Abstract
Some organisms are able to survive the loss of almost all their body water content, entering a latent state known as anhydrobiosis. The sleeping chironomid (Polypedilum vanderplanki) lives in the semi-arid regions of Africa, and its larvae can survive desiccation in an anhydrobiotic form during the dry season. To unveil the molecular mechanisms of this resistance to desiccation, an anhydrobiosis-related Expressed Sequence Tag (EST) database was obtained from the sequences of three cDNA libraries constructed from P. vanderplanki larvae after 0, 12, and 36 h of desiccation. The database contained 15,056 ESTs distributed into 4,807 UniGene clusters. ESTs were classified according to gene ontology categories, and putative expression patterns were deduced for all clusters on the basis of the number of clones in each library; expression patterns were confirmed by real-time PCR for selected genes. Among up-regulated genes, antioxidants, late embryogenesis abundant (LEA) proteins, and heat shock proteins (Hsps) were identified as important groups for anhydrobiosis. Genes related to trehalose metabolism and various transporters were also strongly induced by desiccation. Those results suggest that the oxidative stress response plays a central role in successful anhydrobiosis. Similarly, protein denaturation and aggregation may be prevented by marked up-regulation of Hsps and the anhydrobiosis-specific LEA proteins. A third major feature is the predicted increase in trehalose synthesis and in the expression of various transporter proteins allowing the distribution of trehalose and other solutes to all tissues.
Keywords: Antioxidant, Chaperone Chaperonin, Oxidative Stress, Sugar Transport, Water Channel, Anhydrobiosis, Desiccation Stress, Gene Expression
Introduction
Water is essential for life, and terrestrial organisms have developed various strategies to avoid water loss, which would ultimately be fatal. Small organisms are particularly sensitive to desiccation due to their relatively large body-surface-to-volume ratio (1, 2). Under extreme conditions, such as drought or in arid environments, animals may avoid desiccation in a behavioral manner (by migration or burying deeply in the soil) or physiologically (by entering into diapause, or estivation). Some aquatic insects, which can resist moderate desiccation, secrete a thick cuticle, thus limiting the rate of water loss (3, 4). Alternatively, various organisms have developed the opposite strategy; they allow water loss relatively easily but are able to survive essentially complete desiccation (4). This phenomenon, known as anhydrobiosis, is a specific example of the more general cryptobiosis, which is an ametabolic state induced by various adverse environmental conditions, such as desiccation, low temperature, or lack of oxygen. Cryptobiosis was first defined by Keilin (5), although the observation of bdelloid rotifers resurrecting from dry mud samples had already been described in the early 18th century. Anhydrobiosis represents an extreme case of dormancy during which the water content falls to less than 3% of the body weight and no metabolic activity can be detected (6, 7). However, anhydrobiotic individuals can revive and rapidly resume activity once in contact with water (8). The most familiar cases of anhydrobiosis are plant seeds or fungal spores, but many microscopic organisms, such as terrestrial tardigrades, bdelloid rotifers, or nematodes, can survive in an anhydrobiotic state (5, 8, 9). For plant seeds or the cysts of some crustacean embryos (Artemia, Triops), anhydrobiosis is only observed in early developmental stages (10). In contrast, tardigrades and rotifers enter anhydrobiosis also during postembryonic stages in response to environmental change and can experience anhydrobiosis several times successively (11, 12). This is also the case for the sleeping chironomid (Polypedilum vanderplanki), the largest anhydrobiotic animal known to date. However, in P. vanderplanki, only the larval stages are able to enter anhydrobiosis; the other stages (eggs, pupae, adults) last only a couple of days, and such a rapid development is more adapted to a strategy of desiccation avoidance.
P. vanderplanki lives in small temporary rock pools in the semi-arid regions of the African continent (13). Its larvae are found in small tubular nests in the mud at the bottom of pools, and when the water dries up, they can survive in an almost completely desiccated anhydrobiotic state throughout the dry season, which may last up to 8 months (14, 15). To achieve successful anhydrobiosis, P. vanderplanki larvae need an appropriate desiccation rate and, experimentally, at least 2 days are required for the necessary physiological adjustments; appropriate conditions for successful anhydrobiosis have been reproduced in the laboratory (4, 7, 15). Viably desiccated larvae will recover normal activity within 1 h of contact with water (see Fig. 1). The mechanism of anhydrobiosis induction in P. vanderplanki was found to be independent of any control by the central nervous system or endocrine influence (16), and salt concentration apparently plays an important role in the induction of this phenomenon (7). Intriguingly, anhydrobiosis can even be successfully induced in isolated fat body tissue (17). As observed in many other anhydrobionts (18–20) with the exception of bdelloid rotifers and some tardigrades (21, 22), the accumulation of trehalose in the tissues of P. vanderplanki has a central protective role against desiccation stress, and the induction period of 2 days leading up to successful anhydrobiosis is thus necessary for appropriate levels of trehalose accumulation in the larval body (7, 15, 16). During this induction phase, trehalose is thought first to act as a water replacement molecule and then to form a glassy matrix, which protects cells and their biomolecules against desiccation stress (23).
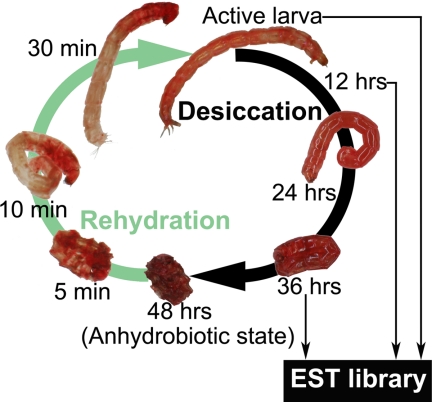
FIGURE 1.
Anhydrobiotic cycle of P. vanderplanki larvae. In laboratory conditions, the process of desiccation lasts 48 h, after active larvae have been removed from water. Water loss accelerates after 24 h of desiccation treatment, at which time trehalose accumulates in the body of larvae. After 48 h of desiccation, the larvae reach the anhydrobiotic state and can maintain this state for several months and even years. Once larvae are immersed in water, the process of rehydration takes place rapidly. The anhydrobiotic larvae quickly absorb water, and muscular contractions can be observed after a few minutes. They recover their original active state about 20 min to 1 h after the beginning of rehydration. In the present study, mRNAs used to construct the anhydrobiosis-related EST library were collected from active larvae and from individuals at 12 and 36 h after the beginning of desiccation treatment.
Apart from the important role of trehalose, the molecular mechanisms of anhydrobiosis are still poorly understood. Molecular studies led to the identification in nematodes and in a rotifer of proteins related to desiccation tolerance, called late embryogenesis abundant (LEA)4 proteins, which were originally known only in plants (22, 24, 25). Anhydrobiosis-related gene expression has been also investigated in a brine shrimp (26, 27) and a rotifer species, allowing the identification of desiccation-responsive genes such as LEA proteins, heat shock proteins, and some antioxidants. However, more information about the molecular mechanisms of desiccation resistance is needed to fully understand the anhydrobiosis phenomenon. The construction of an anhydrobiosis-related EST library in P. vanderplanki (see Fig. 1) has already led to the characterization of genes involved in the desiccation tolerance of this chironomid, such as LEA proteins (28), the trehalose transporter Tret1 (29), or aquaporins (30). The present work describes the construction and general analysis of this anhydrobiosis-related EST database. Many genes involved in the protection of cellular components and biomolecules, such as chaperone proteins and enzymes governing trehalose metabolism, were found to be up-regulated during the process of desiccation, and the dataset also provides evidence that the response to oxidative stress appears to play a central role in anhydrobiosis.
EXPERIMENTAL PROCEDURES
Insect Rearing and Induction of Anhydrobiosis
Anhydrobiotic larvae of P. vanderplanki were collected from temporary rock pools in Nigeria in 2000. The larvae were reared on milk agar under a 13:11-h light:dark photoregime at 27–28 °C, and these insects were kept in the laboratory for successive generations according to procedures described previously (16). The procedure for slow desiccation to induce anhydrobiosis was performed as described previously (15, 16).
EST Library Construction and Sequence Processing
Total RNAs were obtained from the whole body of third instar larvae 0 (hydrated and active larvae), 12, and 36 h after the beginning of the desiccation process (see Fig. 1). Independent cDNA libraries for the three groups (named pvD00, pvD12, and pvD36, respectively) were obtained as described in the supplemental Experimental Procedures. After sequencing each clone in both 5′- and 3′-directions, the resulting ESTs were subjected to processing and clustering as described elsewhere (31). Detailed procedures are provided in supplemental Experimental Procedures.
Gene annotation was performed by sequence similarity comparison, using BLASTX searches against protein sequence databases at the National Center for Biotechnology Information (NCBI), FlyBase, and WormBase. In addition, gene ontology assignments were obtained by scanning EST sequences with InterProScan (supplemental Experimental Procedures).
EST Database Analysis
First, ESTs were classified as a function of their gene ontology, and the most representative gene ontology groups were shown as a percentage of the total EST number in each library (pvD00, pvD12, pvD36). Then, keyword searches in the database for specific gene families or gene functions were performed, and this allowed us to classify gene clusters into each of these keyword groups as a function of their EST representation pattern. Finally, assuming that the number of ESTs for each gene cluster is representative of its expression level, gene clusters were classified as a function of their expression pattern (supplemental Experimental Procedures) in the three time point-specific libraries (pvD00, pvD12, and pvD36), using the Macro function of the Visual Basic Editor of the Excel software (Microsoft Corp.). A ranking of the most strongly induced/inhibited gene clusters was finally obtained.
Real-time Quantitative PCR
Relative expression of selected genes (PvLea1, PvTps, PvAqp1, PvTret1, PvGlobin2, PvHbCTT6, PvTrx2, and PvDip1; primers are listed in supplemental Table S2) was investigated by real-time quantitative PCR in wet and desiccating larvae (at 8, 16, 24, and 48 h after the beginning of desiccation treatment). Results were normalized with P. vanderplanki ribosomal protein L32 (PvRpl32; accession number AB244986) or elongation factor 1-α (PvEf1-α; accession number AB490338) as internal controls (see supplemental Experimental Procedures for details).
Statistical Analysis
The changes in the representation of each gene ontology group and each functional family group throughout the three libraries were statistically analyzed by the χ-square test with a confidence interval of 95%. Analyses were performed with the Prism 4.0 software for Macintosh (GraphPad Software, Inc.).
RESULTS AND DISCUSSION
Anhydrobiosis-related EST Libraries
Three independent EST libraries were constructed during the course of anhydrobiosis induction in P. vanderplanki as follows. ESTs were obtained from wet and active larvae (pvD00), from larvae taken 12 h after the beginning of desiccation treatment (pvD12), and from larvae desiccated for 36 h (pvD36), respectively (Fig. 1). After sequence processing, assembly into gene clusters, grouping related contigs, and gene annotation by BLASTX analysis, an anhydrobiosis-related EST database (NEMURIYUSURIKA (Chironomidae) EST database)5 was constructed on the basis of the clustering results from all three libraries. A total of 22,272 sequences were originally obtained from 10,944 clones (pvD00 comprised 4,800 clones; pvD12 and pvD36 both consisted of 3,072 clones). After trimming and removal of contaminating sequences, the remaining 15,056 ESTs averaged 491 bp in size, with a distribution ranging from 300 to 737 bp (supplemental Fig. S1). ESTs were distributed into 4,807 gene clusters, each of which was given name of the most representative clone and added to the database. ESTs were distributed as follows: 7,186 ESTs (2,795 clusters) for the pvD00 library, 3,760 ESTs (1,675 clusters) for the pvD12 library, and 4,110 ESTs (1,725 clusters) for the pvD36 library.
General Features of the EST Libraries and Gene Ontology Analysis
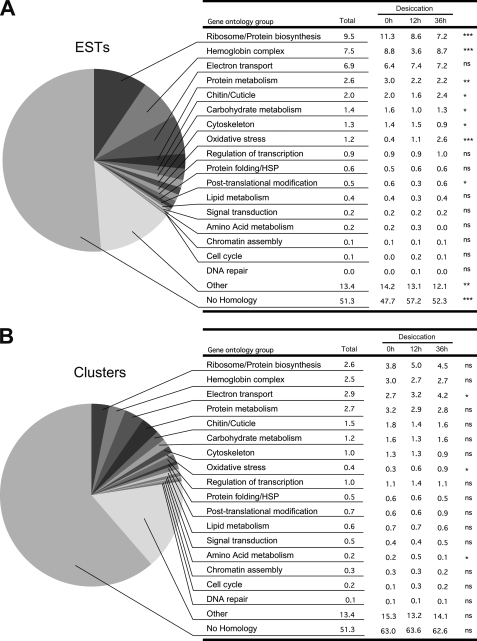
All EST sequences were subjected to similarity search against databases. About one-half of all sequences showed no similarity to any known gene from the three databases investigated (NCBI/nr, FlyBase, WormBase). A substantial proportion of these ESTs may correspond to 3′-UTR sequences. However, the analysis restricted to clusters containing only 5′-ESTs also showed a large proportion (55.8%) of unknown sequences, which may thus correspond to new genes. ESTs and clusters were classified into major groups defined by gene ontology keywords (Fig. 2, A and B, respectively). The three largest groups were related to protein synthesis or ribosome proteins (9.5% of the total number of ESTs), hemoglobin (7.5%), and electron transport (6.9%). These three groups show a large number of ESTs per cluster (Fig. 2A versus Fig. 2B). These data are comparable with other insect EST libraries (32–34), but the high proportion of hemoglobin-related genes is characteristic of chironomid larvae, which often live in oxygen-poor environments (35, 36).
FIGURE 2.
EST clones classified by gene ontology. A and B, general proportions of ESTs (A) and clusters (B) for each ontology group in the whole database. Details of the percentages of ESTs and clusters for each ontology group in the three libraries (desiccation 0, 12, and 36 h) are shown in the tables on the right. Statistical significance (χ-square test) of the proportion changes in the three libraries is shown on the right side (ns, non significant; *, p < 0.05; **, p < 0.001; ***, p < 0.0001).
The relative proportions of each ontology group varied between the three libraries, and groups that showed increasing proportions of ESTs with desiccation time were potentially induced during anhydrobiosis. The oxidative stress response (Fig. 2A) appeared to be the most strongly induced group (p < 0.0001). Significant increase (p = 0.0244) was also observed for this group at the cluster level (Fig. 2B). Representation of the electron transport gene ontology group also increased significantly (from 2.7 to 4.2%, p = 0.0229) at the cluster level during desiccation. The number of ESTs showing no similarity to sequences from available databases also increased significantly on drying, suggesting that genes specific to P. vanderplanki may be involved in the process of anhydrobiosis.
Deduced Gene Expression Patterns during the Induction of Anhydrobiosis
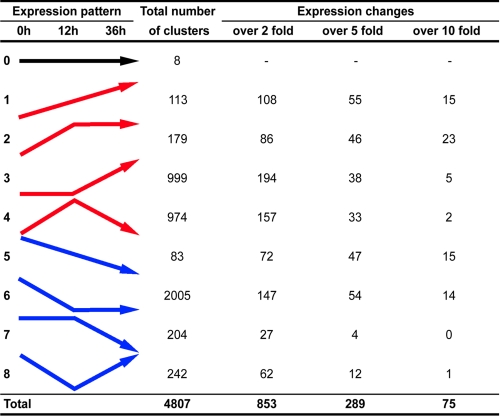
On the basis of the normalized number of ESTs throughout the three libraries, gene clusters were classified into different groups according to expression pattern during desiccation (Fig. 3). Most clusters were only represented by one or two ESTs, and differences in expression less than 2-fold were observed in the majority of cases. Such a poor clone representation did not permit correct assessment of actual expression patterns. Thus, after excluding those clusters with little variation (Fig. 3, over 2 fold), repression of gene expression was predominant during the first 12 h of desiccation treatment (147 clusters), whereas expression changes specific to the later part of desiccation were predominantly represented by up-regulation (194/853 clusters).
FIGURE 3.
General expression patterns. EST clusters were classified on the basis of their putative expression pattern, deduced from the changes in EST numbers for each cluster during desiccation. Nine expression patterns were defined for genes expressed constantly (pattern 0), up-regulated (patterns 1, 2, 3, and 4), or down-regulated (patterns 5, 6, 7, and 8). Further ranking of each expression pattern was established, based on the greatest amplitude of EST number variation, as compared with the value at 0 h (indicated by over 2 fold, over 5 fold, and over 10 fold).
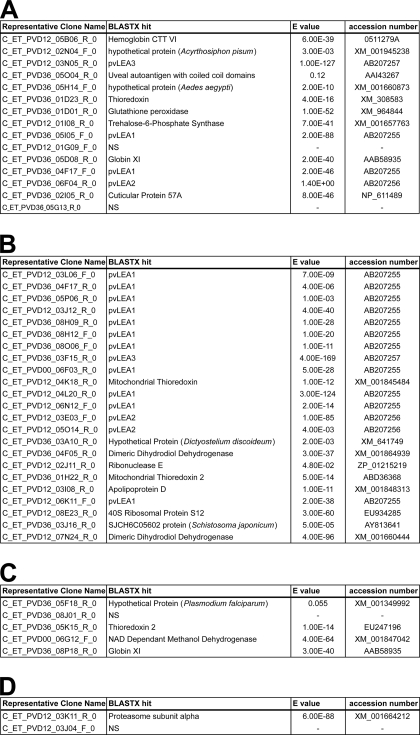
Although some housekeeping genes, such as those encoding actin, ribosomal proteins, globins, or cytochrome oxidases, were mainly down-regulated during the process of desiccation (p = 0.014, p < 0.0001, p < 0.0001 and p < 0.0001, respectively; supplemental Fig. S2, supplemental Table S1, and Fig. 3, patterns 5 and 6), genes involved in the process of anhydrobiosis were more likely to be up-regulated following desiccation. Thus, we focused on the clusters showing an increase in clone representation of over 10-fold (Table 1). Among the clusters showing a constant increase in representation with drying time (Fig. 3, pattern 1), a homolog of hemoglobin CTT VI featured strongly (Table 1A). Many LEA protein ESTs featured also in this category, and they monopolized the upper ranking of the strongly enhanced clusters (Table 1B). Antioxidant genes, such as thioredoxins and glutathione peroxidase, were highly up-regulated (Table 1, A–C). Finally, a non-negligible number of highly up-regulated clusters corresponded to unknown genes, such as the coiled protein (predicted on the Phyre server), which we have named desiccation-induced protein 1 (PvDip1) and which gave only a poor match with a uveal autoantigen sequence (Table 1A; clone C_ET_PVD36_05O04_R_0).
TABLE 1.
Clusters most strongly up-regulated (over 10-fold) during desiccation
All clusters are identified by the name of their representative clone. For each cluster, the closest informative BLASTX hit is given with the corresponding E-value and accession number from NCBI (search performed on June 2008). A, clusters continuously up-regulated throughout desiccation treatment (cf. Fig. 3, pattern 1). B, clusters up-regulated during the first 12 h of desiccation (cf. Fig. 3, pattern 2). C, clusters up-regulated between 12 and 36 h of desiccation (cf. Fig. 3, pattern 3). D, clusters up-regulated during the first 12 h of treatment and then down-regulated (cf. Fig. 3, pattern 4).
Validation of the Expected Expression Patterns for Selected Genes
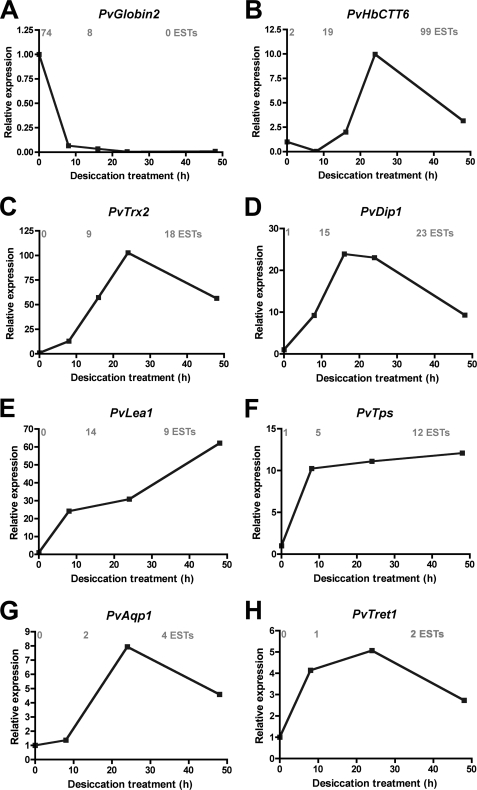
General expression profiles deduced from the database were largely confirmed for most genes investigated. About 87% of the genes for which expression patterns were checked had profiles congruent with the clone distribution in the libraries. The remaining 13% corresponded to genes with poor representation in the database (only one or two ESTs). Although the expression of PvGlobin2 was strongly down-regulated during desiccation (Fig. 4A), genes potentially involved in anhydrobiosis were rapidly induced by desiccation (Fig. 4, B–H), as expected from their EST representation.
FIGURE 4.
Relative expression patterns of selected genes. Expression levels were obtained by real-time quantitative PCR during the first 48 h of desiccation for globin 2 (A), hemoglobin CTT6 (B), thioredoxin 2 (C), desiccation-inducible protein 1 (D), LEA protein 1 (E), trehalose-6-phosphate synthase (F), aquaporin 1 (G), and trehalose transporter 1 (H). The corresponding accession numbers were respectively AB513664, AB513663, AB513662, AB513665, AB207255, AB490332, AB281619, and AB272983. Relative expression levels were calculated relative to elongation factor 1 expression (A–D) or ribosomal protein L32 expression (E–H) and were calibrated using the expression level at 0 h of desiccation treatment as 1.0. The numbers of ESTs at 0, 12, and 36 h of desiccation treatment in the corresponding clusters from the database are written in gray over each graph.
General Expression Patterns of Gene Families or Functional Groups
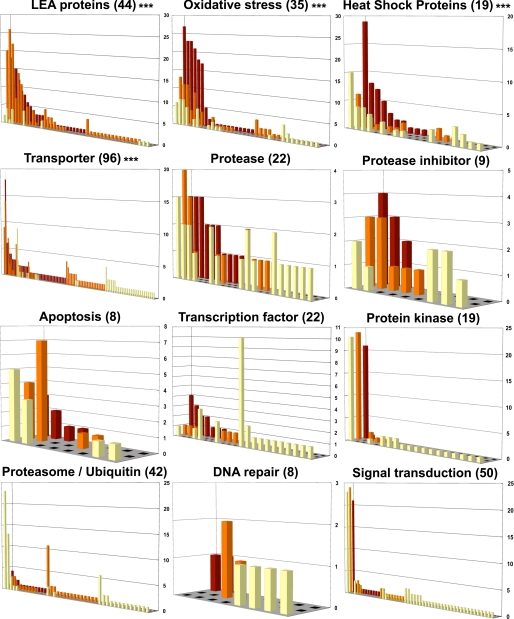
Clusters belonging to specific gene families or functional groups were extracted from the database by keyword search. For each keyword, clusters were classified as a function of their putative expression pattern (number of ESTs). The clusters corresponding to LEA proteins, heat shock proteins (Hsps), genes related to oxidative stress, and transporter proteins showed general induction following desiccation (Fig. 5, p < 0.0001). General gene repression on desiccation was observed for the protein kinase (Fig. 5) actin (p = 0.014) and ribosomal protein (p < 0.0001) groups (supplemental Fig. S2 and supplemental Table S1). For most of the other investigated groups, no clear relation was found between desiccation and the general expression profile (Fig. 5). In the following sections, we focused on the groups showing major transcriptional changes associated with the induction of anhydrobiosis.
FIGURE 5.
Expression profiles of some selected groups of clusters, defined by functional keywords. x-axis, cluster numbers; y-axis, number of ESTs in each cluster; z-axis, expression pattern, represented by the number of ESTs at 0 (yellow bars), 12 (orange bars), and 36 h (red bars) after the beginning of the desiccation treatment. The total number of clusters in each group is indicated in parentheses. Statistical significance (χ-square test) of the proportion changes in the three libraries is shown on the right (***, p < 0.0001).
Oxidative Stress Response
The up-regulation of antioxidant genes during the desiccation process was the most obvious phenomenon observed at all levels in the database (Figs. 2 and 5; Table 1). Desiccation stress is in general accompanied by oxidative stress because the loss of water from any tissue will result in the production of reactive oxygen species (ROS) through enzyme dysfunctions or chemical reactions (37, 38). An excess of ROS in turn generates lipid peroxidation, denaturation of proteins, and nucleic acid damage (38).
In our database, the most strongly induced antioxidant gene was mitochondrial thioredoxin (Fig. 4 and Table 1). Mitochondria represent one of the major sources of ROS production, and the thioredoxin system is the first line of defense against oxidative stress. Reduced thioredoxin can directly scavenge ROS, and the resulting oxidized form of thioredoxin is subsequently reduced by thioredoxin reductase (also induced by desiccation in our database) and NADPH (39, 40). Thioredoxins also prevent damage to proteins, which often occurs during desiccation-associated oxidative stress (38, 41). Interestingly, the strong up-regulation of the proteasome subunit α, observed at 12 h of desiccation (Table 1D), may also be related to the degradation of carbonylated proteins, products of oxidative stress.
The most highly represented antioxidants in the database were glutathione peroxidase homologs, which were also strongly induced by desiccation (Table 1). The glutathione antioxidant system is the most abundant intracellular thiol-based protectant. Toxic hydrogen peroxide (38) and other ROS can be directly neutralized by glutathione through the reaction catalyzed by glutathione peroxidase (40–42). Glutathione peroxidase is especially involved in the protection of biological membranes by reducing the membrane hydroperoxides (43). Several isoforms of glutathione S-transferase (GST) were also present in our anhydrobiosis-related database. This enzyme is generally involved in the detoxification of xenobiotics, but it was also suggested that GST is induced by oxidative stress and helps in the detoxification of end products of lipid peroxidation (38, 44).
Another pathway of ROS detoxification comprises superoxide dismutases, which transform O2-derived free radicals into hydrogen peroxide (H2O2). Hydrogen peroxide will in turn be reduced to a water molecule by catalase (38, 39, 41). Both mitochondrial manganese-superoxide dismutase (one cluster) and cytosolic/extracellular copper/zinc-superoxide dismutase (six clusters) were found in the anhydrobiosis-related EST database. A catalase homolog was also induced about 4-fold during desiccation.
All the antioxidant genes listed here belong to well known ROS detoxification systems, which are widespread in nature. These general antioxidant systems are strongly induced by desiccation in various anhydrobiotes, with different emphasis among species on the superoxide dismutase/catalase pathway or the glutathione system, for example (38, 45). Transcriptomic and proteomic analyses also revealed such antioxidant systems in nematodes and in tardigrades (46, 47). The antioxidant response during desiccation is best characterized in yeast (38), but an EST analysis approach in a desiccation-resistant monogonont rotifer has recently revealed up-regulation of peroxiredoxins, superoxide dismutases, and many GSTs in resting eggs (43). One unique feature of P. vanderplanki anhydrobiosis seems to be involvement of the thioredoxin system, which has not yet been reported in other anhydrobionts to our knowledge (38). Thus, desiccating P. vanderplanki larvae apparently employ a first line of defense against ROS directly in mitochondria and use other general antioxidant systems for the protection of cytoplasmic proteins and phospholipid membranes.
LEA Proteins
Along with antioxidant genes, LEA protein genes were markedly induced during the desiccation of P. vanderplanki larvae (Fig. 5 and Table 1). LEA proteins represented 0.76% of all the clusters in the database, but the corresponding ESTs increased in proportion from 0.67% in wet larvae up to 11.9% of all the ESTs in the database after 12 h of desiccation. Thus, the expression of LEA proteins accounted for a large part of the total amount of transcripts in desiccating larvae.
LEA proteins were first discovered in orthodox plant seeds during the late stages of embryonic development and are associated with desiccation tolerance. They are also found in the tissues of drought-resistant resurrection plants, and they have been suggested to play important roles as molecular chaperones, ion scavengers, or cytoskeletal components (48). LEA proteins were considered for a long time to be plant-specific proteins, but the first animal LEA protein was recently discovered in an anhydrobiotic nematode (24). Subsequently, LEA proteins were also identified in other anhydrobiotic organisms such as brine shrimps (72, 73), bdelloid rotifers, and a monogonont rotifer (43). In P. vanderplanki, we found 44 clusters showing sequence similarity with LEA proteins. From these, three LEA proteins were characterized, and their expression was shown to be enhanced by desiccation (Fig. 4E) or saline stress treatment (28). These proteins were classified as group 3 LEA proteins, as were homologs from other animal anhydrobionts, and did not aggregate even after boiling in vitro, as described elsewhere (28). Desiccation-associated LEA proteins were shown to prevent protein aggregation in vivo through their molecular shield function (49), and they may also be involved in the stabilization of trehalose glasses (50). Each of the three characterized LEA proteins corresponded to only a few clusters (between two and eight) in the database. Three additional LEA proteins sharing 37–47% of amino acid identity with PvLEA1 were also identified.6 According to the large number of clusters in the database, several putative LEA proteins expressed during the anhydrobiosis of P. vanderplanki remain to be characterized, and it is probable that they have several protective functions in desiccating larvae.
Heat Shock Proteins
Desiccation provokes denaturation and aggregation of proteins. Stressed organisms produce chaperone proteins, so-called heat shock proteins (Hsps), to avoid aggregation and refold proteins to their native conformation (51–53). In P. vanderplanki larvae, desiccation also enhanced the expression of Hsps, especially in the late period of desiccation at 36 h (Fig. 5). Several types of Hsps, such as Hsp40, Hsp60, Hsp70, Hsp82, and Hsp90, were found in the database. The most frequently represented Hsps in the database, in terms of clones, were members of the Hsp70 family, which constitute the most classical stress-inducible pathway. Hsp70/Hsc70 chaperones are expressed in the cytosol and promote correct folding of proteins through the stabilization of hydrophobic regions (51, 54). Five clusters corresponding to homologs of Hsp40 (DnaJ) family proteins were also found in the database. DnaJ is thought to act together with Hsp70 to reactivate stress-damaged proteins (51, 54).
Another important family of Hsps is the small Hsps, characterized by their α-crystallin domain (55). At least four clusters corresponding to small Hsps were detected in the database with clear desiccation-induced expression patterns but apparently no constitutive expression in the majority of cases. Small Hsps form large oligomeric complexes to prevent protein aggregation under stress conditions, such as desiccation (51, 52, 55). Interestingly, two clusters showed significant sequence similarity (E-value = 10−25 and 3·10−27) with a small Hsp from the cryoresistant midge Belgica antartica (56). In this species, expression of the small Hsp, but also of Hsp70, was induced during desiccation, which is associated with cold stress in the larvae (56). This connection of small Hsps and Hsp70 with desiccation tolerance was also found in the resting eggs of the rotifer Brachionus plicatilis (43). One possible scenario is that small Hsps prevent desiccation stress-induced aggregation of proteins after which Hsp70 acts together with DnaJ to refold the proteins to their native form when desiccation stress disappears. In the desiccation-tolerant encysted embryos of the brine shrimp, the small Hsp p26 was actually shown to protect proteins from aggregation and denaturation and functioned synergistically with trehalose (57). It is also worth noting that there is a strong correlation between oxidative stress and the expression of these Hsps (58, 59).
Trehalose Metabolism
Trehalose was demonstrated to accumulate in large amounts during desiccation in some anhydrobionts, and it has been suggested to have a protective role against dehydration stress damage (9, 19, 60). Significant production of trehalose was also observed in desiccating larvae of P. vanderplanki (up to 20% of the dry body mass), and vitrification of this disaccharide was demonstrated to be essential for survival of desiccation (7, 16, 23).
A key enzyme for trehalose synthesis, which exploits glycogen reserves, is trehalose-6-phosphate synthase, and the corresponding gene is one of the most strongly up-regulated genes in the database (Table 1A). The marked induction of trehalose-6-phosphate synthase expression during desiccation was verified by real-time quantitative PCR (Fig. 4F) (61). The terminal enzyme in the trehalose synthesis pathway, trehalose-6-phosphatase, was also identified in the database. Four clusters corresponded to trehalose-6-phosphate synthase, and two clusters corresponded to trehalose-6-phosphatase. Another important gene for trehalose metabolism is the trehalose transporter 1 (Tret1), which was first identified in our database. TRET1 is a transmembrane transporter, and further characterization in the laboratory demonstrated that TRET1 was a facilitated diffusion transporter with specific affinity for trehalose (29). Tret1 expression was also strongly up-regulated during the process of desiccation (Fig. 4H). Consequently, the whole gene battery responsible for trehalose production, and accumulation is apparently induced by desiccation; dehydration stress enhances the expression of trehalose-6-phosphate synthase and trehalose-6-phosphatase, thus initiating the production of trehalose in the fat body. At the same time, TRET1 expression is also induced, and trehalose is transported in a gradient-dependent manner to all the tissues of the larvae. Ultimately, vitrification of trehalose generates a glassy matrix (23), which will protect biological membranes and proteins, in association with Hsps and LEA proteins.
Interestingly, one cluster corresponding to trehalase was found in the database with expression detected only after 36 h of desiccation. This suggests that the expression of this trehalose-degrading enzyme would be induced during the desiccation process to allow rapid recycling of the accumulated trehalose just after rehydration.
Carrier Proteins (Transporters)
Many clusters annotated as being involved with transporter function also exhibited general increase of their expression during desiccation (Fig. 5). The onset of anhydrobiosis is likely to be associated with drastic perturbations in homeostatic systems, and significant movements of water, ion, and metabolites are expected during this period.
About 75% of the transporters potentially up-regulated during desiccation correspond to transmembrane transporters. Clusters were annotated as solute carrier transporters from various families or as ATP-binding cassette transporters. The latter are active transporters, requiring the energy of ATP hydrolysis, which transfer various substrates such as ions, carbohydrates, or lipids across membranes (62, 63). In contrast to ATP-binding cassette transporters, solute carriers include transmembrane passive or facilitated diffusion transporters, ion-coupled transporters, and exchangers (62). Among these solute carriers, at least six distinct clusters corresponded to sugar transporters apparently up-regulated in the database, including a facilitated glucose transporter, a sodium/glucose transporter, and the trehalose transporter TRET1, described above. We have already emphasized the importance of trehalose for anhydrobiosis in P. vanderplanki, but the exchange of other sugars such as glucose clearly also plays a fundamental role. Although trehalose is mobilized for biophysical protection of the cells, free glucose should be the main source of energy during preparation for anhydrobiosis. Apart from sugar transporters, other hits for solute carriers corresponded to amino acid transporters, monocarboxylate transporters, sodium-dependent phosphate transporters, and mitochondrial carriers. The presence of a Na-K-Cl cotransporter (BLASTX, E-value = 9·10−47) after 36 h of desiccation highlights the importance of salt stress during the onset of anhydrobiosis (7). Along with these solutes and metabolites, water also requires specific channels to rapidly transit through cell membranes. Two water channels, or aquaporins (PvAqp1 and PvAqp2), were characterized from the database (30). The expression of PvAqp1 was strongly induced by desiccation (Fig. 4G), and this gene was suggested to accelerate water loss from the body of desiccating larvae so as to allow proper vitrification of trehalose and hence effective protection in the dry state (30).
With regard to soluble transporters, a cluster showing similarity to apolipoprotein D (BLASTX, E-value = 1·10−11) was apparently up-regulated by desiccation (Table 1), and a putative cholesterol transporter (BLASTX, E-value = 4·10−13) was identified after 12 h of desiccation. Apolipoproteins are involved in the transport of lipids through invertebrate plasma (64), and the up-regulation of such proteins in P. vanderplanki suggests that lipid metabolism is important for successful anhydrobiosis. A key role for fatty acids and glycerolipids has already been suggested for anhydrobiosis (65), and lipid transporters have also been identified and shown to be differentially regulated during desiccation in the anhydrobiotic nematode Plectus murrayi (66) or expressed in resting eggs of the rotifer B. plicatilis (43).
Finally, we found three clusters with some similarity to α-tocopherol (= vitamin E) transfer protein (Blastn, E-value = 1·10−8), all of which were up-regulated during desiccation. sThis is interesting because this protein regulates α-tocopherol levels in the plasma of mammals (67), and α-tocopherol is known to be a major antioxidant molecule (42), with an important protective role during the anhydrobiosis of resurrection plants (38).
The Specific Case of Hemoglobins
At the gene ontology level, hemoglobins represented one of the largest groups in the database (Fig. 2). A total of 118 clusters, corresponding to globins or hemoglobins (about 2.4% of all clusters), comprised about 7.5% of all the ESTs in the database. Hemoglobins are respiratory proteins that bind molecular O2 and are found in some insect orders. Chironomidae are the only insects that have hemoglobin in their hemolymph, and more than 40 hemoglobin genes were found in the genome of Chironomus tentans (68).
Most hemoglobins in the database (104 clusters) were down-regulated or showed stable expression, whereas only 14 clusters were apparently up-regulated. In Chironomidae, hemoglobins are involved in oxygen transport, as in vertebrates, but they also allow oxygen storage for short periods because Chironomidae live in the sediment of eutrophic waters, which represent a chronically hypoxic environment (68). During desiccation, oxygen storage is probably less important to P. vanderplanki larvae, and hemoglobin expression is largely down-regulated in favor of anhydrobiosis-related genes, which are strongly up-regulated in the same period (Fig. 2). However, a few hemoglobin clusters were up-regulated in the desiccated larvae. These genes may thus encode particular oxygen binding properties adapted to the anhydrous cellular environment, or alternatively, they may play a totally different role. Interestingly, the most strongly up-regulated gene in the database was a hemoglobin similar to the hemoglobin CTT 6 of Chironomus thummi (Table 1A). In terms of total number of ESTs, only one other cluster (one of the LEA protein clusters) exceeded this hemoglobin cluster. Such a strong induction during desiccation suggests that this hemoglobin must play an important role in anhydrobiosis. Some globins carry out enzymatic functions, such as the detoxification of nitric oxide, and a role as ROS scavenger has also been proposed for some hemoglobins (68, 69). Such a function in ROS detoxification for the strongly induced hemoglobin would be consistent with the general mobilization of antioxidant genes during the onset of anhydrobiosis. However that may be, it is clear that hemoglobins are expressed differentially in P. vanderplanki and that at least some of them have been co-opted for anhydrobiosis.
Other Putative Anhydrobiosis-related Genes
However, even in groups showing no obvious general induction following desiccation treatment, such as Signal transduction, Proteasome/Ubiquitin, or DNA repair (Fig. 5), some individual clusters were up-regulated. Among these, clusters showing sequence similarity with Rab7 and Rab11 GTPases were apparently induced at 12 and 36 h. This may reflect an increase in endocytosis and lysosome activity (70), which is congruent with a high degree of transmembrane trafficking and the degradation of aggregated proteins that are expected to occur during desiccation in P. vanderplanki. Some cuticular proteins were also up-regulated during desiccation, probably in relation to a possible modification of the cuticle permeability. Similarly, a putative homolog of the DNA repair protein xeroderma pigmentosum group C-complementing factor was also induced at 12 h, suggesting a possible need for global genome nucleotide excision repair (71). Many other individual clusters corresponding to important genes for anhydrobiosis, such as Tret1 (Fig. 4H), were present in the database but did not appear in the analysis because of poor EST representation. Thus, genes with low expression levels, such as transcription factors or components of the signal transduction pathways, were often represented by a very poor number of ESTs in the database (Fig. 5), and this did not allow us to predict significant expression changes for these genes. A candidate gene approach on the basis of sequence similarities is probably more appropriate for the identification of such important, but poorly represented, clusters.
Conclusions
The general scenario for successful anhydrobiosis emerging from the above data implicates various antioxidant molecules in protection against ROS and a combination of trehalose glasses with LEA proteins and Hsps in the protection of proteins and membranes. However, it appears that substantial damage occurs in desiccating cells despite all this protective machinery.7,8 The next challenge will thus be the identification of the repair systems allowing survival after anhydrobiosis. For8 this, the construction of a new EST library, covering the process of rehydration in anhydrobiotic P. vanderplanki larvae, is now in progress in the laboratory.
Supplementary Material
Acknowledgments
We are grateful to Akihiko Fujita and Yoko Saito for the maintenance and rearing of P. vanderplanki. We also thank Takashi Noda for help with statistics.
This work was supported by the Promotion of Basic Research Activities for Innovative Bioscience (PROBRAIN), a grant-in-aid (Bio Design Program) from the Ministry of Agriculture, Forestry and Fisheries of Japan, and by Grant-in-aid for Scientific Research Number 21688004 from the Ministry of Education, Culture, Sports, Science and Technology of Japan. A part of this study is also the result of the project “Characterization of the Mechanisms Underlying the Radiation Resistance Associated with Cryptobiosis” carried out under the grant “Strategic Promotion Program for Basic Nuclear Research” by the Ministry of Education, Culture, Sports, Science and Technology of Japan.
The nucleotide sequence(s) reported in this paper has been submitted to the DDBJ/GenBankTM/EBI Data Bank with accession number(s) FS595283–FS610333.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, Figs. S1 and S2, and Tables S1 and S2.
http://pvcdna.dna.affrc.go.jp.
R. Cornette, Y. Kanamori, M. Watanabe, Y. Nakahara, O. Gusev, K. Mitsumasu, K. Kadono-Okuda, M. Shimomura, K. Mita, T. Kikawada, and T. Okuda, unpublished data.
K. Iwata, Y. Nakahara, M. Watanabe, R. Cornette, T. Kikawada, and T. Okuda, manuscript in preparation.
O. Gusev, Y. Nakahara, V. Vanyagina, L. Malutina, R. Cornette, T. Sakashita, N. Hamada, T. Kikawada, M. Fujita, Y. Kobayashi, and T. Okuda, manuscript in preparation.
- LEA
- late embryogenesis abundant
- EST
- expressed sequence tag
- Hsp
- heat shock protein
- ROS
- reactive oxygen species.
REFERENCES
- 1.Danks H. V. (2000) J. Insect Physiol. 46, 837–852 [DOI] [PubMed] [Google Scholar]
- 2.Gibbs A. G. (2002) Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 133, 781–789 [DOI] [PubMed] [Google Scholar]
- 3.Suemoto T., Kawai K., Imabayashi H. (2004) Hydrobiologia 57, 107–114 [Google Scholar]
- 4.Nakahara Y., Watanabe M., Fujita A., Kanamori Y., Tanaka D., Iwata K., Furuki T., Sakurai M., Kikawada T., Okuda T. (2008) J. Insect Physiol. 54, 1220–1225 [DOI] [PubMed] [Google Scholar]
- 5.Keilin D. (1959) Proc. R. Soc. Lond. B Biol. Sci. 150, 149–191 [DOI] [PubMed] [Google Scholar]
- 6.Crowe J. H., Hoekstra F. A., Crowe L. M. (1992) Annu. Rev. Physiol. 54, 579–599 [DOI] [PubMed] [Google Scholar]
- 7.Watanabe M., Kikawada T., Okuda T. (2003) J. Exp. Biol. 206, 2281–2286 [DOI] [PubMed] [Google Scholar]
- 8.Sømme L. (1995) in Invertebrates in Hot and Cold Arid Environments, pp. 95–114, Springer-Verlag, New York [Google Scholar]
- 9.Clegg J. S. (2001) Comp. Biochem. Physiol. B 128, 613–624 [DOI] [PubMed] [Google Scholar]
- 10.Hochachka D. W., Guppy M. (1987) Metabolic Arrest and the Control of Biological Time, pp. 146–165, Harvard University Press, Cambridge, Massachusetts and London [Google Scholar]
- 11.Watanabe M., Kikawada T., Fujita A., Forczek E., Adati T., Okuda T. (2004) Eur. J. Entomology 101, 439–444 [Google Scholar]
- 12.Ricci C. (2001) Hydrobiologia 446/447, 1–11 [Google Scholar]
- 13.Hinton H. E. (1951) Proc. Zool. Soc. Lond. 121, 371–380 [Google Scholar]
- 14.Hinton H. E. (1960) J. Insect Physiol. 5, 286–300 [Google Scholar]
- 15.Kikawada T., Minakawa N., Watanabe M., Okuda T. (2005) Integ. Comp. Biol. 45, 710–714 [DOI] [PubMed] [Google Scholar]
- 16.Watanabe M., Kikawada T., Minagawa N., Yukuhiro F., Okuda T. (2002) J. Exp. Biol. 205, 2799–2802 [DOI] [PubMed] [Google Scholar]
- 17.Watanabe M., Kikawada T., Fujita A., Okuda T. (2005) J. Insect Physiol. 51, 727–731 [DOI] [PubMed] [Google Scholar]
- 18.Madin K. A., Crowe J. H. (1975) J. Exp. Zool. 193, 335–342 [Google Scholar]
- 19.Crowe L. M. (2002) Comp. Biochem. Physiol. A Mol. Integr. Physiol. 131, 505–513 [DOI] [PubMed] [Google Scholar]
- 20.Clegg J. S., Campagna V. (2006) Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 145, 119–125 [DOI] [PubMed] [Google Scholar]
- 21.Hengherr S., Heyer A. G., Köhler H. R., Schill R. O. (2008) FEBS J. 275, 281–288 [DOI] [PubMed] [Google Scholar]
- 22.Tunnacliffe A., Lapinski J., McGee B. (2005) Hydrobiologia 546, 315–321 [Google Scholar]
- 23.Sakurai M., Furuki T., Akao K., Tanaka D., Nakahara Y., Kikawada T., Watanabe M., Okuda T. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5093–5098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Browne J., Tunnacliffe A., Burnell A. (2002) Nature 416, 38. [DOI] [PubMed] [Google Scholar]
- 25.Denekamp N. Y., Reinhardt R., Kube M., Lubzens E. (2010) Biol. Reprod. 82, 714–724 [DOI] [PubMed] [Google Scholar]
- 26.Qiu Z., Tsoi S. C., MacRae T. H. (2007) Mech. Dev. 124, 856–867 [DOI] [PubMed] [Google Scholar]
- 27.Chen W. H., Ge X., Wang W., Yu J., Hu S. (2009) BMC Genomics 10, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kikawada T., Nakahara Y., Kanamori Y., Iwata K., Watanabe M., McGee B., Tunnacliffe A., Okuda T. (2006) Biochem. Biophys. Res. Commun. 348, 56–61 [DOI] [PubMed] [Google Scholar]
- 29.Kikawada T., Saito A., Kanamori Y., Nakahara Y., Iwata K., Tanaka D., Watanabe M., Okuda T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11585–11590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kikawada T., Saito A., Kanamori Y., Fujita M., Snigórska K., Watanabe M., Okuda T. (2008) Biochim. Biophys. Acta 1778, 514–520 [DOI] [PubMed] [Google Scholar]
- 31.Suetsugu Y., Minami H., Shimomura M., Sasanuma S., Narukawa J., Mita K., Yamamoto K. (2007) BMC Genomics 8, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eigenheer A. L., Keeling C. I., Young S., Tittiger C. (2003) Gene 316, 127–136 [DOI] [PubMed] [Google Scholar]
- 33.Tagu D., Prunier-Leterme N., Legeai F., Gauthier J. P., Duclert A., Sabater-Muñoz B., Bonhomme J., Simon J. C. (2004) Insect Biochem. Mol. Biol. 34, 809–822 [DOI] [PubMed] [Google Scholar]
- 34.Wu-Scharf D., Scharf M. E., Pittendrigh B. R., Bennett G. W. (2003) Sociobiology 41, 479–490 [Google Scholar]
- 35.Hankeln T., Amid C., Weich B., Niessing J., Schmidt E. R. (1998) J. Mol. Evol. 46, 589–601 [DOI] [PubMed] [Google Scholar]
- 36.Hoback W. W., Stanley D. W. (2001) J. Insect Physiol. 47, 533–542 [DOI] [PubMed] [Google Scholar]
- 37.Pereira Ede J., Panek A. D., Eleutherio E. C. (2003) Cell Stress Chaperones 8, 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.França M. B., Panek A. D., Eleutherio E. C. (2007) Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 146, 621–631 [DOI] [PubMed] [Google Scholar]
- 39.Corona M., Robinson G. E. (2006) Insect Mol. Biol. 15, 687–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maulik N., Das D. K. (2008) Biochim. Biophys. Acta 1780, 1368–1382 [DOI] [PubMed] [Google Scholar]
- 41.Franco R., Sánchez-Olea R., Reyes-Reyes E. M., Panayiotidis M. I. (2009) Mutat. Res. 674, 3–22 [DOI] [PubMed] [Google Scholar]
- 42.Limón-Pacheco J., Gonsebatt M. E. (2009) Mutat. Res. 674, 137–147 [DOI] [PubMed] [Google Scholar]
- 43.Denekamp N. Y., Thorne M. A., Clark M. S., Kube M., Reinhardt R., Lubzens E. (2009) BMC Genomics 10, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freitas D. R., Rosa R. M., Moraes J., Campos E., Logullo C., Da Silva Vaz I., Jr., Masuda A. (2007) Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 146, 688–694 [DOI] [PubMed] [Google Scholar]
- 45.Rizzo A. M., Negroni M., Altiero T., Montorfano G., Corsetto P., Berselli P., Berra B., Guidetti R., Rebecchi L. (2010) Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 156, 115–121 [DOI] [PubMed] [Google Scholar]
- 46.Haegeman A., Jacob J., Vanholme B., Kyndt T., Mitreva M., Gheysen G. (2009) Mol. Biochem. Parasitol. 167, 32–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schokraie E., Hotz-Wagenblatt A., Warnken U., Mali B., Frohme M., Förster F., Dandekar T., Hengherr S., Schill R. O., Schnölzer M. (2010) PLoS One 5, e9502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cuming A. C. (1999) in Seed Proteins (Shewry P. R., Casey R. eds.) pp. 753–780, Kluwer Academic Publishers, The Netherlands [Google Scholar]
- 49.Chakrabortee S., Boschetti C., Walton L. J., Sarkar S., Rubinsztein D. C., Tunnacliffe A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 18073–18078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimizu T., Kanamori Y., Furuki T., Kikawada T., Okuda T., Takahashi T., Mihara H., Sakurai M. (2010) Biochemistry 49, 1093–1104 [DOI] [PubMed] [Google Scholar]
- 51.Parsell D. A., Lindquist S. (1993) Annu. Rev. Genet. 27, 437–496 [DOI] [PubMed] [Google Scholar]
- 52.Berjak P., Farrant J. M., Pammenter N. W. (2007) in Plant Desiccation Tolerance (Jenks M. A., Wood A. J. eds.) pp. 151–192, Blackwell Publishing, Oxford [Google Scholar]
- 53.Hinault M. P., Goloubinoff P. (2007) Adv. Exp. Med. Biol. 594, 47–54 [DOI] [PubMed] [Google Scholar]
- 54.Bukau B., Horwich A. L. (1998) Cell 92, 351–366 [DOI] [PubMed] [Google Scholar]
- 55.Narberhaus F. (2002) Microbiol. Mol. Biol. Rev. 66, 64–93; table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lopez-Martinez G., Benoit J. B., Rinehart J. P., Elnitsky M. A., Lee R. E., Jr., Denlinger D. L. (2009) J. Comp. Physiol. B 179, 481–491 [DOI] [PubMed] [Google Scholar]
- 57.Viner R. I., Clegg J. S. (2001) Cell Stress Chaperones 6, 126–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arrigo A. P. (2007) Adv. Exp. Med. Biol. 594, 14–26 [DOI] [PubMed] [Google Scholar]
- 59.Lopez-Martinez G., Elnitsky M. A., Benoit J. B., Lee R. E., Jr., Denlinger D. L. (2008) Insect Biochem. Mol. Biol. 38, 796–804 [DOI] [PubMed] [Google Scholar]
- 60.Clegg J. S. (1965) Comp. Biochem. Physiol. 14, 135–143 [DOI] [PubMed] [Google Scholar]
- 61.Mitsumasu K., Kanamori Y., Fujita M., Iwata K., Tanaka D., Kikuta S., Watanabe M., Cornette R., Okuda T., Kikawada T. (2010) FEBS J. 277, 4215–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hediger M. A., Romero M. F., Peng J. B., Rolfs A., Takanaga H., Bruford E. A. (2004) Pflugers Arch. 447, 465–468 [DOI] [PubMed] [Google Scholar]
- 63.Tarr P. T., Tarling E. J., Bojanic D. D., Edwards P. A., Baldán A. (2009) Biochim. Biophys. Acta 1791, 584–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Narayanaswami V., Ryan R. O. (2000) Biochim. Biophys. Acta 1483, 15–36 [DOI] [PubMed] [Google Scholar]
- 65.Wharton D. A. (2003) J. Comp. Physiol. B 173, 621–628 [DOI] [PubMed] [Google Scholar]
- 66.Adhikari B. N., Wall D. H., Adams B. J. (2009) BMC Genomics 10, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meier R., Tomizaki T., Schulze-Briese C., Baumann U., Stocker A. (2003) J. Mol. Biol. 331, 725–734 [DOI] [PubMed] [Google Scholar]
- 68.Burmester T., Hankeln T. (2007) J. Insect Physiol. 53, 285–294 [DOI] [PubMed] [Google Scholar]
- 69.Wajcman H., Kiger L., Marden M. C. (2009) C. R. Biol. 332, 273–282 [DOI] [PubMed] [Google Scholar]
- 70.Fan G. H., Lapierre L. A., Goldenring J. R., Richmond A. (2003) Blood 101, 2115–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sugasawa K., Shimizu Y., Iwai S., Hanaoka F. (2002) DNA Repair 1, 95–107 [DOI] [PubMed] [Google Scholar]
- 72.Hand S. C., Jones D., Menze M. A., Witt T. L. (2007) J. Exp. Zool. A Ecol. Genet. Physiol. 307, 62–66 [DOI] [PubMed] [Google Scholar]
- 73.Sharon M. A., Kozarova A., Clegg J. S., Vacratsis P. O., Warner A. H. (2009) Biochem. Cell Biol. 87, 415–430 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.