Abstract
Ceramide is produced by the condensation of a long chain base with a very long chain fatty acid. In Saccharomyces cerevisiae, one of the two major long chain bases is called phytosphingosine (PHS). PHS has been shown to cause toxicity in tryptophan auxotrophic strains of yeast because this bioactive ceramide precursor causes diversion of the high affinity tryptophan permease Tat2 to the vacuole rather than the plasma membrane. Loss of the integral membrane protein Rsb1 increased PHS sensitivity, which was suggested to be due to this protein acting as an ATP-dependent long chain base efflux protein. More recent experiments demonstrated that loss of the genes encoding the ATP-binding cassette transporter proteins Pdr5 and Yor1 elevated PHS tolerance. This increased resistance was suggested to be due to increased expression of RSB1. Here, we provide an alternative view of PHS resistance influenced by Rsb1 and Pdr5/Yor1. Rsb1 has a seven-transmembrane domain topology more consistent with that of a regulatory protein like a G-protein-coupled receptor rather than a transporter. Importantly, an rsb1Δ cell does not exhibit higher internal levels of PHS compared with isogenic wild-type cells. However, tryptophan transport is increased in pdr5Δ yor1 strains and reduced in rsb1Δ cells. Localization and vacuolar degradation of Tat2 are affected in these genetic backgrounds. Finally, internalization of FM4-64 dye suggests that loss of Pdr5 and Yor1 slows normal endocytic rates. Together, these data argue that Rsb1, Pdr5, and Yor1 regulate the endocytosis of Tat2 and likely other membrane transporter proteins.
Keywords: ABC Transporter, Amino Acid Transport, Endocytosis, G-protein-coupled Receptors, Phospholipid
Introduction
Sphingolipids represent one of the major components of the lipid fraction of the eukaryotic plasma membrane. Biosynthesis of these lipids proceeds through production of ceramide that is formed from the linkage of a long chain base (LCB)4 with a very long chain fatty acid. In the yeast Saccharomyces cerevisiae, one of the two LCBs produced in vivo is referred to as phytosphingosine (PHS) (for reviews see Refs.1, 2). PHS is required for sphingolipid production but also has regulatory properties in terms of subcellular localization of proteins. Elevated levels of PHS cause mislocalization of nutrient permeases from the plasma membrane to the vacuole where these proteins are degraded (3, 4). Regulation of PHS levels in the cell is tightly controlled and important to ensure normal metabolism.
One of the best described routes of PHS degradation is provided by the LCB-phosphate lyase Dpl1 (5). This enzyme breaks LCB phosphate into an aldehyde and ethanolamine phosphate limiting accumulation of LCBs. Strains lacking Dpl1 are hypersensitive to PHS. This phenotype was exploited to identify an integral membrane protein designated Rsb1 that, when overproduced, suppressed the PHS hypersensitivity of a dpl1Δ strain (6). Evidence was presented that elevated levels of Rsb1 led to increased LCB efflux. More recent work demonstrated that loss of the multidrug transporters Pdr5 and Yor1 from cells led to a strong increase in PHS tolerance (7). This increase was argued to be a result of increased RSB1 gene expression with accompanying elevation in Rsb1-mediated LCB export activity.
Inspection of the predicted topology of Rsb1 suggests an alternative view to the idea that this membrane protein directly acts on PHS levels in cells. Rsb1 is predicted to have seven transmembrane domains making it a potential member of the family of the seven-transmembrane receptor (7-TM) proteins or G-protein-coupled receptors (recently reviewed in Ref. 8). These membrane receptor proteins are well known to serve as transducers of various signals, but no examples have yet been described in which these proteins possess transporter activity. Rsb1 is localized to the plasma membrane and contains two N-linked glycosylation sites in its N terminus consistent with the topology expected for a 7-TM receptor protein (9).
Work from several laboratories has demonstrated that increased function of the high affinity tryptophan transporter Tat2 influences PHS resistance (3, 4, 10). Because Rsb1 resembles a 7-TM receptor protein, we hypothesized that Rsb1 might influence Tat2 activity, which in turn would stimulate PHS resistance. Screening rsb1Δ strains for phenotypes in addition to their already described PHS sensitivity revealed that these mutants are also sensitive to detergents and a lysophospholipid analogue. Direct measurements of LCB levels in these strains demonstrated that no Rsb1-dependent change in these sphingolipid intermediates was seen. Finally, rsb1Δ cells produced lower levels of plasma membrane-localized tryptophan transport activity. These data are consistent with the view that Rsb1 regulates Tat2 activity rather than levels of intracellular LCBs.
Strains lacking the plasma membrane ATP-binding cassette transporters Pdr5 and Yor1 were hyper-resistant to the same compounds to which rsb1Δ strains were hypersensitive. The pdr5Δ yor1 strains degraded Tat2 more slowly and maintained higher levels of this permease on the plasma membrane than did wild-type strains. These findings suggest that endocytosis in pdr5Δ yor1 strains is slowed compared with wild-type cells. Measuring endocytosis using the fluorescent FM4-64 dye suggested that bulk endocytosis is slowed in pdr5Δ yor1 strains. We interpret these data to implicate Rsb1 and Pdr5/Yor1 as modulators of Tat2 endocytosis.
EXPERIMENTAL PROCEDURES
Yeast Strains and Media
The genotypes of yeast strains used in this study are listed in Table 1. Yeast transformations were performed using the lithium acetate procedure (11). Cells were grown in YPD (1% yeast extract, 2% peptone, 2% glucose) under nonselective conditions or appropriate SC media under selective conditions. Hexadecylphosphocholine (HePC) resistance, SDS, and low tryptophan tolerance was assessed by spot test assays on plates containing different concentrations of HePC (Avanti Polar Lipids, Inc.), SDS, or tryptophan, respectively. PHS resistance was also determined by spot test assays on plates containing varying concentrations of PHS (Avanti Polar Lipids, Inc.) as well as Nonidet P-40 as described previously (9).
TABLE 1.
Strains and plasmids used in this study
| Strain | Genotype | Source/Ref. |
|---|---|---|
| SEY6210 | MATα leu2-3,-112 ura3-52 lys2-801 trp1-901 his3-200 suc2-9 Mel- | Scott Emr |
| SEY6210 pdr5Δ yor1 | SEY6210 pdr5-Δ1::hisG yor1::hisG | Moye-Rowley laboratory collection |
| SEY6210 rsb1Δ | SEY6210 rsb1::natMX4 | 9 |
| SRY5 | SEY6210 pdr5-Δ1::hisG yor1::hisG rsb1:natMX4 | This study |
| SRY149 | SEY6210 bul1::kanMX4 | This study |
| SRY150 | SEY6210 rsb1::natMX4 bul1::kanMX4 | This study |
| SRY156 | SEY6210 pdr5-Δ1::hisG yor1::hisG bul1::kanMX4 | This study |
| Plasmid | Plasmid description | Source/Ref. |
|---|---|---|
| pSLP8 | pRS316-RSB1–3×HA | 9 |
| pSR74 | pRS316-CUP1-TAT2-GFP-3×HA | This study |
| pSR86 | pRS316-RSB1Δ335–382-3×HA | This study |
| pSR87 | pRS316-RSB1Δ360–383-3×HA | This study |
To construct the pdr5Δ yor1 rsb1Δ strain (SRY5), the rsb1::NatMX4 cassette containing 500-bp flanking sequence was removed by digestion from the TOPO 2.1 vector (pSLP2) and transformed into the pdr5Δ yor1 strain and selected on nourseothricin (200 μg/ml). Primers Rsb1 forward 5′-GACAGTGCGGCAATTGATAT-3′ and Nat reverse primer −5′-ACACTGGTGCGGTACCGGTAA-3′ were used to confirm deletion alleles.
SRY149, SRY150, and SRY156 were constructed by amplifying the bul1::KanMX4 cassette from the BY4742 Open Biosystems strain with the forward primer 5′-GCGCCAGCGGCACTGGCGGT-3′ and reverse primer 5′-GCATGCGAGATTTAATCGTT-3′ and transforming SEY6210 wild-type, rsb1Δ, and pdr5Δ yor1 strains. These were selected on G-418 (150 μg/ml). These deletions were confirmed by PCR using the primers Bul1 forward 5′-GCGTATCTGGCACTGGTCAA-3′ with internal Kan B reverse primer 5′-CTGCAGCGAGGAGCCGTAAT-3′ and the Bul1 reverse primer 5′-TGGCAATGCAGCGTAATAAC-3′ with the internal Kan C forward primer 5′-TGATTTTGATGACGAGCGTAAT-3′.
Plasmids
The Tat2-containing plasmid pSR74 was constructed by recombination of TAT2 into the pRS316 plasmid containing the CUP1 promoter, MVB12, and the 3×HA C-terminal tag (provided by Robert Piper), which was digested with EcoRI and BglII to remove MVB12. TAT2 was amplified with forward primer (uppercase letters denote TAT2-specific sequences, and lowercase letters direct recombination to target sites) 5′-tagaagtcatcgaaatagatattaagaaaaacaaactgtaacgaattcataTGACCGAAGACTTTATTTC-3′ and reverse primer 5′-gcccgcatagtcaggaacatcgtatgggtaaaagatgcggcccagatctttACACCAGAAATGGAACTGTC-3′. This amplicon was transformed into SEY6210 wild-type cells along with the digested MVB12 plasmid and selected on CSM -URA media. Plasmids were isolated and confirmed by digestion and sequencing. This plasmid was called pSR71. To facilitate microscopic visualization of Tat2, eGFP was inserted into this plasmid between TAT2 and the 3×HA tag as a single C-terminal eGFP tag. The eGFP cassette was amplified from a previously constructed Tat2-GFP plasmid with forward primer 5′-cctagattcccgtccatggtacgtgagacagttccatttctggtgtaaagatcccTCTAAAGGTGAAGAATTATT-3′ and reverse primer 5′-agggatagcccgcatagtcaggaacatcgtatgggtaaaagatgcggcccagatctttTTTGTACAATTCATCCATAC-3′. This amplicon was transformed into wild-type yeast with pSR71 plasmid digested with BglII (a unique site is present between the ORF and 3×HA tag), and recombinants were selected on CSM −URA media. Plasmids were isolated by standard techniques and confirmed by digestion and sequencing.
To construct pSR86 and pSR87, pSLP8 was digested with PflFI and SmaI to gap this plasmid. The RSB1 ORF from amino acids 41 to 334 or 41 to 359 was amplified with primers Rsb1 forward 5′-TCATTTGGGGTATACTACTG-3′ and Rsb1 reverse 5′-tagtcaggaacatcgtatgggtaaaagatgttaattaacccggggatccgTTCAACATCGTCAGTATGTG-3′ (lowercase letters indicate 50 bp of 3×HA tag and uppercase letters indicate 20 bp of RSB1 ORF starting at amino acid 334) or with primers Rsb1 forward 5′-TCATTTGGGGTATACTACTG-3′ and Rsb1 reverse 5′-tagtcaggaacatcgtatgggtaaaagatgttaattaacccggggatccgCGGGTATTTCATGCTTGCTT-3′ (lowercase letters indicate 50 bp of 3×HA tag and uppercase letters indicate 20 bp of RSB1 ORF starting at amino acid 359), respectively. The PCR products and gapped plasmids were transformed into SEY6210 rsb1Δ and selected for gap repair on CSM −URA. Plasmids were isolated from yeast by standard procedures and electroporated into Escherichia coli. Plasmids isolated from bacteria were confirmed by digestion and sequencing.
Tryptophan Transport Assays
Cells were cultured overnight in 10 ml of CSM + 40 μg/ml tryptophan and diluted to A600 of 0.25 in 30 ml of CSM + 40 μg/ml tryptophan. Cells were grown to an A600 0.5–1.0 and 15 A600 eq were harvested. Cells were washed in 2 ml of 10 mm sodium citrate, pH 4.5, and resuspended in 4.5 ml of assay buffer (10 mm sodium citrate, pH 4.5, plus 2% glucose) at A6000 ∼3. Tritiated tryptophan was added at 2 μCi/ml; samples were mixed, and 1-ml samples were transferred to a 0.45-μm Durapore membrane filter (Millipore), which was equilibrated with 1–2 ml of 10 mm sodium citrate. The filter was washed six times with 1 ml of wash buffer (10 mm sodium citrate, pH 4.5, plus 2 mm tryptophan) per wash. The washed filter was transferred to a scintillation vial and allowed to air-dry overnight. The procedure was repeated at 10-min intervals after addition of radiolabeled tryptophan. Liquid scintillation counting was used for quantification, and values were expressed as counts/min per A600. Tritiated tryptophan was from American Radiolabeled Chemicals, Inc.
Sphingoid Base Measurements
Yeast were grown to mid-log phase in YPD media and harvested by centrifugation. Frozen pellets were resuspended in water, and sphingolipids were extracted by the method of Bligh and Dyer (12), followed by mild alkaline hydrolysis. Extracts were derivatized with o-phthaldialdehyde and resolved by HPLC as described (13) using C17 sphingosine as an internal standard.
Immunological Methods
For Western blot analysis of the Rsb1 truncation mutants, cells were grown to an A600 of 0.8–1 in CSM −URA, and whole cell extracts were prepared using the TWIRL buffer (8 m urea, 5% SDS, 10% glycerol, 50 mm Tris, pH 6.8, 5% β-mercaptoethanol) extraction method. Briefly, 2 A600 units of cells were resuspended in 60 μl of TWIRL buffer and vortexed for 1 min with glass beads at 4 °C and centrifuged for 5 min at 12,000 rpm in an Eppendorf microcentrifuge at 4 °C. Samples were electrophoresed on SDS-PAGE, transferred to nitrocellulose, and probed using monoclonal anti-HA antibody (1:1000) (Covance). The membrane was stripped and re-probed for the plasma membrane H+-ATPase, Pma1 (Invitrogen). Pulse-chase immunoprecipitation analyses were carried out essentially as described previously (14) following copper induction of the CUP1-TAT2-eGFP-3×HA fusion gene. Lysates were immunoprecipitated with the anti-HA antibody described above. Immunoprecipitated Tat2 was detected by phosphorimage scanning of dried gels using a Cyclone PhosphorImager and quantitated using Optiquant software. Each immunoprecipitation was performed at least twice and a representative autoradiogram is shown.
Fluorescence Microscopy
GFP:SEY6210 wild-type and SEY6210 pdr5Δ yor1 strains transformed with pSR74 or empty vector control were grown overnight in CSM −URA and subcultured to an A600 of 0.15 in CSM −URA containing 4 μg/ml tryptophan to starve the cells. After 6 h of growth in low tryptophan conditions, a 500-μl sample was harvested, washed in water, and resuspended in SC-azide (SC medium containing 0.13% w/v sodium azide) buffer. Cells were visualized for GFP fluorescence and Nomarski optics using an Olympus (Tokyo, Japan) BX-60 microscope with a 100× oil objective. Images were captured using a Hamamatsu (Shizuoka, Japan) ORCA charge-coupled device camera.
FM 4–64 Chase Analysis
Overnight cultures were diluted to an A600 of 0.15 in 5 ml of YPD and grown to an A600 of 0.8–1.0. Cultures were harvested and resuspended at 20 absorbance units/ml in YPD. These samples were chilled on ice for 20 min. FM4-64 dye was added to these chilled samples, which were then rotated at 4 °C for 45 min. Samples were washed with cold water and resuspended at 10 A600 units/ml in pre-warmed YPD and kept at 30 °C. At indicated time points, samples were harvested, washed in cold SC-azide buffer, and visualized as described above for GFP analysis. The number of pixels per area on vacuoles and whole cells were calculated using ImageJ (rsbweb.nih.gov) and expressed as a percentage of pixels on the vacuole and as compared with the whole cell. A minimum of 50 cells were analyzed for each time point shown.
RESULTS
Tryptophan Phenotypes Correlate with Rsb1 Function
S. cerevisiae expresses a single high affinity plasma membrane tryptophan transporter called Tat2 (15, 16). Because many standard laboratory S. cerevisiae strains are tryptophan auxotrophs, this places a premium on Tat2 because this protein now serves as the primary route for obtaining this amino acid. This important role for Tat2 has been exploited to identify factors involved in controlling its subcellular localization (17–21). The toxicity of the LCB PHS in S. cerevisiae has been demonstrated to be caused in large part by PHS-induced mislocalization of Tat2 to the vacuole (3, 4).
Overexpression of the 7-TM protein Rsb1 was demonstrated to strongly elevate PHS resistance (6). Evidence was presented in this study that Rsb1 acted as a PHS translocase, which was believed to lower the intracellular concentration of this toxic LCB. Interestingly, PHS resistance was induced in cells lacking genes encoding the plasma membrane ATP-binding cassette (ABC) transporters Pdr5 and Yor1 (7). This increase was suggested to be due to induction of Rsb1 expression with attendant elevation in LCB translocase activity.
We were prompted to investigate the role of Rsb1 in PHS resistance for two reasons. First, loss of Pdr5 and Yor1 did not lead to increased RSB1 expression in our standard wild-type strain, although PHS tolerance was increased (9). Second, the 7-TM topology of Rsb1 is more reminiscent of a signaling protein like a G-protein-coupled receptor than a transporter. We wondered if Rsb1 might increase PHS resistance through an effect on Tat2 rather than acting directly on PHS. Because this indirect model of Rsb1 action would require a link with tryptophan prototrophy, we screened a collection of different compounds to evaluate their ability to influence cell growth by potentially interfering with tryptophan transport. Our standard laboratory wild-type strain was transformed with low copy number plasmids containing either the URA3 or TRP1 genes. Transformants were then compared for the ability to grow on medium containing the compounds indicated in Fig. 1. Although many compounds were screened, data for two are presented here as follows: the strong detergent SDS and HePC or miltefosine (22).
FIGURE 1.

Multiple phenotypes are affected by tryptophan prototrophy. Wild-type cells transformed with either a low copy URA3 plasmid (pRS316) or a low copy TRP1 plasmid (pRS314) were grown in selective minimal media to mid-log phase and spotted onto YPD plates containing the indicated drug concentration or YPD alone. Trp+ (pRS314) strains were more resistant to PHS, SDS, and HePC as compared with their Trp− (pRS316) counterparts, indicating that tryptophan status influences multiple phenotypes.
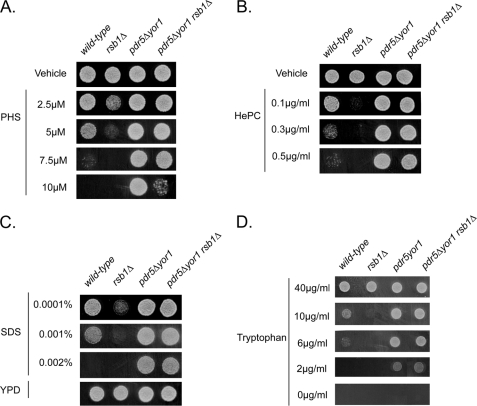
The Trp+ strain was better able to grow in the presence of these compounds than the Ura+ transformant. Because all these compounds behaved in a manner similar to PHS (less effective on Trp+ strains), we wanted to determine whether mutants with known PHS phenotypes would behave similarly in response to challenge with these other compounds. Wild-type, rsb1Δ, pdr5Δ yor1, and pdr5Δ yor1 rsb1Δ cells were grown to mid-log phase and placed on media containing various concentrations of PHS, SDS, or HePC (Fig. 2).
FIGURE 2.
Bypass of rsb1Δ by pdr5Δ yor1. Wild-type and isogenic derivatives lacking the indicated genes were grown in YPD to mid-log phase and spotted onto YPD plates containing the indicated concentrations of PHS (A), HePC (B), SDS (C), or YPD alone. D, these same strains were spotted onto minimal media plates containing the indicated concentrations of tryptophan. Strains lacking Rsb1 are sensitive to all three compounds, whereas strains lacking both Pdr5 and Yor1 show increased resistance.
Loss of RSB1 caused hypersensitivity to all three compounds, whereas disruption of PDR5 and YOR1 led to pronounced resistance as reported previously for PHS susceptibility (7). Importantly, the sensitivity caused by loss of Rsb1 could be strongly suppressed by removal of Pdr5 and Yor1. Because loss of Rsb1 still reduced the PHS resistance seen in a pdr5Δ yor1 strain, we suggest that Rsb1 and the two ABC transporter proteins act in opposition to influence the resistance to these different compounds. Rsb1 positively regulates resistance, whereas Pdr5 and Yor1 act negatively. These data support the view that Rsb1 acts generally to influence tolerance to these three compounds rather than as a PHS-specific resistance determinant and are not consistent with Pdr5 and Yor1 acting upstream of Rsb1.
Previous experiments both here and elsewhere (6) require the addition of an exogenous compound to the medium (PHS, SDS, or HePC) to cause a phenotype dependent on Rsb1 levels. To assess the likelihood that the different phenotypes seen for rsb1Δ and pdr5Δ yor1 cells could be simply explained by changes in tryptophan transport, we tested the ability of these same strains to grow on progressively limiting concentrations of tryptophan in the medium. Note that no foreign compounds are present in the media, and the only variable is the level of tryptophan supplementation. Strains were grown as above but placed on minimal medium containing the indicated levels of tryptophan (Fig. 2D).
At high levels of tryptophan, all strains grew equally. However, as the tryptophan concentration in the medium was reduced, rsb1Δ cells failed to grow at 10 μg/ml, a concentration tolerated by the other strains. Further lowering the tryptophan concentration to 6 μg/ml caused a major growth defect in wild-type cells, whereas the pdr5Δ yor1 strains, containing or lacking Rsb1, were not significantly inhibited. These data support the view that a major contributor to the phenotypes seen in strains lacking Rsb1 or Pdr5/Yor1 is altered tryptophan transport. To directly test this hypothesis, tryptophan uptake was measured in these same strains.
Changes in Tryptophan Transport Correlate with the Observed Tryptophan Sensitivity of rsb1Δ and pdr5Δ yor1 Strains
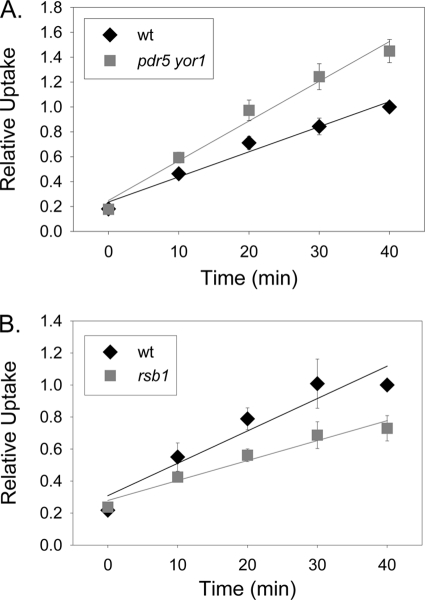
The data above suggests that rsb1Δ cells have less ability to take up tryptophan from the media, as they are unable to grow on low levels of exogenous tryptophan. This could be explained by a reduction in levels of Tat2 at the plasma membrane. To test this idea, wild-type and rsb1Δ strains were grown in minimal media to mid-log phase, and tryptophan transport assays were performed. The rsb1Δ strain showed a decreased rate of transport at 0.012 relative tryptophan uptake per min as compared with wild type, which had relative tryptophan uptake per min of 0.02 (Fig. 3B). The pdr5Δ yor1 strain showed a significant increase in transport with a relative tryptophan uptake per min of 0.032 as compared with the wild type that had a relative uptake per min of 0.02 (Fig. 3A). These data suggest that the pdr5Δ yor1 strain may have increased levels of the high affinity tryptophan transporter, Tat2, at the plasma membrane. The presence of Pdr5 and Yor1 could reduce the levels of Tat2 in the plasma membrane, whereas Rsb1 would act to increase plasma membrane content of this tryptophan permease. Genetically, these experiments argue that Rsb1 acts as a positive regulator of tryptophan transport, whereas Pdr5 and Yor1 serve as negative regulators of uptake of this amino acid.
FIGURE 3.
Tryptophan uptake is increased in the pdr5Δ yor1 strain and decreased in the rsb1Δ strain. A, wild-type (diamonds) and pdr5Δ yor1 (squares) strains were grown to mid-log phase and harvested. Cells were subjected to tryptophan uptake assays as described above. Time points were taken at 10-min intervals for 40 min. B, wild-type (diamonds) and rsb1Δ (squares) strains were grown and assayed as in A.
PHS Levels Are Not Altered in the Absence of Rsb1 or Pdr5 and Yor1
Although the data above indicate that changes in tryptophan transport may be sufficient to explain the phenotypes caused by loss of Rsb1 and Pdr5/Yor1, it is also possible that loss of these proteins influences endogenous PHS levels. For example, if rsb1Δ cells had an elevated intracellular PHS pool, then this in turn could cause mistargeting of Tat2 with the attendant effects on tryptophan transport. Similarly, loss of Pdr5 and Yor1 might lead to lowered intracellular PHS, which could stimulate Tat2 delivery to the plasma membrane.
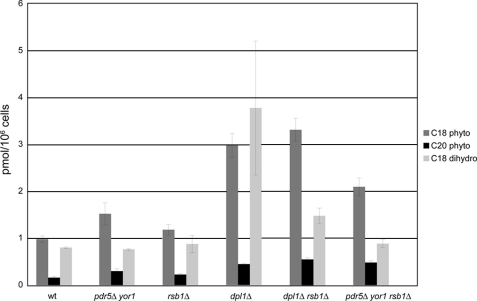
To directly assess this alternative possibility, we measured levels of intracellular LCBs in these strains by HPLC. A model in which Rsb1 acts to efflux LCBs predicts that these lipids should accumulate in mutants lacking Rsb1 and be lowered in pdr5Δ yor1 strains. The data in Fig. 4 show no such correlation. Deletion of RSB1 had no significant effect on constitutive levels of C18 or C20 phytosphingosine, or C18 dihydrosphingosine (C20 dihydrosphingosine was undetectable in this background strain). Similarly, deletion of PDR5 and YOR1 did not lower sphingoid base levels either in the presence or absence of RSB1. In fact, loss of Pdr5 and Yor1 led to an elevation in C18 phytosphingosine, in direct opposition to a mechanism involving induction of an efflux activity upon loss of these ABC transporters. These data argue strongly against a role for these genes in regulation of intracellular sphingoid base levels.
FIGURE 4.
Endogenous PHS levels are not significantly elevated in either the absence of Rsb1 or the absence of Pdr5 and Yor1. Wild-type, rsb1Δ, rsb1Δ dpl1Δ, dpl1Δ, pdr5Δ yor1, and pdr5Δ yor1 rsb1Δ strains were grown to mid-log phase and processed for HPLC analysis.
A role for Rsb1 in sphingoid base efflux from cells was previously suggested (6). Thus, although deletion of RSB1 from otherwise wild-type cells did not increase sphingoid bases, as would be expected given this former hypothesis, it might be possible that a role in regulating sphingoid base levels might be more easily detected under conditions that aberrantly increase sphingoid base levels. To rule out any difficulties caused by LCB degradation or problems with low endogenous LCB levels, we utilized a strain deleted in the sphingoid base phosphate lyase, DPL1. Because this strain lacks a key enzyme in sphingolipid degradation, it constitutively accumulates high LCB levels (5). This was confirmed in our hands (Fig. 4). However, deletion of RSB1 in this strain did not change phytosphingosine levels and showed only a modest decrease in dihydrosphingosine levels. We conclude that regulation of endogenous sphingoid base levels does not seem to be a key role for Rsb1 because its absence has no significant influence on normal or elevated LCB levels. Its effect on Tat2 and tryptophan transport likely occur through other mechanisms.
Turnover of Tat2 Is Stimulated in rsb1Δ Strain and Decreased in the pdr5Δ yor1 Strain
Together, the data above support the view that Rsb1 and Pdr5/Yor1 regulate Tat2-mediated tryptophan transport activity. Tat2 regulation can occur at the transcriptional as well as post-translational level (23). To simplify analysis of Tat2 regulation, we constructed a gene fusion in which the TAT2 coding sequences were placed under regulation of the well studied CUP1 promoter. CUP1 transcription can be induced with the addition of copper to the medium, providing a means to study post-translational regulation of Tat2 without interference from potential transcriptional regulatory inputs. Additionally, Tat2 was fused to eGFP that in turn contained an 3×HA epitope tag at the C-terminal end of the fusion protein. This hybrid protein allowed facile detection of subcellular distribution of Tat2 by fluorescence microscopy and efficient detection via the epitope tags. The chimeric Tat2 protein was functional for tryptophan transport as it was able to elevate PHS resistance of wild-type cells when overproduced (data not shown).
Previous work has shown that Tat2 follows the typical itinerary of a plasma membrane nutrient permease (21, 24). Under conditions of low substrate, in this case tryptophan, Tat2 is delivered to the plasma membrane where it acts to facilitate tryptophan transport. When tryptophan levels are high, Tat2 is targeted to the vacuole for degradation. To determine whether Tat2 turnover was influenced by Rsb1 and/or Pdr5/Yor1, we carried out a pulse-chase immunoprecipitation analysis using the CUP1-driven Tat2-eGFP-3×HA protein. Appropriate transformants of wild-type, rsb1Δ, or pdr5Δ yor1 cells were grown to mid-log phase and then labeled with [35S]methionine. Next, an excess of unlabeled methionine was added, and samples were removed at specified time points. These samples were processed for immunoprecipitation using an anti-HA antibody as described previously (14). Immunoprecipitates were electrophoresed through SDS-PAGE and Tat2-eGFP-HA was detected (Fig. 5).
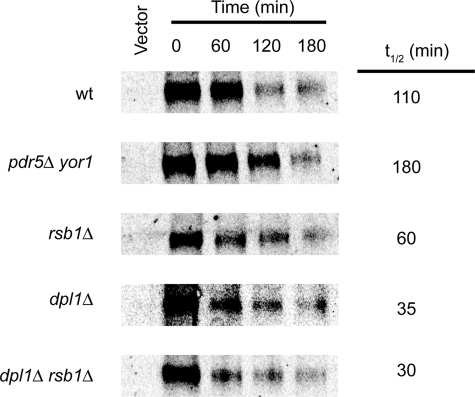
FIGURE 5.
Tat2 stability correlates with PHS resistance. The indicated strains were transformed with either pRS316-CUP1-Tat2-GFP-3×HA (pSR74) or empty vector (pRS316). Transformants were grown to mid-log phase and labeled with [35S]methionine for 10 min. A large excess of unlabeled methionine was added, and samples were withdrawn at the indicated times. Cells were lysed and processed for immunoprecipitation using an anti-HA antibody as described previously (14). Immunoprecipitates were resolved on SDS-PAGE, and levels of Tat2-eGFP-HA were determined using a phosphorimager.
Tat2 was degraded with a half-life of 110 min in the SEY6210 wild-type strain. Loss of Pdr5 and Yor1 increased this half-life to more than 180 min. Finally, the rsb1Δ strain produced Tat2 that was less stable than in wild-type cells, with a t½ of only 60 min. These data fit well with the view that internalization of Tat2 is slowed in pdr5Δ yor1 cells, whereas loss of Rsb1 accelerates turnover of this permease.
We used this same approach to evaluate the turnover of Tat2 in cells lacking the LCB lyase Dpl1 and a double mutant strain containing the dpl1Δ and rsb1Δ alleles. Our analysis of the LCB levels indicated that these ceramide precursors were elevated in the dpl1Δ but not the rsb1Δ background (Fig. 4). To determine whether loss of Dpl1 and/or Rsb1 had a similar effect on Tat2 stability, pulse-chase immunoprecipitation was carried out as above.
Loss of Dpl1 had the most dramatic effect on Tat2 degradation in our strain background, reducing the t½ to only 35 min. Introducing an rsb1Δ allele into the dpl1Δ mutant strain had only a minor effect on Tat2 turnover, suggesting that loss of Dpl1 was epistatic to loss of Rsb1. The relationship between Dpl1, Rsb1, PHS, and Tat2 turnover will be considered below.
Plasma Membrane Localization of Tat2 Is Increased in the pdr5Δ yor1 Strain
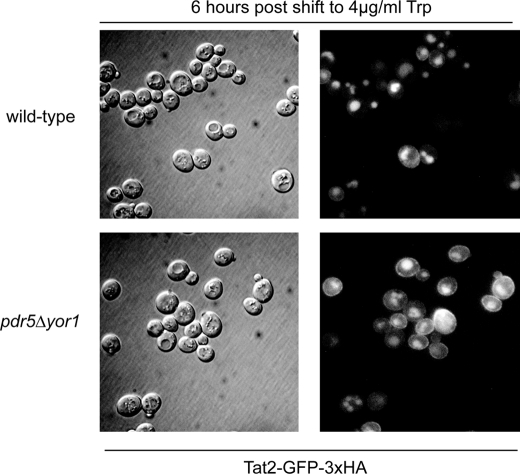
The above data support the idea that Tat2 is stabilized at the plasma membrane for a prolonged period of time in the absence of Pdr5 and Yor1, thereby explaining both the enhanced growth on low levels of tryptophan and increased Tat2 stability. To test this prediction, wild-type and pdr5Δ yor1 strains were transformed with a plasmid expressing Tat2-eGFP-3×HA or empty vector control. Transformants were grown in minimal media containing a high level of tryptophan (>20 μg/ml) to mid-log phase. These cultures were then shifted to media containing a low concentration of tryptophan (4 μg/ml). Six hours after the shift, cells were washed and visualized for GFP fluorescence (Fig. 6).
FIGURE 6.
Tat2 is present at the plasma membrane longer in the absence of Pdr5 and Yor1. Wild-type and pdr5Δ yor1 strains were transformed with pRS316-Cup1-Tat2-GFP-3×HA (pSR74), and transformants were grown overnight in CSM −URA media. These cultures were shifted to CSM −URA media containing 4 μg/ml tryptophan, grown for 6 h, and visualized by Nomarski optics and fluorescence microscopy.
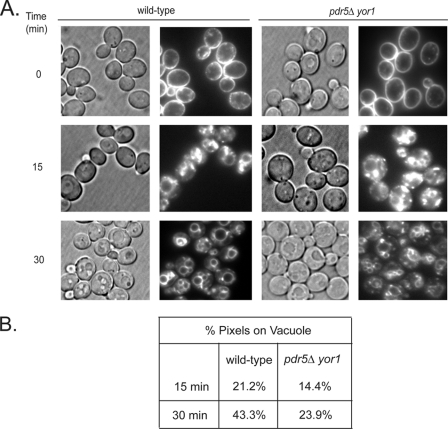
Fig. 6 shows an increased amount of Tat2-GFP at the plasma membrane in the pdr5Δ yor1 strain as compared with the wild-type strain. This analysis provides further evidence that loss of the plasma membrane ABC transporters Pdr5 and Yor1 inhibit the endocytosis of Tat2. To determine whether this decrease in turnover was specific to Tat2 or a more general slowing of internalization from the plasma membrane, we used a well characterized probe of yeast endocytosis, FM4-64 (25). Wild-type and pdr5Δ yor1 strains were labeled with FM4-64 for 45 min at 4 °C, and excess dye was removed by washing the cells and then chased with fresh media at 30 °C for the indicated times. Distribution of the dye was assessed by fluorescence microscopy (Fig. 7). We compared the intensity of fluorescence of the vacuoles compared with the entire cell through the use of ImageJ software. Pixels were quantitated inside a circle corresponding to the vacuole and compared with the total fluorescence detected for a circle enclosing the cell to generate the vacuolar pixel fraction (Fig. 7B).
FIGURE 7.
Bulk internalization is slowed in the pdr5Δ yor1 strain. A, wild-type and pdr5Δ yor1 strains were grown to mid-log phase and chilled on ice. FM4-64 dye was added to pre-chilled cells, and cells were incubated at 4 °C for 45 min. Samples were harvested and resuspended in pre-warmed YPD and incubated at 30 °C for the indicated time points. At the designated time points, samples were washed with cold SC-azide buffer and visualized by Nomarski optics (left panels) or fluorescence microscopy (right panels). B, quantification of the percentage of pixels (fluorescence) on the vacuolar membrane as compared with the whole cell was determined as described under “Experimental Procedures.”
Within 30 min, more than 40% of FM4-64 fluorescence was detectable on the vacuolar limiting membrane of wild-type cells. In cells lacking Pdr5 and Yor1, only 24% of FM4-64 was found on vacuoles. We carried out this same quantitation for the Tat2-GFP protein and found 40% of the GFP signal seen in wild-type vacuoles, although only 33% was present in the vacuole in pdr5Δ yor1 cells. These data provide supporting evidence that these ABC transporter proteins are required for normal bulk endocytic rates and not only internalization of Tat2. Because we are measuring localization to the vacuole, we cannot exclude the possibility that endocytic rates are normal, but intracellular trafficking is slowed in the pdr5Δ yor1 strain. We carried out this same analysis using the rsb1Δ strain but were unable to visualize any detectable difference in vacuolar fluorescence of FM4-64 (data not shown).
Interaction of Rsb1 and Pdr5/Yor1 with the Ubiquitin Ligase Adaptor Protein Bul1
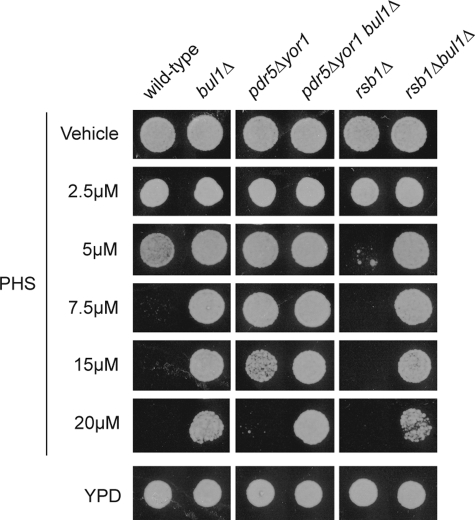
Previous experiments have implicated the Rsp5 adaptor protein Bul1 in controlling the endocytosis of Tat2 and other nutrient permeases (17, 26). Loss of Bul1 along with other Rsp5 adaptor proteins, also called arrestins (reviewed in Ref. 27), led to a pronounced retention of Tat2 on the plasma membrane (28). To determine whether Bul1 regulation of Tat2 endocytosis might interact with the effects of Pdr5/Yor1 and/or Rsb1 on Tat2 described above, the BUL1 gene was deleted from these genetic backgrounds. Representative mutant strains were grown to mid-log phase and tested for the ability to tolerate PHS challenge by placing equal numbers of cells on medium containing various concentrations of this LCB. Plates were allowed to develop at 30 °C and then photographed (Fig. 8).
FIGURE 8.
Interaction of Bul1 with Rsb1 and Pdr5/Yor1. Wild-type or isogenic mutants lacking the indicated genes were grown to mid-log phase and spotted onto YPD plates containing varying concentrations of PHS.
Loss of Bul1 caused a dramatic increase in PHS resistance. This effect has been seen before for both PHS resistance and also tolerance to the immunosuppressant FTY720 (10). Combining a bul1Δ allele with the pdr5Δ yor1 mutant background increased PHS resistance to a level greater than either starting mutant strain alone. This finding suggested that the effects of Bul1 and Pdr5/Yor1 are nonequivalent and define two separate pathways controlling activity of Tat2. Finally, the PHS sensitivity caused by the rsb1Δ allele could be fully suppressed by deleting BUL1. This result suggests that Rsb1 may act through Bul1 to negatively regulate Tat2 endocytosis.
Role of the C Terminus of Rsb1 in Control of PHS Resistance
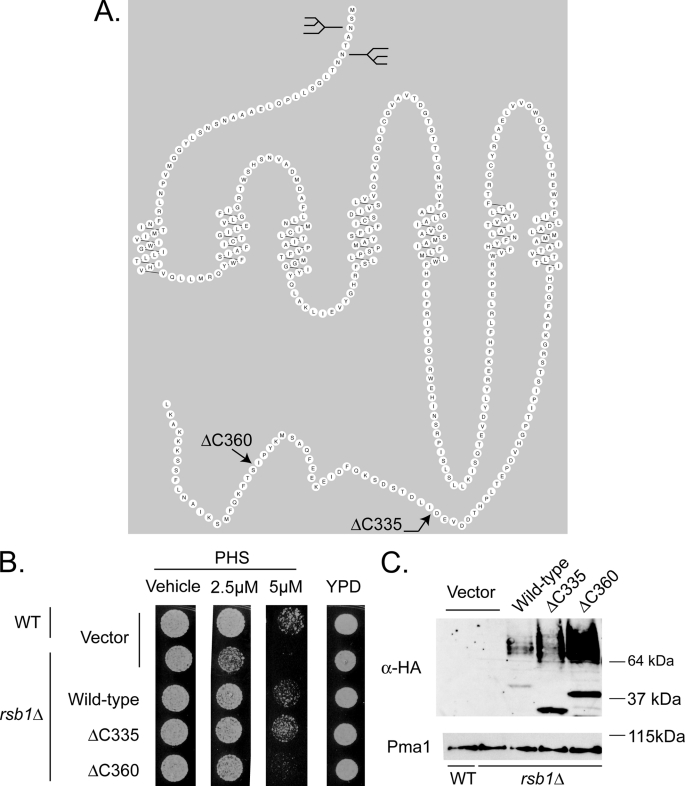
Earlier models suggested that Rsb1 might act as a translocase or transporter (6). Our in silico analyses of Rsb1 provided a different possibility. Topological predictions by programs such as Griffin or RbDe led to structural models for Rsb1 resembling the well known 7-TM receptor superfamily, most often associated with G-protein-coupled receptor proteins (Fig. 9A). Because there are no examples of 7-TM receptor proteins behaving as direct mediators of transport to our knowledge, we hypothesized that Rsb1 might carry out a regulatory role. Furthermore, 7-TM receptor proteins are known to often be regulated by modulation of their cell surface residence after ubiquitination on their cytoplasmic C-terminal domains.
FIGURE 9.
C-terminal truncations of Rsb1 show differential expression and PHS tolerance. A, predicted topology of Rsb1. A prediction for the organization of Rsb1 made by the program RbDe is shown. The locations of previously determined N-linked glycosylation sites in the luminal N terminus (9) are graphically represented by the diagrams. The positions of the C-terminal truncation mutations are indicated by arrows. B, low copy plasmids containing the above forms of Rsb1, along with an empty vector control, were transformed into wild-type and rsb1Δ strains. These transformants were grown to mid-log phase in selective minimal media and spotted onto YPD plates containing varying concentrations of PHS as described above. C, these same cultures from A were extracted using the TWIRL method (as described under “Experimental Procedures”), and equal amounts of total protein were run on SDS-PAGE, transferred to nitrocellulose, and probed using an anti-HA epitope antibody. These membranes were stripped and re-probed for Pma1.
To begin the structure-function analysis of Rsb1, we constructed two different C-terminal truncation mutant variants. Wild-type Rsb1, which contains 382 amino acids, was shortened to either 360 or 335 amino acids. These proteins, termed Rsb1ΔC360 or Rsb1ΔC335, were expressed with an 3×HA epitope tag inserted at their new C termini. Low copy number plasmids expressing each of these truncation mutants, along with wild-type and empty vector controls, were transformed into a rsb1Δ strain. A wild-type yeast strain was transformed with the empty vector to serve as a control for the wild-type chromosomal copy of RSB1. Transformants were grown to mid-log phase, placed on medium containing various concentrations of PHS, and analyzed by Western blotting using an anti-HA antibody (Fig. 9).
Removal of the C-terminal 22 residues of Rsb1 to form the Rsb1ΔC360 protein reduced but did not eliminate function as this protein was still able to complement the PHS sensitivity of the rsb1Δ strain. Interestingly, the steady-state level of this mutant protein was significantly greater than that of the wild-type polypeptide. Rsb1ΔC360 exhibited both the glycosylated (>64 kDa) and the unglycosylated species (∼37 kDa) that were present in elevated amounts compared with the wild-type protein. This same behavior was seen for the Rsb1ΔC335 mutant, although the glycosylated form of this mutant was less abundant than that of the Rsb1ΔC360 protein.
These data implicate the C terminus of Rsb1 as a regulator of the steady-state levels of this protein. The increased levels of Rsb1 in mutants lacking the complete C terminus are consistent with a role for this protein domain in the endocytosis and eventual turnover of this plasma membrane protein. Further work is required to substantiate this idea.
DISCUSSION
The identification of Rsb1 as a determinant of PHS resistance led to the understandable focus on the role of this membrane protein as a potential transporter of this LCB. PHS is a naturally occurring intermediate in sphingolipid biosynthesis but can be toxic at high levels. An informative feature of this PHS toxicity is its dependence on tryptophan auxotrophy. Loss of tryptophan prototrophy dramatically increases sensitivity to PHS (3). This type of dependence on tryptophan prototrophy has been noticed before for a range of phenotypes (reviewed in Ref. 29) and led us to consider that there might be a relationship between Rsb1 and control of tryptophan transport, rather than directly on efflux of LCBs. We argue that Rsb1 serves a regulatory function in Tat2-dependent tryptophan uptake based on the findings reported here. First, rsb1Δ cells have a range of phenotypes other than PHS sensitivity. Second, loss of Rsb1 caused no detectable accumulation of PHS in cells. Third, the topology of Rsb1 is most consistent with the structure of a 7-TM receptor rather than a transporter. Strikingly, although the 7-TM receptor family of membrane proteins is one of the largest known in biology, no protein with this topology has been shown to serve as a transporter/translocase. Together, these observations are more consistent with a role for Rsb1 as a regulator of permease endocytosis rather than a direct determinant of lipid distribution.
Loss of the plasma membrane phospholipid floppase activities of the ABC transporter proteins Pdr5 and Yor1 was previously described to strongly elevate PHS tolerance (7). Although previous experiments argued that this elevation proceeded exclusively through Rsb1, these studies used strains that lack the long chain base-phosphate lyase Dpl1. This enzyme plays a major role in degradation of LCB metabolites (5). Our analysis of the epistasis of Rsb1 and Pdr5/Yor1 indicates that, although Rsb1 is a key contributor to PHS resistance, loss of Pdr5/Yor1 still increased PHS tolerance, even if the rsb1Δ allele was present (Fig. 2). We interpret these observations to indicate that Rsb1 and Pdr5/Yor1 influence PHS tolerance via different pathways.
The demonstration that both Rsb1 and Pdr5/Yor1 activities modulate levels of Tat2 transport provides a simplifying theme for an important role of these proteins. We propose that Rsb1 acts as a negative regulator of Tat2 internalization, although Pdr5 and Yor1 positively influence this process. The exquisite sensitivity of tryptophan auxotrophs to levels of Tat2 function, which has been commented on before (17), makes tryptophan transport a faithful reporter of the membrane trafficking control of Tat2 determined by Rsb1 and Pdr5/Yor1. The finding that internalization of the FM4-64 dye probe is also delayed by loss of Pdr5 and Yor1 indicates that the effects of these ABC transporter proteins likely extend beyond control of this tryptophan transporter. Tat2 provides an especially useful phenotype to follow here. We have no evidence that Rsb1 influences membrane trafficking beyond its role in control of Tat2 internalization.
Our data also suggest a potential relationship between the influences of Dpl1 and Rsb1 on PHS resistance. Only loss of Dpl1 elicits an increase in intracellular PHS, whereas both Dpl1 and Rsb1 influence Tat2 function. One interpretation of these data is that loss of either Dpl1 or Rsb1 destabilizes Tat2 (Fig. 5) but by different mechanisms. Rsb1 acts to negatively regulate Tat2 internalization from the plasma membrane, whereas Dpl1 acts at the endoplasmic reticulum to limit accumulation of LCBs in the cell (5, 30). Loss of both Rsb1 and Dpl1 from the cell did not destabilize Tat2 more than the dpl1Δ single mutant. We interpret this to indicate that the high LCB levels in the dpl1Δ strain cause a defect in Tat2 early in its biosynthesis, similar to the previously described Tat2 destabilization seen in particular sterol biosynthetic mutants (21). The majority of Tat2 may never reach the plasma membrane, which would render any possible action of Rsb1 of minor importance.
One of the defining features of the large family of 7-TM receptors is the signaling between these integral membrane receptors and downstream heterotrimeric G-proteins (for review see Ref. 31). S. cerevisiae contains two α-subunit-encoding genes, GPA1 and GPA2, that have been linked to signaling via the G-protein-coupled receptor homologues Ste2/3 (for review see Ref. 32) or Gpr1 (for recent review see Ref. 33). Ste2/3 represent the mating pheromone receptors in S. cerevisiae, whereas Gpr1 is involved in glucose signaling. Disruption of GPA2 has no effect on PHS resistance (data not shown). Recent analyses of downstream signaling from Ste2/3 have demonstrated a link between these pheromone-induced G-protein-coupled receptors and the endosomal function that converges on the kinase Vps15 (34). However, because the effect of Gpa1 signaling is restricted to Vps15 (and its partner Vps34 (35)), although many VPS genes are involved in endosomal function, we doubt that this pathway is involved in Rsb1 signaling. Experiments to directly test this claim are underway.
Because no strong evidence can be derived from the literature that classical G-protein signaling is involved in downstream function of Rsb1, we hypothesize that this protein might act to control endocytosis through an arrestin-mediated mechanism. Arrestins act to deliver ubiquitin ligases to target membrane proteins (for review see Ref. 36). The finding that loss of the Bul1 adaptor protein both elevated PHS resistance alone and suppressed the PHS sensitivity of a rsb1Δ strain is consistent with Rsb1 acting upstream of this adaptor protein. We hypothesize that Rsb1 acts to antagonize Bul1 function, and it is this antagonism that leads to the observed phenotypic effects caused by loss of Rsb1 or Bul1, respectively. The simplest interpretation of the interaction of Rsb1 and Bul1 is that these two proteins act in the same pathway. This is in contrast to the behavior of Rsb1, Bul1, and Pdr5/Yor1. Loss of Bul1 or Pdr5/Yor1 increased PHS tolerance, but the triple mutant combination was more resistant to LCB exposure than either strain alone. This additive behavior is most consistent with Bul1 and Pdr5/Yor1 defining two separate pathways that act to control Tat2 plasma membrane localization.
G-protein-independent signaling as a result of activation of 7-TM receptor proteins is emerging as an important theme in the downstream function of these regulatory proteins (37). 5-Hydroxytryptamine receptors have been found to couple to the nonreceptor tyrosine kinase Src (38) or calmodulin (39) and signal in a fashion that is G-protein-independent. Interestingly, calmodulin binding acts to recruit β-arrestin proteins to the 5-hydroxytryptamine2C receptor (38). Earlier work on the β2-adrenergic receptor demonstrated that this 7-TM receptor protein could activate the ERK MAPK pathway even in the absence of normal levels of the Gα protein or β-arrestin (for review see Ref. 40). Together, these data indicate that 7-TM receptor proteins can couple to a variety of downstream signaling systems other than the classical G-protein pathways. Identification of the regulators of Rsb1-dependent signaling will provide important new information on the links between lipid composition and regulation of endocytosis.
Acknowledgment
We thank Dr. Rob Piper for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants GM75120 and GM49825 (to W. S. M.-R.).
- LCB
- long chain base
- 7-TM
- seven-transmembrane receptor protein
- HePC
- hexadecylphosphocholine
- PHS
- phytosphingosine
- eGFP
- enhanced GFP
- ABC
- ATP-binding cassette.
REFERENCES
- 1.Dickson R. C., Sumanasekera C., Lester R. L. (2006) Prog. Lipid Res. 45, 447–465 [DOI] [PubMed] [Google Scholar]
- 2.Cowart L. A., Obeid L. M. (2007) Biochim. Biophys. Acta 1771, 421–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skrzypek M. S., Nagiec M. M., Lester R. L., Dickson R. C. (1998) J. Biol. Chem. 273, 2829–2834 [DOI] [PubMed] [Google Scholar]
- 4.Chung N., Mao C., Heitman J., Hannun Y. A., Obeid L. M. (2001) J. Biol. Chem. 276, 35614–35621 [DOI] [PubMed] [Google Scholar]
- 5.Saba J. D., Nara F., Bielawska A., Garrett S., Hannun Y. A. (1997) J. Biol. Chem. 272, 26087–26090 [DOI] [PubMed] [Google Scholar]
- 6.Kihara A., Igarashi Y. (2002) J. Biol. Chem. 277, 30048–30054 [DOI] [PubMed] [Google Scholar]
- 7.Kihara A., Igarashi Y. (2004) Mol. Biol. Cell 15, 4949–4959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenbaum D. M., Rasmussen S. G., Kobilka B. K. (2009) Nature 459, 356–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panwar S. L., Moye-Rowley W. S. (2006) J. Biol. Chem. 281, 6376–6384 [DOI] [PubMed] [Google Scholar]
- 10.Welsch C. A., Roth L. W., Goetschy J. F., Movva N. R. (2004) J. Biol. Chem. 279, 36720–36731 [DOI] [PubMed] [Google Scholar]
- 11.Ito H., Fukuda Y., Murata K., Kimura A. (1983) J. Bacteriol. 153, 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bligh E. G., Dyer W. J. (1959) Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 13.Merrill A. H., Jr., Wang E., Mullins R. E., Jamison W. C., Nimkar S., Liotta D. C. (1988) Anal. Biochem. 171, 373–381 [DOI] [PubMed] [Google Scholar]
- 14.Katzmann D. J., Epping E. A., Moye-Rowley W. S. (1999) Mol. Cell. Biol. 19, 2998–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X. H., Xiao Z., Fitzgerald-Hayes M. (1994) Mol. Gen. Genet. 244, 260–268 [DOI] [PubMed] [Google Scholar]
- 16.Schmidt A., Hall M. N., Koller A. (1994) Mol. Cell. Biol. 14, 6597–6606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abe F., Iida H. (2003) Mol. Cell. Biol. 23, 7566–7584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daicho K., Makino N., Hiraki T., Ueno M., Uritani M., Abe F., Ushimaru T. (2009) FEMS Microbiol. Lett. 298, 218–227 [DOI] [PubMed] [Google Scholar]
- 19.Hiraki T., Abe F. (2010) FEBS Lett. 584, 55–60 [DOI] [PubMed] [Google Scholar]
- 20.Khozoie C., Pleass R. J., Avery S. V. (2009) J. Biol. Chem. 284, 17968–17974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Umebayashi K., Nakano A. (2003) J. Cell Biol. 161, 1117–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanson P. K., Malone L., Birchmore J. L., Nichols J. W. (2003) J. Biol. Chem. 278, 36041–36050 [DOI] [PubMed] [Google Scholar]
- 23.Didion T., Regenberg B., Jørgensen M. U., Kielland-Brandt M. C., Andersen H. A. (1998) Mol. Microbiol. 27, 643–650 [DOI] [PubMed] [Google Scholar]
- 24.Beck T., Schmidt A., Hall M. N. (1999) J. Cell Biol. 146, 1227–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vida T. A., Emr S. D. (1995) J. Cell Biol. 128, 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helliwell S. B., Losko S., Kaiser C. A. (2001) J. Cell Biol. 153, 649–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeWire S. M., Ahn S., Lefkowitz R. J., Shenoy S. K. (2007) Annu. Rev. Physiol. 69, 483–510 [DOI] [PubMed] [Google Scholar]
- 28.Nikko E., Pelham H. R. (2009) Traffic 10, 1856–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abe F. (2007) Int. Congr. Ser. 1304, 75–84 [Google Scholar]
- 30.Grote E., Vlacich G., Pypaert M., Novick P. J. (2000) Mol. Biol. Cell 11, 4051–4065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilman A. G. (1987) Annu. Rev. Biochem. 56, 615–649 [DOI] [PubMed] [Google Scholar]
- 32.Dohlman H. G., Thorner J. W. (2001) Annu. Rev. Biochem. 70, 703–754 [DOI] [PubMed] [Google Scholar]
- 33.Gancedo J. M. (2008) FEMS Microbiol. Rev. 32, 673–704 [DOI] [PubMed] [Google Scholar]
- 34.Heenan E. J., Vanhooke J. L., Temple B. R., Betts L., Sondek J. E., Dohlman H. G. (2009) Biochemistry 48, 6390–6401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slessareva J. E., Routt S. M., Temple B., Bankaitis V. A., Dohlman H. G. (2006) Cell 126, 191–203 [DOI] [PubMed] [Google Scholar]
- 36.Léon S., Haguenauer-Tsapis R. (2009) Exp. Cell Res. 315, 1574–1583 [DOI] [PubMed] [Google Scholar]
- 37.Sun Y., McGarrigle D., Huang X. Y. (2007) Mol. Biosyst. 3, 849–854 [DOI] [PubMed] [Google Scholar]
- 38.Barthet G., Framery B., Gaven F., Pellissier L., Reiter E., Claeysen S., Bockaert J., Dumuis A. (2007) Mol. Biol. Cell 18, 1979–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Labasque M., Reiter E., Becamel C., Bockaert J., Marin P. (2008) Mol. Biol. Cell 19, 4640–4650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lefkowitz R. J., Shenoy S. K. (2005) Science 308, 512–517 [DOI] [PubMed] [Google Scholar]