΔL Genes
The construction of the bzdR3, bzdR4, and bzdR5 genes was achieved by using directed PCR mutagenesis with the pECOR7 plasmid (Table 1) as template. The bzdR gene was first amplified with the oligonucleotide 5HisReg (5′-CGGGATCCTTTCCAACGATGAGAACTCATCAC-3′, engineered BamHI is underlined) that anneals at the 5′-end of bzdR start codon, and the 3′E111R (5′-CCGGCGCGCCCATCTATTGCGTCTTG-3′; bzdR3), 3′R114E (5′-CTGGAGCAGCCGTTCCGCCCATTCATG-3′; bzdR4), or 3′E111R/R114E (5′-CTGGAGCAGCCGTTCCGCCCATCTATGCGTCTT-3′; bzdR5) oligonucleotides that anneal at the bzdR sequence to be mutated. The PCR amplification renders fragment A that spans from the start codon of BzdR to the sequence to be mutated. In a second round of amplifications, the bzdR gene was amplified using oligonucleotides 5′E111R (5′-CAAGACGCATAGATGGGCGCGCCGG-3′; bzdR3), 5′R114E (CATGAATGGGCGGAACGGCTGCTCCAG-3′; bzdR4), or 5′E111R/R114E (5′-AAGACGCATAGATGGGCGGAACGGCTGCTCCAG-3′; bzdR5) that anneal at the bzdR sequence to be mutated, and the oligonucleotide 3HISReg (5′-GGGAAGCTTTCAGCGTGCCAGGACTTCGAGG-3′, engineered HindIII is underlined) that anneals at the 3′-end of bzdR. This second round of amplification renders fragment B that spans from the sequence to be mutated to the stop codon of the bzdR gene. The third PCR amplification round uses as templates the A and B fragments and the 5HisReg and 3HISReg oligonucleotides that anneal at the start and stop codon, respectively, of each bzdR mutant variant. The bzdR3, bzdR4, and bzdR5 genes obtained in the third amplification step were BamHI- and HindIII-restricted and cloned into the pQE32 vector (Table 1) producing plasmids pQE32-His6BzdR3, pQE32-His6BzdR4, and pQE32-His6BzdR5, respectively (Table 1). The incorporation of the mutations in the genes was corroborated by DNA sequencing.
The construction of the bzdRΔL gene was achieved by PCR amplification of the NbzdR fragment (nucleotides 1–270 of bzdR gene) using the oligonucleotides 5HisReg (5′-CGGGATCCTTTCCAACGATGAGAACTCATCAC-3′, engineered BamHI is underlined) and new-N1kim (5′-GGGGTACCCTCCGCCTCCTCGCGCACG-3′, engineered KpnI is underlined) and the CbzdR fragment (nucleotides 391–897 of bzdR gene) using the oligonucleotides 5′C1N1 (5′-GGGGTACCTTCAGCGCATCGCCTTCATCGGC-3′, engineered KpnI is underlined) and 3′C1N1 (5′-AACTGCAGTCAGCGTGCCAGGACTTCGAGG-3′, engineered PstI is underlined). The NbzdR fragment was purified and cloned into the pGEM-Teasy vector using the BamHI and KpnI restriction sites. The plasmid obtained was used to clone the CbzdR fragment using the restriction sites KpnI and PstI, generating plasmid pGEMT-BzdRΔL (Table 1). The bzdRΔL gene was BamHI/PstI double-digested in pGEMT-BzdRΔL and subcloned into BamHI/PstI double-digested pQE32 vector, producing plasmid pQE32-His6-BzdRΔL (Table 1).
Overproduction and Purification of His6-BzdR, His6-BzdR3, His6-BzdR4, His6-BzdR5, and His6-BzdRΔL
The recombinant plasmids pQE32-His6BzdR, pQE32-His6BzdR3, pQE32-His6BzdR4, pQE32-His6BzdR5, and pQE32-His6BzdRΔL (Table 1) harbor the bzdR, bzdR3, bzdR4, bzdR5, and bzdRΔL genes, respectively, without the ATG start codon and with a His6 tag coding sequence (MRGSHHHHHHGIL), at their 5′-end, under control of the T5 promoter and two lac operator boxes. The His-tagged proteins were overproduced in E. coli M15 strain harboring also plasmid pREP4 (Table 1), and they were purified following protocols previously established (7).
Analytical Ultracentrifugation Methods
Sedimentation velocity and equilibrium were performed to determine the state of association of His6-BzdR and its complex with DNA. The PN DNA fragment used was obtained as described previously (7). The analytical ultracentrifugation analysis was performed using several protein concentrations (from 1 to 10 μm) and two DNA concentrations (0.1 and 0.2 μm). All samples were equilibrated in 50 mm NaH2PO4, 300 mm KCl, 100 mm imidazole, pH 8.0, buffer. The sedimentation velocity runs were carried out at 43,000 rpm and 20 °C in an XL-I analytical ultracentrifuge (Beckman-Coulter Inc.) equipped with UV-Visible optics detection system, using an An50Ti rotor and 12-mm double sector centerpieces. Sedimentation profiles were registered every 1–5 min at 260 and 275 nm. The sedimentation coefficient distributions were calculated by least squares boundary modeling of sedimentation velocity data using the c(s) method (30), as implemented in the SEDFIT program. These s values were corrected to standard conditions (water, 20 °C, and infinite dilution; Ref. 31) using the SEDNTERP program (32) to get the corresponding standard s values (s20,w). Sedimentation velocity of the purified His6-BzdR (10 μm) in the presence of benzoyl-CoA (0.5, 1, or 2 mm) was also done. In the case of the protein/DNA mixtures, the effect of protein concentration on the s value of the DNA component was used as the first assay to monitor the complex formation. Sedimentation equilibrium assays were carried out at speeds ranging from 5,000 to 15,000 rpm (depending upon the samples analyzed) and at several wavelengths (260, 280, and 290 nm) with short columns (85–95 μl), using the same experimental conditions and instrument as in the sedimentation velocity experiments. After the equilibrium scans, a high speed centrifugation run (40,000 rpm) was done to estimate the corresponding base-line offsets. The measured low speed equilibrium concentration (signal) gradients of His6-BzdR and DNA alone and the protein/DNA mixtures were fit by the equation that characterizes the equilibrium gradient of an ideally sedimenting solute (using a MATLAB program, kindly provided by Dr. Allen Minton, National Institutes of Health) to get the corresponding buoyant signal average molecular weights. The average molecular weight of the different macromolecular solutes were determined from the experimental buoyant molecular weight values using the following partial specific volumes as follows: 0.737 ml/g for His6-BzdR (calculated from the amino acid composition using the SEDNTERP program; Ref. 32) and 0.55 ml/g for 253-bp-PN (taken from an estimated literature value for nucleic acids, Ref. 33).
Sedimentation equilibrium was also used to determine the DNA binding capacity of His6-BzdR. The differences in size between the proteins and PN, together with the fact that most of extinction at 260 nm of the mixture corresponded to the DNA (more than 90% of the total signal at the highest protein concentrations used), allow the design of low speed sedimentation equilibrium experiments (5,000–6,000 rpm) to discriminate the gradients of DNA (free and bound) in the presence of the proteins without the requirement of DNA labeling. The sedimentation equilibrium data of the mixtures were analyzed assuming no significant volume changes upon complex formation (34, 35). Therefore, the buoyant molecular weight of the complex (bMr,C) is the composition weighted sum of the buoyant molecular weights of the individual components as follows: i (bMr,A) + j (bMr,B), where i and j are the number of molecules of A (i.e. PN) and B (protein), respectively; and, bMr,A and bMr,B are the buoyant molecular weights of pure A and pure B, respectively. To further characterize the DNA binding process (in terms of stoichiometry, affinity, and degree of cooperativity), the dependence of the degree of the protein-DNA complex formation upon protein concentration can be described by an empirical Hill function: B(L) = Bmax (L/L50)n/(1 + (L/L50)n),where B(L) is the amount of protein bound to the DNA at a given free concentration of protein (L), and L50 is equal to half of the maximal binding capacity Bmax, and n is a cooperativity parameter (36).
Electron Microscopy Studies
The purified His6-BzdR protein was applied to carbon-coated grids, negatively stained with 2% uranyl acetate, and observed in a JEOL 1230 electron microscope operating at 100 kV. Micrographs were recorded at a magnification of 50,000 under low dose conditions and scanned using a Minolta Dimage Scan Multi Pro scanner at 2,400 dpi. The contrast transfer function of each micrograph was estimated using the program CTFFIND3 (37) and corrected with the program Bsoft (38) by flipping phases. 5,741 individual molecules were selected using “boxer” from the image analysis package EMAN (39) and averaged to a final 4.2 Å/pixel. These images were processed using XMIPP (40) to obtain reference-free class averages. Additionally, a three-dimensional reconstruction was obtained by angular refinement methods using EMAN. Several references were used as initial templates for refinement; some were constructed as Gaussian blobs using commands found in EMAN, whereas others were built after reference-free image classification, averaging, and using common lines. When 2-fold symmetry was applied during the final steps of the refinement, the most likely axis for symmetrization was selected based on the visual inspection of the reconstruction obtained in the absence of symmetry, although the precise orientation of the symmetry axis could not be determined computationally.
The resolution of the final reconstruction was estimated to be 25 Å using the Fourier Shell Correlation method with a cutoff at a correlation of 0.5. The volume was rendered assuming a density for the protein of 1.35 g/ml, with UCSF Chimera (41). The handedness of the structure could not be defined, and one hand was arbitrarily selected for representation. Fitting of the atomic models into the three-dimensional reconstruction of BzdR was performed by using the program COLORES as implemented in the platform SITUS (42, 43). The comparison between the high resolution models and the low resolution EM structure was evaluated by cross-correlation, and the use of a Laplacian filter to enhance the boundaries of the complex.
Cross-linking Assays
Purified His6-BzdR protein at concentrations ranging from 2 to 10 μm was incubated at different concentrations (5–50 mm) of the cross-linking agent glutaraldehyde at 20 °C for 20 min in the protein purification buffer (50 mm NaH2PO4, 300 mm KCl, 100 mm imidazole, pH 8.0), as described previously (44). All reactions were finished by the addition of the stop buffer (62.5 mm Tris-HCl, pH 6.8, 2% SDS, 5% β-mercaptoethanol, 10% glycerol, and 0.005% bromphenol blue). Samples were heated at 100 °C for 10 min and analyzed by SDS-PAGE (27).
Limited Proteolysis Assays
Purified His6-BzdR (10 μm) was incubated with trypsin (0.02 mg/ml, Sigma) in 20 μl of reaction buffer (25 mm Tris-HCl, pH 7.5, 10 mm CaCl2) in the absence or presence of 2 mm benzoyl-CoA for 0–60 min at 37 °C (45). The reactions were finished by the addition of the stop buffer. The samples were boiled and analyzed by SDS-PAGE (27).
Modeling of Proteins
The three-dimensional model of BzdR was generated by using the LOOPP program (46) and visualized with the PyMol program (DeLano Scientific) as described previously (7).
Fluorescence Measurements of Ligand Binding to BzdR and BzdRΔL Proteins
Binding of benzoyl-CoA to BzdR or BzdRΔL was determined by monitoring the change in protein fluorescence upon ligand addition. Measurements were done on an SML-Amico 8000 spectrophotometer using quartz cuvettes with a 0.2-cm excitation and 1-cm emission path lengths (λexcitation = 275 nm and λemission 326 nm; slit widths = 5 nm). Titrations were performed at 25 °C by addition of stock solutions of benzoyl-CoA to 0.4 ml of 20 mm Tris-HCl, pH 8.0, 100 mm KCl containing BzdR or BzdRΔL (1 μm initial concentration). Final ligand concentrations varied from 0 to 300 μm. Corrections were made for dilution of the protein, for background signal, and also for the instrumental response as indicated by the manufacturer. Control titrations with buffer alone did not produce any change in emission signal after correction. The Kd values for benzoyl-CoA for both BzdR and BzdRΔL regulators were calculated using GraphPad Prism software (GraphPad software, San Diego) with the data fit to the equation corresponding to a one-site saturation model, ([L]b/[P]t) = ([L]free/Kd + [L]free); where [L]b is the concentration of bound ligand; [P]t is the total protein concentration, and [L]free is the concentration of free ligand. Fraction of bound ligand was calculated as (Io − I)/(o − Imin); where Io is the maximum BzdR fluorescence intensity (no ligand added); I is the fluorescence intensity after addition of an aliquot of ligand, and Imin is the fluorescence intensity at saturating concentration of ligand. This last value was estimated by nonlinear regression to a single hyperbolic decay curve. A minimum of three experimental trials was averaged for calculation of the dissociation constants.
Gel Retardation Assays
The PN DNA probe was obtained as described previously (7).
The mI probe (145 bp) was obtained by PCR amplification of DNA fragment spanning position −61 to +79 with respect to the transcription start site of the PN promoter from plasmid pECOR7 (Table 1) by using oligonucleotides BLINE (5′-CGTGCCTGACATTTGACTTAGATC-3′) and 3IVTPN (5′-CGGAATTCCATCGAACTATCTCCTCTGATG; the engineered EcoRI site is underlined). The mII probe (80 bp) was obtained by PCR amplification of DNA fragment spanning position −112 to −38 with respect to the transcription start site of PN promoter from plasmid pECOR7 by using oligonucleotides PNII (5′-CAAGAAAGATTGCAGTTTTCCATG-3′) and NIK (5′-CGGAATTCGATCTAAGTCAAATGTCAGGCACG-3′; the engineered EcoRI site is underlined). The mIII probe (79 bp) was obtained by PCR amplification of DNA fragment spanning position −174 to −101 with respect to the transcription start site of PN promoter from plasmid pECOR7 by using oligonucleotides RET1 (5′-CCGAGCCTCGCGTTTTACTGC-3′) and NIU PN-III (5′-CGGAATTCCAATCTTTCTTGCCGCACACGC-3′; the engineered EcoRI site is underlined). The mI, mII, and mIII DNA fragments digested with EcoRI and labeled by filling in the overhanging EcoRI-digested end with [α-32P]dATP (6000 Ci/mmol; Amersham Biosciences) and the Klenow fragment of E. coli DNA polymerase I as described previously (7).
The probes were mixed with the purified proteins at the concentrations indicated in each assay, following procedures previously established (7). The retardation reactions mixtures contained 20 mm Tris-HCl, pH 7.5, 10% glycerol, 2 mm β-mercaptoethanol, 50 mm KCl, 0.05 nm DNA probe, 250 μg/ml bovine serum albumin, and purified His6-BzdR protein in a 9-μl final volume.
DNase I Footprinting Assays
The DNA fragment used for DNase I footprinting assays was the same PN probe as that reported for the gel retardation assays (see above). For the assays, the reaction mixture contained 2 nm DNA probe, 1 mg/ml bovine serum albumin, and purified protein in 15 μl of FP buffer (7). This mixture was incubated for 20 min at 37 °C, after which 3 μl (0.05 unit) of DNase I (Amersham Biosciences) (prepared in 10 mm CaCl2, 10 mm MgCl2, 125 mm KCl, and 10 mm Tris-HCl, pH 7.5) was added, and the incubation was continued at 37 °C for 20 s. The reaction was stopped by addition of 180 μl of solution containing 0.4 m sodium acetate, 2.5 mm EDTA, 50 μg/ml calf thymus DNA, and 0.3 μg/ml glycogen. After phenol extraction, DNA fragments were analyzed as described previously (7). A + G Maxam and Gilbert reactions (47) were carried out with the same fragments and loaded on the gels along with the footprinting samples. The gels were dried on Whatman 3MM paper and exposed to Hyperfilm MP (Amersham Biosciences).
In Vitro Transcription Experiments
Transcription assays were performed as published previously (48). Plasmid pJCD-PN (Table 1) was used as a supercoiled PN template. Reactions (50-μl mixtures) were performed in a buffer consisting of 50 mm Tris-HCl, pH 7.5, 50 mm KCl, 10 mm MgCl2, 0.1 mm BSA, 10 mm dithiothreitol, and 1 mm EDTA. Each DNA template (0.5 mm of supercoiled plasmid pJCD- PN) was premixed with 50 nm σ70-containing E. coli RNA polymerase (Amersham Biosciences), 20 nm of purified His6-Fnr* (a constitutively active Fnr mutant protein that carries a D154A substitution and is able to form a dimer and to bind DNA in the presence of oxygen) (6, 49), and different amounts of purified His6-BzdR or His6-BzdRΔL and 1 mm benzoyl-CoA. For multiple rounds assays, transcription was then initiated by adding a mixture of 500 nm (each) ATP, CTP, and GTP; 50 mm UTP; and 2.5 μCi of [α-32P]UTP (3,000 mCi mmol−1; 111 GBq mmol−1). After incubation for 15 min at 37 °C, the reactions were stopped with an equal volume of a solution containing 50 mm EDTA, 350 mm NaCl, and 0.5 mg of carrier tRNA ml−1. The mRNA produced was then precipitated with ethanol, dissolved in loading buffer (7 m urea, 1 mm EDTA, 0.6 m glycerol, 0.9 mm bromphenol blue, and 1.1 mm xylene cyanol), electrophoresed on a denaturing 7 m urea, 4% polyacrylamide gel, and visualized by autoradiography.
RESULTS AND DISCUSSION
BzdR Is a Homodimer in Solution
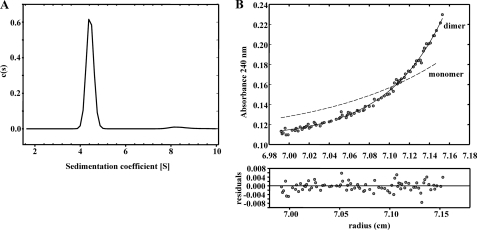
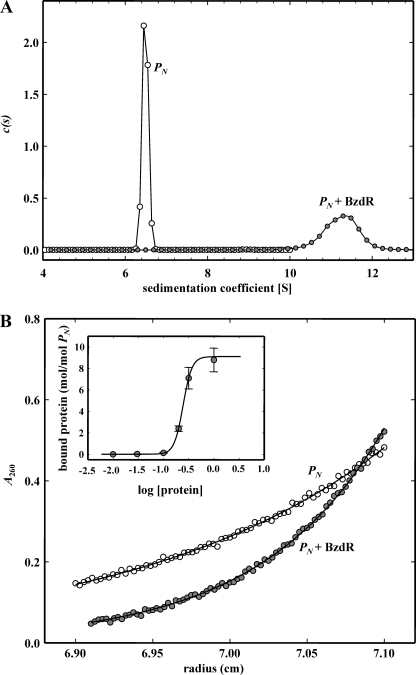
To study the native conformation of the BzdR regulator, we performed analytical ultracentrifugation experiments with the purified His6-BzdR protein. Sedimentation velocity experiments were carried out at different concentrations of BzdR (1–30 μm) and analyzed in terms of distribution of sedimentation coefficients, allowing an evaluation of protein homogeneity and self-association. Fig. 1A shows the sedimentation velocity data for 10 μm BzdR in solution, demonstrating that under these conditions BzdR sediments as a unique species with an s value of 4.4 ± 0.1 S. The molecular weight of the 4.4 S species measured by sedimentation equilibrium was compatible with the mass of the protein dimer (BzdR monomer theoretical mass of 34,895; Fig. 1B). From the combined analytical centrifugation results, we can conclude that under the experimental conditions used, BzdR forms stable dimers with a gross shape (frictional ratio f/f0 of 1.4) that slightly deviates from that expected for a globular protein. In addition, the analytical centrifugation results are in agreement with the glutaraldehyde cross-linking analysis (see the “Experimental Procedures”), that reveal the formation of a band corresponding to the dimeric form of BzdR (data not shown).
FIGURE 1.
Study of the oligomerization state of the BzdR protein in solution. A, sedimentation coefficient distribution c(s) corresponding to the sedimentation velocity of purified His6-BzdR. The distribution pattern of concentration of the protein (c(s)) is represented in front of the sedimentation coefficient (S). B, sedimentation equilibrium data (gray dots) and the best fit analysis assuming a theoretical protein dimer (solid line) and monomer (broken line) species. The lower panel shows residuals between estimated values and experimental data for protein dimer.
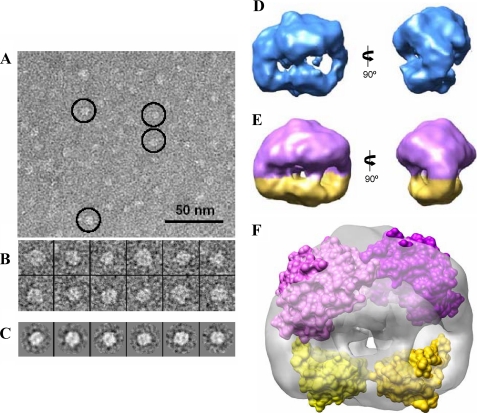
To obtain additional information about the native structure of the BzdR protein, we performed EM studies that have been used successfully before to resolve the structure of many proteins, including some prokaryotic transcriptional regulators (50, 51). Because the molecular weight of the BzdR protein was small compared with what is usual in EM studies, we used negative staining with uranyl acetate to increase contrast. Despite this size limitation, BzdR was clearly detected in the micrographs (Fig. 2A), obtaining a total of 5,741 images of individual molecules (Fig. 2B). First, the data were analyzed by classification and averaging using reference-free methods. Two-dimensional averages revealed that many images complied with an overall rectangular shape displaying a rough 2-fold symmetry (Fig. 2C). Images were then refined into a three-dimensional reconstruction using angular refinement methods without assuming any symmetry (Fig. 2D). This reconstruction revealed that BzdR was composed of a large and a small region with an apparent 2-fold rotational symmetry. This structure was then further refined imposing this symmetry into a 2-fold symmetrized reconstruction at a resolution of 25 Å (Fig. 2E). Calculation of the protein mass accounted for this reconstruction at a threshold adequate to enclose the structure and assuming the average density of proteins (1.35 g/ml) was compatible with a dimer. Therefore, BzdR could be built as a dimeric structure where each monomer includes a large (Fig. 2E, purple color) and a small (Fig. 2E, yellow color) region. The previously obtained high resolution three-dimensional models of the N- and C-terminal domains of BzdR (7) were fitted into the medium resolution EM map (see the “Experimental Procedures”), and we found that the EM structure enclosed sufficient volume as to accommodate a dimer of BzdR (Fig. 2F). The precise orientation of the protein domains within the EM structure could not be determined unambiguously due to the low resolution of the EM map. Therefore, we have represented a low resolution filtered version of the atomic models, represented as a surface, just to reveal the occupancy of the EM density, fitted into the EM density (Fig. 2F), to point out that the three-dimensional structure can accommodate two copies of each of the N- and C-terminal domains of BzdR (Fig. 2F). In summary, both analytical ultracentrifugation and electron microscopy studies demonstrate that BzdR is a dimeric protein, which is in agreement with the reported dimeric conformation observed in many regulators of the XRE family.
FIGURE 2.
Three-dimensional structure of BzdR using negative staining electron microscopy. A, electron microscopy field of purified His6-BzdR protein. Some representative molecules have been highlighted within circles. Scale bar, 50 nm. B, gallery of 12 selected molecules from the data set of 5,741 molecules. C, some reference-free class averages obtained using XMIPP (38). D, two views of the three-dimensional structure of BzdR reconstructed without assuming any symmetry. E, two views of the three-dimensional structure of BzdR reconstructed after imposing 2-fold symmetry. The big (violet) and the small (yellow) domains might correspond to the C-BzdR and N-BzdR domains, respectively. F, two copies of the structural models of C-BzdR (violet) and N-BzdR (yellow) can be fitted into the three-dimensional structure obtained by electron microscopy (shown as a gray transparent density), indicating that BzdR has sufficient volume to accommodate two copies of each domain. The precise orientation of the domains cannot be determined at the resolution of this three-dimensional map, and the two atomic models have been represented as a surface just to reveal the occupancy of the EM density.
Binding of BzdR to the DNA Is Cooperative
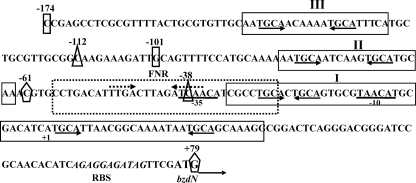
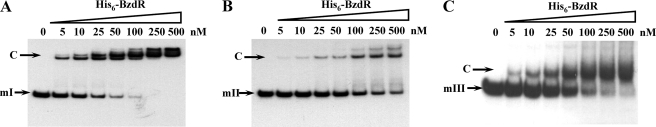
As indicated in the Introduction, it has been described before that the BzdR protein binds to the PN promoter at three operator regions (7). To better understand the interaction of BzdR with PN, three different DNA probes were generated that cover each of the operator boxes already described (Fig. 3). The three probes were termed as mI (from position −61 to +79 with respect to the transcription start site, and it includes the operator region I of PN), mII (from −112 to −38, and it includes the operator region II of PN), and mIII (from −174 to −101, it includes the operator region III of PN). The interaction of BzdR with the three DNA probes was studied by gel shift experiments as described under “Experimental Procedures.” As shown in Fig. 4, purified BzdR retarded the three operator boxes but with different affinities. Thus, whereas BzdR shows the higher affinity for the mI probe (Kd in the range of 10 nm), the affinities for the mIII and mII probes decreased to 50 and 250 nm, respectively. The higher affinity of BzdR for the mI probe could reflect the existence of two palindromic sequences in the long operator region I, which may account for the existence of at least two retardation complexes, but only one palindromic region in the short operators II and III (Fig. 3). Interestingly, the concentration of BzdR needed to totally shift the PN probe (extending from positions −174 to +79 and containing the three operator regions) was similar to that needed to retard the mI probe but lower than that needed to retard the mII or the mIII probes, suggesting that BzdR binds cooperatively to the PN promoter.
FIGURE 3.
Scheme of the PN promoter from Azoarcus sp. CIB. The nucleotide sequence from positions −174 to +79 is presented. The transcription start site (+1) and the inferred −10 and −35 boxes of the PN promoter are indicated. The ribosome-binding site (RBS) and the ATG start codon of the bzdN gene are also shown in italic and boldface type, respectively. The BzdR-binding regions I–III (operators) are boxed. The palindromic regions containing the TGCA repeats within the operator regions are marked with arrows below the sequence. The FNR (AcpR)-binding site (FNR) is boxed with broken lines, and the inverted repeats of the consensus FNR-binding sequence are marked with convergent broken arrows above the sequence. The ends of the mI (−61 to +79), mII (−112 to −38), and mIII (−174 to −101) DNA probes are indicated as pentagons, triangles, and rectangles, respectively.
FIGURE 4.
In vitro binding of His6-BzdR to the mI, mII, and mIII probes. Gel retardation analyses of mI (A), mII (B), and mIII (C) were performed as indicated under “Experimental Procedures.” The concentration of purified His6-BzdR used is indicated on the top. The free DNA probes (mI, mII, or mIII) and the protein-DNA complexes (C) are indicated by arrows.
To elucidate the number of molecules of BzdR protein that interact with the PN promoter, we performed analytical ultracentrifugation experiments of the complex PN::BzdR as described under “Experimental Procedures.” The binding of His6-BzdR to PN was studied by means of sedimentation velocity analysis of a fixed amount of DNA (0.2 μm) in the absence or in the presence of increasing concentrations of His6-BzdR up to a 20-fold protein molar excess (10:0.2 μm). The DNA alone (open circles in Fig. 5A) sediments as a single species with an s value of 6.5 S. The interaction leads to the formation of a complex with an s value of 11.5 ± 0.8 S (closed circles in Fig. 5A) at a protein/DNA molar ratio of 10:1. Upon increasing the protein concentration of the mixture, larger aggregates were formed that sediment out of the cell window (data not shown). The corresponding sedimentation equilibrium gradient of the 11.5 S species is shown in Fig. 5B (closed circles). A single species model with buoyant molecular weight of 230,000 ± 8,000 can account for the experimental gradient. This buoyant value is compatible with the presence of 8–9 molecules of His6-BzdR in this complex. The analysis of the dependence of the complex formation with protein concentration confirms that eight protein molecules interact with the PN promoter in a cooperative manner (Hill coefficient >2) to form the nucleoprotein complex, with a concentration for semi-saturation of 0.24 ± 0.1 μm (Fig. 5B, inset).
FIGURE 5.
Analysis of complex formation in the interaction of protein His6-BzdR with DNA 253-bp-PN. A, sedimentation coefficient distribution c(s) corresponding to the sedimentation velocity of purified DNA (open circles) and protein-DNA complex (closed circles) at a protein/DNA molar ratio of 10:1. B, corresponding sedimentation equilibrium gradients. Inset represents the variation of bound protein (mol/mol DNA) with protein concentration.
It is tempting to speculate that a BzdR dimer binds to each of the four palindromic regions harboring the TGCA sequence within PN, thus being two bound dimers (four molecules) at operator region I, one bound dimer (two molecules) at operator region II, and another dimer at operator region III. This arrangement of short palindromic sequences spread into three operator boxes, and the cooperative binding to the target DNA may ensure a local concentration of BzdR to guarantee a tough control of the activity of the PN promoter preventing the expression of the bzd genes when benzoate is not present in the growth media.
Conformational Changes in BzdR upon Benzoyl-CoA Binding
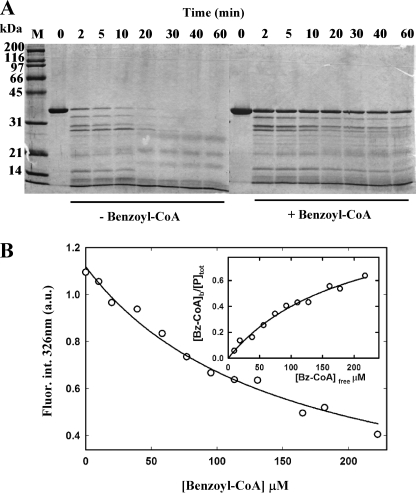
Limited proteolysis has been successfully used to detect conformational changes of regulators upon interaction with their cognate inducers (45). Therefore, to monitor potential conformational changes in the BzdR regulator induced by benzoyl-CoA binding, we first accomplished a limited proteolysis experiment as detailed under “Experimental Procedures.” Fig. 6A shows that the BzdR protein is more accessible to trypsin digestion in the absence of benzoyl-CoA, suggesting that the presence of inducer protects BzdR against the trypsin cleavage and supporting the hypothesis of a conformational change in the regulator induced by its interaction with benzoyl-CoA.
FIGURE 6.
Conformational change of BzdR induced by benzoyl-CoA. A, limited proteolysis assays of purified His6-BzdR. SDS-12.5% PAGE show the results of the trypsin-mediated digestion of purified His6-BzdR (10 μm) in the absence (−Benzoyl-CoA) or in the presence (+Benzoyl-CoA) of 2 mm benzoyl-CoA as detailed under “Experimental Procedures.” Bz, benzoyl. Lanes 0–60, incubations were performed at 37 °C for 0, 2, 5, 10, 20, 30, 40, 60 min, respectively. Lane M represents the molecular mass markers (in kDa). B, intrinsic fluorescence of BzdR as a function of benzoyl-CoA concentration. Data points represent the decrease of the BzdR (7.5 μm) fluorescence emission maximum (326 nm) expressed in arbitrary units (a.u.) upon excitation at 275 nm in the presence of increasing concentrations of benzoyl-CoA. Inset, fitting of benzoyl-CoA binding to BzdR to a single site model.
As shown above, BzdR is a homodimer species in solution (Fig. 1). To check whether the interaction with benzoyl-CoA promotes a change in the oligomeric state of BzdR, additional analytical ultracentrifugation experiments were performed. Sedimentation velocity results showed that in the presence of benzoyl-CoA, the s value of BzdR was not modified in the presence of the inducer (data not shown). This result indicated that BzdR bound to benzoyl-CoA still behaves as a dimeric protein.
To study further the interaction of BzdR with its cognate inducer molecule, fluorescence spectroscopy was used taking advantage of its intrinsic sensitivity to changes in the microenvironment of aromatic residues of the protein (52, 53). BzdR has two tryptophan residues located at positions 112 and 229, and its fluorescence emission spectrum shows a maximum at 326 nm upon excitation at 275 nm. Addition of benzoyl-CoA quenches the fluorescence emission signal of BzdR (Fig. 6B), allowing us to carry out a fluorescence titration assay. Titration of the protein with increasing amounts of benzoyl-CoA yields a binding isotherm with a dissociation constant (Kd) of 157 ± 58 μm, a value that is within the range of that obtained with other transcriptional regulators that control aromatic degradation pathways (54). This analysis assumes a single binding site for benzoyl-CoA that should be located at the C-terminal domain of BzdR, as deduced from the structural model of the protein (7). Interestingly, similar binding assays using the noninducer molecules benzoate, phenylacetyl-CoA, or CoA revealed the absence of significant changes in the fluorescence signal of BzdR (data not shown), suggesting that the quenching effects caused by benzoyl-CoA are highly specific. Furthermore, benzoyl-CoA binding to BzdR results in a red shift of the tryptophan emission, from a maximum at 326 nm in the absence of benzoyl-CoA to a maximum at 332 nm in the presence of the inducer benzoyl-CoA (data not shown). Because a similar red shift of the tryptophan emission maximum and fluorescence quenching induced by benzoyl-CoA addition is observed with a mutant BzdR protein (BzdRΔL) lacking residue Trp112 (see below), it appears reasonable to assume that Trp229 is responsible for the fluorescence emission of BzdR, and therefore ligand binding would result in a change in its microenvironment. In this regard, the structural model proposed for C-BzdR (7) indicates that the Trp229 side chain is almost buried, and therefore the observed red-shifted emission of BzdR upon benzoyl-CoA binding would be consistent with a conformational change in which the polarity of the Trp229 side chain has increased.
Taken as a whole, these results support the idea that benzoyl-CoA binding to the BzdR protein, which most probably occurs through its C-terminal domain, causes a conformational change in the regulator that, however, does not lead to the modification of its dimeric conformation.
Effect of Benzoyl-CoA on the BzdR-mediated Repression of the PN Promoter
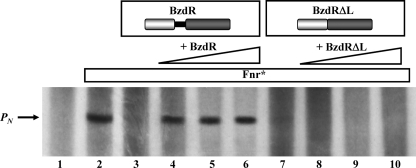
As shown above, the binding of benzoyl-CoA induces a conformational change in C-BzdR. This conformational change in C-BzdR would then trigger a structural change at the DNA binding domain (N-BzdR), rendering a regulator unable to inhibit the activity of the PN promoter. Although the BzdR-dependent regulation of the PN promoter has been observed in vivo (7), this regulatory circuit has not been confirmed in vitro so far. To accomplish this goal, we have used a previously reported in vitro transcription assay using the purified Fnr* activator (an E. coli homolog of the AcpR activator from Azoarcus sp. CIB), the E. coli RNA polymerase, and the plasmid pJCD-PN, which contains the PN promoter as a supercoiled DNA template (6). Whereas the formation of the expected 184-nucleotide transcript due to the activity of the PN promoter was observed in the absence of BzdR (Fig. 7, lane 2), this transcript was missing in the presence of purified BzdR protein (Fig. 7, lane 3), confirming the repressor role of this regulator. The addition of the inducer benzoyl-CoA restored the appearance of the 184-nucleotide transcript (Fig. 7, lanes 4–6) indicating the activation of the PN promoter and therefore the abolition of the repression due to BzdR. Therefore, these data and the results shown above confirm that benzoyl-CoA causes a conformational change in BzdR that prevents its repression effect and leads to activation of the PN promoter in the presence of the Fnr* activator.
FIGURE 7.
Effect of BzdR or BzdRΔL and benzoyl-CoA on in vitro transcription from PN. Multiple round in vitro transcription reactions were performed as indicated under “Experimental Procedures” by using pJCD-PN, a plasmid template that produces a 184-nucleotide mRNAs from PN, 50 nm E. coli RNA polymerase, and 20 nm Fnr* activator. The transcription reactions were carried out in the absence of repressor (lane 2) or in the presence of 25 nm (lanes 3 and 4), 50 nm (lane 5), or 100 nm (lane 6) of purified His6-BzdR or 25 nm (lanes 7 and 8), 50 nm (lane 9), or 100 nm (lane 10) of purified His6-BzdRΔL. Benzoyl-CoA was added at 1 mm (lanes 4–6 and 8–10). Lane 1, control assay without RNA polymerase.
Role of the BzdR Linker Region
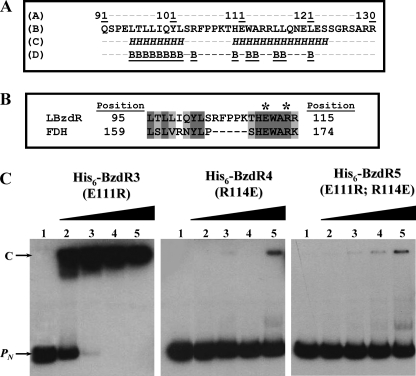
A detailed analysis of the primary structure of BzdR revealed the existence of a linker region (LBzdR), from residue Gln91 to Arg130, connecting the N-BzdR and C-BzdR domains (Fig. 8A). Secondary structure prediction of LBzdR using the JPred3 software (29) identified two possible α-helices spanning residues Leu95–Leu102 and His110–Ser123, respectively, which would not be solvent-exposed in the protein surface (Fig. 8A). Conversely, a BLAST search against the Protein Data Bank revealed that the sequence stretch of LBzdR, including residues Leu95 to Arg115, showed high similarity (42% identity) to the segment making up the α5-helix (residues Leu159 to Lys174) of the homodimeric NAD+-dependent formate dehydrogenase from Pseudomonas sp. 101 (Fig. 8B) (Protein Data Bank code 2NAD; Refs. 55, 56). Notably, analysis of the surface areas at the interface between subunits of the latter enzyme with the PISA server (57) revealed two important issues as follows: first, the above-mentioned α5-helix of formate dehydrogenase is located at the main contacting interface between subunits, and second, some residues of formate dehydrogenase, which are strictly conserved in LBzdR, directly participate in symmetry-related intersubunit contacts, as is the case for Glu170 and Arg173 that form the unique salt bridge between the two α5-helices (Fig. 8B) (56). To experimentally verify the assumption that LBzdR participates in maintaining the dimeric state of BzdR similarly to the α5-helix in formate dehydrogenase, three mutant variants of BzdR were constructed. Two single substitutions, E111R (BzdR3) and R114E (BzdR4), should prevent the formation of the predicted salt bridge, presumably affecting the dimeric state of BzdR, and the double mutant E111R/R114E (BzdR5) should restore the putative salt bridge maintaining the homodimeric conformation of BzdR. The BzdR mutant proteins were constructed with a His6 tag at their N-terminal end, and they were overproduced in E. coli and purified as described under “Experimental Procedures.” The three mutant proteins were produced with a similar yield and presented comparable solubility to that of the wild-type BzdR protein (data not shown). Interestingly, analytical ultracentrifugation experiments revealed that whereas BzdR3 showed a sedimentation coefficient compatible with a dimer (s value of 4.2 ± 0.1), BzdR4 and BzdR5 were compatible with protein with monomeric proteins (s values of 2.7 ± 0.1 and 2.9 ± 0.1, respectively).
FIGURE 8.
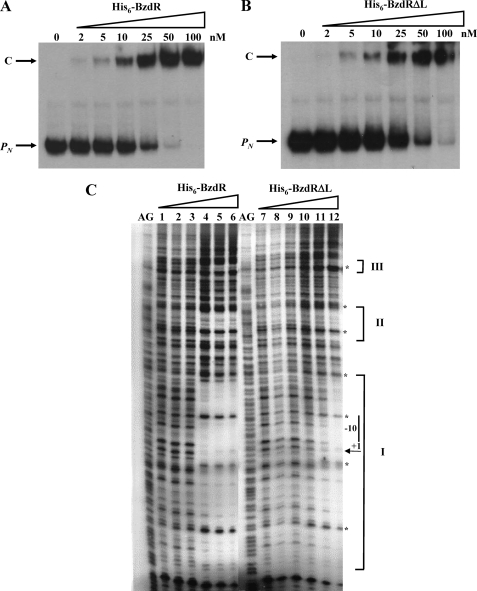
Analysis of the linker region of BzdR. A, amino acid sequence analysis of LBzdR. The position of the LBzdR residues in the primary structure of BzdR is indicated. The sequence of the linker was analyzed using the JPred3 program that predicts residues involved in α-helices, indicated as italic H, and those that have a probability lower than 25% of being located at the protein surface, indicated as underlined B. B, alignment of a region of LBzdR (residues 95–115) with a significantly similar region (residues 159–174) of the formate dehydrogenase from Pseudomonas sp. 101. The dark gray and light gray shadows represent identical residues and conservative substitutions, respectively. The position of the Glu and Arg residues involved in a saline bridge in formate dehydrogenase and the equivalent residues in LBzdR are indicated with an asterisk. C, in vitro analysis of the interaction of BzdR mutants with the PN promoter. Gel retardation analysis of His6-BzdR3, His6-BzdR4, and His6-BzdR5 mutant regulators binding to the PN promoter was performed as indicated in “Experimental Procedures.” Lane 1, free PN probe; lanes 2–5, retardation assays containing 25, 50, 100, or 200 nm, respectively, of purified protein (indicated on the top). The PN probe (PN) and the PN-protein complex (C) are indicated by arrows.
Therefore, despite that substitution R114E disturbs the dimerization of the protein and the fact that the single substitution E111R did not and the double replacement E111R/R114E could not restore such dimeric conformations allow us to discard the predicted salt bridge in the dimerization process. In this sense, although LBzdR seems not to be crucial in BzdR dimerization by the formation of a salt bridge, the monomeric conformation caused by the R114E substitution, which represents a decrease in the total of positive charges in the linker area, suggests that the surface charge distribution in LBzdR may influence the dimerization of BzdR.
Notably, gel shift experiments showed that whereas the BzdR3 (E111R) protein was able to bind to the PN promoter in a concentration-dependent manner and with an affinity similar to that of the parental BzdR regulator (Figs. 8C and 9A), the BzdR4 (R114E) and BzdR5 (E111R and R114E) proteins did not bind efficiently to the PN promoter (Fig. 8C). These results support the idea that the dimeric state of BzdR is necessary for binding to the target DNA, a feature also reported for other regulators of the XRE family such as the λCI repressor (58, 59).
FIGURE 9.
In vitro binding of His6-BzdR and His6-BzdRΔL to the PN probe. A and B, gel retardation analyses of PN were performed as indicated under “Experimental Procedures.” The concentration of purified His6-BzdR (A) and His6-BzdRΔL (B) used is indicated on the top. The free DNA probes (PN) and the protein-DNA complexes (C) are indicated by arrows. C, DNase I footprinting analyses of the interaction of purified His6-BzdR and His6-BzdRΔL with the PN promoter region. The DNase I footprinting experiments were carried out using the PN probe labeled as indicated under “Experimental Procedures.” Lanes 1 and 7 show footprinting assays in the absence of His6-BzdR or His6-BzdRΔL. Lanes 2–6 show footprinting assays containing 5, 25, 200, 400, and 800 nm purified His6-BzdR, respectively. Lanes 8–12 show footprinting assays containing 5, 25, 200, 400, and 800 nm purified His6-BzdRΔL, respectively. Lanes AG show the A + G Maxam and Gilbert sequencing reaction. Protected regions (I, II, and III) are marked by brackets, and the phosphodiester bonds hypersensitive to DNaseI cleavage are indicated by asterisks. The −10 box and the transcription initiation site (+1) of the PN promoter are also shown.
To further investigate the role of LBzdR, we constructed the BzdRΔL mutant regulator that lacks the complete LBzdR linker sequence (from Gln91 to Arg130) as detailed under “Experimental Procedures.” Interestingly, the BzdRΔL protein still retains the ability to bind to the target DNA with an efficiency slightly lower than that shown by the parental BzdR regulator (Fig. 9B), and protecting also the same three operator regions within the PN promoter (Fig. 9C). Moreover, BzdRΔL represses the transcription from PN with similar efficiency to that observed for the BzdR protein as demonstrated by in vitro transcription experiments (Fig. 7, lane 7). These results show that LBzdR does not influence the binding of BzdR to the target DNA, and they suggest that BzdRΔL should be a dimeric protein because BzdR is only able to bind to the PN promoter as a dimer. To demonstrate this last assumption, we performed analytical ultracentrifugation experiments with purified BzdRΔL, a protein that showed a sedimentation coefficient compatible with a dimer in solution (s value of 3.7 ± 0.1), and thus confirmed that LBzdR is dispensable in the dimerization of BzdR.
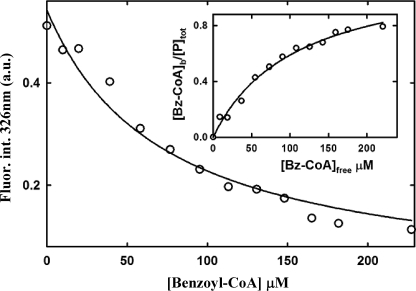
Remarkably, the inducer molecule benzoyl-CoA was not able to release the repressor effect of BzdRΔL on the PN promoter, as demonstrated by in vitro transcription assays (Fig. 7, lanes 8–10). To rule out that the observed effect could be due to the lack of interaction of BzdRΔL with the inducer molecule, fluorescence titration assays were carried out with purified BzdRΔL and benzoyl-CoA. As shown in Fig. 10, BzdRΔL retains the ability to bind benzoyl-CoA, with an affinity (Kd = 107 ± 15 μm) similar to that of wild-type BzdR (see above). This result strongly suggests that the BzdRΔL protein lacking the linker interacts with the inducer, ruling out that LBzdR can be essential in the interaction of BzdR with the inducer.
FIGURE 10.
Fluorescence titration of benzoyl-CoA binding to BzdRΔL. Variation of the fluorescence intensity (Fluor.int.) of BzdRΔL at 326 nm upon excitation at 275 nm expressed in arbitrary units (a.u.), with increasing concentrations of benzoyl-CoA (Bz-CoA). Experimental data were fitted by nonlinear regression to a single hyperbolic decay curve. The inset graph shows the fitting of the latter data to an equation corresponding to a one-site saturation model (see “Experimental Procedures”).
All these results taken together demonstrate that BzdRΔL binds DNA and interacts with the inducer molecule as the wild-type BzdR protein, but in contrast to the latter, the mutant regulator lacking LBzdR is not able to channel the conformational change induced by benzoyl-CoA and necessary to release the regulator from the target DNA, thus behaving as a super-repressor of the PN promoter. Therefore, we suggest that the major role of the linker region would be to facilitate the correct communication between the C-terminal domain of BzdR, the presumed target site for benzoyl-CoA binding, and the N-terminal domain involved in DNA binding.
Because the linker region of BzdR has an amino acid composition quite similar to that of Q-linkers that are present in several transcriptional regulators (60, 61), i.e. high abundance of glutamine (3), arginine (6), glutamic acid (4), serine (5), and proline (3) residues, it could be also considered as a Q-linker. Despite that some Q-linkers are just a connection between protein domains without a detectable function (61), other Q-linkers, such as that of the OmpR regulator, participate actively in the transmission of conformational change between domains (62). In this sense, LBzdR appears to behave as the linkers that directly participate in the transmission of the signal between the two domains of the regulatory protein.
Model of Action of the BzdR Regulatory Protein
As indicated previously, C-BzdR shows high identity to shikimate kinases, both at the sequence and structural levels (7). The shikimate kinases are enzymes that belong to the structural family of the nucleoside monophosphate (NMP) kinases and catalyze the conversion of shikimic acid to shikimate 3-phosphate using ATP as a co-substrate in the chorismate biosynthesis pathway (63). A characteristic feature of the NMP kinases is that they undergo large conformational changes during substrate binding and catalysis (24, 64). Previous studies have shown that the conformational changes are promoted by the LID region of the shikimate kinase that flaps over the active site upon substrate binding (24, 25). In the proposed three-dimensional structural model of C-BzdR, a LID-like region that spans from residues Arg240 to Gly252 can be identified. According to the mechanism of action of the shikimate kinase, we postulate that whereas the LID-like region of C-BzdR is maintained in an “open position” when the benzoyl-CoA is not present after the interaction with benzoyl-CoA, the LID-like region “closes” over the inducer. This movement at the LID-like region could lead a significant conformational change of C-BzdR that would then be transmitted to N-BzdR through the linker region, thus triggering a structural modification at the DNA-binding surfaces of the protein that would release the regulator from the target DNA allowing the activity of the PN promoter. More experiments are currently in progress to demonstrate experimentally this model.
In summary, the biochemical studies of BzdR presented in this work constitute an interesting step forward in the characterization of this new subfamily of regulators and paves the way for further biochemical studies with orthologous proteins (BoxR regulators) that are predicted to control the aerobic degradation of benzoate via benzoyl-CoA in many bacteria.
Acknowledgments
The technical work of A. Valencia is greatly appreciated. The help of Secugen S.L. with sequencing is gratefully acknowledged.
This work was supported in part by Comisión Interministerial de Ciencia y Tecnología Grants BIO2006-05957, BIO2009-10438, BIO2008-04478-C03-03, BFU2007-67404/BMC, SAF2008-00451, and CSD2007-00005 and Comunidad Autónoma de Madrid Grants P-AMB-259-0505 and CAM S-BIO-0214-2006.
- XRE
- xenobiotic response element.
REFERENCES
- 1.Gibson J. S., Harwood C. (2002) Annu. Rev. Microbiol. 56, 345–369 [DOI] [PubMed] [Google Scholar]
- 2.Lovley D. R. (2003) Nat. Rev. Microbiol. 1, 35–44 [DOI] [PubMed] [Google Scholar]
- 3.Carmona M., Zamarro M. T., Blázquez B., Durante-Rodríguez G., Juárez J. F., Valderrama J. A., Barragán M. J., García J. L., Díaz E. (2009) Microbiol. Mol. Biol. Rev. 73, 71–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harwood C. S., Burchhardt G., Herrmann H., Fuchs G. (1999) FEMS Microbiol. Rev. 22, 439–458 [Google Scholar]
- 5.López Barragán M. J., Carmona M., Zamarro M. T., Thiele B., Boll M., Fuchs G., García J. L., Díaz E. (2004) J. Bacteriol. 186, 5762–5774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durante-Rodríguez G., Zamarro M. T., García J. L., Díaz E., Carmona M. (2008) Microbiology 154, 306–316 [DOI] [PubMed] [Google Scholar]
- 7.Barragán M. J., Blázquez B., Zamarro M. T., Mancheño J. M., García J. L., Díaz E., Carmona M. (2005) J. Biol. Chem. 280, 10683–10694 [DOI] [PubMed] [Google Scholar]
- 8.Wintjens R., Rooman M. (1996) J. Mol. Biol. 262, 294–313 [DOI] [PubMed] [Google Scholar]
- 9.Ptashne M., Backman K., Humayun M. Z., Jefferey A., Maurer R., Meyer B., Sauer R. T. (1976) Science 194, 156–161 [DOI] [PubMed] [Google Scholar]
- 10.Roberts T. M., Shimatake H., Brady C., Rosenberg M. (1977) Nature 270, 274–275 [DOI] [PubMed] [Google Scholar]
- 11.Sauer R. T., Yocum R. R., Doolittle R. F., Lewis M., Pabo C. O. (1982) Nature 298, 447–451 [DOI] [PubMed] [Google Scholar]
- 12.Gaur N. K., Oppenheim J., Smith I. (1991) J. Bacteriol. 173, 678–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGeehan J. E., Streeter S. D., Papapanagiotou, Fox G. C., Kneale G. G. (2005) J. Mol. Biol. 346, 689–701 [DOI] [PubMed] [Google Scholar]
- 14.Tao T., Bourne J. C., Blumenthal R. M. (1991) J. Bacteriol. 173, 1367–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis R. J., Brannigan J. A., Offen W. A., Smith I., Wilkinson A. J. (1998) J. Mol. Biol. 283, 907–912 [DOI] [PubMed] [Google Scholar]
- 16.Koudelka G. B. (1998) Nucleic Acids Res. 26, 669–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffman E. C., Reyes H., Chu F. F., Sander F., Conley L. H., Brooks B. A., Hankinson O. (1991) Science 252, 954–958 [DOI] [PubMed] [Google Scholar]
- 18.LeFevre K. R., Cordes M. H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2345–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cervin M. A., Lewis R. J., Brannigan J. A., Spiegelman G. B. (1998) Nucleic Acids Res. 26, 3806–3812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vogels G. D., Van der Drift C. (1976) Bacteriol. Rev. 40, 403–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gan J., Gu Y., Li Y., Yan H., Ji X. (2006) Biochemistry 45, 8539–8545 [DOI] [PubMed] [Google Scholar]
- 22.Krell T., Coggins J. R., Lapthorn A. J. (1998) J. Mol. Biol. 278, 983–997 [DOI] [PubMed] [Google Scholar]
- 23.Saraste M., Sibbald P. R., Wittinghofer A. (1990) Trends Biochem. Sci. 15, 430–434 [DOI] [PubMed] [Google Scholar]
- 24.Gu Y., Reshetnikova L., Li Y., Wu Y., Yan H., Singh S., Ji X. (2002) J. Mol. Biol. 319, 779–789 [DOI] [PubMed] [Google Scholar]
- 25.Gescher J., Zaar A., Mohamed M., Schägger H., Fuchs G. (2002) J. Bacteriol. 184, 6301–6315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J., Rusell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 27.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 28.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 29.Cole C., Barber J. D., Barton G. J. (2008) Nucleic Acids Res. 36, W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuck P. (2000) Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Holde K. E. (1985) Physical Biochemistry, 2nd Ed., Prentice-Hall, Englewood Cliffs, NJ [Google Scholar]
- 32.Laue T. M., Shah B. D., Ridgeway T. M., Pelletier S. L. (1992) Analytical Ultracentrifugation in Biochemistry and Polymer Science, pp. 90–125, Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 33.Howlett G. J., Davidson B. E. (2000) Methods Enzymol. 323, 231–254 [DOI] [PubMed] [Google Scholar]
- 34.Cole J. L. (2004) Methods Enzymol. 384, 212–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rivas G., Stafford W., Minton A. P. (1999) Methods 19, 194–212 [DOI] [PubMed] [Google Scholar]
- 36.Hatters D. M., Minton A. P., Howlett G. J. (2002) J. Biol. Chem. 277, 7824–7830 [DOI] [PubMed] [Google Scholar]
- 37.Mindell J. A., Grigorieff N. (2003) J. Struct. Biol. 142, 334–347 [DOI] [PubMed] [Google Scholar]
- 38.Heymann J. B., Belnap D. M. (2007) J. Struct. Biol. 157, 3–18 [DOI] [PubMed] [Google Scholar]
- 39.Ludtke S. J., Baldwin P. R., Chiu W. (1999) J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 40.Sorzano C. O., Marabini R., Velázquez-Muriel J., Bilbao-Castro J. R., Scheres S. H., Carazo J. M., Pascual-Montano A. (2004) J. Struct. Biol. 148, 194–204 [DOI] [PubMed] [Google Scholar]
- 41.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 42.Wriggers W., Milligan R. A., McCammon J. A. (1999) J. Struct. Biol. 125, 185–195 [DOI] [PubMed] [Google Scholar]
- 43.Chacón P., Wriggers W. (2002) J. Mol. Biol. 317, 375–384 [DOI] [PubMed] [Google Scholar]
- 44.Russell A. D., Hopwood D. (1976) Prog. Med. Chem. 13, 271–301 [DOI] [PubMed] [Google Scholar]
- 45.Schujman G. E., Guerin M., Buschiazzo A., Schaeffer F., Llarrull L. I., Reh G., Vila A. J., Alzari P. M., de Mendoza D. (2006) EMBO J. 25, 4074–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teodorescu O., Galor T., Pillardy J., Elber R. (2004) Proteins 54, 41–48 [DOI] [PubMed] [Google Scholar]
- 47.Maxam A. M., Gilbert W. (1980) Methods Enzymol. 65, 499–560 [DOI] [PubMed] [Google Scholar]
- 48.Carmona M., Magasanik B. (1996) J. Mol. Biol. 261, 348–356 [DOI] [PubMed] [Google Scholar]
- 49.Kiley P. J., Reznikoff W. S. (1991) J. Bacteriol. 173, 16–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vénien-Bryan C., Schertler G. F., Thouvenin E., Courty S. (2000) J. Mol. Biol. 296, 863–871 [DOI] [PubMed] [Google Scholar]
- 51.Larquet E., Schreiber V., Boisset N., Richet E. (2004) J. Mol. Biol. 343, 1159–1169 [DOI] [PubMed] [Google Scholar]
- 52.Lakowicz J. R., Kierdaszuk B., Callis P., Malak H., Gryczynski I. (1995) Biophys. Chem. 56, 263–271 [DOI] [PubMed] [Google Scholar]
- 53.Manso I., Torres B., Andreu J. M., Menéndez M., Rivas G., Alfonso C., Díaz E., García J. L., Galán B. (2009) J. Biol. Chem. 284, 21218–21228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clark T. J., Phillips R. S., Bundy B. M., Momany C., Neidle E. L. (2004) J. Bacteriol. 186, 1200–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tishkov V. I., Galkin A. G., Egorov A. M. (1991) Dokl. Akad. Nauk. SSSR 317, 745–748 [PubMed] [Google Scholar]
- 56.Lamzin V. S., Aleshin A. E., Strokopytov B. V., Yukhnevich M. G., Popov V. O., Harutyunyan E. H., Wilson K. S. (1992) Eur. J. Biochem. 206, 441–452 [DOI] [PubMed] [Google Scholar]
- 57.Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 58.Weiss M. A., Pabo C. O., Karplus M., Sauer R. T. (1987) Biochemistry 26, 897–904 [DOI] [PubMed] [Google Scholar]
- 59.Burz D. S., Beckett D., Benson N., Ackers G. K. (1994) Biochemistry 33, 8399–8405 [DOI] [PubMed] [Google Scholar]
- 60.Argos P. (1990) J. Mol. Biol. 211, 943–958 [DOI] [PubMed] [Google Scholar]
- 61.Wootton J. C., Drummond M. H. (1989) Protein Eng. 2, 535–543 [DOI] [PubMed] [Google Scholar]
- 62.Mattison K., Oropeza R., Kenney L. J. (2002) J. Biol. Chem. 277, 32714–32721 [DOI] [PubMed] [Google Scholar]
- 63.Ely B., Pittard J. (1979) J. Bacteriol. 138, 933–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vonrhein C., Schlauderer G. J., Schulz G. E. (1995) Structure 3, 483–490 [DOI] [PubMed] [Google Scholar]