Abstract
The lipase activity of most phospholipases C (PLCs) is basally repressed by a highly degenerate and mostly disordered X/Y linker inserted within the catalytic domain. Release of this auto-inhibition is driven by electrostatic repulsion between the plasma membrane and the electronegative X/Y linker. In contrast, PLC-γ isozymes (PLC-γ1 and -γ2) are structurally distinct from other PLCs because multiple domains are present in their X/Y linker. Moreover, although many tyrosine kinases directly phosphorylate PLC-γ isozymes to enhance their lipase activity, the underlying molecular mechanism of this activation remains unclear. Here we define the mechanism for the unique regulation of PLC-γ isozymes by their X/Y linker. Specifically, we identify the C-terminal SH2 domain within the X/Y linker as the critical determinant for auto-inhibition. Tyrosine phosphorylation of the X/Y linker mediates high affinity intramolecular interaction with the C-terminal SH2 domain that is coupled to a large conformational rearrangement and release of auto-inhibition. Consequently, PLC-γ isozymes link phosphorylation to phospholipase activation by elaborating upon primordial regulatory mechanisms found in other PLCs.
Keywords: Phosphatidylinositol Signaling, Phospholipase C, Phosphotyrosine Signaling, Protein Conformation, SH2 Domains
Introduction
Phospholipase C (PLC)2 isozymes hydrolyze phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) into the second messengers diacylglycerol and inositol 1,4,5-trisphosphate (1). Diacylglycerol and inositol 1,4,5-trisphosphate subsequently activate PKC isozymes and liberate intracellular calcium stores, respectively, thereby initiating and propagating numerous cellular events including fertilization, proliferation, differentiation, and chemotaxis (2).
The mammalian PLC family contains 13 isozymes subdivided into six subtypes (β, δ, γ, ϵ, η, and ζ) based on structural homology (1). The highest sequence similarity among PLCs lies within the catalytic triose phosphate isomerase barrel comprised of two halves (X and Y boxes) separated by a highly degenerate X/Y linker. The active site is essentially invariant in PLC enzymes and ligates a calcium ion cofactor necessary for deprotonation of the PtdIns(4,5)P2 substrate and stabilization of the transition state (3).
Using structural and biochemical information, we recently showed that the X/Y linker in PLC-β, -δ, and -ϵ isozymes mediates auto-inhibition of phospholipase activity (4). The X/Y linker of these PLCs is highly negatively charged, mostly disordered, and prevents PtdIns(4,5)P2 access to the active site by a combination of steric exclusion and electrostatic repulsion of negatively charged membranes. Consequently, we proposed a general mechanism of interfacial activation of these isozymes in which any upstream input that preferentially orients the active site toward PtdIns(4,5)P2-containing membranes would remove the X/Y linker to allow access of substrate for hydrolysis (4).
In contrast to all other PLCs, the PLC-γ isozymes (PLC-γ1 and -γ2) possess highly elaborated X/Y linkers containing multiple domains: a split PH domain, two SH2 domains, and an SH3 domain (see Fig. 1A). Nevertheless, some reports suggest that PLC-γ isozymes are also auto-inhibited via their X/Y linkers. For example, deletion (5) or proteolysis (6–8) of the X/Y linker modestly (2–5-fold) enhanced PLC-γ1 activity. This potential auto-inhibition has been neither rigorously confirmed nor evaluated. Moreover, the X/Y linker of PLC-γ isozymes lacks the high density of negative charge critical for interfacial activation of other PLCs, and because these isozymes are directly activated by phosphorylation, their mode of activation must be mechanistically distinct.
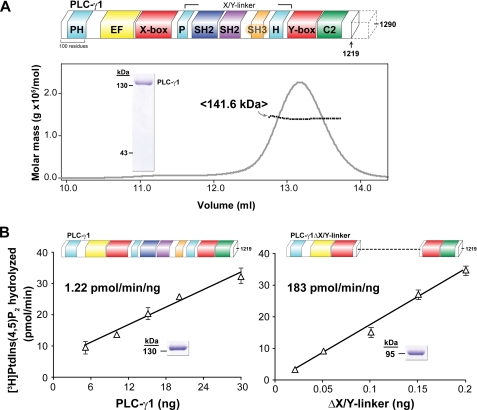
FIGURE 1.
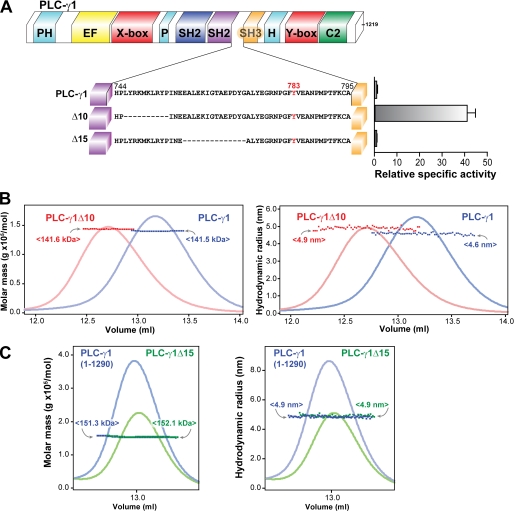
Purified PLC-γ1 is monomeric and auto-inhibited by its X/Y linker. A, purified PLC-γ1 is monomeric. Schematic representation of full-length rat PLC-γ1 (upper panel). PLC isozymes contain a conserved core consisting of an N-terminal PH domain (cyan), an array of four EF hands (yellow), a catalytic TIM barrel composed of X and Y boxes (red), and a C-terminal C2 domain (green). PLC-γ isozymes are unique in containing multiple domains within the X/Y linker: a split PH domain (cyan), two SH2 domains (navy and purple), and an SH3 domain (gold). The C terminus (dotted outline) of PLC-γ is degenerate; unless otherwise noted, studies reported here used PLC-γ1 truncated at residue 1219. Lower panel, elution of PLC-γ1 from a size exclusion column and monitored by absorbance (gray line) was simultaneously analyzed with multi-angle light scattering (black line) to provide a mean molecular mass of 141.6 kDa consistent with a monomer (calculated molecular mass of 140.1 kDa). Purity of PLC-γ1 protein (2 μg, inset) was assessed by SDS-PAGE analysis followed by staining with Coomassie Brilliant Blue. B, removal of the X/Y linker constitutively activates purified PLC-γ1. Lipase activity of purified PLC-γ1 (left panel) or PLC-γ1ΔX/Y linker (right panel) was measured at the indicated protein concentrations using mixed detergent-phospholipid micelles. The indicated specific activities are the means of at least three independent experiments. Purity (2 μg, inset) of PLC-γ1 proteins was assessed by SDS-PAGE analysis followed by Coomassie Brilliant Blue staining. See also supplemental Fig. S1.
Although it has been recognized for 20 years that activation of PLC-γ isozymes downstream of numerous receptor tyrosine kinases (RTKs) and cytosolic tyrosine kinases (2) involves membrane recruitment through the binding of the N-terminal SH2 (nSH2) within the X/Y linker and subsequent tyrosine phosphorylation of PLC-γ isozymes (9–11), the molecular mechanism linking phosphorylation to activation remains poorly defined.
In this report, we used highly purified proteins and robust in vitro assays to discover that PLC-γ isozymes have modified the common mechanism of interfacial activation present in most PLCs to couple phosphorylation of a highly elaborated X/Y linker with release of auto-inhibition and consequential activation.
EXPERIMENTAL PROCEDURES
Molecular Constructs
Standard PCR-mediated mutagenesis (Stratagene; QuikChange site-directed mutagenesis manual) was used to introduce deletions or substitutions into the open reading frames of rat PLC-γ1 and human PLC-γ2 (kind gifts from Dr. Graham Carpenter, Vanderbilt University, Nashville, TN) in modified pFastBacHT, pcDNA3.1, or pPICZ vectors (Invitrogen). See also supplemental “Experimental Procedures.”
The kinase domain of FGFR2 (residues 458–778), FGFR2K, was PCR-amplified from the full-length FGFR2 (provided by Dr. Channing Der, University of North Carolina at Chapel Hill) (12) and subcloned into a modified pET15b vector (Novagen). The constitutively active mutation (E565A) (13) was generated using PCR-mediated mutagenesis (Stratagene; QuikChange site-directed mutagenesis manual). All of the mutant genes were verified by automated DNA sequencing of the entire open reading frame. PLC-γ1-derived proteins were purified to homogeneity as described in great detail in the supplemental “Experimental Procedures.”
Measurement of PLC-γ Activity in Mammalian Cells
The accumulation of [3H]inositol phosphates was measured in transiently transfected HEK293 or HEK293T cells as previously described (4). Western blotting was performed to confirm the expression of each PLC construct in HEK cells using a monoclonal antibody directed toward the HA epitope (Covance). β-Actin (Sigma) antibody was used as a loading control.
In Vitro Reconstitution Assays to Measure [3H]PtdIns(4,5)P2 Hydrolysis
Two different procedures were utilized for measuring phospholipase activity of PLC enzymes. Initial experiments measuring maximum enzymatic activity were carried out in the presence of a detergent substrate mixture. The second assay employed phospholipid vesicles. See also supplemental “Experimental Procedures.”
In Vitro Kinase Assay
Equimolar concentrations (35 μm) of FGFR2K E565A and PLC-γ1 were incubated in a buffer containing 20 mm HEPES (pH 7.4), 50 mm NaCl, 50 μm ATP, 10 mm MnCl2, 0.2 mm Na3VO4, 2 mm DTT, 50 ng/ml fatty acid-free BSA, and 1 μl of [γ-32P]ATP (10 mCi/ml; PerkinElmer Life Sciences) on ice in a final volume of 70 μl. After 1 h, 10 μl was removed and used in the phospholipase assay described above. Another 10 μl was subjected to SDS-PAGE analysis followed by Coomassie Brilliant Blue staining. After extensive destaining, the bands corresponding to PLC-γ1 proteins were excised, washed with acetonitrile solvent, and dried using a SpeedVac (Thermo Scientific). Recovery of radioactivity from gel pieces was determined by Cherenkov counting.
Isothermal Titration Calorimetry
ITC studies were carried out on a VP-ITC (MicroCal; GE Healthcare) at 25 °C. In general, macromolecules were dialyzed in 50 mm HEPES (pH 7.5) and 50 mm NaCl, and peptides were dissolved in the same buffer prior to use. Peptides (450 μm to 1 mm) or split PH domain (1 mm) were titrated into macromolecules (isolated SH2 domains (35–50 μm) or X/Y linker (45–70 μm)) in a 1.45-ml sample cell. Titrations were performed, while samples were stirred at 300 rpm. An initial 2-μl injection was followed by 30 injections of 10 μl each with a 4-min delay between injections to allow for signal to return to base line. The data were fit with a single independent site binding model using Origin software (MicroCal). Background reaction enthalpy caused by titrant dilution or buffer dilution of macromolecule was determined from the last five to ten injections, and data were corrected for background heat released prior to curve fitting. See also supplemental “Experimental Procedures.”
Size Exclusion Chromatography-Multi-angle Light Scattering (SEC-MALS)
Light scattering measurements were made with a Wyatt DAWN EOS light scattering instrument (Wyatt Optilab refractometer and Wyatt dynamic light scattering module) coupled to an AKTA FPLC (GE Healthcare). A Superdex 200 10/300 GL (GE Healthcare) pre-equilibrated with 20 mm Tris (pH 8.0), and 200 mm NaCl was loaded with 250 μl of PLC-γ1 proteins at ∼4.0 mg/ml. Data collection and analysis were performed with ASTRA software version 4.90.08 (Wyatt Technologies).
Circular Dichroism
Purified isolated X/Y linker proteins, either wild type or Δ10, at a concentration of 0.125 mg/ml in 20 mm sodium phosphate buffer (pH 7.5), were analyzed by circular dichroism at 25 °C using a Chirascan circular dichroism spectrophotometer (Applied Photophysics) with a 10-mm path length. The data were plotted as mean residue ellipticity as a function of wavelength.
RESULTS
Purified PLC-γ1 Is Monomeric and Inhibited by Its X/Y Linker
Full-length rat PLC-γ1, purified to homogeneity after heterologous expression in insect cells, was predominantly monomeric with a minor aggregate component based on its light scattering properties after elution from a size exclusion column (supplemental Fig. S1A). The majority of the C-terminal region following the C2 domain is degenerate in PLC-γ isozymes, and aggregation was eliminated with PLC-γ1 lacking its last 71 amino acids (residues 1–1219; Fig. 1A). Consequently, unless otherwise noted, all of the work described here with purified PLC-γ1 used this truncated form. The absolute molecular masses for both monomeric species of purified PLC-γ1 measured using size exclusion chromatography coupled to multi-angle light scattering (SEC-MALS) were almost identical to their calculated molecular masses based on amino acid composition (Fig. 1A and supplemental Fig. S1A). In addition, the specific activities of both forms of purified PLC-γ1 determined with 50 μm [3H]PtdIns(4,5)P2 were ∼1 pmol/min/ng (Fig. 1B and data not shown), consistent with specific activities reported for endogenous (14) and recombinant (15) PLC-γ1. As discussed below, it was necessary to use the yeast, Pichia pastoris, for expression of certain very active mutants of PLC-γ1. Therefore, we also compared the properties of wild-type PLC-γ1 purified after expression in yeast (P. pastoris) versus insect cells (Trichoplusia ni). The two samples of purified PLC-γ1 exhibited identical specific activities, molecular masses, and hydrodynamic radii measured by SEC-MALS (supplemental Fig. S1B), indicating that the intrinsic physical properties of PLC-γ1 are independent of expression system.
We recently showed that deletion of the entire X/Y linker of PLC-β2, -δ1, or -ϵ increased basal phospholipase activity 10–100-fold in transfected cells (4). Given these observations, we examined potential auto-inhibitory roles for the elaborated X/Y linker within PLC-γ isozymes. Thus, PLC-γ1 lacking its entire X/Y linker (residues 471–938; PLC-γ1ΔX/Y linker) was purified to homogeneity after expression in P. pastoris. Pichia was utilized for expression of this mutant because its expression in insect cells prevented baculovirus replication, presumably because of highly elevated phospholipase activity. The specific activity of purified PLC-γ1ΔX/Y linker (∼180 pmol/min/ng) was ∼150-fold greater than PLC-γ1 with an intact X/Y linker (Fig. 1B), indicating that PLC-γ1 is dramatically auto-inhibited by its X/Y linker. It is unclear why a previous study reported only modest (4-fold) enhancement of phospholipase activity for a mutant of PLC-γ1 that roughly corresponds to PLC-γ1ΔX/Y linker (5).
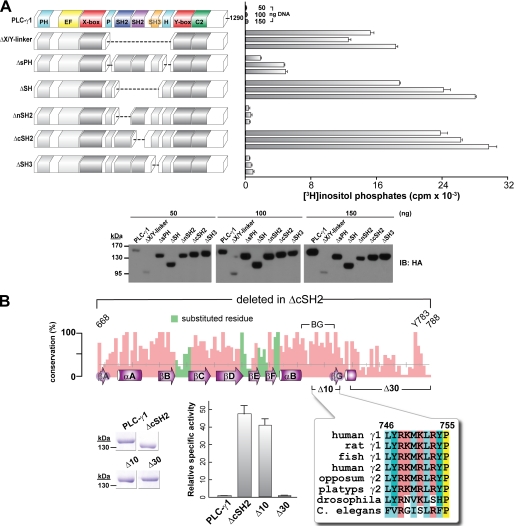
A Minimal Inhibitory Motif of PLC-γ1 Maps to the BG Loop of the C-terminal SH2 Domain
A series of deletions were introduced to delimit the minimal auto-inhibitory portion of the X/Y linker of PLC-γ1, and the activities of these mutants were determined after transient expression in HEK293 cells (Fig. 2A). Removal of the entire X/Y linker, the SH array, or the C-terminal SH2 (cSH2) domain produced robust constitutive activation of PLC-γ1. In contrast, removal of the split pleckstrin homology (sPH) domain modestly elevated phospholipase activity, and loss of either the nSH2 domain or the SH3 domain did not substantially affect phospholipase activity. These results suggest that the cSH2 domain within the X/Y linker of PLC-γ1 is the main determinant mediating auto-inhibition of phospholipase activity.
FIGURE 2.
Deletion mapping delineates a 10-residue span within the X/Y linker of PLC-γ1 critical for auto-inhibition. A, systematic deletion of the domains in the X/Y linker implicate the C-terminal SH2 domain in mediating auto-inhibition. HEK293 cells were transfected with the indicated amounts of DNA encoding either wild-type PLC-γ1 or the indicated deleted forms (left) prior to quantification of [3H]inositol phosphates (right). Vector alone was subtracted from all of the measurements. The data shown are the means ± S.D. of triplicate samples and are representative of three or more independent experiments. Western blotting (bottom) of cell lysates confirmed expression of PLC-γ1 deletion constructs. B, high resolution mutational mapping of the C-terminal SH2 domain highlights 10 residues required for auto-inhibition. Top panel, bar chart plotting the percentage of identity per position of a multiple sequence alignment of 22 PLC-γ orthologs spanning the region deleted (residues 668–788) in rat PLC-γ1ΔcSH2. Also shown on this chart are the secondary structure elements (purple) of the crystal structure of the cSH2 domain (Protein Data Bank entry 3GQI) with the nomenclature proposed by Harrison and co-workers (37); two deletions (Δ10, red; Δ30, blue) introduced into PLC-γ1; and Tyr783 required to be phosphorylated during receptor-mediated lipase activation. Green bars highlight sites of substitution to alanine in PLC-γ1 yielding no change in [3H]inositol phosphate accumulation after transfection of mutant DNAs into HEK293 cell (data not shown). In contrast, purified PLC-γ1 proteins (bottom left panel) indicate that PLC-γ1ΔcSH2 and PLC-γ1Δ10 have comparable constitutive activation (bottom middle panel). A subset of the multiple sequence alignment spanning Δ10 is shown in the bottom right panel. Specific activities are the means ± S.E. of at least three independent experiments using phospholipid vesicles and normalized to wild-type activity. Purified proteins (2 μg; bottom left panel) were assessed by SDS-PAGE followed by staining with Coomassie Brilliant Blue. See also supplemental Fig. S2.
The mutant forms of PLC-γ1 did not express to identical levels (Fig. 2A). In particular, the PLC-γ1ΔX/Y linker was poorly expressed over the range of concentrations (50–150 ng) of transfected expression vectors tested. To more accurately compare the phospholipase activities of PLC-γ1ΔX/Y linker and PLC-γ1ΔcSH2, the corresponding expression vectors were transfected at lower concentrations (supplemental Fig. S2A). The expression levels of the two PLC-γ1 mutants were more comparable under these conditions, and the corresponding phospholipase assays indicate that PLC-γ1ΔX/Y linker and PLC-γ1ΔcSH2 are both similarly active (within a factor of 2), further substantiating the idea that the cSH2 domain of PLC-γ1 is the main determinant for auto-inhibition of phospholipase activity.
The series of PLC-γ1 mutants harboring deletions within the X/Y linker subsequently were purified to homogeneity and tested for phospholipase activity (supplemental Fig. S2B). Consistent with the previous transfection experiments, deletion of the cSH2 domain, but not the nSH2 domain or the SH3 domain, substantially (∼50-fold) elevated the phospholipase activity of PLC-γ1. Surprisingly, the phospholipase activity of purified PLC-γ1ΔsPH was essentially equivalent to the phospholipase activity of purified PLC-γ1ΔcSH2 despite the fact that PLC-γ1ΔsPH exhibited only modest increases in basal phospholipase activity after transient expression in HEK293 cells. These results suggest that the sPH domain might stabilize the auto-inhibitory conformation of the cSH2 domain and that this stabilization is more pronounced within a cellular milieu. However, we were unable to detect an interaction between the purified sPH and cSH2 domains using ITC (supplemental Fig. S2C), suggesting that any potential stabilization of the auto-inhibited state of PLC-γ1 involves, at best, weak direct interactions between the two domains.
The cSH2 domain of PLC-γ1 was subjected to more extensive mutagenesis to define potential regions responsible for auto-inhibition of phospholipase activity (Fig. 2B). The recent crystal structure of the tandem SH2 domain of PLC-γ1 bound to the kinase domain of fibroblast growth factor receptor 1 (FGFR1) highlights a set of loops within the nSH2 domain that, in addition to the canonical phosphate-binding pocket, are needed for engagement of FGFR1 (16). Reasoning that the equivalent loops in the cSH2 domain might participate in intramolecular interactions needed for auto-inhibition, we introduced a series of single substitutions into specific loops (BC, DE, and EF) of the cSH2 domain of PLC-γ1, and the mutant isozymes were tested for altered phospholipase activity in HEK293 cells. In all cases, the mutants of PLC-γ1 isozymes exhibited phospholipase activities indistinguishable from wild-type PLC-γ1 (data not shown).
Two small deletions (Δ10 and Δ30) were subsequently introduced into PLC-γ1 near the end of the cSH2 domain (Fig. 2B). The Δ10 (residues 746–755) deletion results in removal of 10 residues that encompass the highly mobile BG loop and the short βG strand not found in all SH2 domains; the Δ30 (residues 759–788) deletion removes 30 residues encompassing Tyr783, which is critical for phosphorylation-mediated activation of PLC-γ1. The two deletion mutants of PLC-γ1 were purified to homogeneity, and their specific activities were compared (Fig. 2B). The specific activity of PLC-γ1Δ30 was indistinguishable from that of wild-type PLC-γ1. In contrast, the specific activities of purified PLC-γ1Δ10 and PLC-γ1ΔcSH2 were essentially equivalent, and enzyme activity was elevated ∼40-fold relative to wild-type PLC-γ1. Therefore, some portion of the cSH2 domain encompassing the BG loop and subsequent βG strand is critical for auto-inhibition of PLC-γ isozymes. The trivial explanation that deletion of the BG loop and βG strand promotes unfolding of the cSH2 domain can be dismissed because the amount and type of secondary structure of the purified X/Y linker of PLC-γ1 and the equivalent portion lacking these 10 residues are essentially identical based on circular dichroism spectroscopy (supplemental Fig. S2D). Also, Tyr783 and the surrounding sequence, although essential for phosphorylation-induced activation of PLC-γ1, does not directly participate in auto-inhibition because its deletion did not affect basal phospholipase activity.
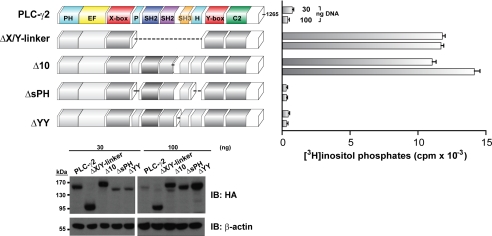
Mechanism of Auto-inhibition Is Conserved in PLC-γ2
PLC-γ1 and -γ2 possess identical domain architecture, 50% sequence identity, and 77% sequence similarity. Therefore, we hypothesized that PLC-γ1 and -γ2 are similarly auto-inhibited. To test this idea explicitly, a series of deletions were individually introduced into the X/Y linker of PLC-γ2, and the phospholipase activity of the resulting mutant isozymes was determined after expression in HEK293T cells (Fig. 3). As expected based on the earlier studies with PLC-γ1, PLC-γ2ΔX/Y linker exhibited robust lipase activity relative to wild-type PLC-γ2. More informatively, however, PLC-γ2Δ10 also had essentially the same high level of phospholipase activity as PLC-γ2ΔX/Y linker, and thus, PLC-γ2, like PLC-γ1, is strongly auto-inhibited by a mechanism that requires the region encompassing the BG loop of the cSH2 domain. PLC-γ2ΔYY harbors a deletion centered on Tyr759, which is analogous to Tyr783 in PLC-γ1 and is deleted in PLC-γ1Δ30. Like PLC-γ1Δ30, PLC-γ2ΔYY exhibited low basal phospholipase activity, supporting the conclusion that this region, although required for activation upon its phosphorylation, does not directly participate in auto-inhibition. The phospholipase activity of PLC-γ2ΔsPH and wild-type PLC-γ2 were essentially indistinguishable, supporting the idea that the split PH domain is not critical for auto-inhibition of phospholipase activity. In total, these analyses indicated that PLC-γ1 and PLC-γ2 share identical modes of auto-inhibition that require a portion of the region encompassed by the BG loop and the βG strand of the cSH2 domain.
FIGURE 3.
PLC-γ1 and -γ2 are auto-inhibited by analogous portions of the X/Y linker. HEK293T cells were transfected with the indicated amount of DNA encoding either wild-type PLC-γ2 or the deleted forms of PLC-γ2 (left panel) prior to quantification of [3H]inositol phosphates (right panel). Vector alone was subtracted from all measurements. The data are the means ± S.D. of triplicate samples and are representative of three or more independent experiments. Western blotting of HEK293T cell lysates confirmed expression of PLC-γ2 constructs.
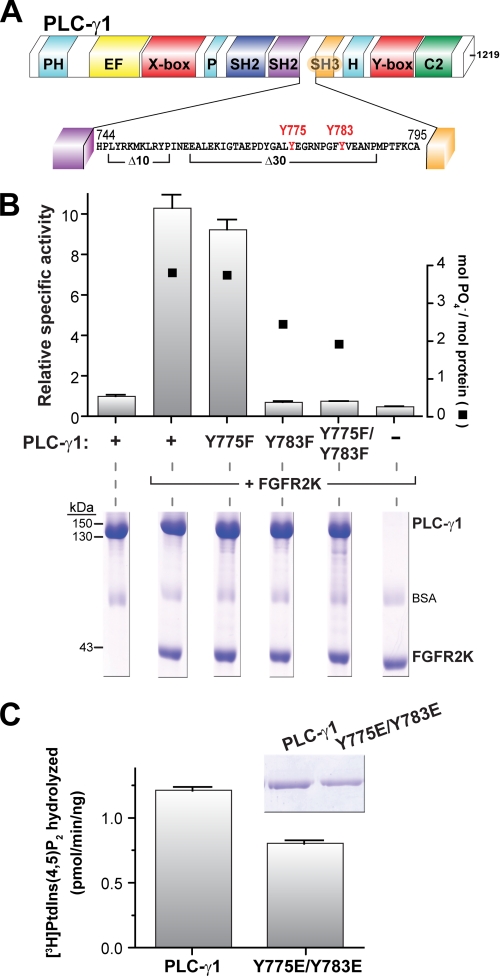
PLC-γ1 Activity Is Dependent on Tyr783 Phosphorylation
Phosphorylation of Tyr775 or Tyr783 has been shown to be essential for activation of PLC-γ1 downstream of the B-cell receptor (11) and RTKs (10); phosphorylation of the analogous tyrosines (at positions 753 and 759) in PLC-γ2 is also essential for its activation by tyrosine kinases (17–20). As shown above, deletion of residues that surround these tyrosines had no impact on auto-inhibition of lipase activity of either PLC-γ2 (ΔYY) or PLC-γ1 (Δ30) (Fig. 4A). To begin to place phosphorylation of these tyrosines into a mechanistic framework for release of auto-inhibition of PLC-γ isozymes, Tyr775 and Tyr783 of PLC-γ1 were substituted for phenylalanine, either individually or in combination, and the resulting mutant PLC-γ1 isozymes were purified to homogeneity. Phospholipase activity was quantified after phosphorylation of the purified mutants with the constitutively active kinase domain of fibroblast growth factor receptor 2 (FGFR2K) (Fig. 4B), a RTK known to phosphorylate and activate PLC-γ1 (21). Approximately 4 mol of phosphate were incorporated into purified wild-type PLC-γ1 under these conditions, and phospholipase activity was accelerated ∼10-fold. Mutation of Tyr775 did not substantively affect the level of FGFR2K-promoted phosphorylation or phospholipase activity, indicating that phosphorylation of Tyr775 is not essential to relieve auto-inhibition. In contrast, mutation of Tyr783, either alone or in combination with Y775F, reduced FGFR2K-promoted phosphorylation of PLC-γ1 and completely abrogated kinase-stimulated enhancement of phospholipase activity. Thus, phosphorylation of Tyr783 is crucial for relief of auto-inhibition. Phosphorylation of Tyr771 also was suggested to regulate lipase activity of PLC-γ1 (10). However, this tyrosine is not conserved in PLC-γ2 (supplemental Fig. S3A), and its mutation affected neither the degree of FGFR2K-promoted phosphorylation nor the lipase activity of purified PLC-γ1 (supplemental Fig. S3B).
FIGURE 4.
Phosphorylation, but not phosphomimetic mutation, of tyrosine 783 enhances the lipase activity of purified PLC-γ1. A, expanded view of PLC-γ1 sequence between the C-terminal SH2 and SH3 domains. Tyrosines previously implicated in phosphorylation-dependent activation of the lipase are highlighted in red. Also marked are deletions (Δ10 and Δ30) described in Fig. 2. B, phosphorylation of tyrosine 783, but not tyrosine 775, activates PLC-γ1. Equimolar concentrations of purified PLC-γ1 or the indicated phospho-deficient forms were incubated with purified, constitutively active kinase domain of FGFR2 (FGFR2K) and [γ-32P]ATP in BSA-containing buffer. Aliquots of reaction mixtures were subsequently tested for: (i) lipase activity in phospholipid vesicles (left y axis) and (ii) incorporation of [γ-32P]phosphate into PLC-γ1 (right y axis) after SDS-PAGE followed by staining with Coomassie Brilliant Blue (lower panel). Specific activities are the means ± S.E. of three or more independent experiments and normalized to basal activity of wild-type PLC-γ1. C, phosphomimetic mutations of tyrosines 775 and 783 are insufficient to enhance the lipase activity of PLC-γ1. The specific activity of either purified PLC-γ1 or the doubly mutated (Y775E/Y783E) form was measured in mixed detergent-phospholipid micelles. The data are the means ± S.D. of triplicate samples and are representative of three or more independent experiments. The purity (2 μg, inset) of PLC-γ1 proteins was assessed by SDS-PAGE followed by staining with Coomassie Brilliant Blue. See also supplemental Fig. S3.
Tyrosine phosphorylation often regulates protein function by introducing a highly localized negative charge that can be mimicked by substitution of the phosphorylated tyrosines with glutamic acid. However, this is not the case for PLC-γ1, because tandem substitution of Tyr775 and Tyr783 with glutamic acid failed to increase the phospholipase activity of PLC-γ1 (Fig. 4C).
Interaction between Phosphorylated Tyr783 and the C-terminal SH2 Domain Is Required for Phosphorylation-mediated Activation of PLC-γ1
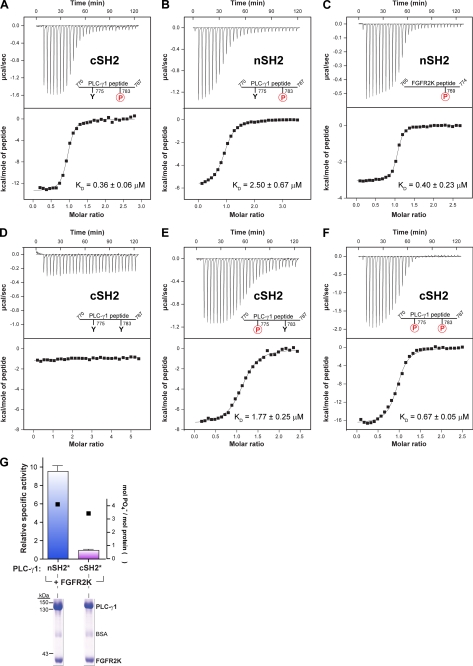
A previous report suggested that phosphorylated Tyr783 of PLC-γ1 engages its cSH2 domain to enhance phospholipase activity of partially purified PLC-γ1 (22). Aspects of this idea were tested directly using purified proteins (Fig. 5). A peptide encompassing phosphorylated Tyr783 of PLC-γ1 bound with high affinity (KD = ∼0.4 μm) to the purified cSH2 domain of PLC-γ1 (Fig. 5A) as measured by ITC. The phosphorylated peptide showed a clear preference for the cSH2 domain of PLC-γ1 (Fig. 5B) because the affinity of complex formation was ∼8-fold lower (KD ∼2.5 μm) if the peptide was titrated with a purified nSH2 domain under identical conditions. Conversely, the nSH2 domain of PLC-γ1 preferentially bound (KD = ∼0.4 μm) a relevant phosphorylated peptide of FGFR2 as opposed to the phosphorylated peptide of PLC-γ1 (Fig. 5C).
FIGURE 5.
Phosphorylated tyrosine 783 must interact with the C-terminal SH2 domain for activation of purified PLC-γ1. A–F, phosphorylation of Tyr783 mediates high affinity interaction with the C-terminal SH2 domain of PLC-γ1. Heats evolved from titration of individual SH2 domains of PLC-γ1 (nSH2 or cSH2) with indicated peptides of PLC-γ1 or FGFRK2 were measured with isothermal titration calorimetery and used to derive associated dissociation constants (KD). The range of residues are indicated for the peptides as well as phosphorylated (circled red P) and nonphosphorylated tyrosines (Y). The top panels show the base line-corrected heats of titration; the bottom panels show the integrated heat released as a function of the molar ratio of peptide titrated into the sample cell. The data were corrected for the heat of dilution of the peptide and subsequently fit to a one-site model using a nonlinear least squares algorithm to obtain the associated KD values. Measured thermodynamic parameters are given in supplemental Table S1. G, mutations within the phosphotyrosine-binding pocket of the C-terminal SH2 domain of PLC-γ1 prevent receptor-mediated lipase activation. PLC-γ1 was mutated to prevent binding of phosphotyrosine to either the N-terminal (nSH2*) or C-terminal (cSH2*) SH2 domain, purified, and incubated with an equimolar concentration of purified FGFR2K and [γ-32P]ATP. Aliquots of the reaction mixtures were subsequently quantified for: (i) lipase activity using phospholipid vesicles (left y axis) and (ii) incorporation of [γ-32P]phosphate in PLC-γ1 (right y axis) after SDS-PAGE and staining with Coomassie Brilliant Blue (lower panel). Specific activities are the means ± S.E. for three or more independent experiments and are normalized to wild-type activity. See also supplemental Table S1.
Phosphorylation of Tyr783 was required for association because the equivalent nonphosphorylated peptide exhibited no affinity for purified cSH2 domain under identical conditions (Fig. 5D). The equivalent peptide phosphorylated on Tyr775 (Fig. 5E) also bound to the cSH2 domain, but with an affinity (KD = ∼1.8 μm) 4–5-fold lower than observed with the peptide phosphorylated at Tyr783.
The isolated cSH2 domain of PLC-γ1 has the capacity to bind dually phosphorylated peptides as highlighted in the crystal structure of this domain bound to a dually phosphorylated peptide from the Syk tyrosine kinase (23). Although it is uncertain whether Syk engages PLC-γ1 in this manner under physiological conditions, a peptide of PLC-γ1 dually phosphorylated on Tyr775 and Tyr783 bound to the purified cSH2 domain of PLC-γ1 with an affinity (KD = ∼0.7 μm) intermediate between the singly phosphorylated forms of the peptide (Fig. 5F).
The results from these titration experiments are consistent with occurrence of an intramolecular association with the cSH2 domain of PLC-γ1 driven by tyrosine phosphorylation. This idea was directly tested using purified forms of PLC-γ1 mutated in either the N- or C-terminal SH2 domain to prevent the binding of phosphorylated tyrosines (Fig. 5G). The constitutively active kinase domain of FGFR2 enhanced the phospholipase activity of PLC-γ1 harboring a defective nSH2 domain by ∼10-fold, which is similar to the rate of enhancement shown earlier for wild-type PLC-γ1. In contrast, PLC-γ1 with a defective cSH2 domain was refractory to stimulation by the kinase domain of FGFR2. These results are consistent with the idea that phosphorylation of Tyr775 or Tyr783 prompts intramolecular association of this phosphorylated region with the cSH2 domain and that this association is required for enhanced phospholipase activity.
The peptide of PLC-γ1 containing phosphorylated Tyr783 bound to purified X/Y linker of PLC-γ1 with an affinity (KD = ∼3.5 μm) ∼10-fold lower than observed in equivalent titrations with the cSH2 domain (supplemental Fig. S4A). This result suggests that the interface within the cSH2 domain needed for engagement of the phosphorylated peptide is at least partially occluded within the larger X/Y linker. This conclusion is supported by the fact that it was not possible to use ITC to measure the affinity of the X/Y linker of PLC-γ1 for the corresponding peptide phosphorylated on Tyr775 (supplemental Fig. S4B).
Substantial Conformational Rearrangement of PLC-γ1 Is Linked to Phospholipase Activation
Poulin et al. (22) previously suggested that engagement of the cSH2 domain of PLC-γ1 by phosphorylated Tyr783 leads to a conformational rearrangement within PLC-γ1, but this proposal was based solely on differential retention of phosphorylated forms of PLC-γ1 during chromatography with a heparin matrix. To directly examine this concept, we applied SEC-MALS to measure potential conformational changes in PLC-γ1 upon its activation.
Phosphorylation of PLC-γ1 in vitro likely would produce a heterogeneous preparation containing isozyme with differing amounts and sites of phosphorylation that would confound biophysical analyses. Consequently, purified PLC-γ1Δ10 was assumed to recapitulate potential conformational changes within phosphorylated and activated PLC-γ1 and therefore was studied as a reasonable surrogate (Fig. 6). PLC-γ1 and PLC-γ1Δ10 were monodisperse with experimentally measured molecular masses equal to their calculated molecular masses (Fig. 6B), as expected for two proteins differing by less than 0.1% of total mass. In contrast, the hydrodynamic radius of PLC-γ1Δ10 was ∼6.5% larger than the equivalent value for PLC-γ1 (4.9 versus 4.6 nm), indicating that constitutively active PLC-γ1Δ10 exists in a substantively different conformation than auto-inhibited PLC-γ1.
FIGURE 6.
PLC-γ1 adopts an extended conformation upon activation. A, specific activity of either purified PLC-γ1 or the indicated deleted forms was measured in phospholipid vesicles. The relative specific activities are the means ± S.E. for three or more independent experiments and are normalized to wild-type activity. Note that the data for PLC-γ1 and PLC-γ1Δ10 are repeated from Fig. 2 and are shown here for comparative purposes. B and C, SEC-MALS analysis of either PLC-γ1Δ10 (B) or PLC-γ1Δ15 (C) compared with wild-type PLC-γ1. Molar mass (left panels; dark colored squares) and hydrodynamic radius (right panels; dark colored squares) of the indicated proteins were assessed by multi-angle light scattering as a function of elution volume (light colored curves; relative absorbance) after application to a Superdex 200 size exclusion column. The mean molecular masses (amounts in angle brackets) and hydrodynamic radii for elutions are color-coded to the indicated proteins. Note that in C, PLC-γ1Δ15 was generated within the context of full-length PLC-γ1 (1290 residues) and therefore compared with it.
Interestingly, the intact and Δ10 versions of the X/Y linker of PLC-γ1 retain identical hydrodynamic radii (and similar molecular masses) (supplemental Fig. S5), suggesting that the conformational rearrangement in PLC-γ1Δ10 relative to PLC-γ1 does not reflect gross rearrangement within the X/Y linker itself. Rather, the alteration of conformation derives from changes in the spatial rearrangement of the X/Y linker relative to the remainder of the protein. This result is also consistent with the previous circular dichroism studies indicating no change in the overall secondary structure of the X/Y linker upon introduction of Δ10 (supplemental Fig. S2D). PLC-γ1Δ15 lacks 15 residues adjacent to the deletion found in PLC-γ1Δ10 (Fig. 6A). However, PLC-γ1Δ15 remains auto-inhibited, and its hydrodynamic radius was unchanged relative to PLC-γ1 (Fig. 6C). Overall, these data strongly argue that the regulated phospholipase activity of PLC-γ1 is tightly linked to a substantial conformational rearrangement of PLC-γ1 with a transition from a “closed” basally inhibited state to an “open” activated state.
DISCUSSION
Model for the Activation of PLC-γ Isozymes
Although previous groups sought to study this mechanism using purified, recombinant PLC-γ isozymes, these studies were hampered by several limitations, including: (i) low yield (24) and specific activity (19) of purified PLC-γ isozymes after expression in E. coli; (ii) substoichiometric tyrosine phosphorylation of purified PLC-γ1 expressed in insect cells (15); and (iii) a predicted molecular mass based on gel exclusion chromatography of PLC-γ1 partially purified from eukaryotic cells that was approximately half the calculated mass (22). In contrast, we utilized highly purified proteins with accurate molecular mass and specific activity as well as robust biochemical and biophysical assays to delineate a definite and comprehensive model for the activation of PLC-γ isozymes.
We have convincingly identified the cSH2 domain within the highly elaborated X/Y linker as the primary structural determinant in the regulated auto-inhibition of PLC-γ isozymes. Moreover, specific deletion of 10 residues encompassing the highly mobile BG loop of the cSH2 domain is sufficient to constitutively activate PLC-γ isozymes (Figs. 2 and 3). In contrast, the other domains within the X/Y linker are ancillary to auto-inhibition of phospholipase activity (Fig. 2A). Activation is associated with an increase in the hydrodynamic radius of the full-length protein that cannot be recapitulated by introduction of an identical deletion into the isolated X/Y linker (Fig. 6B and supplemental Fig. S5). In a similar vein, the capacity of the cSH2 domain to bind phosphotyrosine peptides must be maintained to relieve this auto-inhibition (Fig. 5G), and phosphomimetic substitution of the physiologically relevant tyrosines within PLC-γ1 is insufficient to drive activation (Fig. 4C).
A simple model that fits these data posits a compact and basally inhibited state of PLC-γ isozymes mediated by the cSH2 domain directly engaging the phospholipase active site to block entrance of PtdIns(4,5)P2 substrate (Fig. 7). PLC-γ isozymes are recruited to membranes through docking of the nSH2 domain to a phosphotyrosine-containing region of a RTK or cytoplasmic tyrosine kinase, but simple recruitment is insufficient to release auto-inhibition and promote phospholipase activity. Instead, activation is coupled to phosphorylation, by the docked kinase, of specific tyrosines within the PLC-γ isozyme (e.g. Tyr783) that engage the cSH2 domain to disrupt its interactions with the catalytic TIM barrel and concomitantly free the active site for substrate entry. Disruption of interactions between the cSH2 domain and active site is associated with a large conformation change within the full-length PLC-γ isozyme that most likely reflects an alteration in the spatial disposition of the entire X/Y linker with respect to the remainder of the enzyme.
FIGURE 7.
Mechanism for phosphorylation-stimulated increase in PLC-γ1 lipase activity. In the absence of stimulus, the catalytic domain (gray doughnut) of PLC-γ1 is basally auto-inhibited by the cSH2 domain (purple cylinder). Activation of RTKs (gold) provides a phosphotyrosine (red dot) docking site for the nSH2 domain (blue cylinder) of PLC-γ1, thereby translocating PLC-γ1 from the cytosol to the plasma membrane. RTKs then phosphorylate PLC-γ1 at a specific tyrosine, which promotes direct interactions with the cSH2 domain. Engagement of the cSH2 domain by phosphorylated tyrosine induces structural rearrangements of the cSH2 domain with respect to the catalytic domain, leading to release of auto-inhibition and subsequent substrate hydrolysis.
The small GTPase, Rac2, directly binds the split PH domain of PLC-γ2 to enhance its phospholipase activity, and this activation does not require tyrosine phosphorylation of PLC-γ2 (25–27). Although deletion of the split PH domain of PLC-γ2 did not enhance its phospholipase activity in cells (Fig. 3), the equivalent deletion modestly activated PLC-γ1 in cells (Fig. 2A), and this effect was more pronounced with purified PLC-γ1 (Fig. 2B). Thus, the split PH domain is not essential for auto-inhibition of PLC-γ isozymes but apparently buttresses the position of the cSH2 domain for auto-inhibition. If true, Rac2 likely promotes activation of PLC-γ2 by influencing the position of the cSH2 domain through its interactions with the split PH domain.
The SHP phosphatases possess tandem SH2 domains that share substantial sequence homology (30% identity) with the equivalent portion of the PLC-γ isozymes. Indeed, detailed structural analysis of the SHP phosphatases (28, 29) highlights striking similarities to the model proposed here for the regulation of PLC-γ isozymes. The nSH2 domain of SHP phosphatases blocks the active site, leading to repression of phosphatase activity. Engagement of this domain by phosphotyrosine-containing regions within substrates and docking proteins causes structural changes within the nSH2 domain that concomitantly destabilize interactions with the active site to elevate phosphatase activity up to 10-fold (30). The two SH2 domains can simultaneously engage the same target substrate through separate phosphotyrosine-containing regions, in which case phosphatase activity is elevated greater than 100-fold (30). Therefore, the tandem SH2 domains of PLC-γ isozymes and SHP phosphatases perform dual functions: (i) they repress intrinsic catalytic activity and (ii) they integrate phosphorylation of specific tyrosines to the relief of auto-inhibition, leading to high catalytic activity that is preferentially localized in the vicinity of substrates.
Critical Role of the cSH2 Domain
The interface of the nSH2 domain of SHP-2 with its phosphatase domain is centered on the DE and FB loops of the SH2 domain (28). However, targeting the equivalent loops within the cSH2 domain of PLC-γ1 failed to identify sites critical for auto-inhibition of phospholipase activity, indicating that the two SH2 domains do not share the identical determinants necessary for auto-inhibition.
In contrast, we identified the BG loop of the cSH2 domain as a critical determinant for regulated auto-inhibition of PLC-γ isozymes (Figs. 2 and 3). Similar to many SH2 domains, the BG loop of the cSH2 domain of PLC-γ1 forms part of the groove needed to bind phosphotyrosine-containing peptides (31). Perhaps not surprisingly then, the BG loop of the cSH2 domain of PLC-γ1 alters its conformation upon peptide engagement, and different peptides stabilize distinct conformations of the BG loop (23, 32, 33). An attractive possibility is that changes in the conformation of the BG loop in response to binding phosphotyrosine-containing peptides are key to the release of auto-inhibition. In the simplest form of this scenario, the BG loop directly engages the catalytic TIM barrel to occlude the active site, and this occlusion is destabilized upon peptide engagement by the cSH2 domain. This idea is supported by the fact that removal of the BG loop induces a large conformational change within full-length PLC-γ1 that results in full constitutive activation of the phospholipase (Figs. 2 and 6). This conformational change is not observed if the BG loop is deleted within the context of the isolated X/Y linker, suggesting that conformational alterations upon the activation of full-length PLC-γ1 arise because of en bloc movement of the X/Y linker relative to the remainder of the enzyme (supplemental Fig. S5). This model does not preclude the possibility that other portions of the cSH2 domain participate in interactions with the TIM barrel to contribute to auto-inhibition.
Homma et al. (34) showed that fragments corresponding to the SH array of PLC-γ1 could inhibit several PLC isoforms (PLC-β1, -δ1, -γ1, and -γ2) and that this inhibition was noncompetitive using PLC-γ1. Further studies delimited a series of short peptides that required the BG loop for inhibition of PLC-γ1 in vitro and in vivo (35). These results were interpreted to suggest that the peptides directly interacted with the active site of the PLC isozymes. Our results are consistent with the possibility of the BG loop directly engaging the active site of PLC-γ isozymes, but it is difficult to envision how peptides of the BG loop of PLC-γ1 could inhibit multiple PLC isoforms unless the site of interaction is highly conserved among all PLCs.
Differential Phosphorylation
The differential roles of tyrosines 775 and 783 of PLC-γ1 in regulating phosphorylation-induced activation are superficially perplexing. That is, multiple lines of evidence indicate that both tyrosines (11), or the equivalent sites (positions 753 and 759) in PLC-γ2 (17–20), can be functionally redundant in phosphorylation-dependent activation of PLC-γ isozymes. Indeed, peptides of PLC-γ1 phosphorylated at either site bound to the isolated cSH2 domain of PLC-γ1, albeit with a 5-fold difference in affinity (Fig. 5). However, when the kinase domain of FGFR2 was used to phosphorylate PLC-γ1, substitution of Tyr783, but not Tyr775, resulted in a PLC-γ1 mutant refractory to activation (Fig. 4B). One possible explanation for these results is that physiologically relevant kinases exhibit a differential capacity to phosphorylate Tyr775 and Tyr783 in PLC-γ1. Indeed, Tyr775 of PLC-γ1, or its equivalent in PLC-γ2 (Tyr753), has been implicated in regulating PLC-γ isozymes downstream of immune receptor signaling controlled by cytosolic tyrosine kinases. To the best of our knowledge, neither Tyr775 of PLC-γ1 nor Tyr753 of PLC-γ2 has been shown to be necessary for activation downstream of RTKs, including FGFR2.
The possibility that multiple tyrosines within a PLC-γ isozyme fulfill important physiological functions should not be entirely dismissed. That is, we illustrated that a peptide from PLC-γ1 phosphorylated at both Tyr775 and Tyr783 bound essentially as well to the cSH2 domain of PLC-γ1 as the equivalent peptide phosphorylated at Tyr783 alone (Fig. 5), and this dual phosphorylation might be expected to have substantial regulatory ramifications. For example, a dually phosphorylated peptide of Syk bound to the cSH2 domain of PLC-γ1 elicited large changes in the BG loop of the cSH2 domain (23) in comparison with this domain alone or the domain bound to a singly phosphorylated peptide of PDGF (32, 33). These results suggest that dual tyrosine phosphorylation might elicit differential levels of phospholipase activation because of differential structural responses. Alternatively, the SH2 domain of the adapter protein, APS, binds dually phosphorylated regions of certain kinases to sustain signaling through enhanced protection to dephosphorylation (36). Similar alterations in PLC-γ activity might also depend on the sites and number of phosphorylations within the X/Y linker.
Core Regulation
We previously proposed a general model for the regulated activation of PLC isozymes auto-inhibited by mostly disordered X/Y linkers (4). In contrast, the PLC-γ isozymes, with their highly elaborated X/Y linkers containing multiple domains, suggest a fundamentally different form of regulation. Nevertheless, these structural differences mask fundamental auto-inhibition by the X/Y linker preserved within the entire PLC family. However, despite shared auto-inhibition, the PLC-γ isozymes differ dramatically in their mechanism of activation. This understanding is reinforced with the knowledge that PLC-γ isozymes, unlike other PLCs, are activated upon phosphorylation of specific tyrosines within their X/Y linkers. Therefore, PLC-γ isozymes evolved complex X/Y linkers to preserve basal auto-inhibition while coupling phospholipase activation to tyrosine phosphorylation.
Supplementary Material
Acknowledgments
We thank Dr. Janeen L. Vanhooke for generating a protocol for protein expression in P. pastoris, Mary Onorati for assistance with alanine scanning mutagenesis, as well as members from the Sondek and Harden laboratory for many helpful discussions and suggestions. We are indebted to Eric Zimmerman and Dr. Lee Graves for assistance with the in vitro kinase assay and Dr. Ashutosh Tripathy, director of the Macromolecular Interaction Facility at the University of North Carolina at Chapel Hill for technical expertise in the ITC, SEC-MALS, and CD instruments.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-GM57391 (to J. S. and T. K. H.). This work was also supported by American Heart Association Pre-doctoral Fellowship AHA 0715332U (to A. G.).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Table S1, and Figs. S1–S5.
- PLC
- phospholipase C
- PtdIns(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- TIM
- triose phosphate isomerase
- FGFR
- fibroblast growth factor receptor
- SEC-MALS
- size exclusion chromatography-multi angle light scattering
- ITC
- isothermal titration calorimetry
- RTK
- receptor tyrosine kinase
- nSH2
- N-terminal SH2
- cSH2
- C-terminal SH2
- sPH
- split pleckstrin homology.
REFERENCES
- 1.Harden T. K., Sondek J. (2006) Annu. Rev. Pharmacol. Toxicol. 46, 355–379 [DOI] [PubMed] [Google Scholar]
- 2.Rhee S. G. (2001) Annu. Rev. Biochem. 70, 281–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Essen L. O., Perisic O., Cheung R., Katan M., Williams R. L. (1996) Nature 380, 595–602 [DOI] [PubMed] [Google Scholar]
- 4.Hicks S. N., Jezyk M. R., Gershburg S., Seifert J. P., Harden T. K., Sondek J. (2008) Mol. Cell 31, 383–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horstman D. A., Chattopadhyay A., Carpenter G. (1999) Arch. Biochem. Biophys. 361, 149–155 [DOI] [PubMed] [Google Scholar]
- 6.Fernald A. W., Jones G. A., Carpenter G. (1994) Biochem. J. 302, 503–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horstman D. A., DeStefano K., Carpenter G. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 7518–7521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones G. A., Wu Y. (2000) Arch. Biochem. Biophys. 375, 229–239 [DOI] [PubMed] [Google Scholar]
- 9.Wahl M. I., Nishibe S., Kim J. W., Kim H., Rhee S. G., Carpenter G. (1990) J. Biol. Chem. 265, 3944–3948 [PubMed] [Google Scholar]
- 10.Kim H. K., Kim J. W., Zilberstein A., Margolis B., Kim J. G., Schlessinger J., Rhee S. G. (1991) Cell 65, 435–441 [DOI] [PubMed] [Google Scholar]
- 11.Serrano C. J., Graham L., DeBell K., Rawat R., Veri M. C., Bonvini E., Rellahan B. L., Reischl I. G. (2005) J. Immunol. 174, 6233–6237 [DOI] [PubMed] [Google Scholar]
- 12.Cha J. Y., Maddileti S., Mitin N., Harden T. K., Der C. J. (2009) J. Biol. Chem. 284, 6227–6240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen H., Ma J., Li W., Eliseenkova A. V., Xu C., Neubert T. A., Miller W. T., Mohammadi M. (2007) Mol. Cell 27, 717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryu S. H., Suh P. G., Cho K. S., Lee K. Y., Rhee S. G. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 6649–6653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horstman D. A., Ball R., Carpenter G. (1995) Protein Expr. Purif. 6, 278–283 [DOI] [PubMed] [Google Scholar]
- 16.Bae J. H., Lew E. D., Yuzawa S., Tomé F., Lax I., Schlessinger J. (2009) Cell 138, 514–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez R., Matsuda M., Perisic O., Bravo J., Paul A., Jones N. P., Light Y., Swann K., Williams R. L., Katan M. (2001) J. Biol. Chem. 276, 47982–47992 [DOI] [PubMed] [Google Scholar]
- 18.Watanabe D., Hashimoto S., Ishiai M., Matsushita M., Baba Y., Kishimoto T., Kurosaki T., Tsukada S. (2001) J. Biol. Chem. 276, 38595–38601 [DOI] [PubMed] [Google Scholar]
- 19.Ozdener F., Dangelmaier C., Ashby B., Kunapuli S. P., Daniel J. L. (2002) Mol. Pharmacol. 62, 672–679 [DOI] [PubMed] [Google Scholar]
- 20.Humphries L. A., Dangelmaier C., Sommer K., Kipp K., Kato R. M., Griffith N., Bakman I., Turk C. W., Daniel J. L., Rawlings D. J. (2004) J. Biol. Chem. 279, 37651–37661 [DOI] [PubMed] [Google Scholar]
- 21.Mohammadi M., Honegger A. M., Rotin D., Fischer R., Bellot F., Li W., Dionne C. A., Jaye M., Rubinstein M., Schlessinger J. (1991) Mol. Cell. Biol. 11, 5068–5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poulin B., Sekiya F., Rhee S. G. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4276–4281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groesch T. D., Zhou F., Mattila S., Geahlen R. L., Post C. B. (2006) J. Mol. Biol. 356, 1222–1236 [DOI] [PubMed] [Google Scholar]
- 24.Koblan K. S., Schaber M. D., Edwards G., Gibbs J. B., Pompliano D. L. (1995) Biochem. J. 305, 745–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piechulek T., Rehlen T., Walliser C., Vatter P., Moepps B., Gierschik P. (2005) J. Biol. Chem. 280, 38923–38931 [DOI] [PubMed] [Google Scholar]
- 26.Walliser C., Retlich M., Harris R., Everett K. L., Josephs M. B., Vatter P., Esposito D., Driscoll P. C., Katan M., Gierschik P., Bunney T. D. (2008) J. Biol. Chem. 283, 30351–30362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bunney T. D., Opaleye O., Roe S. M., Vatter P., Baxendale R. W., Walliser C., Everett K. L., Josephs M. B., Christow C., Rodrigues-Lima F., Gierschik P., Pearl L. H., Katan M. (2009) Mol. Cell 34, 223–233 [DOI] [PubMed] [Google Scholar]
- 28.Hof P., Pluskey S., Dhe-Paganon S., Eck M. J., Shoelson S. E. (1998) Cell 92, 441–450 [DOI] [PubMed] [Google Scholar]
- 29.Yang J., Liu L., He D., Song X., Liang X., Zhao Z. J., Zhou G. W. (2003) J. Biol. Chem. 278, 6516–6520 [DOI] [PubMed] [Google Scholar]
- 30.Eck M. J., Pluskey S., Trüb T., Harrison S. C., Shoelson S. E. (1996) Nature 379, 277–280 [DOI] [PubMed] [Google Scholar]
- 31.Kaneko T., Huang H., Zhao B., Li L., Liu H., Voss C. K., Wu C., Schiller M. R., Li S. S. (2010) Sci. Signal 3, ra34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., Kay L. E. (1994) Biochemistry 33, 5984–6003 [DOI] [PubMed] [Google Scholar]
- 33.Pascal S. M., Singer A. U., Gish G., Yamazaki T., Shoelson S. E., Pawson T., Kay L. E., Forman-Kay J. D. (1994) Cell 77, 461–472 [DOI] [PubMed] [Google Scholar]
- 34.Homma Y., Takenawa T. (1992) J. Biol. Chem. 267, 21844–21849 [PubMed] [Google Scholar]
- 35.Homma M. K., Homma Y., Yamasaki M., Ohmi-Imajoh S., Yuasa Y. (1996) Cell Growth Differ. 7, 281–288 [PubMed] [Google Scholar]
- 36.Hu J., Liu J., Ghirlando R., Saltiel A. R., Hubbard S. R. (2003) Mol. Cell 12, 1379–1389 [DOI] [PubMed] [Google Scholar]
- 37.Eck M. J., Shoelson S. E., Harrison S. C. (1993) Nature 362, 87–91 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.