Abstract
Spermatogenic cells exhibit a lower spontaneous mutation frequency than somatic tissues in a lacI transgene and many base excision repair (BER) genes display the highest observed level of expression in the testis. In this study, uracil-DNA glycosylase-initiated BER activity was measured in nuclear extracts prepared from tissues obtained from each of three mouse strains. Extracts from mixed spermatogenic germ cells displayed the greatest activity followed by liver then brain for all three strains, and the activity for a given tissue was consistent among the three strains. Levels of various BER proteins were examined by western blot analyses and found to be consistent with activity levels. Nuclear extracts prepared from purified Sertoli cells, a somatic component of the seminiferous epithelium, exhibited significantly lower activity than mixed spermatogenic cell-type nuclear extracts, thereby suggesting that the high BER activity observed in mixed germ cell nuclear extracts was not a characteristic of all testicular cell types. Nuclear extracts from thymocytes and small intestines were assayed to assess activity in a mitotically active cell type and tissue. Overall, the order of tissues/cells exhibiting the greatest to lowest activity was mixed germ cells > Sertoli cells > thymocytes > small intestine > liver > brain.
INTRODUCTION
Endogenous sources of spontaneous DNA damage continuously challenge the integrity of genetic material (1). Unrepaired DNA damage can have a variety of biological ramifications including inhibition of transcription and/or replication, apoptosis, mutagenesis and carcinogenesis. Base damage is thought to represent a significant proportion of spontaneous damage (2) and can result in cytotoxicity or mutagenesis if not repaired. The base excision repair (BER) pathway is the major mechanism for ameliorating spontaneous base damage (3).
In general, BER involves the action of specific DNA glycosylases that catalyze hydrolysis of the N-glycosylic bond leaving apurinic/apyrimidinic (AP) sites in DNA. These lesions are further processed by an AP endonuclease (APE) (3,4). A DNA polymerase fills in the resulting gap, the 5′-terminus is processed to leave a 5′ phosphate group and a DNA ligase completes the reaction (2,5). Over the past few years multiple BER pathways have been defined such that short- and long-patch BER pathways are recognized. DNA polymerase β (β-pol) performs short-patch BER and a single nucleotide is incorporated into the strand undergoing repair (6–8). During long-patch BER, 2–6 nt are excised in the strand undergoing repair (9,10). Long-patch repair (≥4 nt) is carried out in a PCNA-dependent manner (9,11), and utilizes DNA polymerase δ or ɛ (11).
Effort has also been directed towards describing BER activity in mammalian tissues. Steady-state levels of BER gene transcripts have been examined with the consistent finding that the highest observed levels of expression were detected in testis (12–19). For a limited number of BER genes, it has been specifically shown that male germ cells are the source of the highest observed expression. Furthermore, some genes have been shown to vary in expression through spermatogenesis such that the highest detected level of expression occurs in pachytene spermatocytes and round spermatids (12,14–18,20).
Gene knockouts have been developed for some BER genes in part to better understand the biological significance of this pathway. Inactivation of hydroxymethyluracil-DNA glycosylase did not result in an increased toxicity by 5-hydroxymethyl-2′-deoxyuridine (21), whereas inactivation of 3-methyladenine (m3A)-DNA glycosylase (alkylpurine-DNA-N-glycosylase) resulted in increased cell killing by the simple alkylating agent methyl methanesulfonate (MMS) (22,23) and an increase in induced mutations by MMS (24). Uracil-DNA glycosylase (UDG)-deficient mice exhibit increased steady-state levels of incorporated uracil with only a slight increase in mutation frequency, but lack an overt pathological phenotype (25). Mice, in which the Ogg1 gene has been inactivated, accumulate spontaneous 8-hydroxyguanine at an accelerated rate, and to higher levels, with a corresponding increase in spontaneous mutation frequency (26,27). Homozygous null mice for Ape, β-pol, Xrcc-1 and Lig I were embryonic lethals (28–32); thus the effects of gene inactivation on biological endpoints such as mutagenesis and carcinogenesis in specific tissues are not available.
Despite intense investigation into BER, the relative activity among mammalian tissues and cell types remains largely undetermined. Therefore, to quantitatively assess BER activity in various mammalian tissues, an in vitro BER assay was modified and used to specifically quantify UDG-initiated BER (UDG-BER) activity in nuclear extracts prepared from tissues and cell types of mice. Three different commonly used mouse strains were examined to determine if there were variations in activity among these strains. Variation in activity among tissues but not among strains of mice was observed.
MATERIALS AND METHODS
Animals
Four- to six-month-old male C57BL/6J mice were obtained from The Jackson Laboratory, or from in-house breeding. Male CD1 mice (5- to 8-day-old and 4- to 6-month-old) were obtained from Charles River (Wilmington, MA). B6D2F1 (4- to 6-month-old) male mice were obtained from Harlan Sprague Dawley (Indianapolis, IN). The animals were housed in an animal facility accredited by the American Association for the Accreditation of Laboratory Animal Care and fed standard mouse laboratory chow and water ad libitum. Mice were specific-pathogen-free. Mice were humanely killed at the appropriate ages, organs rapidly removed and used immediately for mixed germ cell and Sertoli cell isolations or nuclear extract (liver, brain, small intestine and thymocyte) preparations.
Mixed germ cell, Sertoli cell and thymocyte isolations
Testes were collected from three to four male mice (4- to 6-month-old), pooled for each preparation of mixed germ cells and enriched as described previously (33,34). The approximate composition of the mixed germ cell preparations was 4% spermatogonia, 22% spermatocytes, 71% spermatids and <3% Sertoli cells (35). Sertoli cells were prepared from 200 male 5- to 8-day-old CD1 mice at ~85% purity by using a Sta Put gradient system as described previously (33,34). Thymocytes were isolated from four sets of 50 male 5- to 6-day-old CD1 mice. Thymuses were removed and teased apart in homogenization buffer, cells were dissociated by gentle passage through a 16-guage needle and thymocyte-enriched preparations obtained by passage through Nytex mesh.
Nuclear extracts
Three volumes of homogenization buffer [10 mM HEPES pH 8.0, 1.5 mM MgCl2, 10 mM NaCl, 0.5 mM dithiothreitol (DTT), 0.5 mM PMSF, 1 µg/ml pepstatin A and 10 mM sodium metabisulfite] were added to ∼300 mg of minced tissue or ≥109 cells (36). Tissue/cells were homogenized with six full strokes in a motor-driven glass-Teflon homogenizer then centrifuged at 10 000 g for 10 min. The resulting supernatant was removed and the nuclear pellet washed with 3 vol of homogenization buffer. Approximately 1.5 vol of lysis buffer (homogenization buffer with 1 M NaCl and 2.5 ng/µl luciferase), relative to the starting volume of tissue or cell pellet, was added to the nuclear pellet fraction, homogenized with three strokes and then centrifuged at 100 000 g for 1 h. Supernatant was transferred to a 10 000 MWC dialysis cassette (Pierce) and dialyzed against 500 ml of buffer A (20 mM Tris–HCl pH 8.0, 10 mM NaCl, 0.1 mM DTT, 0.1 mM PMSF, 1 µg/ml pepstatin A and 10 mM sodium metabisulfite) for 90 min. Precipitate was removed by centrifugation at 10 000 g for 10 min. Solid (NH2)2SO4 was added to the supernatant to 40% saturation and mixed gently for 30 min. Precipitate was collected by centrifugation at 20 000 g for 20 min, dissolved in a minimal volume of buffer B (identical to buffer A except it contained 100 mM KCl rather than NaCl) and dialyzed in a 10 000 MWC cassette (Pierce) against 500 ml of buffer B for 90 min. The resulting dialysate was centrifuged at 10 000 g for 10 min to remove insoluble material. All procedures were performed at 4°C. Protein concentration was determined using the Bradford assay with immunoglobin protein standard (Bio-Rad). Samples were routinely diluted to 10 mg/ml, separated into single use aliquots and stored at –80°C until use.
Due to the small size of Sertoli cell and thymocyte samples, minor modifications were incorporated into the preparation of nuclear extracts for these cell types. Nuclei were disrupted by sonication with three, 5 s intervals. (NH2)2SO4 precipitation was omitted and after dialysis against buffer A, the dialysate was centrifuged at 10 000 g for 10 min before transferring the supernatant to a 10 000 MWC Microcon filter (Millipore) to reduce the volume by at least 50%. The remainder of the nuclear extract preparation protocol was identical to that described above.
Prior to use in UDG-BER activity assays or western blot analysis, the amount of luciferase recovered in each aliquot of nuclear extract was determined by adding 2 µl of nuclear extract to 400 µl luciferase buffer (60 mM Tris-acetate pH 7.5, 2.5 mM EDTA, 12 mM Mg-acetate, 60 mM DTT, 5 mM ATP, 0.075% BSA and 150 µM luciferin). Relative light units were measured on a Lumat LB 9501 (Berthold) luminometer and compared to a luciferase standard curve (37,38). The amount of luciferase recovered was used to correct for differences in protein recovered between samples.
Oligonucleotide substrate
The double-strand oligonucleotide (Integrated DNA Technologies) utilized for measuring UDG-BER in vitro was:
5′-GCTTGCATGCCTGCAGGTCTGAUTCTAGAGGATCCCCGGGTACCGAGCTCGA-3′
3′-CGAACGTACGGACGTCCAGACTGAGATCTCCTAGGGGCCCATGGCTCGAGCT-5′
as described by Singhal et al. (6). The guanine residue at the 5′-end of the U-containing strand was considered base 1 and was 5′-end-labeled with fluorescein (Integrated DNA Technologies) thereby facilitating oligonucleotide recovery measurements.
UDG-BER assay
Reaction conditions for measuring BER were similar to those reported by Singhal et al. (6). Standard reaction mixtures (25 µl) contained 100 mM Tris–HCl pH 7.5, 5 mM MgCl2, 1 mM DTT, 0.1 mM EDTA, 2 mM ATP, 0.5 mM NAD, dATP, dGTP and dTTP at 20 µM each, 5 mM ditrisphosphocreatine, 10 U of creatine phosphokinase, 3 pmol of fluorescein 5′-end-labeled duplex oligonucleotide, 20 nM of dCTP, 20 µCi of [α-33P]dCTP (3000 Ci/mmol) and 10–40 µg nuclear extract. Except for experiments to assess dNTP incorporation, all reactions were carried out in the presence of [α-33P]dCTP. After incubation for 10 min at 37°C, reactions were stopped by placing on ice and the addition of 4.5 µl of 50 mM EDTA, 0.3 M NaCl and 80% formamide. Samples were heated to 65°C for 3 min and the entire volume subjected to electrophoresis on a 12% polyacrylamide gel containing 7 M urea in 90 mM Tris-base, 90 mM boric acid and 2 mM EDTA pH 8.8. Standards consisting of serially diluted fluorescein-labeled oligonucleotide from 0.25 to 4 pmol, encompassing the linear range of fluorescent quantification, were loaded with every assay to quantify recovery of oligonucleotide for each sample using a ChemiImager 4400 (Alpha Innotech). Radionucleotide incorporation was measured using a GS-363 Molecular Imager System (Bio-Rad). Incorporation of dATP, dGTP and dTTP in the reaction were each quantified as described for dCTP using the appropriate radiolabeled dNTP and mixture of cold dNTPs.
Western blot analysis
Nuclear protein extracts prepared from brain and mixed germ cells (50 µg) and liver (200 µg) were separated using SDS–PAGE on a 10% gel (acrylamide:bis-acrylamide 29:1), followed by electroblotting onto Trans-Blot Transfer Medium (Bio-Rad). The blot was cut into three sections based on molecular mass to facilitate detection of specific antigens. β-pol and Ape/Ref-1 were detected using rabbit polyclonal anti-hβpol (S.Wilson, NIEHS, Research Triangle Park, NC) and rabbit anti-hAPE/ref-1 polyclonal antibody (Novus Biologicals), respectively. Xrcc-1 was detected with rabbit anti-hXRCC-1 polyclonal antibody (Serotec). DNA ligases I and III were detected by rabbit polyclonal anti-ligase I and anti-ligase III antibodies (A.Tomkinson, University of Texas Health Science Center at San Antonio, San Antonio, TX), respectively. Purified β-pol and Ape/Ref-1 (Trevigene) and DNA ligases I and III (A.Tomkinson) were included as standards and controls. Primary antibodies were followed by horseradish peroxidase-conjugated goat anti-rabbit antibody (Pierce) and signal generated with enhanced chemiluminescence (Pierce). Chemiluminescence intensity of visualized bands was measured as an integrated density value (IDV) using a ChemiImager 4400 (Alpha Innotech).
Statistical analysis
The data were analyzed using analysis of variance. Comparisons among means were Bonferroni adjusted. In vitro UDG-BER data were log-transformed prior to statistical analysis. We present P-values from analysis of log-transformed data whereas we present means and standard errors computed from untransformed data. P-values <0.05 were considered significant.
RESULTS
Linearity of the assay
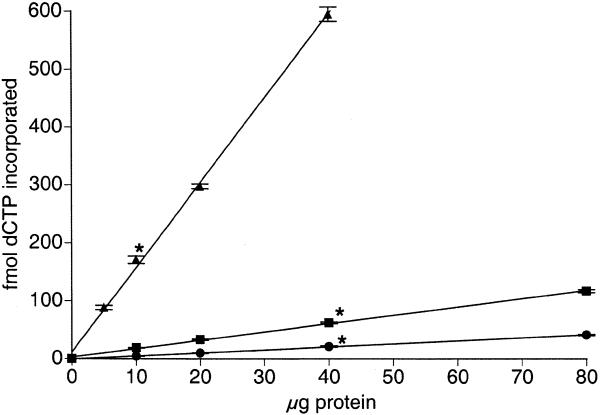
To quantify in vitro BER activity in nuclear extracts prepared from multiple tissues and cell types, an assay described by Singhal et al. (6) was modified. Oligonucleotide recovery measurements were facilitated with a fluorescein tag on the 5′ end of the uracil-containing strand. Optimal reaction conditions were determined to be 3 pmol oligonucleotide incubated with nuclear extract for 10 min thereby yielding ≥95% of the recovered oligonucleotide in the 51mer form. Figure 1 shows a fluorescent image observed after running the BER assay (top panel), with the corresponding radiographic image resulting from [α-33P]dCTP incorporation during repair synthesis (bottom panel). Various amounts of nuclear extract were examined to ensure the BER assays were being performed in the linear range. Samples to which no nuclear extract was added did not incorporate detectable levels of [α-33P]dCTP. Increasing amounts of nuclear extract prepared from brain, liver and mixed germ cells showed a linear increase in incorporation of α-33P (Fig. 2; test of non-linear trend: brain, P = 0.77; liver, P = 0.20; mixed germ cells, P = 0.12). Although the actual limits of linearity were not identified, between 5 and 40 µg of protein isolated from mixed germ cells and between 10 and 80 µg protein isolated from brain and liver resulted in linear increases in [α-33P]dCTP incorporation. Subsequently, assays were performed with 10 µg protein obtained from mixed germ cells and 40 µg protein obtained from brain, liver, Sertoli cells, small intestine and thymocytes.
Figure 1.
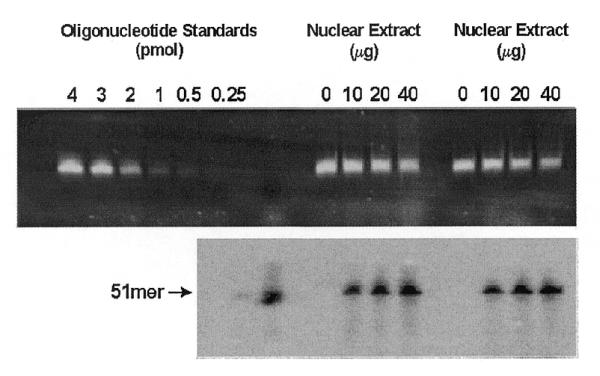
Fluorescent (top) and autoradiographic visualization (bottom) of recovered oligonucleotide from the UDG initiated-in vitro BER assay. Oligonucleotide standards were loaded spanning the linear range of fluorescent quantification for generation of a standard curve and as a 51 bp size standard (top). A 5′-radiolabeled 51 bp size standard was loaded for autoradiographic visualization.
Figure 2.
Linearity of the UDG-BER in vitro assay. Nuclear extracts were prepared from tissues of male 4- to 6-month-old C57BL/6J mice. Data are expressed as means (±SEM) from three replicate assays for each of three nuclear extract preparations obtained from brain (circles), liver (squares) and mixed germ cells (triangles). The amounts of protein selected for subsequent assays are indicated by an asterisk.
dNTP incorporation
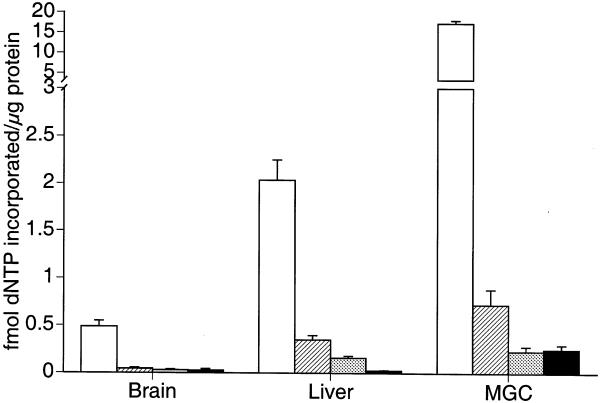
Each of the four dNTPs were independently assessed for incorporation during repair of the G:U mismatch in the oligonucleotide. As shown in Figure 3, nucleotide incorporation results for nuclear extracts from three different tissues indicate that dCTP is preferentially incorporated (P = 0.003). dATP was the second most commonly incorporated nucleotide, but it represented only 9% of the total incorporation for all four dNTPs in liver, 7% of incorporation for all four dNTPs in brain and 3% incorporation for all four dNTPs in mixed germ cells. In brain, dGTP accounted for 5% of the incorporation of all four dNTPs. Similarly, dTTP accounted for 5% incorporation of all four dNTPs in brain. In liver dGTP and dTTP accounted for 6 and 1% of all four dNTPs incorporated, respectively. In mixed germ cells dGTP and dTTP each accounted for only 1% of all four dNTPs incorporated. Thus, [α-33P]dCTP was predominantly incorporated during repair synthesis by all tested nuclear extracts. These results were consistent with the results of others studying short-patch BER using this oligonucleotide substrate (6).
Figure 3.
Incorporation of various [α-33P]dNTPs during UDG-BER in vitro. Nuclear extracts were prepared from tissues and cell types obtained from male 4- to 6-month-old C57BL/6J mice. Results are expressed as means (±SEM) for three replicate assays for each of five different nuclear extract preparations. (dCTP, open bars; dATP, hatched bars; dGTP, gray bars; dTTP, closed bars)
In vitro BER activity
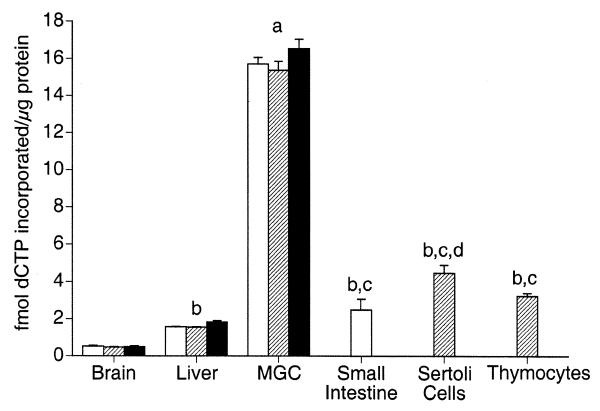
The level of UDG-BER in vitro in nuclear extracts prepared from select tissues for each of three strains of mice was measured (Fig. 4). Nuclear extracts prepared from liver, brain and mixed germ cells differed significantly in UDG-BER for each strain tested. Nuclear extracts prepared from mixed germ cells exhibited an ~30- and 10-fold higher UDG-BER activity than nuclear extracts from brain and liver, respectively. Liver exhibited ~3-fold higher UDG-BER activity than brain. However, no significant differences were detected for a given tissue among an inbred strain (C57BL/6J), an out-bred strain (CD1) and a hybrid strain (B6D2F1) of mouse.
Figure 4.
UDG-BER in vitro activities for nuclear extracts prepared from select tissues and cell types. Results are presented as means (±SEM) of three replicate assays for each of three nuclear protein extract preparations. Nuclear extracts were prepared from C57Bl/6 (open bars), CD1 (hatched bars) and B6D2F1 (closed bars) male mice. (a) Significantly higher UDG-BER activity than all other tissues and cell types. (b) Significantly higher UDG-BER activity than brain. (c) Significantly higher UDG-BER activity than liver. (d) Significantly higher UDG-BER activity than small intestine and thymocytes.
To determine whether high UDG-BER activity in mixed germ cells is a characteristic of germ cells or of testis as a whole, UDG-BER activity of nuclear extracts prepared from Sertoli cells, a somatic cell type present in the testis, was determined. Sertoli cell nuclear extracts from an out-bred mouse strain (CD1) exhibited a 3-fold higher UDG-BER activity than nuclear extracts prepared from liver and a 9-fold higher activity than nuclear extracts prepared from brain. Notably, the UDG-BER activity of nuclear extracts prepared from mixed germ cells was 3.5-fold higher than nuclear extracts prepared from Sertoli cells.
Unlike brain and liver, many spermatogenic cell types undergo cell division at reasonably high rates in conjunction with spermatogenesis. To determine whether somatic cells with high division rates had similarly high UDG-BER activities, nuclear extracts were prepared from small intestine (C57BL/6J) and thymocytes (CD1). Small intestine nuclear extracts had significantly higher UDG-BER activity compared to liver and brain. Likewise, thymocyte nuclear extracts had moderately high UDG-BER activity, which was significantly greater than that determined for brain and liver. Compared to Sertoli cells, small intestine and thymocyte nuclear extracts had significantly lower activity, however, this difference was <2-fold. Notably, nuclear extracts prepared from mixed germ cells exhibited a 5-fold higher activity than thymocytes and a 6-fold higher activity than small intestine.
BER protein composition of nuclear extracts
The amounts of BER proteins in nuclear extracts prepared from brain, liver and mixed germ cells were determined by western blot (Fig. 5). Quantification of chemiluminescence from western blots shows a greater amount of all tested BER proteins present within nuclear extracts prepared from mixed germ cells compared to those from brain and liver (Table 1). DNA ligase I and Ape1 were detected at significantly greater levels in liver than in brain, while DNA ligase III, Xrcc-1 and β-pol were found at similar levels in brain and liver.
Figure 5.
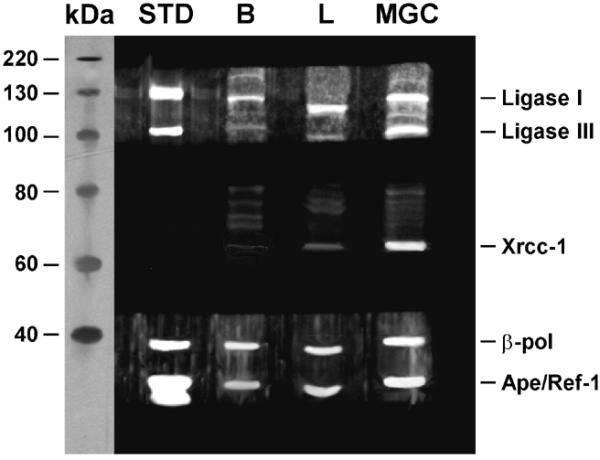
Western blot analysis of BER proteins. A total of 50 µg of nuclear extract was loaded for brain (B) and mixed germ cells (MGC) and 200 µg of nuclear extract was loaded for liver (L). Bands corresponding to DNA ligase I (130 kDa), DNA ligase III (93 kDa), Xrcc-1 (69 kDa), β-pol (39 kDa) and Ape/Ref-1 (37 kDa) proteins were visualized. A molecular mass protein standard and purified DNA ligases I and III, β-pol and Ape are shown for comparison. Faint bands at ~75 and 80 kDa are due to background binding of secondary antibody.
Table 1. Abundance of BER proteins in brain, liver and mixed germ cells.
| Protein |
Brain |
Liver |
Mixed germ cells |
| DNA ligase I | 784 ± 139a | 1229 ± 91 | 2088 ± 178 |
| (42.4%)b | (43.8%) | (24.5%) | |
| DNA ligase III | 215 ± 31 | 143 ± 29 | 2022 ± 56 |
| (11.7%) | (5.1%) | (23.7%) | |
| Xrcc-1 | 44 ± 10 | 38 ± 3 | 858 ± 81 |
| (2.4%) | (1.4%) | (10.1%) | |
| β-pol | 527 ± 94 | 747 ± 105 | 2265 ± 117 |
| (28.5%) | (26.6%) | (26.6%) | |
| Ape/Ref1 | 279 ± 28 | 648 ± 95 | 1283 ± 176 |
| (15.1%) | (23.1%) | (15.1%) |
aValues are expressed as (IDV/µg protein) ± SEM.
bPercent of total chemiluminescence for a tissue calculated by: (IDV/µg protein) × 100%.
When the proportions of various BER proteins within tissues were compared, the proportion of DNA ligase I was lower in nuclear extracts prepared from mixed germ cells compared to brain and liver. In contrast, the proportion of DNA ligase III was higher in nuclear extracts prepared from mixed germ cells compared to brain and liver. The proportion of Xrcc-1 was also higher in nuclear extracts prepared from mixed germ cells compared to brain and liver. The proportion of β-pol was similar in all tested nuclear extracts. Compared to nuclear extracts prepared from brain and mixed germ cells, nuclear extracts prepared from liver exhibited a modestly higher level of Ape1.
DISCUSSION
Mouse strains are known to vary in many phenotypes. For example, differences between strains in the activity of m3A-DNA glycosylase, another glycosylase involved in BER, have been identified (39). Accordingly, because this was the first study to quantitatively examine the levels of UDG-BER activity in mouse tissues, we examined three different commonly used mouse strains to determine if there were significant differences among strains. No differences in UDG-BER activity were observed. However, there was significant variation among tissues for all three strains. The highest UDG-BER activity was observed in nuclear extracts prepared from mixed germ cells. Although nuclear extracts prepared from liver exhibited higher UDG-BER activity than nuclear extracts from brain, both somatic tissues had significantly lower UDG-BER activity compared to mixed germ cells. This is consistent with the observation that mixed germ cells exhibit a relatively low spontaneous mutation frequency compared to somatic tissues (40–42).
After observing the high UDG-BER activity of mixed germ cells, nuclear extracts were prepared from Sertoli cells, a somatic component of the testis, and were compared with those from mixed germ cell preparations to determine if the high UDG-BER activity was a characteristic of testicular cells in general. The activity observed for Sertoli cell nuclear extracts was substantially lower than that for the mixed germ cells, thereby suggesting the high activity detected for the mixed germ cell preparations was not a characteristic of all testicular cell types. The higher activity observed in Sertoli cell nuclear extracts compared to brain and liver is probably due to minor contamination (maximally 15%) of Sertoli cell samples with spermatogonia.
Spermatogonia and spermatocytes, representing 4 and 22%, respectively, of the cells present in mixed germ cell preparations made from adult mice (35) continue to undergo cell division while the remainder of the cell types present, spermatids (71%) and Sertoli cells (<3%), do not undergo additional cell division events. Because 25% of the cells present in mixed germ cell preparations undergo cell division, additional cell and tissue types were examined to determine if the relatively high UDG-BER activity detected in mixed germ cell nuclear extracts was unique to germ cells or a characteristic of cells and tissues that continue to divide. Sertoli cell preparations obtained from 5- to 8-day old rodents continue to undergo cell division although at diminishing levels compared to earlier stages (43). The significantly lower UDG-BER activity observed for nuclear extracts prepared from these cells suggest that the high UDG-BER activity in germ cell nuclear extracts is not simply due to cell division activity. In addition, nuclear extracts from small intestine and mitotically active thymocyte were prepared and found to have substantially lower UDG-BER activity than mixed germ cell nuclear extracts. Notably, the activity in the nuclear extracts from these mitotically active cells/tissues was higher than extracts prepared from brain and liver. These results suggest that UDG-BER may be modestly elevated in cells/tissues that continue to undergo division, but that the large elevation in mixed germ cell nuclear extracts cannot be explained simply by high division rates. Interestingly, spontaneous mutation frequencies for Sertoli cells, thymus and small intestine in LacI transgenic mice are similar to those for brain and liver (42,44) and are significantly higher than that reported for male germ cells (40,42).
The high UDG-BER activity observed for mixed germ cell extracts is consistent with the western blot analyses. Accordingly, BER proteins were detected in greatest amounts in mixed germ cell nuclear extracts followed by liver then brain. The high UDG-BER activity and levels of BER proteins in mixed germ cell nuclear extracts observed in this study are consistent with previous studies that demonstrated higher levels of expression of BER genes in testis (12–14,16–19,45) and significantly lower spontaneous mutation frequencies (40–42) for mixed germ cells compared to somatic tissues.
Consistent with the observation that rapidly dividing cells contain higher levels of DNA ligase I than non-proliferating cells (12,46,47), DNA ligase I was detected at significantly higher levels in nuclear extracts prepared from mixed germ cells than in those from brain or liver. In contrast, the proportion of DNA ligase I to other BER proteins in mixed germ cells was half that detected in brain or liver. This is presumably due to the high amounts of DNA ligase III and Xrcc-1 present in mixed germ cell nuclear extracts in comparison to their extremely low abundance in brain and liver. While Xrcc-1 and DNA ligase III are believed to participate in short-patch BER (8,48), the precise contributions of Xrcc-1 and DNA ligase III to BER within germ cells is not clear. Two forms of DNA ligase III are found in spermatogenic cells with DNA ligase IIIα requiring Xrcc-1 for stability while DNA ligase IIIβ is not dependent upon Xrcc-1 (16,49,50). Expression of α and β forms of DNA ligase III peak in pachytene spermatocytes and round spermatids (16), and it has been suggested that DNA ligase IIIβ may play an important role in meiotic recombination (12,16,51).
Xrcc-1 has also been implicated in single-strand break repair (52–57) and similar to DNA ligase III, Xrcc-1 transcripts are most abundant in pachytene spermatocytes and round spermatids (18). Thus, the data suggest that DNA ligase III and Xrcc-1 may be involved in multiple pathways. Other DNA repair proteins have been shown to participate in multiple pathways. For example β-pol has been implicated in short- and long-patch BER (6,58,59), XPG, a component of nucleotide excision repair, has been implicated in repair of oxidized bases in BER (60) and XPB and XPD participate in transcription and nucleotide excision repair (61,62). Therefore, it seems possible that DNA ligase III and Xrcc-1 participate in multiple pathways.
In conclusion, a high UDG-BER activity was observed in nuclear extracts prepared from adult mixed germ cells and this activity appears to be a unique characteristic of spermatogenic cells that cannot be explained simply by their division rates. In addition, UDG-BER activity was found to vary significantly among somatic tissues and the relative proportions of BER proteins varied among tissues. Whether or not the variation in proteins among tissues impacts the ability to repair various lesions remains undetermined. Finally, the high UDG-BER activity measured in nuclear extracts prepared from mixed germ cells, and the high BER protein content in mixed germ cells, compared to that in somatic tissues suggests that BER plays an important role in maintaining the integrity of the germline genome during the process of spermatogenesis.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Dr Sam Wilson for supplying antibody against DNA polymerase β and Dr Alan Tomkinson for supplying purified DNA ligases I and III and antibody against DNA ligases I and III. This publication was made possible by grant numbers ESO9136, AG13560, AG14674 and AG00205 from the NIEHS and NIA, NIH, the VA Environmental Hazards Center, STVHCS and the Nutritional and Interventional Gerontology Training Program. The contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH or the VHCS.
References
- 1.Mullaart E., Lohman,P.H.M., Berends,F. and Vijg,J. (1990) DNA damage metabolism and aging. Mutat. Res., 237, 189–210. [DOI] [PubMed] [Google Scholar]
- 2.Glassner B.J., Posnick,L.M. and Samson,L.D. (1998) The influence of DNA glycosylases on spontaneous mutation. Mutat. Res., 400, 33–44. [DOI] [PubMed] [Google Scholar]
- 3.Lindahl T. (1995) Recognition and processing of damaged DNA. J. Cell Sci., 19 (Suppl.), 73–77. [DOI] [PubMed] [Google Scholar]
- 4.Demple B., Herman,T. and Chen,D.S. (1991) Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc. Natl Acad. Sci. USA, 88, 11450–11454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dianov G. and Lindahl,T. (1994) Reconstitution of the DNA base excision-repair pathway. Curr. Biol., 4, 1069–1076. [DOI] [PubMed] [Google Scholar]
- 6.Singhal R.K., Prasad,R. and Wilson,S.H. (1995) DNA polymerase β conducts the gap-filling step in uracil-initiated base excision repair in a bovine testis nuclear extract. J. Biol. Chem., 270, 949–957. [DOI] [PubMed] [Google Scholar]
- 7.Sobol R.W., Horton,J.K., Kuhn,R., Gu,H., Singhal,R.K., Prasad,R., Rajewsky,K. and Wilson,S.H. (1996) Requirement of mammalian DNA polymerase-β in base-excision repair. Nature, 379, 183–186. [DOI] [PubMed] [Google Scholar]
- 8.Kubota Y., Nash,R.A., Klungland,A., Schar,P., Barnes,D.E. and Lindahl,T. (1996) Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase β and the XRCC1 protein. EMBO J., 15, 6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 9.Frosina G., Fortini,P., Rossi,O., Corrazzino,F., Raspaglio,G., Cox,L.S., Lane,D.P., Abbandandolo,A. and Dogliotti,E. (1996) Two pathways for base excision repair in mammalian cells. J. Biol. Chem., 271, 9573–9578. [DOI] [PubMed] [Google Scholar]
- 10.Klungland A. and Lindahl,T. (1997) Second pathway for completion of DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J., 16, 3341–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsumoto Y., Kim,K. and Bogenhagen,D.F. (1994) Proliferating cell nuclear antigen-dependent abasic site repair in Xenopus laevis oocytes: an alternative pathway of base excision DNA repair. Mol. Cell. Biol., 14, 6187–6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J., Tomkinson,A.E., Ramos,W., Mackey,Z.B., Danehower,S., Walter,C.A., Shultz,R.A., Besterman,J.M. and Husain,I. (1995) Mammalian DNA ligase III: Molecular cloning, chromosomal location and expression in spermatocytes undergoing meiotic recombination. Mol. Cell. Biol., 15, 5412–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelward B.P., Boosalis,M.S., Chen,B.J., Deng,Z., Siciliano,M.J. and Samson,L.D. (1993) Cloning and characterization of a mouse 3-methyladenine/7-methylguanine/3-methylguanine DNA glycosylase cDNA whose gene maps to chromosome 11. Carcinogenesis, 14, 175–181. [DOI] [PubMed] [Google Scholar]
- 14.Hirose F., Hotta,Y., Yamaguchi,M. and Matsukage,A. (1989) Difference in the expression level of DNA polymerase β among mouse tissues: high expression in the pachytene spermatocytes. Exp. Cell Res., 181, 169–180. [DOI] [PubMed] [Google Scholar]
- 15.Alcivar A.A., Hake,L.E. and Hecht,N.B. (1992) DNA polymerase-β and poly(ADP)ribose polymerase mRNAs are differentially expressed during the development of male germinal cells. Biol. Reprod., 46, 201–207. [DOI] [PubMed] [Google Scholar]
- 16.Mackey Z.B., Ramos,W., Levin,D.S., Walter,C.A., McCarrey,J.R. and Tomkinson,A.E. (1997) An alternative splicing event which occurs in mouse pachytene spermatocytes generates a form of DNA ligase III with distinct biochemical properties that may function in meiotic recombination. Mol. Cell. Biol., 17, 989–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walter C.A., Lu,J., Bhakta,M., Zhou,Z.Q., Thompson,L.H. and McCarrey,J.R. (1994) Testis and somatic Xrcc-1 DNA repair gene expression. Somat. Cell Mol. Genet., 20, 451–461. [DOI] [PubMed] [Google Scholar]
- 18.Walter C.A., Trolian,D.A., McFarland,M.B, Street,K.A. and McCarrey,J.R. (1996) Xrcc-1 expression during male meiosis. Biol. Reprod., 55, 630–635. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Z.Q. and Walter,C.A. (1995) Expression of the DNA repair gene XRCC1 in baboon tissues. Mutat. Res., 348, 111–116. [DOI] [PubMed] [Google Scholar]
- 20.Wilson T.M., Rivkees,S.A., Deutsch,W.A. and Kelley,M.R. (1996) Differential expression of the apurinic/apyrimidinic endonuclease (APE/ref-1) multifunctional DNA base excision repair gene during fetal development and in adult rat brain and testis. Mutat. Res., 362, 237–248. [DOI] [PubMed] [Google Scholar]
- 21.Mi L.J., Mahl,E., Chaung,W. and Boorstein,R.J. (1997) Lack of phenotypic alteration of hmUra-DNA glycosylase-deficient hamster cells exposed to DNA-damaging agents. Mutat. Res., 374, 287–295. [DOI] [PubMed] [Google Scholar]
- 22.Engelward B.P., Dreslin,A., Christensen,J., Huszar,D., Kurahara,C. and Samson,L. (1996) Repair-deficient 3-methyladenine DNA glycosylase homozygous mutant mouse cells have increased sensitivity to alkylation-induced chromosome damage and cell killing. EMBO J., 15, 945–952. [PMC free article] [PubMed] [Google Scholar]
- 23.Engelward B.P., Weeda,G., Wyatt,M.D., Broekhof,J.L.M., De Wit,J., Donker,I., Allan,J.M., Gold,B., Hoeijmakers,J.H. and Samson,L.S. (1997) Base excision repair deficient mice lacking the Aag alkyladenine DNA glycosylase. Proc. Natl Acad. Sci. USA, 94, 13087–13092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elder R.H., Jansen,J.G., Weeks,R.J., Willington,M.A., Deans,B., Watson,A.J., Mynett,K.J., Bailey,J.A., Cooper,D.P., Rafferty,J.A., Heeran,M.C., Wijnhoven,S.W.P., van Zeeland,A.A. and Margison,G.P. (1998) Alkylpurine-DNA-glycosylase knockout mice show increased susceptibility to induction of mutations by methyl methanesulfonate. Mol. Cell. Biol., 18, 5828–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nilsen H., Roswell,I., Robins,P., Skjelbred,C.F., Andersen,S., Slupphaug,G., Daly,G., Krokan,H.E., Lindahl,T. and Barnes,D.E. (2000) Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of enzyme during DNA replication. Mol. Cell, 5, 1059–1065. [DOI] [PubMed] [Google Scholar]
- 26.Klungland A., Rosewell,I., Hollenbach,S., Larsen,E., Daly,G., Epe,B., Seeberg,E., Lindahl,T. and Barnes,D.E. (1999) Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl Acad. Sci. USA, 96, 13300–13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minowa O., Arai,T., Hirano,M., Monden,Y., Nakai,S., Fukuda,M., Itoh,M., Takano,H., Hippou,Y., Aburatani,H., Masumura,K., Nohmi,T., Nishimura,S. and Noda,T. (2000) Mmh/Ogg1 gene inactivation results in accumulation of 8-hydroxyguanine in mice. Proc. Natl Acad. Sci. USA, 97, 4156–4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xanthoudakis S., Smeyne,R.J., Wallace,J.D. and Curran,T. (1996) The redox/DNA repair protein, Ref-1 is essential for early embryonic development in mice. Proc. Natl Acad. Sci. USA, 93, 8919–8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ludwig D.L., MacInnes,M.A., Takiguchi,Y., Purtymun,P.E., Henrie,J., Flannery,M., Maneses,J., Pedersen R.A. and Chen,D.J. (1998) A murine AP-endonuclease gene-targeted deficiency with post-implantation embryonic progression and ionizing radiation sensitivity, Mutat. Res., 409, 17–29. [DOI] [PubMed] [Google Scholar]
- 30.Gu H., Marth,J.D., Orban,P.C., Mossmann,H. and Rajewsky,K. (1994) Deletion of a DNA polymerase β gene segment in T cells using cell type-specific gene targeting. Science, 265, 103–106. [DOI] [PubMed] [Google Scholar]
- 31.Tebbs R.S., Flannery,M.L., Meneses,J.J., Hartmann,A., Tucker,J.D., Thompson,L.H., Cleaver,J.E. and Pedersen,R.A. (1999) Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev. Biol., 208, 513–529. [DOI] [PubMed] [Google Scholar]
- 32.Bentley D.J., Selfridge,J., Millar,J.K., Samuel,K., Hole,N., Ansell,J.D. and Melton,D.W. (1996) DNA ligase I is required for fetal liver erythropoiesis but not essential for mammalian cell viability. Nature Genet., 13, 489–491. [DOI] [PubMed] [Google Scholar]
- 33.Bellve A.R., Cavicchia,J.C., Millette,C.F., O’Brien,D.A., Bhatnagar,Y.M. and Dym,M. (1977) Spermatogenic cells of the prepuberal mouse. J. Cell Biol., 74, 68–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romrell L.J., Bellve,A.R. and Fawcett,D.W. (1976) Separation of mouse spermatogenic cells by sedimentation velocity. A morphological characterization. Dev. Biol., 49, 119–131. [DOI] [PubMed] [Google Scholar]
- 35.Bellve, A.R. (1993) Purification, culture and fractionation of spermatogenic cells. Methods Enzymol., 225, 84–113. [DOI] [PubMed] [Google Scholar]
- 36.Widen S.G. and Wilson,S.H. (1991) Mammalian β-polymerase promoter: large-scale purification and properties of ATF/CREB palindrome binding protein from bovine testis. Biochemistry, 30, 6296–6305. [DOI] [PubMed] [Google Scholar]
- 37.De Wet J.R., Wood,K.V., Helinski,D.R. and DeLuca,M. (1985) Cloning of firefly luciferase cDNA and the expression of active luciferase in Escherichia coli. Proc. Natl Acad. Sci. USA, 82, 7870–7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Wet J.R., Wood,K.V., DeLuca,M., Helinski,D.R. and Subramani,S., (1987) Firefly luciferase gene: Structure and expression in mammalian cells. Mol. Cell. Biol., 7, 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Washington W.J., Dunn,W.C.,Jr, Generoso,W.M. and Mitra,S. (1988) Tissue-specific variation in repair activity for 3-methyladenine in DNA in two stocks of mice. Mutat. Res., 207, 165–169. [DOI] [PubMed] [Google Scholar]
- 40.Kohler S.W., Provost,G.S., Fieck,A., Kretz,P.L., Bullock,W.O., Sorge,J.A., Putman,D.L. and Short,J.M. (1991) Spectra of spontaneous and mutagen-induced mutations in the lacI gene in transgenic mice. Proc. Natl Acad. Sci. USA, 88, 7958–7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dycaico M.J., Provost,G.S., Kretz,P.L., Ransom,S.L., Moores,J.C. and Short,J.M. (1994) The use of shuttle vectors for mutation analysis in transgenic mice and rats. Mutat. Res., 307, 461–478. [DOI] [PubMed] [Google Scholar]
- 42.Walter C.A., Intano,G.W., McCarrey,J.R., McMahan,C.A. and Walter,R.B. (1998) Mutation frequency declines during spermatogenesis in young mice but increases in old mice. Proc. Natl Acad. Sci. USA, 95, 10015–10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orth J.M. (1982) Proliferation of Sertoli cells in fetal and postnatal rats: a quantitative autoradiographic study. Anat. Rec., 203, 485–492. [DOI] [PubMed] [Google Scholar]
- 44.Andrew S.E., Reitmair,A.H., Fox,J., Hsiao,L., Francis,A., McKinnon,M., Mak,T.W. and Jirik,F.R., (1997) Base transitions dominate the mutational spectrum of a transgenic reporter gene in MSH2 deficient mice. Oncogene, 15, 123–129. [DOI] [PubMed] [Google Scholar]
- 45.Menegazzi M., Grassi-Zucconi,G., Carcerero De Prati,A., Ogura,T., Poltronieri,P., Nyunoya,H., Shiratori-Nyunoya,Y., Miwa,M. and Suzuki,H. (1991) Differential expression of poly(ADP-ribose) polymerase and DNA polymerase beta in rat tissues. Exp. Cell Res., 197, 66–74. [DOI] [PubMed] [Google Scholar]
- 46.Elder R.H. and Rossignol,J.-M. (1990) DNA ligases from rat liver. Purification and partial characterization of two molecular forms. Biochemistry, 29, 6009–6017. [DOI] [PubMed] [Google Scholar]
- 47.Chan J.Y.H. and Becker,F.F. (1985) DNA ligase activities during hepatocarcinogenesis induced by N-acetylaminofluorene. Carcinogenesis, 6, 1275–1277. [DOI] [PubMed] [Google Scholar]
- 48.Cappelli E., Taylor,R., Cevasco,M., Abbondandolo,A., Caldecott,K. and Frosina,G. (1997) Involvement of XRCC1 and DNA ligase III gene products in DNA base excision repair. J. Biol. Chem., 272, 23970–23975. [DOI] [PubMed] [Google Scholar]
- 49.Caldecott K.W., Tucker,J.D., Stanker,L.H. and Thompson,L.H. (1995) Characterization of the XRCC1–DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res., 23, 4836–4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nash R.A., Caldecott,K.W., Barnes,D.E. and Lindahl,T. (1997) XRCC1 protein interacts with one of two distinct forms of DNA ligase III. Biochemistry, 36, 5207–5211. [DOI] [PubMed] [Google Scholar]
- 51.Husain I., Tomkinson,A.E., Burkhart,W.A., Moyer,M.B., Ramos,W., Mackey,Z.B., Besterman,J.M. and Chen,J. (1995) Purification and characterization of DNA ligase III from bovine testes. J. Biol. Chem., 270, 9683–9690. [DOI] [PubMed] [Google Scholar]
- 52.Thompson L.H., Brookman,K.W., Jones,N.J., Allen,S.A. and Carrano,A.V. (1990) Molecular cloning of the human XRCC1 gene, which corrects defective DNA strand break repair and sister chromatid exchange. Mol. Cell. Biol., 10, 6160–6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caldecott K.W., Aoufouchi,S., Johnson,P. and Shall,S. (1996) XRCC1 polypeptide interacts with DNA polymerase β and possibly poly (ADP-ribose) polymerase and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res., 24, 4387–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Masson M., Niedergang,C., Schreiber,V., Muller,S., Menissier-De Murcia,J. and De Murcia,G. (1998) XRCC1 is specifically associated with poly (ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol., 18, 3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marintchev A., Mullen,M.A., Maciejewski,M.W., Pan,B., Gryk,M.R. and Mullen,G.P. (1999) Solution structure of the single-strand break repair protein XRCC1 N-terminal domain. Nature Struct. Biol., 6, 884–893. [DOI] [PubMed] [Google Scholar]
- 56.Taylor R.M., Moore,D.J., Whitehouse,J., Johnson,P. and Caldecott,K.W. (2000) A cell cycle-specific requirement for the XRCC1 BRCT II domain during mammalian DNA strand break repair. Mol. Cell. Biol., 20, 735–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moore D.J., Taylor,R.M., Clements,P. and Caldecott,K.W. (2000) Mutation of a BRCT domain selectively disrupts DNA single-strand break repair in noncycling Chinese hamster ovary cells. Proc. Natl Acad. Sci. USA, 97, 13649–13654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilson S.H. (1998) Mammalian base excision repair and DNA polymerase β. Mutat. Res., 407, 203–215. [DOI] [PubMed] [Google Scholar]
- 59.Dianov G.L., Prasad,R., Wilson,S.H. and Bohr,V.A. (1999) Role of DNA polymerase β in the excision step of long patch mammalian base excision repair. J. Biol. Chem., 274, 13741–13743. [DOI] [PubMed] [Google Scholar]
- 60.Klungland A., Hoss,M., Gunz,D., Constantinou,A., Clarkson,S.G., Doetsch,P.W., Bolton,P.H., Wood,R.D. and Lindahl,T. (1990) Base excision repair of oxidative DNA damage activated by XPG protein. Mol. Cell, 3, 33–42. [DOI] [PubMed] [Google Scholar]
- 61.Schaeffer L., Roy,R., Humbert,S., Moncollin,V., Vermeulen,W., Hoeijmakers,J.H.H., Chambon,P. and Egly,J.-M. (1993) DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science, 260, 58–63. [DOI] [PubMed] [Google Scholar]
- 62.Drapkin R., Reardon,J.T., Ansarl,A., Huang,J-C., Zawel,L., Ahn,K., Sancar,A. and Reinberg,D. (1994) Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature, 368, 769–772. [DOI] [PubMed] [Google Scholar]