Abstract
MyD88 is an essential adaptor molecule for Toll-like receptors (TLRs) and interleukin (IL)-1 receptor. MyD88 is thought to be present as condensed forms or aggregated structures in the cytoplasm, although the reason has not yet been clear. Here, we show that endogenous MyD88 is present as small speckle-like condensed structures, formation of which depends on MyD88 dimerization. In addition, formation of large aggregated structures is related to cytoplasmic accumulation of sequestosome 1 (SQSTM1; also known as p62) and histone deacetylase 6 (HDAC6), which are involved in accumulation of polyubiquitinated proteins. A gene knockdown study revealed that SQSTM1 and HDAC6 were required for MyD88 aggregation and exhibited a suppressive effect on TLR ligand-induced expression of IL-6 and NOS2 in RAW264.7 cells. SQSTM1 and HDAC6 were partially involved in suppression of several TLR4-mediated signaling events, including activation of p38 and JNK, but they hardly affected degradation of IκBα (inhibitor of nuclear factor κB). Biochemical induction of MyD88 oligomerization induced recruitment of SQSTM1 and HDAC6 to the MyD88-TRAF6 signaling complex. Repression of SQSTM1 and HDAC6 enhanced formation of the MyD88-TRAF6 complex and conversely decreased interaction of the ubiquitin-specific negative regulator CYLD with the complex. Furthermore, ubiquitin-binding regions on SQSTM1 and HDAC6 were essential for MyD88 aggregation but were not required for interaction with the MyD88 complex. Thus, our study reveals not only that SQSTM1 and HDAC6 are important determinants of aggregated localization of MyD88 but also that MyD88 activates a machinery of polyubiquitinated protein accumulation that has a modulatory effect on MyD88-dependent signal transduction.
Keywords: Histone Deacetylase, Inflammation, Innate Immunity, Intracellular Processing, MyD88, Protein Translocation, Signal Transduction, Toll-like Receptors (TLR), TRAF, Ubiquitination
Introduction
MyD88 was originally identified as an inducible protein during terminal differentiation of M1 myeloleukemic cells upon interleukin (IL)-6 stimulation (1). The essential function of MyD88 was later revealed to be a universal adaptor molecule for type 1 IL-1 receptor (IL-1R)2 and Toll-like receptors (TLRs) (2–4). MyD88 is composed of three distinct regions, an N-terminal death domain, an intermediary domain, and a Toll/IL-1 homology domain at the C terminus (5). After receptor ligation, MyD88 interacts with the Toll/IL-1 homology domain of IL-1R/TLRs and then activates the signaling pathway through dimerization and utilizing death domain-containing IL-1R-associated kinases (2, 6, 7). This pathway is further activated through the ubiquitin E3 ligase TRAF6 (tumor necrosis factor (TNF) receptor-associated factor 6) that works together with a ubiquitin-conjugating enzyme complex consisting of UBC13 and UEV1A to catalyze Lys63-linked polyubiquitination, which then activates the TAK1 kinase. TAK1 activates IκB kinases and cascades of mitogen-activated protein kinases (MAPKs), ultimately leading to early phase activation of nuclear factor (NF)-κB and AP-1 and the transcription of genes encoding various proinflammatory mediators, such as TNF, NOS2 (nitric-oxide synthase 2), and IL-6 (6, 7).
In contrast to many findings in the signaling function of MyD88, the cellular distribution of MyD88 has still been unclear. Most studies on the subcellular localization have shown that MyD88 is present as condensed forms, such as discrete foci, fibrillar aggregates, and inclusion bodies, in the cytoplasm (8–11). Nishiya et al. (10) found that aggregated structures of MyD88 have irregular morphologies and do not reside in known particular organelles. They also showed that the entire molecule except the Toll/IL-1 homology domain was required for forming the structures. However, because overexpressed MyD88 automatically induces death domain-dependent activation of downstream signaling pathways (4, 10, 12, 13), it is still not known whether such condensed distribution is an artificial observation ascribed to overexpression.
Here we show that endogenous MyD88 is present as a dimerized form to exhibit small speckle-like structures. In addition, the aggregated distribution is associated with the mechanism for protein accumulation by sequestosome 1 (SQSTM1; also known as p62) and histone deacetylase 6 (HDAC6). Both SQSTM1 and HDAC6 are important molecules for accumulation of Lys63-linked polyubiquitinated proteins to form protein aggregates, which ultimately leads to lysosomal degradation by autophagy (14). Our results indicate that oligomerized MyD88 recruits SQSTM1 and HDAC6 to the signaling complex, which may be an important step for formation of aggregated structures. In addition, SQSTM1 and HDAC6 are involved in down-regulation of formation of the MyD88-TRAF6 complex and have a suppressive effect on the signal transduction. Thus, our results may provide novel information to elucidate the enigmatic subcellular localization and complex functioning of MyD88.
EXPERIMENTAL PROCEDURES
Reagents, Antibodies, and Cell Culture
The Streptomyces product coumermycin A1 was purchased from Sigma. Novobiocin was obtained from Fluka. Highly purified Escherichia coli LPS and the synthetic bacterial lipopeptide Pam3CSK4 were described previously (15, 16). CpG oligodeoxynucleotide (ODN) 1826 was purchased from Invivogen. Anti-FLAG M2 monoclonal antibody (F3165) was obtained from Sigma. Antibodies to SQSTM1 (sc-28359 and sc-25575), HDAC6 (sc-28386 and sc-11420), MyD88 (sc-11356), TRAF6 (sc-7221), cylindromatosis 1 (CYLD; sc-25779), GAPDH (sc-25778), and Akt1/2/3 (sc-8312) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Anti-MyD88 rabbit polyclonal antibody (ab2064) was obtained from Abcam. Phosphorylation-specific antibodies to p38 (4631), JNK (4671), ERK (4376), IκBα (9246), Akt Thr308 (4056), and Akt Ser473 (9271) and antibodies to p38 (9212), JNK (9258), ERK (9102), and IκBα (9242) were obtained from Cell Signaling Technology. FK2 monoclonal antibody (MFK-004) was obtained from Nippon Biotest. RAW264.7 mouse macrophages and human embryonic kidney 293T cells were maintained at 37 °C in a humidified atmosphere of 5% CO2 in DMEM supplemented with 10% FBS, penicillin G (100 units/ml), and streptomycin (100 μg/ml). The cells were stimulated with coumermycin and TLR ligands in antibiotic-free DMEM supplemented with 5% FBS.
DNA Constructs
The expression plasmid for N-terminal FLAG-tagged MyD88 was described previously (15). The DNA construct for MyD88 C-terminally fused to enhanced green fluorescent protein (EGFP) (17) was a gift from Margaret K. Offermann. The expression plasmid for the E. coli B subunit of DNA gyrase (GyrB) (18) was a gift from Michael A. Farrar. The expression plasmid (pcDNA3; Invitrogen) encoding 3×FLAG-tagged MyD88 fused to GyrB was originally constructed and provided by Häcker et al. (13). The cDNA of (1×)FLAG-MyD88-GyrB fusion protein was amplified by PCR and then subcloned into pcDNA3.1/V5-His (Invitrogen). Constructs encoding N-terminally hemagglutinin (HA) epitope-tagged SQSTM1 (HA-SQSTM1) and HA-HDAC6 were generated by amplifying RAW264.7 cDNA and by subcloning them into pcDNA3.1/V5-His (Invitrogen). Constructs encoding UBA domain-deleted HA-SQSTM1 (HA-SQSTM1 ΔUBA), BUZ domain-deleted HA-HDAC6 (HA-HDAC6 ΔBUZ), and an enzyme-inactive mutant of HA-HDAC6 (His215 and His613 substituted to Ala) were generated using a QuikChange II site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. Transfection of plasmids into RAW264.7 cells and 293T cells was performed using Lipofectamine 2000 reagent (Invitrogen). RAW264.7 cells stably expressing FLAG-MyD88-GyrB fusion protein were maintained in DMEM supplemented with 10% FBS and 1 mg/ml G418 (Roche Applied Science).
Small Interference RNA
ON-TARGET plus SMARTpool small interference RNAs (siRNAs) against mouse Hdac6 (L-043456-00-0005) and mouse Sqstm1 (L-047628-01-0005) were purchased from Dharmacon as well as the control ON-TARGET plus non-targeting siRNA pool (D-001810-01). ON-TARGET plus SMARTpool siRNAs consist of four distinct RNA oligoduplexes per target gene. The ON-TARGET plus non-targeting siRNA pool also consists of four distinct RNA oligoduplexes. For the transfection of siRNAs, RAW264.7 cells were washed once with Opti-MEM I medium (Invitrogen), and then transfection of siRNAs (100 nm) was performed with Lipofectamine RNAi MAX reagent (Invitrogen) as instructed by the manufacturer (1 μl per 20 pmol of siRNA was used). After 12 h of incubation, culture media were changed to DMEM supplemented with 5% FBS, and incubation was continued further for 12 h. ON-TARGET plus SMARTpool siRNAs have a guaranteed silencing effectiveness of at least 75% at the mRNA level. Indeed, more than 85% of suppressive effects on the respective gene expression could be confirmed by quantitative reverse transcription-coupled polymerase chain reaction (qRT-PCR) compared with the control transfection (data not shown).
Microscopy and Image Analysis
Images of live 293T cells expressing MyD88-EGFP were obtained using a KEYENCE BZ-8000 fluorescent microscope. For immunofluorescence microscopy, cells were seeded on Lab-Tek chamber 8-well Permanox slides (Nunc) and fixed at −20 °C with methanol for 20 min. Double immunostaining was then carried out using anti-FLAG M2 monoclonal antibody (Sigma) and Alexa488-conjugated anti-mouse IgG antibody (Invitrogen) and then with anti-SQSTM1 or anti-HDAC6 rabbit polyclonal antibody (Santa Cruz Biotechnology, Inc.) and Alexa564-conjugated anti-rabbit IgG antibody (Invitrogen). Stained cells were embedded in Mowiol 4–88 (Calbiochem) in the presence of the Prolong Gold Antifade reagent (Invitrogen). Fluorescent images were obtained as described previously (16, 19).
Quantification of Cellular Condensed Structures
Cells seeded on Lab-Tek chamber 8-well Permanox slides were stimulated with coumermycin, novobiocin, or TLR ligands. Immunofluorescent staining of cells was carried out with anti-FLAG, anti-SQSTM1, or anti-HDAC6 antibody and then with Alexa 488-conjugated secondary antibody. The percentage of cells with small speckle-like MyD88 condensed structures or MyD88 inclusion body-like structures of more than 1 μm in diameter or with SQSTM1 or HDAC6 condensed structures was quantified by counting cells on three or five images of microscopic fields including at least 30 cells scored. Results are representative of three separate experiments and expressed as the mean ± S.E. (n = 3 or 5).
RNA Isolation and qRT-PCR
Total RNA was prepared from cells using a GenElute mammalian total RNA miniprep kit (Sigma). One μg of total RNA was reverse-transcribed using ReverTraAce reverse transcriptase (TOYOBO) with both an oligo21dT primer and random hexamer primers. qRT-PCR was performed using SYBR Premix Ex Taq (TaKaRa) on a thermal cycler dice real-time system TP800 (TaKaRa), according to the manufacturer's instructions. All of the primer sets used in this study were obtained from TaKaRa. We confirmed that there was no critical difference between the values normalized to the levels of each of three different housekeeping genes, Gapdh, Ppia1, and Hprt1. Results shown were normalized to the level of Gapdh.
Immunoprecipitation
For immunoprecipitation of FLAG-MyD88-GyrB, cells seeded on 6-well plates were lysed with 600 μl of lysis buffer (20 mm HEPES buffer (pH 7.4) containing 0.5% Triton X-100, 150 mm sodium chloride, 12.5 mm β-glycerophosphate, 1.5 mm magnesium chloride, 10 mm sodium fluoride, 2 mm DTT, 1 mm sodium orthovanadate, 2 mm EGTA, 1 mm phenylmethylsulfonyl fluoride, EDTA-free “Complete” protease inhibitor mixtures (Roche Applied Science) and “PhosSTOP” phosphatase inhibitor mixtures (Roche Applied Science)) at 4 °C for 15 min. After clarification by centrifugation at 15,000 × g for 10 min, cell lysates were immunoprecipitated using 25 μl of anti-FLAG M2-agarose (Sigma) for 1.5 h at 4 °C on a rotating platform. For immunoprecipitation of endogenous SQSTM1, HDAC6, or GAPDH, cells seeded on 100-mm dishes were lysed with 750 μl of the lysis buffer. After clarification, cell lysates were immunoprecipitated using 50 μl of Dynabeads Protein A (Invitrogen; treated beforehand with 10 μg of antibody to SQSTM1, HDAC6, or GAPDH) for 1 h at 4 °C on a rotating platform. The beads were washed four times with 1 ml of lysis buffer, boiled with SDS sample buffer containing 2-mercaptoethanol, and subjected to immunoblotting using the indicated antibodies.
Immunoblotting and Densitometry Analysis
Cell lysates were prepared with lysis buffer (20 mm HEPES buffer (pH 7.4) containing 1% Triton X-100, 0.5% Nonidet P-40, 150 mm sodium chloride, 12.5 mm β-glycerophosphate, 1.5 mm magnesium chloride, 10 mm sodium fluoride, 2 mm DTT, 1 mm sodium orthovanadate, 2 mm EGTA, 1 mm phenylmethylsulfonyl fluoride, protease inhibitor mixtures, and phosphatase inhibitor mixtures) at 4 °C for 15 min. Lysates were boiled with SDS sample buffer, separated on 5–20% gradient SDS-PAGE, and transferred to Immobilon PVDF transfer membranes (Millipore). The membranes were blocked in 10% skim milk in PBS. Immunoreactive bands were detected using the antibodies described above. The ECL Plus Western blotting detection system (Amersham Biosciences) was used to visualize the blots on an ECL minicamera (Amersham Biosciences) with the instant black and white film FP-3000B (Fuji Films). Densitometric quantification of the immunoblot bands was performed using an Epson ES-H7200 scanner and ImageJ densitometry software (Version 1.6, National Institutes of Health). Values of the bands of phosphorylated proteins were normalized to the levels of respective proteins and expressed as -fold increase by taking the control values as 1.
Statistical Analysis
Data are expressed as the mean ± S.E. (Figs. 1 (D–F), 2 (B and D), 3A, 6, and 8C) or as the mean ± S.D. (Fig. 5). p values were calculated by Student's t test and one-way analysis of variance and were considered significant at a value of 0.05 or 0.01.
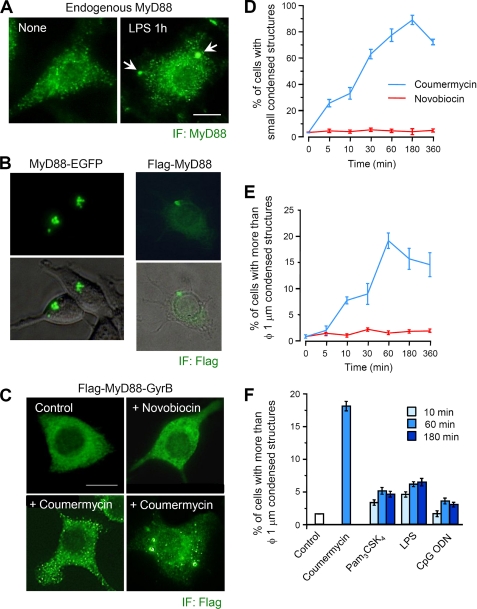
FIGURE 1.
Dimerization of MyD88 induces formation of condensed structures. A, immunofluorescent images of endogenous MyD88 in RAW264.7 cells. Cells were left untreated or treated with 1 μg/ml E. coli LPS for 60 min. Immunofluorescent staining (IF) of cells was carried out with anti-MyD88 antibody (obtained from Abcam) and Alexa 488-conjugated anti-mouse IgG antibody. Images are representative of three independent experiments. Arrowheads, MyD88 aggregated structures; Scale bar, 10 μm. B, fluorescent images of live 293T cells expressing MyD88-EGFP (left) and RAW264.7 cells stably expressing FLAG-MyD88 (right). Morphology of live 293T cells was determined by differential interference contrast microscopy. Immunofluorescent staining of RAW264.7 cells was carried out with anti-FLAG and Alexa 488 anti-mouse IgG antibodies. C, immunofluorescent images of RAW264.7 cells stably expressing FLAG-MyD88-GyrB. Cells were treated with 1 μm coumermycin for 10–60 min or 10 μm novobiocin for 30 min. Immunofluorescent staining of cells was carried out with anti-FLAG and Alexa 488 anti-mouse IgG antibodies. Images are representative of three independent experiments. Scale bar, 10 μm. D and E, quantification of the percentage of RAW264.7 cells stably expressing FLAG-MyD88-GyrB with small speckle-like condensed structures (D) and with structures of more than 1 μm in diameter (E). Cells were treated with 1 μm coumermycin or 10 μm novobiocin for the indicated periods. Immunofluorescent staining of cells was carried out with anti-FLAG and Alexa 488 anti-mouse IgG antibodies. Results, shown as the mean ± S.E. (n = 3), were obtained from three images of microscopic fields including at least 30 cells and are representative of three independent experiments. F, quantification of the percentage of RAW264.7 cells stably expressing FLAG-MyD88-GyrB with structures of more than 1 μm in diameter. Cells were stimulated with 1 μm coumermycin, 1 μg/ml Pam3CSK4, 100 ng/ml LPS, or 10 μm CpG ODN 1826 for the indicated periods. Immunofluorescent staining of cells was carried out with anti-FLAG and Alexa 488 anti-mouse IgG antibodies. Results, shown as the mean ± S.E. (error bars) (n = 3), were obtained from three images of microscopic fields including at least 30 cells and are representative of three independent experiments.
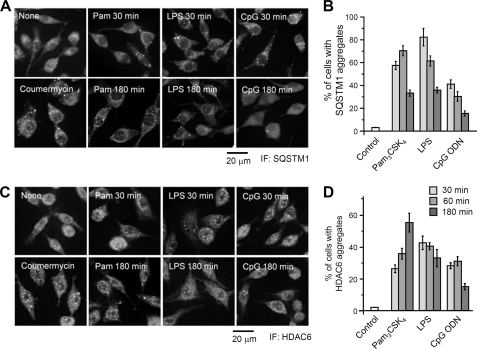
FIGURE 2.
SQSTM1 and HDAC6 are condensed in the cytoplasm after TLR stimulation. A and C, parental RAW264.7 cells were left untreated or stimulated with 1 μg/ml Pam3CSK4, 100 ng/ml LPS, or 10 μm CpG ODN 1826 for 30 or 180 min. RAW264.7 cells stably expressing FLAG-MyD88-GyrB were stimulated with 1 μm coumermycin for 30 min. Cells were then fixed and stained with anti-SQSTM1 antibody (A) or anti-HDAC6 antibody (C) and with Alexa488-conjugated anti-rabbit IgG antibody. Scale bar, 20 μm. B and D, quantification of the percentage of parental RAW264.7 cells with condensed structures of SQSTM1 or HDAC6. Cells were stimulated with 1 μg/ml Pam3CSK4, 100 ng/ml LPS, or 10 μm ODN 1826 for the indicated periods. Immunofluorescent staining (IF) of cells was carried out with anti-SQSTM1 antibody (B) or anti-HDAC6 antibody (D) and with Alexa488-conjugated anti-rabbit IgG antibody. Results, shown as the mean ± S.E. (error bars) (n = 5), were obtained from five images of microscopic fields including at least 30 cells and are representative of three independent experiments.
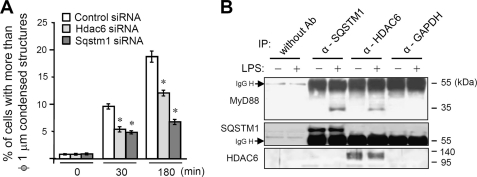
FIGURE 3.
SQSTM1 and HDAC6 are involved in formation of MyD88 aggregated structures. A, quantification of the percentage of cells with structures of more than 1 μm in diameter in RAW264.7 cells stably expressing FLAG-MyD88-GyrB. Cells were transfected with siRNA pools directed against Sqstm1 and Hdac6 or with control siRNA. After 24 h, cells were treated with 1 μm coumermycin for the indicated periods. Immunofluorescent staining of cells was carried out with anti-FLAG and Alexa 488 anti-mouse IgG antibodies. Results, shown as the mean ± S.E. (IP) (n = 3), were obtained from three images of microscopic fields including at least 30 cells and are representative of three independent experiments. *, p < 0.01 for comparison with the control siRNA group. B, immunoblot analysis of endogenous MyD88 coimmunoprecipitated with SQSTM1 and HDAC6. RAW264.7 cells were left unstimulated or stimulated with 1 μg/ml LPS for 1 h. Then immunoprecipitation (IP) using Dynabeads protein A with anti-SQSTM1, anti-HDAC6 or anti-GAPDH antibody was carried out with clarified cell lysates, followed by immunoblotting with anti-MyD88, anti-SQSTM1, and anti-HDAC6 antibodies. Results are representative of three independent experiments.
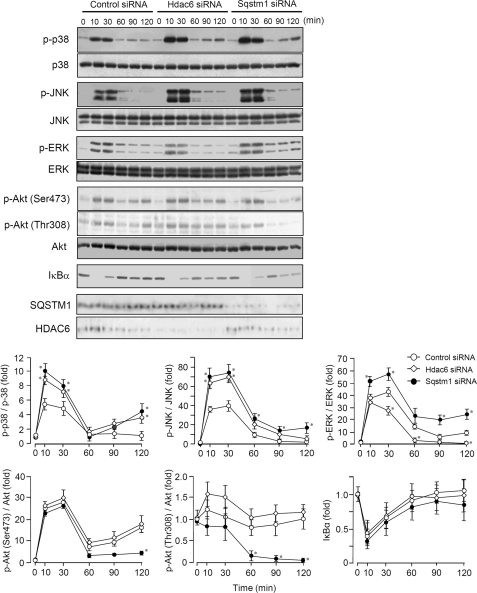
FIGURE 6.
Effects of SQSTM1 and HDAC6 on signaling events activated by LPS. Immunoblot analysis of parental RAW264.7 cells transfected with siRNA against Sqstm1 or Hdac6. Cells were stimulated with 100 ng/ml LPS for the indicated periods. Whole cell lysates were analyzed by immunoblotting with antibodies to phosphorylated (p-) JNK, p38, ERK, and Akt and IκBα. Total JNK, p38, ERK, Akt, SQSTM1, and HDAC6 served as loading controls. Results are representative of three independent experiments. Densitometric quantification was performed on all of the immunoblot bands obtained by three independent experiments. Each value of phospho-p38, phospho-JNK, phospho-ERK, phospho-Akt (Ser473), and phospho-Akt (Thr308) was normalized to each level of total p38, total JNK, total ERK, total Akt, and total Akt, respectively. Results were expressed as the mean ± S.E. (error bars) of three independent experiments and as -fold increase by taking the control values (control siRNA, 0 min) as 1. *, p < 0.05, for comparison with the control group.
FIGURE 8.
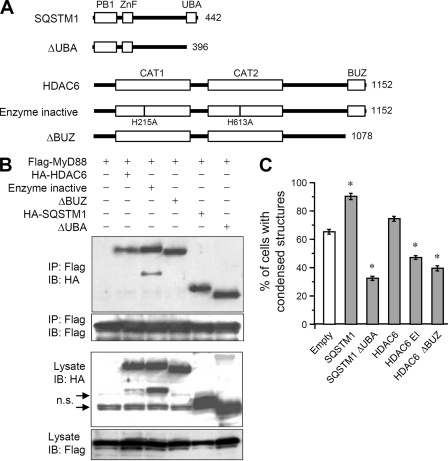
Roles of the ubiquitin-binding domain of SQSTM1 and HDAC6. A, schematic diagram of SQSTM1, HDAC6, and their mutant constructs. CAT, deacetylase catalytic domain. B, immunoblot analysis of interaction of MyD88 with SQSTM1, HDAC6, and their mutant proteins. 293T cells were transfected with FLAG-MyD88 together with HA-SQSTM1, HA-SQSTM1 ΔUBA, HA-HDAC6, enzyme-inactive HA-HDAC6, or HA-HDAC6 ΔBUZ. Immunoprecipitation (IP) with anti-FLAG-agarose was carried out with clarified cell lysates, followed by immunoblotting (IB) with anti-FLAG and anti-HA antibodies. n.s., nonspecific bands. C, quantification of the percentage of RAW264.7 cells stably expressing FLAG-MyD88-GyrB with condensed structures. Cells were transfected with HA-SQSTM1, HA-SQSTM1 ΔUBA, HA-HDAC6, enzyme-inactive HA-HDAC6, or HA-HDAC6 ΔBUZ. After 24 h, cells were stimulated with 0.1 μm coumermycin for 30 min. Immunofluorescent staining of cells was carried out with anti-FLAG and Alexa 488 anti-mouse IgG antibodies. Results, shown as the mean ± S.E. (error bars) (n = 3), were obtained from three images of microscopic fields including at least 30 cells and are representative of three independent experiments. *, p < 0.01 for comparison with the empty vector group.
FIGURE 5.
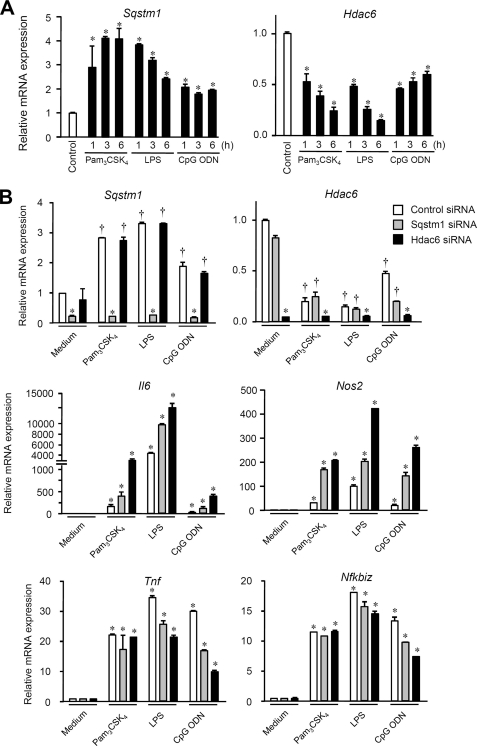
Effects of SQSTM1 and HDAC6 on proinflammatory gene expression induced by TLR stimulation. A, expression levels of Sqstm1 and Hdac6 in parental RAW264.7 cells. Cells were stimulated with 1 μg/ml Pam3CSK4, 100 ng/ml LPS, or 10 μm ODN 1826 for the indicated periods. The expression levels of Sqstm1 and Hdac6 in cells were determined by qRT-PCR. Each value is expressed as the mean ± S.D. (error bars) of three independent experiments by taking the control group as 1 (*, p < 0.01; †, p < 0.05, for comparison with the control groups). B, RAW264.7 cells transfected with siRNA against Sqstm1 or Hdac6 or with control siRNA were stimulated with 1 μg/ml Pam3CSK4, 100 ng/ml LPS, or 10 μm ODN 1826 for 6 h. The expression levels of Sqstm1, Hdac6, Il6, Nos2, Tnf, and Nfkbiz in cells were determined by qRT-PCR. Each value is expressed as the mean ± S.D. of three independent experiments by taking the control group as 1 (*, p < 0.01; †, p < 0.05, for comparison with the control groups).
RESULTS
MyD88 Dimerization Induces Formation of Condensed Structures
MyD88 is thought to be physiologically present as condensed forms in the cytoplasm. Immunofluorescent staining of RAW264.7 cells revealed that endogenous MyD88 is present as small speckle-like condensed forms scattered throughout the cytoplasm (Fig. 1A, left). In addition, after LPS stimulation, large aggregated structures of more than 1 μm in diameter could be found in a small number of the cells (Fig. 1A, right), indicating that MyD88 has the potential to form small condensed and large aggregated structures.
In contrast to endogenous MyD88, overexpression of C-terminally EGFP-tagged MyD88 showed an aggregated feature in all types of cells that we tested (Fig. 1B, left) (data not shown). The similar aggregated feature was obtained for overexpression of N-terminally FLAG-tagged MyD88 (Fig. 1B, right), indicating that the property to form aggregated structures depends on MyD88 itself and does not depend on the tag. However, it is still unclear whether the formation of such aggregated structures is an artificial observation attributable to overexpression.
To further analyze the properties of MyD88, we applied a biochemical approach utilizing MyD88 C-terminally fused to GyrB, which is known to dimerize upon binding with the Streptomyces product coumermycin with a stoichiometry of 2:1, whereas a related monomeric antibiotic, novobiocin, binds GyrB as a 1:1 complex (18). Häcker et al. (13) found that the MyD88-GyrB fusion protein does not exhibit nonspecific activation unless cells are exposed to coumermycin or to IL-1R/TLR stimulation. In cells expressing MyD88-GyrB, they also found that coumermycin can activate signaling pathways similar to TLR signaling pathways through recruitment of TRAF6 and TRAF3 to dimerized or oligomerized MyD88-GyrB (13). We investigated the cytoplasmic localization of MyD88-GyrB in RAW264.7 cells. Of note, in contrast to the condensed or aggregated localization of endogenous MyD88, MyD88-GyrB was diffusely present in the cytoplasm despite overexpression (Fig. 1C). Identical results were obtained in other types of cells that we tested (data not shown). Intriguingly, treatment of cells with coumermycin resulted in formation of a large number of small speckle-like condensed structures scattered throughout the cytoplasm (Fig. 1C), which were very similar to endogenous MyD88 shown in Fig. 1A (left). Treatment with novobiocin, which would bind monomeric MyD88-GyrB to inhibit dimerization, did not exhibit such an effect (Fig. 1C). After coumermycin treatment, some cells formed relatively large aggregated structures of more than 1 μm in diameter (Fig. 1C). Such large structures were detected as polyubiquitinated proteins by FK2 antibody, whereas this antibody could not detect any small speckle-like condensed structures (data not shown). Cells containing small speckle-like structures increased by 25% within 5 min of coumermycin treatment, followed by a further increase by ∼90% (Fig. 1D). In contrast to small speckle-like structures promptly formed after coumermycin treatment, large aggregated structures gradually increased (Fig. 1E).
We next investigated the formation of MyD88 aggregated structures after physiological TLR stimulation. RAW264.7 cells stably expressing MyD88-GyrB were stimulated with Pam3CSK4 (for TLR2), LPS (for TLR4), and CpG ODN 1826 (for TLR9), all of which are known to activate the MyD88-dependent signaling pathway. All of the TLR ligands had the ability to induce formation of relatively large MyD88 aggregated structures of more than 1 μm in diameter, although the activity was modest compared with coumermycin (Fig. 1F).
These results collectively suggest that 1) endogenous MyD88 is normally present as a dimerized form to exhibit inactive small speckle-like structures; 2) monomeric MyD88 is able to exist diffusely in the cytoplasm; and 3) the aggregated structures of MyD88 may be a result of activation of dimerized MyD88 to be switched to further oligomerized forms or to “Myddosome” that consists of six MyD88, four IRAK-4, and four IRAK-2 (20).
Involvement of SQSTM1 and HDAC6 in the Formation of MyD88 Aggregated Structures
We were very interested in why dimerized MyD88 could have the potential to be aggregated in the cytoplasm. One important possibility is that MyD88 signaling may activate a machinery of intracellular protein accumulation. Generally, mechanisms of protein accumulation have been considered to be linked to cellular management of misfolded (or unnecessary) proteins. These proteins interact with specific ubiquitin E3 ligases after recognition by heat shock protein family members, by which accumulation of polyubiquitinated misfolded proteins is promoted in the cytoplasm (14). Polymerization of the polyubiquitinated misfolded proteins results in the formation of microscopically visible structures known as inclusion bodies and aggresomes (14). SQSTM1 has been found to serve as an important adaptor molecule to recognize the polyubiquitinated misfolded proteins, leading to sequestration of misfolded proteins for the formation of inclusion bodies or sequestosomes (21–23). The microtuble-associated cytoplasmic deacetylase HDAC6 is known to be an important molecule for retrograde transportation of ubiquitinated aggregated proteins to the microtubule-organizing center to form aggresomes (14, 24, 25).
We examined whether TLR stimulation alters the subcellular localization of SQSTM1 and HDAC6 in parental RAW264.7 cells. After stimulation with Pam3CSK4, LPS, and CpG ODN for 30 min, SQSTM1 was observed as condensed structures in the cytoplasm (Fig. 2A). Cells containing SQSTM1 aggregates increased by ∼40–80% within 30 min of stimulation, followed by a decrease over a period of 3 h (Fig. 2B). HDAC6 was also observed as condensed structures in the cytoplasm and nucleus after stimulation with TLR ligands (Fig. 2C). Cells containing HDAC6 aggregates increased by more than 25% within 30 min of TLR stimulation (Fig. 2D). Similar results of formation of condensed structures of SQSTM1 and HDAC6 were observed in RAW264.7 cells stably expressing MyD88-GyrB stimulated with coumermycin (Fig. 2, A and C). Thus, MyD88-dependent TLR stimulation is able to induce cytoplasmic accumulation of SQSTM1 and HDAC6.
To determine whether formation of MyD88 aggregated structures is linked to the mechanisms involving SQSTM1 and HDAC6, we investigated the effect of repression of Sqstm1 and Hdac6 on the formation of MyD88 aggregated structures in cells stably expressing MyD88-GyrB by using siRNA. Of note, the number of cells containing aggregated structures of more than 1 μm in diameter after coumermycin treatment was decreased by repression of Sqstm1 or Hdac6 (Fig. 3A). Repression of Sqstm1 exhibited a more obvious effect than that of Hdac6 on reduction of the number of cells containing aggregated structures (Fig. 3A). These data indicate that both SQSTM1 and HDAC6 participate in the processes of formation of aggregated structures of MyD88. We therefore investigated whether endogenous MyD88 can interact with SQSTM1 and HDAC6. Immunoprecipitation analysis revealed that both SQSTM1 and HDAC6 were recruited to MyD88 only after LPS stimulation of the cells (Fig. 3B and supplemental Fig. 1).
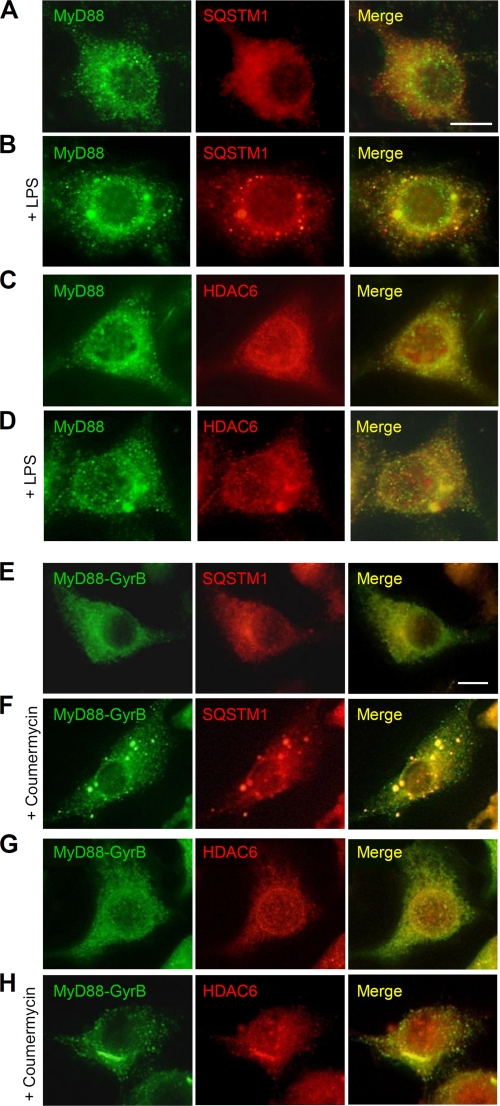
We also examined the subcellular localization of endogenous MyD88 and SQSTM1/HDAC6. In unstimulated cells, MyD88 was found as small speckle-like structures, and SQSTM1 was diffusely present in the cytoplasm (Fig. 4A). Upon LPS stimulation, SQSTM1 formed inclusion body-like structures with an elliptic or a circular shape throughout the cytoplasm, which partly colocalized with large MyD88 aggregated structures (Fig. 4B). HDAC6 was present throughout the cytoplasm and around the nuclear membrane in unstimulated cells (Fig. 4C). After LPS stimulation, HDAC6 was condensed around the juxtanucleus to form aggresome-like structures, which colocalized with MyD88 aggregated structures (Fig. 4D). We further examined the subcellular localization of MyD88-GyrB and SQSTM1/HDAC6. In unstimulated cells stably expressing MyD88-GyrB, SQSTM1 was diffusely present in the cytoplasm (Fig. 4E). Upon MyD88-GyrB dimerization by coumermycin treatment, SQSTM1 formed inclusion body-like structures throughout the cytoplasm, which colocalized with large MyD88-GyrB aggregated structures (Fig. 4F). HDAC6 was present throughout the cytoplasm and around the nuclear membrane similarly to that found in parental RAW264.7 cells (Fig. 4G). After MyD88-GyrB dimerization, HDAC6 was condensed around the juxtanucleus to form aggregated structures with an elongated shape, which colocalized with MyD88-GyrB aggregated structures (Fig. 4H). Coumermycin itself did not have any effect on formation of aggregates of SQSTM1 and HDAC6 in parental RAW264.7 cells (data not shown). Collectively, given the fact that both SQSTM1 and HDAC6 have an ability to form intracellular protein aggregates, our results strongly suggest that SQSTM1 and HDAC6 are involved in the formation of MyD88 aggregated structures.
FIGURE 4.
SQSTM1 and HDAC6 are colocalized with MyD88 aggregated structures. A–D, RAW264.7 cells were left untreated (A and C) or stimulated with 1 μg/ml LPS for 1 h (B and D). Cells were then fixed and stained with anti-MyD88 rabbit polyclonal antibody and Alexa488-conjugated anti-rabbit IgG antibody and then with anti-SQSTM1 mouse monoclonal antibody (A and B) or anti-HDAC6 mouse monoclonal antibody (C and D) and Alexa564-conjugated anti-mouse IgG antibody. Scale bar, 10 μm. E–H, RAW264.7 cells stably expressing FLAG-MyD88-GyrB were left untreated (E and G) or stimulated with 1 μm coumermycin for 30 min (F and H). Cells were then fixed and stained with anti-FLAG mouse monoclonal antibody and Alexa488-conjugated anti-mouse IgG antibody and then with anti-SQSTM1 rabbit polyclonal antibody (E and F) or anti-HDAC6 rabbit polyclonal antibody (G and H) and Alexa564-conjugated anti-rabbit IgG antibody. Scale bar, 10 μm.
SQSTM1 and HDAC6 Regulate the MyD88-dependent Signaling Pathway
In addition to the alteration of the cytoplasmic state of SQSTM1 and HDAC6 by TLR ligands (Fig. 2, A and C), we found that TLR ligands exhibit a certain extent of regulatory effects on gene expression levels of Sqstm1 and Hdac6 (Fig. 4A). Within 1 h, Sqstm1 expression was increased 2–5-fold by TLR ligand stimulation in RAW264.7 cells (Fig. 5A, left). Elevated expression of Sqstm1 was maintained at least for a period of 6 h (Fig. 4A, left). On the other hand, Hdac6 expression was decreased 1.5–2-fold by TLR ligand stimulation (Fig. 5A, right). In Pam3CSK4- and LPS-stimulated parental RAW264.7 cells, Hdac6 expression was gradually decreased during a period of 6 h (Fig. 5A, right). Therefore, we speculated that SQSTM1 and HDAC6 play some physiological roles in regulation of TLR-mediated signaling pathways.
To test this, we investigated the effect of repression of Sqstm1 and Hdac6 on TLR ligand-induced proinflammatory gene expression. In RAW264.7 cells, Pam3CSK4, LPS, and CpG ODN could induce expression of Tnf, Nos2, Il6, and Nfkbiz (Fig. 5B). Induction of Nfkbiz expression is known to be specific to the MyD88-dependent signaling pathway (26). Compared with the control, repression of Sqstm1 and Hdac6 exerted an up-regulatory effect on the TLR ligand-induced expression of Il6 and Nos2 but did not affect the expression of Tnf and Nfkbiz (Fig. 5B). Repression of Sqstm1 and Hdac6 had a suppressive effect on the CpG ODN-induced expression of Tnf and Nfkbiz (Fig. 5B). These results suggest that both SQSTM1 and HDAC6 serve as negative regulators for several proinflammatory gene expression induced by the MyD88-dependent pathway, although the effects are limited.
To further test whether SQSTM1 and HDAC6 affect the MyD88-dependent signaling pathway, we investigated phosphorylation of three MAPKs and Akt and degradation of IκB under the condition of repression of Sqstm1 and Hdac6 in LPS-stimulated RAW264.7 cells. Compared with the control, LPS-induced activation of p38 and JNK was enhanced under the condition of HDAC6 repression (Fig. 6). On the other hand, compared with the control, Sqstm1 repression enhanced LPS-induced activation of p38, JNK, ERK, and IκBα and suppressed later phase activation of Akt (Fig. 6). Intriguingly, repression of Hdac6 and Sqstm1 hardly affected LPS-induced degradation of IκBα (Fig. 6), suggesting that SQSTM1 and HDAC6 do not have a regulatory effect on NF-κB signaling. These results indicate that SQSTM1 and HDAC6 differentially regulate the TLR-mediated signaling pathways. However, both molecules at least had suppressive regulatory effects on activation of p38 and JNK.
Role of SQSTM1 and HDAC6 in Regulation of MyD88-dependent Signaling
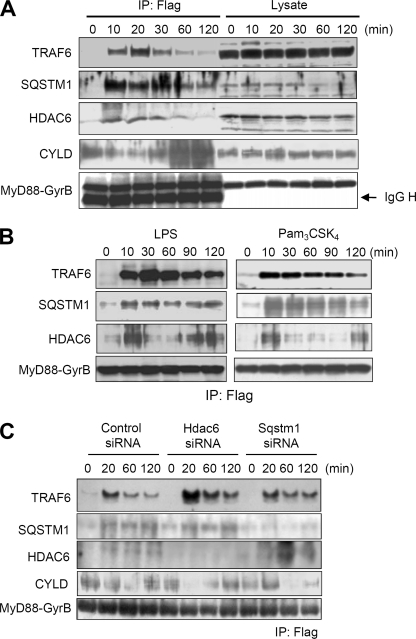
We sought to understand how SQSTM1 and HDAC6 exert the regulatory effect. To examine whether SQSTM1 and HDAC6 were recruited to the MyD88 signaling complex, we analyzed components coimmunoprecipitated with the MyD88-GyrB protein complex formed upon coumermycin treatment. Consistent with the results of a previous study (13), TRAF6 was coimmunoprecipitated with MyD88-GyrB after coumermycin treatment (Fig. 7A). Importantly, both SQSTM1 and HDAC6 were coimmunoprecipitated in a coumermycin treatment-dependent manner (Fig. 7A). Interestingly, the ubiquitination-specific TLR signaling regulator CYLD was coimmunoprecipitated with MyD88-GyrB and found to dissociate from the MyD88 complex after coumermycin treatment (Fig. 7A). Our results for recruitment of SQSTM1 and CYLD to the MyD88 signaling complex are consistent with the results of previous studies showing that SQSTM1 directly interacts with TRAF6 (23, 27, 28) and CYLD (23). Also, the results for recruitment of HDAC6 to the complex may be consistent with the results showing that the MyD88-interacting molecule nucleoredoxin can interact with HDAC6 (29). Recruitment of SQSTM1 and HDAC6 to the MyD88-GyrB complex was also observed when cells were stimulated with LPS and Pam3CSK4 (Fig. 7B).
FIGURE 7.
Recruitment of SQSTM and HDAC6 to the MyD88 signaling complex and their regulatory effect on CYLD. A, immunoblot analysis of components of the MyD88 dimerization-induced signaling complex in RAW264.7 cells stably expressing FLAG-MyD88-GyrB. Cells were treated with 1 μm coumermycin for the indicated periods. Then immunoprecipitation (IP) with anti-FLAG-agarose was carried out with clarified cell lysates, followed by immunoblotting with anti-TRAF6, anti-SQSTM1, anti-HDAC6, anti-CYLD, and anti-FLAG antibodies. Results are representative of three independent experiments. B, immunoblot analysis of TRAF6, SQSTM1, and HDAC6 in the MyD88 dimerization-induced signaling complex in RAW264.7 cells stably expressing FLAG-MyD88-GyrB. Cells were stimulated with 100 ng/ml LPS (left) and 2.5 μg/ml Pam3CSK4 (right) for the indicated periods. Immunoprecipitation with anti-FLAG-agarose was carried out with clarified cell lysates. Results are representative of three independent experiments. C, RAW264.7 cells stably expressing FLAG-MyD88-GyrB were transfected with siRNA pools directed against Sqstm1 or Hdac6 or with control siRNA. The cells were treated with 1 μm coumermycin for the indicated periods. Immunoprecipitation with anti-FLAG-agarose was carried out with clarified cell lysates, followed by immunoblot analysis of TRAF6, CYLD, SQSTM1, HDAC6, and FLAG-MyD88-GyrB. Results are representative of three independent experiments.
To examine whether SQSTM1 and HDAC6 affect MyD88 signaling complex, we immunoprecipitated dimerized MyD88-GyrB to analyze coimmunoprecipitated TRAF6 and CYLD under the condition of repression of each expression of Sqstm1 and Hdac6 by using siRNA. Compared with the control, repression of Hdac6 did not alter coimmunoprecipitated SQSTM1 upon coumermycin treatment (Fig. 7C). Interestingly, coimmunoprecipitated TRAF6 and CYLD upon coumermycin treatment were obviously increased and decreased by Hdac6 repression, respectively, suggesting that HDAC6 functions as a modulator of the early phase recruitment of TRAF6 and CYLD to the MyD88 signaling complex. In contrast, compared with the control, repression of Sqstm1 did not abrogate but on the contrary increased coimmunoprecipitated HDAC6 upon coumermycin treatment (Fig. 7C). Repression of Sqstm1 suppressed dissociation of TRAF6 from the MyD88 complex and interaction of CYLD with the complex (Fig. 7C), suggesting that SQSTM1 functions as a modulator of the later phase recruitment of TRAF6 and CYLD to the MyD88 signaling complex. Thus, SQSTM1 and HDAC6 at least separately and negatively affect the formation of the MyD88-TRAF6 signaling complex possibly through regulation of CYLD interaction with the MyD88 complex.
SQSTM1 is composed of an N-terminal Phox and Bem1p domain, a zinc finger domain, and the C-terminal UBA domain (Fig. 8A). The UBA domain of SQSTM1 can bind both Lys48-linked and Lys63-linked ubiquitin chains but with a higher affinity for Lys63-linked chains (23). HDAC6 is composed of two distinct deacetylase catalytic domains and the C-terminal ubiquitin-binding BUZ domain (Fig. 8A). The BUZ domain of HDAC6 preferentially binds Lys63-linked ubiquitin chains (24). It has recently been shown that SQSTM1 and HDAC6 participate in accumulation of misfolded proteins through recognition of the ubiquitin chain by UBA and BUZ, respectively (14, 30). To determine whether the interaction of these molecules with the MyD88 complex is mediated through ubiquitin chain recognition, we performed coimmunoprecipitation analysis using ubiquitin-binding domain-deleted mutants of SQSTM1 and HDAC6 (Fig. 8B). An enzyme-inactive mutant of HDAC6 was also used. Interestingly, not only intact SQSTM1 and HDAC6 but also their deletion mutants could interact with MyD88 (Fig. 8B), suggesting that ubiquitin-binding domains are not required for interaction with the MyD88 signaling complex. In addition, the HDAC6 enzyme inactive mutant could also interact with the complex (Fig. 8B).
To determine whether formation of MyD88 condensed structures by SQSTM1 and HDAC6 is mediated through ubiquitin recognition, these ubiquitin-binding domain-deleted mutants were expressed in cells stably expressing MyD88-GyrB. The numbers of cells containing condensed structures formed upon coumermycin treatment were increased by intact SQSTM1 and HDAC6 (Fig. 8C). On the other hand, ubiquitin-binding domain-deleted mutants greatly reduced the numbers of cells containing condensed structures (Fig. 8C). In addition, the HDAC6 enzyme inactive mutant also had a suppressive effect (Fig. 8C). These data indicate that SQSTM1 and HDAC6 interact with the MyD88 signaling complex in a ubiquitination-independent manner. However, their functions in the formation of condensed structures are dependent on recognition of protein ubiquitination.
DISCUSSION
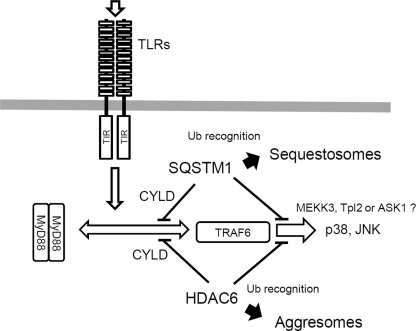
Our results suggest that MyD88 is physiologically present as a dimerized form and the formation of aggregated structures of MyD88 depends on its potential to activate machineries of protein accumulation through SQSTM1 and HDAC6. MyD88 can activate downstream signaling through the ubiquitin E3 ligase TRAF6, which is currently known to induce Lys63-linked polyubiquitination of TRAF6 itself and target proteins (31). In accordance with this, overexpressed TRAF6 forms aggregated structures in the cytoplasm (23, 28). SQSTM1 and HDAC6 have been found to be important molecules for selective recognition of Lys63-linked polyubiquitination and their accumulation in the cytoplasm. Recent findings have provided important evidence that Lys63-linked polyubiquitination is a signaling event for the selective removal of misfolded proteins by autophagy (14, 30). SQSTM1 mediates accumulation of polyubiquitinated proteins for formation of sequestosomes (inclusion bodies), whereas HDAC6 mediates formation of aggresomes (14, 30). Although the biological importance of the formation of sequestosomes and aggresomes for cell signal transduction has not generally been understood, the present study revealed that SQSTM1 and HDAC6 suppress the formation of the MyD88-TRAF6 complex through regulation of CYLD recruitment and limit activation of several signaling events, especially activation of p38 and JNK. However, these molecules did not have a distinct regulatory effect on NF-κB signaling. Therefore, it is possible that the suppressive effects are more preferentially exerted through inhibition of MAPKKKs downstream of TRAF6, such as MEKK3, Tpl-2, and ASK1, than through inhibition of TAK1 that mediates NF-κB signaling. The functions of SQSTM1 and HDAC6 are summarized in Fig. 9. Such functions of these molecules may be useful to protect cells from strong extracellular stimuli or cytotoxic signaling because the MyD88-dependent signaling pathway is cytotoxic under certain cellular circumstances (12, 32, 33).
FIGURE 9.
Schematic of MyD88-induced protein accumulation pathways through SQSTM1 and HDAC6. See “Discussion” for details. Ub, ubiquitin.
Our results indicate that the ability of TLRs to form MyD88 condensed structures is not so high. Although TLRs are thought to activate signaling pathways through MyD88 recruitment, it is possible that TLRs only transiently interact with MyD88, which may allow recycling of MyD88 to avoid aggregation (through SQSTM1 and HDAC6) and to induce further signal transduction. Alternatively, it is possible that MyD88 is rapidly degraded after TLR signal transduction, in which a specific ubiquitin E3 ligase may rapidly ubiquitinate MyD88. Recently, a novel ubiquitin E3 ligase, Nrdp1, was revealed to directly bind MyD88 for Lys48-linked polyubiquitination, then allowing its degradation (34). Lys48-linked ubiquitination is known to target substrates for proteasomal degradation rather than for autophagic degradation (35).
It is increasingly becoming evident that the MyD88-dependent signaling pathway is intricately regulated by protein ubiquitination and deubiquitination. TRAF6 advances the signaling processes itself through autoubiquitination. A20 serves as a ubiquitin-editing enzyme through removing Lys63-linked polyubiquitin chains from TRAF6 and then ligating Lys48-linked polyubiquitin chains, leading to restriction of TLR responses (36, 37). TRIM30α, a ubiquitin E3 ligase, limits TLR signaling by targeting TAB2 and TAB3, causing their degradation (38). CYLD, a member of the ubiquitin-specific protease family, negatively regulates TLR-induced inflammatory responses by deubiquitination of TRAF6 (39, 40). In this study, we found that the Lys63-linked polyubiquitin-binding molecules SQSTM1 and HDAC6 separately serve as modulators for the MyD88-dependent signaling pathway through binding with the MyD88 signaling complex that contains TRAF6 and CYLD. Our result shown in Fig. 6C suggests that SQSTM1 is more preferentially recruited to the MyD88 complex than HDAC6, probably through its higher affinity, because SQSTM1 repression enhanced HDAC6 recruitment to the complex. We found that HDAC6 regulates the early phase recruitment of TRAF6 and CYLD to the MyD88 signaling complex. It has recently been suggested that HDAC6 is rapidly translocated to the actin-enriched membrane ruffles after growth factor stimulation and subsequently associates with endocytic vesicles (macropinosomes) in an Hsp90-dependent manner (41). For HDAC6 regulation of formation of the MyD88 signaling complex, such a mechanism may be deeply associated with the case of MyD88-dependent TLR signal transduction. Indeed, initiation of the MyD88 signaling has been shown to be induced in the site of membrane ruffles and endocytic vesicles in an actin-dependent manner (9). SQSTM1 seemed to function as a modulator of TRAF6 dissociation from the MyD88 signaling complex (Fig. 7C). Recently, SQSTM1 has been shown to be involved not only in binding with TRAF6 and CYLD but also in increased polyubiquitination and destabilization of IRF8 (23, 42), which ultimately results in attenuation of inflammatory responses. Several results of our study may support such conclusions.
Protein aggregates are widely found in diseases in both the brain and the liver. These include Lewy bodies in Parkinson disease, neurofibrillary tangles in Alzheimer disease, and huntingtin aggregates. In the liver, Mallory bodies in steatohepatitis, hyaline bodies in hepatocellular carcinoma, and α1-antitrypsin aggregates are known. Importantly, all of these aggregates are attributed to accumulation of polyubiquitinated proteins, and polyubiquitinated protein-sequestering molecules, including SQSTM1 and HDAC6, have been found in the aggregates (21, 24, 25, 30, 43, 44). Given that MyD88 has a capability to form aggregates through SQSTM1 and HDAC6, dysregulation of MyD88 expression may lead to development of diseases associated with polyubiquitinated protein aggregates.
Supplementary Material
Acknowledgments
We thank Margaret K. Offermann (Emory University School of Medicine), Michael A. Farrar (University of Minnesota), and Hans Häcker (Department of Infectious Diseases, St. Jude Children's Research Hospital) for providing DNA constructs used in this study.
This work was supported by the Science Research Promotion Fund (2010) (to T. I.) from the Promotion and Mutual Aid Corporation for Private Schools of Japan and a grant from the Miyata Science Research Foundation in Asahi University (to T. I.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- IL-1R
- IL-1 receptor
- EGFP
- enhanced green fluorescent protein
- ODN
- oligodeoxynucleotide
- Pam3CSK4
- the synthetic bacterial lipopeptide N-palmitoyl-(S)-dipalmitoylglyceryl-CSKKKK
- qRT-PCR
- quantitative reverse transcription-coupled PCR
- TLR
- Toll-like receptor
- UBA
- ubiquitin-associated
- BUZ
- bound to ubiquitin zinc finger.
REFERENCES
- 1.Lord K. A., Abdollahi A., Hoffman-Liebermann B., Liebermann D. A. (1990) Cell. Growth Differ. 1, 637–645 [PubMed] [Google Scholar]
- 2.Muzio M., Ni J., Feng P., Dixit V. M. (1997) Science 278, 1612–1615 [DOI] [PubMed] [Google Scholar]
- 3.Wesche H., Henzel W. J., Shillinglaw W., Li S., Cao Z. (1997) Immunity 7, 837–847 [DOI] [PubMed] [Google Scholar]
- 4.Medzhitov R., Preston-Hurlburt P., Kopp E., Stadlen A., Chen C., Ghosh S., Janeway C. A., Jr. (1998) Mol. Cell. 2, 253–258 [DOI] [PubMed] [Google Scholar]
- 5.Hardiman G., Jenkins N. A., Copeland N. G., Gilbert D. J., Garcia D. K., Naylor S. L., Kastelein R. A., Bazan J. F. (1997) Genomics 45, 332–339 [DOI] [PubMed] [Google Scholar]
- 6.Akira S., Uematsu S., Takeuchi O. (2006) Cell 124, 783–801 [DOI] [PubMed] [Google Scholar]
- 7.O'Neill L. A., Bowie A. G. (2007) Nat. Rev. Immunol. 7, 353–364 [DOI] [PubMed] [Google Scholar]
- 8.Kawai T., Sato S., Ishii K. J., Coban C., Hemmi H., Yamamoto M., Terai K., Matsuda M., Inoue J., Uematsu S., Takeuchi O., Akira S. (2004) Nat. Immunol. 5, 1061–1068 [DOI] [PubMed] [Google Scholar]
- 9.Kagan J. C., Medzhitov R. (2006) Cell 125, 943–955 [DOI] [PubMed] [Google Scholar]
- 10.Nishiya T., Kajita E., Horinouchi T., Nishimoto A., Miwa S. (2007) FEBS Lett. 581, 3223–3229 [DOI] [PubMed] [Google Scholar]
- 11.Jaunin F., Burns K., Tschopp J., Martin T. E., Fakan S. (1998) Exp. Cell. Res. 243, 67–75 [DOI] [PubMed] [Google Scholar]
- 12.Aliprantis A. O., Yang R. B., Weiss D. S., Godowski P., Zychlinsky A. (2000) EMBO J. 19, 3325–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Häcker H., Redecke V., Blagoev B., Kratchmarova I., Hsu L. C., Wang G. G., Kamps M. P., Raz E., Wagner H., Häcker G., Mann M., Karin M. (2006) Nature 439, 204–207 [DOI] [PubMed] [Google Scholar]
- 14.Ding W. X., Yin X. M. (2008) Autophagy 4, 141–150 [DOI] [PubMed] [Google Scholar]
- 15.Into T., Inomata M., Nakashima M., Shibata K., Häcker H., Matsushita K. (2008) Mol. Cell. Biol. 28, 1338–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Into T., Kanno Y., Dohkan J., Nakashima M., Inomata M., Shibata K., Lowenstein C. J., Matsushita K. (2007) J. Biol. Chem. 282, 8134–8141 [DOI] [PubMed] [Google Scholar]
- 17.Kaiser W. J., Offermann M. K. (2005) J. Immunol. 174, 4942–4952 [DOI] [PubMed] [Google Scholar]
- 18.Farrar M. A., Alberol-Ila, Perlmutter R. M. (1996) Nature 383, 178–181 [DOI] [PubMed] [Google Scholar]
- 19.Fujita M., Into T., Yasuda M., Okusawa T., Hamahira S., Kuroki Y., Eto A., Nisizawa T., Morita M., Shibata K. (2003) J. Immunol. 171, 3675–3683 [DOI] [PubMed] [Google Scholar]
- 20.Lin S. C., Lo Y. C., Wu H. (2010) Nature 465, 885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., Hamazaki J., Nishito Y., Iemura S., Natsume T., Yanagawa T., Uwayama J., Warabi E., Yoshida H., Ishii T., Kobayashi A., Yamamoto M., Yue Z., Uchiyama Y., Kominami E., Tanaka K. (2007) Cell 131, 1149–1163 [DOI] [PubMed] [Google Scholar]
- 22.Moscat J., Diaz-Meco M. T. (2009) Cell 137, 1001–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wooten M. W., Geetha T., Babu J. R., Seibenhener M. L., Peng J., Cox N., Diaz-Meco M. T., Moscat J. (2008) J. Biol. Chem. 283, 6783–6789 [DOI] [PubMed] [Google Scholar]
- 24.Kawaguchi Y., Kovacs J. J., McLaurin A., Vance J. M., Ito A., Yao T. P. (2003) Cell 115, 727–738 [DOI] [PubMed] [Google Scholar]
- 25.Olzmann J. A., Li L., Chudaev M. V., Chen J., Perez F. A., Palmiter R. D., Chin L. S. (2007) J. Cell. Biol. 178, 1025–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto M., Yamazaki S., Uematsu S., Sato S., Hemmi H., Hoshino K., Kaisho T., Kuwata H., Takeuchi O., Takeshige K., Saitoh T., Yamaoka S., Yamamoto N., Yamamoto S., Muta T., Takeda K., Akira S. (2004) Nature 430, 218–222 [DOI] [PubMed] [Google Scholar]
- 27.Moscat J., Diaz-Meco M. T., Wooten M. W. (2007) Trends Biochem. Sci. 32, 95–100 [DOI] [PubMed] [Google Scholar]
- 28.Sanz L., Diaz-Meco M. T., Nakano H., Moscat J. (2000) EMBO J. 19, 1576–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayashi T., Funato Y., Terabayashi T., Morinaka A., Sakamoto R., Ichise H., Fukuda H., Yoshida N., Miki H. (2010) J. Biol. Chem. 285, 18586–18593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirkin V., McEwan D. G., Novak I., Dikic I. (2009) Mol. Cell. 34, 259–269 [DOI] [PubMed] [Google Scholar]
- 31.Deng L., Wang C., Spencer E., Yang L., Braun A., You J., Slaughter C., Pickart C., Chen Z. J. (2000) Cell 103, 351–361 [DOI] [PubMed] [Google Scholar]
- 32.Lehnardt S., Wennekamp J., Freyer D., Liedtke C., Krueger C., Nitsch R., Bechmann I., Weber J. R., Henneke P. (2007) J. Immunol. 179, 6134–6143 [DOI] [PubMed] [Google Scholar]
- 33.Peck-Palmer O. M., Unsinger J., Chang K. C., Davis C. G., McDunn J. E., Hotchkiss R. S. (2008) J. Leukoc. Biol. 83, 1009–1018 [DOI] [PubMed] [Google Scholar]
- 34.Wang C., Chen T., Zhang J., Yang M., Li N., Xu X., Cao X. (2009) Nat. Immunol. 10, 744–752 [DOI] [PubMed] [Google Scholar]
- 35.Sun S. C. (2008) Nat. Rev. Immunol. 8, 501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boone D. L., Turer E. E., Lee E. G., Ahmad R. C., Wheeler M. T., Tsui C., Hurley P., Chien M., Chai S., Hitotsumatsu O., McNally E., Pickart C., Ma A. (2004) Nat. Immunol. 5, 1052–1060 [DOI] [PubMed] [Google Scholar]
- 37.Wertz I. E., O'Rourke K. M., Zhou H., Eby M., Aravind L., Seshagiri S., Wu P., Wiesmann C., Baker R., Boone D. L., Ma A., Koonin E. V., Dixit V. M. (2004) Nature 430, 694–699 [DOI] [PubMed] [Google Scholar]
- 38.Shi M., Deng W., Bi E., Mao K., Ji Y., Lin G., Wu X., Tao Z., Li Z., Cai X., Sun S., Xiang C., Sun B. (2008) Nat. Immunol. 9, 369–377 [DOI] [PubMed] [Google Scholar]
- 39.Yoshida H., Jono H., Kai H., Li J. D. (2005) J. Biol. Chem. 280, 41111–41121 [DOI] [PubMed] [Google Scholar]
- 40.Trompouki E., Hatzivassiliou E., Tsichritzis T., Farmer H., Ashworth A., Mosialos G. (2003) Nature 424, 793–796 [DOI] [PubMed] [Google Scholar]
- 41.Gao Y. S., Hubbert C. C., Lu J., Lee Y. S., Lee J. Y., Yao T. P. (2007) Mol. Cell. Biol. 27, 8637–8647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim J. Y., Ozato K. (2009) J. Immunol. 182, 2131–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. (2005) J. Cell. Biol. 171, 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kirkin V., Lamark T., Sou Y. S., Bjørkøy G., Nunn J. L., Bruun J. A., Shvets E., McEwan D. G., Clausen T. H., Wild P., Bilusic I., Theurillat J. P., Øvervatn A., Ishii T., Elazar Z., Komatsu M., Dikic I., Johansen T. (2009) Mol. Cell. 33, 505–516 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.