Abstract
Non-proteolytic activities of matrix metalloproteinases (MMPs) have recently been shown to impact cell migration, but the precise mechanism remains to be understood. We previously demonstrated that the hemopexin (PEX) domain of MMP-9 is a prerequisite for enhanced cell migration. Using a biochemical approach, we now report that dimerization of MMP-9 through the PEX domain appears necessary for MMP-9-enhanced cell migration. Following a series of substitution mutations within the MMP-9 PEX domain, blade IV was shown to be critical for homodimerization, whereas blade I was required for heterodimerization with CD44. Blade I and IV mutants showed diminished enhancement of cell migration compared with wild type MMP-9-transfected cells. Peptides mimicking motifs in the outermost strands of the first and fourth blades of the MMP-9 PEX domain were designed. These peptides efficiently blocked MMP-9 dimer formation and inhibited motility of COS-1 cells overexpressing MMP-9, HT-1080, and MDA-MB-435 cells. Using a shRNA approach, CD44 was found to be a critical molecule in MMP-9-mediated cell migration. Furthermore, an axis involving a MMP-9-CD44-EGFR signaling pathway in cell migration was identified using antibody array and specific receptor tyrosine kinase inhibitors. In conclusion, we dissected the mechanism of pro-MMP-9-enhanced cell migration and developed structure-based inhibitory peptides targeting MMP-9-mediated cell migration.
Keywords: Cell Migration, Hemopexin, Matrix Metalloproteinase, Peptides, Signal Transduction, CD44, Dimer Formations, Signaling Pathway
Introduction
Human matrix metalloproteinases (MMPs)2 make up a family of 23 Zn2+-dependent proteinases that degrade and remodel multiple components of the extracellular matrix including collagens, fibronectin, laminin, hyaluronan, proteoglycans, and elastin (1, 2). The biological importance of MMPs has been described in multiple cellular processes including proliferation, angiogenesis, migration, host defense, cancer invasion, and metastasis (3, 4). In the 1990s, several broad-spectrum MMP inhibitors were evaluated in clinical trials involving cancer, arthritis, and later, heart failure. The absence of clinical efficacy of these drug trials and the conviction that MMPs are critical players in disease processes lead to a more thorough investigation of the biological roles of individual MMPs (3, 5).
Crystal structures of various domains have been solved for several MMPs including MMP-1, -2, -3, -9, -13, and -14. Bridging biochemical information with in vitro and in vivo experiments has been helpful in better understanding the specific roles of individual MMPs. Because the catalytic sites of most MMPs are highly homologous, leading to difficulty producing non-cross-reactive inhibitors, intense scrutiny of other MMP domains has followed. The substrate binding function of the hemopexin (PEX) domain is recognized to play an important role in MMP function (4). With the exception of MMP-7, -23, and -26, which lack the PEX domain, all other MMPs form a propeller structure composed of four blades; each blade consists of one α-helix and four anti-parallel β-strands.
Among secreted MMPs, only MMP-9 is capable of forming a homodimer; the precise role of this homodimerization has yet to be elucidated. Solving the crystal structure of MMP-9 demonstrated that the homodimer is formed through blade IV of the PEX domain (6). In contrast to all other secreted MMPs, pro-MMP-9 and pro-MMP-2 bind TIMP-1 and TIMP-2, respectively, through their PEX domain. Other MMPs require activation for TIMP to bind to their catalytic domain.
MMP-9 has been shown to bind to several cell surface receptors including CD44, LRP-1, LRP-2, Ku, and β1-integrin (7–10). CD44, a cell surface glycoprotein involved in cell-cell and cell-matrix interactions, has been associated with the ability to regulate cell migration and cell shape by association with actin microfilaments (11, 12). CD44 has an extracellular domain that binds hyaluronic acid and promotes intracellular signaling involving ERK and Rho (13, 14).
Because MMPs are involved in multiple diseases, it has been proposed that better understanding of MMP domains might reveal crucial information for specific and novel inhibitory drug design (3, 5, 15). Based on less homology between PEX domains as compared with catalytic domains of different MMPs (homology sequence alignment: Clustalw2), targeting the PEX domain has been proposed as an option to inhibit a specific MMP.
In contrast to general concepts regarding the requirement for activation of pro-MMPs to generate biological activity, we recently demonstrated that pro-MMP-9 enhances COS-1 cell migration independent of its proteolytic activity (16). In this report we investigated biochemical and biological properties of MMP-9 dimerization and dissected the signaling pathways involved in MMP-9-mediated cell migration. Using structure-functional analysis, inhibitory peptides targeting MMP-9-induced cell migration were designed and assessed. This novel structure-based peptide approach serves as a proof of principle for design of the next generation of MMP inhibitors.
EXPERIMENTAL PROCEDURES
Reagents
Oligo primers were purchased from Operon (Huntsville, AL). The pcDNA3.1-myc expression vectors were purchased from Invitrogen. Anti-Myc and anti-HA antibodies were purchased from Roche Applied Science. MMP-9 antibody was described previously (17). Anti-tubulin, anti-AKT, anti-pAKT, anti-ERK, anti-pERK, anti-pEGFR, and anti-EGFR antibodies were purchased from Cell Signaling Technology (Davers, MA). Anti-FAK and anti-pFAK antibodies were purchased from BioSource (Camarillo, CA). Anti-TIMP-1 and anti-TIMP-2 antibodies were purchased from Calbiochem (Cambridge, MA). Anti-CD44 antibodies were purchased from Novus Biologicals (Littleton, CO). Genistein, PP2 (SRC), AG490 (JAK-2), AG1296 (PDGFR), and AG1478 (EGFR) were purchased from Calbiochem (Cambridge, MA). AG1024 (IGFR), PD173074 (FGFR and VEFGR), and PHA665752 (c-Met) were purchased from EMD Chemicals (Gibbstown, NJ). Peptides were synthesized from EZBiolab (Carmel, IN) and purity was verified by HPLC.
Cell Culture, Transfection, and Peptides Treatment
COS 1 monkey kidney epithelial, human fibrosarcoma HT-1080, and breast cancer MDA-MB-435 cell lines were purchased from ATCC (Manassas, VA) and maintained in Dulbecco's modified Eagle's medium (Invitrogen). Transfection of plasmid DNA in COS-1 cells was achieved using polyethylenimine (Polysciences) or TransfectinTM reagents (Bio-Rad) and the transfected cells were incubated for 48 h at 37 °C followed by biochemical and biological assays. Peptides and inhibitors were incubated with cells for 24 h prior to co-immunoprecipitation and transwell chamber migration assays. Final concentrations of the peptides ranged from 1 μm to 1 mm, as indicated in the figure legends.
Construction of Plasmids
MMP-9 with a carboxyl-terminal Myc tag (MMP-9/Myc) was generated by using the pcDNA3.1 expression vector (Invitrogen). The MMP-9 cDNA containing the open reading frame of MMP-9 was amplified by a PCR approach using the primers sets: forward primer 1315, 5′-CGGAATTCCGCCAACATGAGCCTCTGGCAGCCCCT-3′ and reverse primer 2507, 5′-GGAAGATCTCTAGTCCTCAGGGCACTGCAGGATGTC-3′. The resultant PCR fragment was then cloned into the pcDNA3.1 vector to generate MMP-9/Myc chimeric cDNA.
To generate HA (human influenza hemagglutinin)-tagged MMP-9 chimeric cDNA, the HA tag was placed between the propeptide and catalytic domains of MMP-9 by a site-directed mutagenesis approach (QuikChange Site-directed Mutagenesis kit, Stratagene) using wild type MMP-9 as a template with mutagenesis primers containing the HA sequence (forward primer 2512, 5′-GGGGTCCCAGACCTGGGCAGATACCCCTACGACGTGCCCGACTCGCCTCCAAACCTTTGAGGGCGAC-3′ and reverse primer 2513, 5′-GTCGCCCTCAAAGGTTTGGAAGGCGTAGTCGGGCACGTCGTAGGGGTATCTGCCCAGGTCTGGGACCCC-3′).
To determine the role of the PEX domain of MMP-9, a substitution mutation was engineered by replacing the PEX domain of MMP-9 with that of MMP-2. A modified two-step PCR was employed as previously described (18). In brief, MMP-9 signal peptide/propeptide/catalytic/hinge domains (fragment A) (primer sets: forward primer 1315 and reverse primer 2534, 5′-TACAATGTCCTGTTTGCAGATCTCGTCCACCGGACTCAAAGGCAC-3′), and MMP-2 PEX domain (fragment B) (primer sets: forward primer 2535, 5′-GAGATCTGCAAACAGGACATTGTATTTGAT-3′, and reverse primer 1356, 5′-CCCAAGCTTCTAGCAGCCTAGCCAGTCGGATTT-3′) were first amplified by PCR, respectively. Using the resultant PCR products as templates (fragment A and B), the signal/propeptide/catalytic/hinge region of MMP-9 was then fused together with the MMP-2 PEX domain by PCR using forward primer 1315 and reverse primer 1356. The resultant PCR fragments were then inserted into the pcDNA3.1 vector (Invitrogen) to generate MMP9/PEXMMP2/Myc chimeric cDNAs.
Similarly, we generated substitution mutations of MMP-9/blade I, II, III, and IV by replacing the outermost β-strand of each blade with the corresponding sequence of MMP-2. Primers used to generate these chimeric cDNAs were designed as follow: 1) MMP-9/blade I, fragment A, 1315 and 2557 (5′-GGGAGCCGGCCGGGGCCCCTGCTGATCGCCGAC-3′),and fragment B, 2558 (5′-GTCGGCGATCAGCAGGGGCCCCGGCCGGCTCCC-3′) and 2507; 2) MMP-9/blade II, fragment A, 1315, and 2596 (5′-GTGTACACAGGCGCGACCTTGGAGCGAGGGTACCCCAAGCCACTGGACAAGCTGGGC-3′), and fragment B, 2597 (5′-CACATGTGTCCGCGCTGGAACCTCGCTCCCATGGGGTTCGGTGACCTGTTCGACCCG-3′), and 2507; 3) MMP-9/blade III, fragment A, 1315 and 2540 (5′-GTTCGACGTGAAGGCGAAGAAAATGGATCCTGGCTTCCCCAAGCTCGTGGACCGGATGTTCC-3′), and fragment B, 2541 (5′-GGAACATCCGGTCCACGAGCTTGGGGAAGCCAGGATCCATTTTCTTCGCCTTCACGTCGAAC-3′) and 2507; and 4) MMP-9/blade IV: fragment A, 1315 and 2538 (5′-GTTCCCGGAGTGAGTTGAAGAGCGTGAAGTTTGGAAGCGTGACCTATGACATCCTG-3′), and fragment B, 2539 (5′-CAGGATGTCATAGGTCACGCTTCCAAACTTCACGCTCTTCAACTCACTCCGGGAAC-3′), and 2507. All the constructs were confirmed by DNA sequencing.
Construction of Short Hairpin RNA Vectors and Retroviral Infection
Small interfering oligonucleotides specific for human and monkey CD44 and control luciferase to express short hairpin RNA were designed using a Worldwide Web-based online software system (Block-iT RNAi Designer, Invitrogen) for mammalian RNA interference. Two specific 21-nucleotide sequences spanning positions 173–193 (CD44shRNA-1) and 678–698 (CD44-shRNA-2) of the human CD44 gene (GenBankTM accession number L05424) were synthesized. The sense and antisense template oligonucleotides encoding a hairpin structure were annealed and cloned into the RNAi-Ready pSIREN-Retro Q vector (Clontech). As a control, a luciferase protein from firefly Pyrocoelia pectoralis as a target gene was employed as previously reported (19). A retroviral supernatant was obtained by co-transfection of a vector encoding the envelope gene (pAmphotropic) and a retroviral expression vector containing the CD44 shRNA, or luciferase shRNA control into human embryonic kidney GP2–293 packaging cells (Clontech) according to the manufacturer's protocol. COS-1 cells were infected with the supernatant containing retrovirus, and the cells were then selected with 4 μg/ml of puromycin for 1–2 weeks. The effects of shRNA on gene expression were evaluated by real time RT-PCR using RNA of the pooled resistant cells. The most effective stable CD44 knockdown cell lines were selected.
Immunofluorescence
Cultured cells were fixed with 4% paraformaldehyde/phosphate-buffered saline (PBS) followed by blocking with 3% bovine serum albumin (BSA)/PBS. CD44 was detected with anti-CD44 antibody (Novus Biologicals, CO) followed by secondary antibodies conjugated with Alexa 568 (Invitrogen). Nuclei were counterstained with DAPI (Invitrogen).
Flow Cytometry
1 × 106 cells/ml of cells suspended in Dulbecco's modified Eagle's medium containing 2% BSA were incubated with CD44 primary monoclonal antibody for 60 min at 4 °C followed by incubation with anti-mouse FITC-labeled (1:1000) secondary antibody for 60 min at 4 °C. After extensive washing, the cells were stained with propidium iodide to determine viability of the cells. CD44 expression was measured using a FACS Calibur flow cytometer (BD Biosciences).
Antibody Microarray and Protein Phosphorylation Assay (KinexTM Antibody Microarray KAM-1.2 and Phospho-Site 1.3 Kinetworks Screen)
Cell lysates were prepared as recommended by Kinexus Bioinformatics Corporation (Vancouver, BC, Canada). COS-1 cells were transiently transfected with vector or MMP-9 cDNA. Forty-eight hours after transfection, total cellular proteins were extracted using ice-cold lysis buffer containing detergent and protease inhibitor mixture and subjected to a customer antibody microarray analysis (Kinexus Bioinformatics Corp.). Over 500 pan-specific and 300 phospho-site-specific antibodies were screened against both vector control and MMP-9 samples. Sixteen different proteins that are known to link cell migration and displayed over 2-fold difference between control and MMP-9-transfected cells were further analyzed by a semi-quantitative immunoblotting examination (Kinexus Bioinformatics Corp.).
Co-immunoprecipitation
Twenty-four hours following transfection, the conditioned medium in the absence of serum was collected from the transfected COS-1 cells. The cells were then lysed in lysis buffer containing 150 mm NaCl, 25 mm HEPES (pH 7.4), 1% CHAPS, and protease inhibitors mixture (Sigma). Both the conditioned medium and the cell lysates were incubated with specific antibodies for 24 h at 4 °C. Antigen-antibody complexes were precipitated with protein A-agarose beads followed by a brief centrifugation, washing, and then electrophoresis in a 10% SDS-polyacrylamide gel. Western blotting was then followed using a corresponding antibody.
Preparation of Plasma Membrane-enriched Cell Fractions
Cell membranes were prepared from MMP-9-transfected COS-1 cells following nitrogen cavitation as described previously (20). The post-nuclear supernatant (770 × g for 10 min) was collected, and heavy organelles were removed by centrifugation at 6,000 × g for 15 min. This supernatant was centrifuged at 100,000 × g for 1 h at 4 °C to recover the plasma membrane-enriched lighter cell organelles in the pellet.
Fast Protein Liquid Chromatography (FPLC)
Conditioned medium of COS-1 cells transfected with MMP-9 cDNA was isolated and clarified by centrifugation. Cell-free conditioned medium was subjected to gelatin-Sepharose 4B column chromatography as previously described (21). The DMSO eluted fractions were dialyzed against buffer (150 mm NaCl, 10 mm Tris, 0.03% sodium azide, pH 7.4), concentrated, and subjected to a Hiload 16/60 Superdex 200 prep grade column (GE Healthcare) mounted on an FPLC apparatus (GE Pharmacia Aktapurifier, GE Healthcare, Piscataway, NJ). One ml of sample was injected and the flow rate was 1 ml min−1. One-ml fractions were collected and 10 μl of each fraction was examined by gelatin zymogram.
Statistical Analysis
Data are expressed as the mean ± S.E. of triplicates. Each experiment was repeated as least 3 times. Student's t test and analysis of variants were used to assess differences with p < 0.05 considered to be significant.
Procedures for Gelatin Substrate Zymography, Western Blotting, Transwell Chamber Cell Migration, and Phagokinetic Cell Migration Assays
Basic protocols for these techniques have been described previously (16, 22).
RESULTS
Dimerization of MMP-9 in Transfected COS-1 Cells
We previously demonstrated that the PEX domain of pro-MMP-9 is required for enhanced cell migration independent of its proteolytic activity (16). To dissect the mechanism underlying MMP-9-induced cell migration, the PEX domain of MMP-9 was examined using biochemical and molecular approaches.
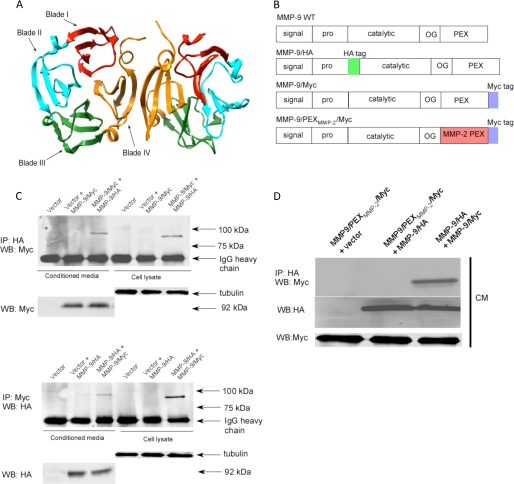
Among all secreted MMPs, only MMP-9 has been found to form a homodimer. Employing proteins purified from an Escherichia coli expression system (Fig. 1A, ribbon diagram based on PDB file 1ITV), Cha et al. (6) showed that MMP-9 homodimer formation requires an interaction of the fourth blade of adjacent PEX domains. Because proteins purified from a bacterial system lack posttranslational modifications, e.g. glycosylation and phosphorylation, which can impact the function of most mammalian proteins (23), it is essential to test if the dimerization of MMP-9 also occurs in a mammalian cell expression system. To this end, we fused HA and Myc tags into the MMP-9 cDNA, to generate MMP-9/HA and MMP-9/Myc chimeras, respectively (Fig. 1B), followed by transfection of the cDNAs into COS-1 monkey kidney epithelial cells. Insertion of either HA tag between the propeptide domain and catalytic domain or Myc tag at the end of the PEX domain of MMP-9 does not interfere with the overall properties of wild type MMP-9 as evidenced by gelatin zymography and Western blotting (supplemental Fig. S1, A and B). Immunoprecipitation with either HA or Myc antibodies resulted in identifying MMP-9 in both the cell lysates and conditioned medium of transfected COS-1 cells (supplemental Fig. S1C).
FIGURE 1.
MMP-9 homodimerizes through its PEX domain. A, ribbon diagram of the MMP-9 PEX domain (PDB code 1ITV). Homodimerization of MMP-9 is through the fourth blade of the MMP-9 PEX domain. B, schematic diagram of wild type MMP-9, MMP-9/HA, MMP-9/Myc, and MMP9/PEXMMP2/Myc. Five typical domains of MMP-9 from the N terminus to C terminus are the signal peptide (signal), propeptide (pro), catalytic domain (catalytic), hinge region (OG), and hemopexin-like domain (PEX). HA and Myc tags were inserted as shown. The PEX domain of MMP-9 was replaced by MMP-2 to generate MMP-9/PEXMMP-2/Myc. C, MMP-9 forms a homodimer in the COS-1 cells transfected with MMP-9 cDNAs. COS-1 cells were transfected with a combination of cDNAs as indicated. The conditioned medium (CM) and cell lysates were examined by a co-immunoprecipitation (IP) assay (upper panel) and a reciprocal co-immunoprecipitation assay (lower panel). 20 μg of total cell lysates or 20 μl of the conditioned medium were used as loading controls by anti-α/β-tubulin antibody for cell lysates and anti-Myc or anti-HA antibodies for the conditioned medium. D, replacement of the MMP-9 PEX domain with the corresponding region of MMP-2 prohibited dimerization with wild type MMP-9 as examined through a co-immunoprecipitation assay. The conditioned medium of COS-1 cells transfected with a combination of cDNAs as indicated was examined by a co-immunoprecipitation assay. 20 μl of the conditioned medium were examined by Western blotting (WB) using anti-Myc antibody for monitoring expression of MMP-9/Myc and MMP9/PEXMMP2/Myc.
COS-1 cells co-transfected with HA- and Myc-tagged MMP-9 cDNAs were then employed to examine homodimer formation using a co-immunoprecipitation approach. The HA-tagged MMP-9 proteins in the conditioned medium of transfected COS-1 cells were immunoprecipitated with anti-HA antibodies followed by a Western blotting probed by anti-Myc antibodies. This approach revealed the identification of Myc-tagged MMP-9 in the complex immunoprecipitated by anti-HA antibody, suggesting that MMP-9 forms homodimers in cell-conditioned medium (Fig. 1C). To further confirm this observation, the reciprocal co-immunoprecipitation was performed. Anti-Myc antibodies were employed to precipitate Myc-tagged MMP-9 complexes in the conditioned medium of co-transfected COS-1 cells followed by Western blotting using anti-HA antibodies. This reciprocal approach confirms that MMP-9 does form a homodimer in the condition medium of transfected COS-1 cells. To further explore if homodimer formation of MMP-9 occurs within the cell or following secretion from the cell, the cell lysate of transfected COS-1 cells was also examined by the co-immunoprecipitation method (Fig. 1C). MMP-9 homodimer formation was demonstrated in the cell lysate, suggesting that MMP-9 dimerization occurs during MMP-9 secretion.
Prerequisite of MMP-9 Dimerization in Pro-MMP-9-induced Cell Migration
We previously demonstrated that the PEX domain of pro-MMP-9 plays an important role in enzymatic activity-independent cell migration (16). To test if dimerization of the MMP-9 PEX domain is a prerequisite for MMP-9-induced cell migration, we generated MMP-9 PEX domain mutations. Because MMP-2 does not form homodimers (24), the PEX domain of MMP-9 was replaced by that of MMP-2 to generate MMP9/PEXMMP-2 chimeric cDNA using a two-step PCR approach (22). The resultant PCR product containing MMP9/PEXMMP-2 was then inserted into a pcDNA3.1/Myc vector to generate a final construct of MMP9/PEXMMP-2/Myc chimeric cDNA (Fig. 1B). Substitution of the PEX domain of MMP-9 with MMP-2 failed to couple with the wild type MMP-9 in co-transfected COS-1 cells not only in the conditioned medium (Fig. 1D) but also in the cell lysate (data not shown). The failure of the dimerization of the PEX domain mutation was not due to the loss of protein synthesis, trafficking, or proteolytic activity as evidenced by Western blotting and gelatin zymography (supplemental Fig. S1D).
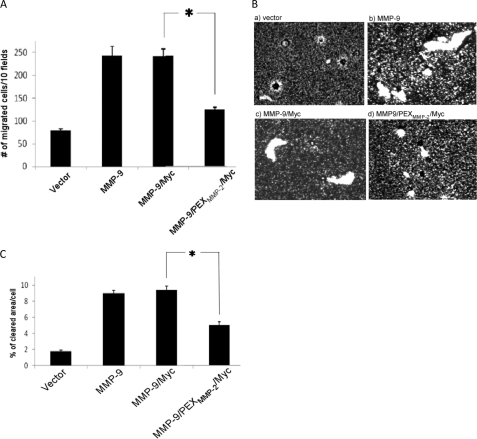
To determine whether the loss of homodimerization of this MMP-9 PEX domain mutation affects MMP-9-induced cell migration, a transwell chamber migration assay was employed (16). COS-1 cells transfected with the PEX mutant MMP-9 (MMP-9/PEXMMP-2/Myc) failed to enhance cell migration to the extent of wild type MMP-9, suggesting a role of MMP-9 homodimer in cell migration (Fig. 2A). To further confirm the biological role of MMP-9 homodimers in cell migration, wild type and mutant MMP-9-transfected cells were subjected to phagokinetic migration analysis, which permits quantification by clearance of colloidal gold particles within the cell migratory pathway (Fig. 2B). Similarly to the data from the transwell migration assay (Fig. 2A), cells transfected with MMP-9/Myc cDNA displayed enhanced migration as compared with vector-transfected cells determined by NIH ImageJ software (Fig. 2C). Enhanced cell migration did not occur in cells transfected with MMP-9/PEXMMP-2/Myc. These data indicate that homodimerization through the MMP-9 PEX domain plays an important role in MMP-9-induced cell migration.
FIGURE 2.
MMP-9 homodimer is required for MMP-9-enhanced cell migration. COS-1 cells transfected with wild type and mutant MMP-9 cDNAs were examined by a transwell chamber migration assay (A) and phagokinetic assay (B). *, p < 0.05. Migratory ability of cells using the phagokinetic assay was quantitatively determined using Image J software (C). *, p < 0.05.
Inhibition of MMP-9 Dimerization by TIMP-1 Resulting in Diminished Cell Migration
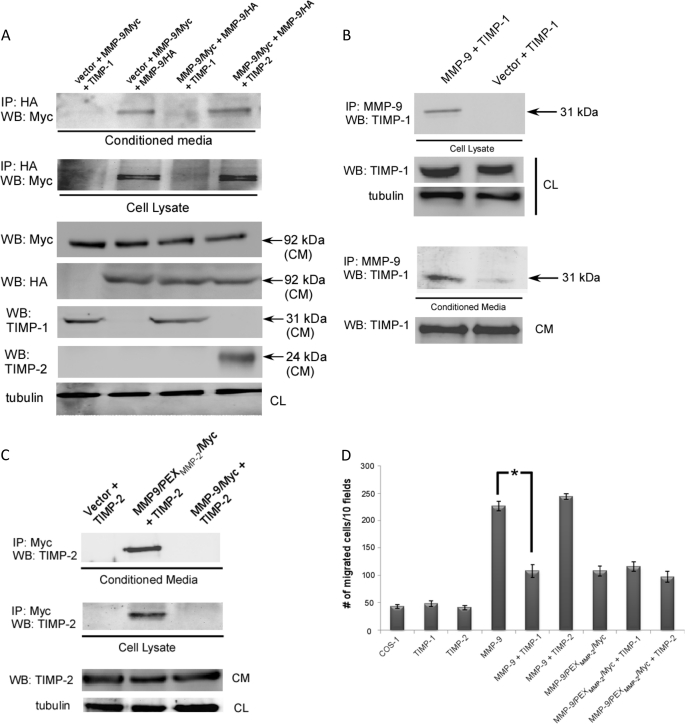
The role of TIMP-1 in interfering with MMP-9 homodimer formation remains controversial (25, 26). This discrepancy led us to re-evaluate the specific interference of TIMP-1 on pro-MMP-9 homodimer formation using the biochemical approach described above. Because MMP-9 homodimer formation is a prerequisite for enhancing cell migration (Fig. 2A), and TIMP-1 blocks MMP-9-induced cell migration (16), we hypothesized that binding of TIMP-1 to the PEX domain might prevent pro-MMP-9 homodimerization, thus, interfering with MMP-9-induced cell migration. To test this hypothesis, COS-1 cells transfected with both pro-MMP-9/Myc and pro-MMP-9/HA cDNAs along with TIMP-1 or TIMP-2 cDNAs were examined by the co-immunoprecipitation assay, as described above. As shown in Fig. 3A, co-expression of TIMP-1 with pro-MMP-9 cDNAs resulted in blocking MMP-9 homodimer formation both in the conditioned medium and cell lysate of transfected cells. As demonstrated by co-immunoprecipitation studies, abrogation of pro-MMP-9 homodimer formation by TIMP-1 coincided with heterodimer formation between pro-MMP-9 and TIMP-1 (Fig. 3B and supplemental Fig. S1E). In contrast, TIMP-2 has no effect on pro-MMP-9 homodimer formation (Fig. 3A), which is in agreement with previous observations that TIMP-2 only binds to the PEX domain of MMP-2, but not MMP-9 (Fig. 3C) (24).
FIGURE 3.
TIMP-1 interferes with MMP-9 homodimerization. A, TIMP-1, but not TIMP-2 interferes with MMP-9 dimerization in transfected COS-1 cells examined by a co-immunoprecipitation (IP) assay for conditioned medium (CM) and cell lysates (upper and middle panels). Western blotting (WB) using an aliquot of the conditioned medium and cell lysates (CL) was performed using anti-Myc, HA, TIMP-1, TIMP-2, and α/β-tubulin antibodies (lower panel, loading control). B, TIMP-1 co-precipitated with MMP-9 in both the cell lysate and the conditioned medium of transfected COS-1 cells examined by a co-immunoprecipitation assay. The reciprocal co-immunoprecipitation experiment is shown in supplemental Fig. 1E. The conditioned medium and cell lysate were examined by Western blotting using anti-TIMP-1 and α/β-tubulin antibodies for control of protein expression. C, MMP-9/PEXMMP-2/Myc, but not wild type MMP-9, co-precipitates with TIMP-2. COS-1 cells were co-transfected with a combination of cDNAs as indicated. The conditioned medium and cell lysate were examined by a co-immunoprecipitation assay. Aliquots of conditioned medium and cell lysate were examined by Western blotting using anti-TIMP-2 and MMP-9 antibody, respectively, for control of protein expression. D, TIMP-1, but not TIMP-2, interferes with MMP-9-induced cell migration. COS-1 cells transfected with corresponding cDNAs, as indicated, were subjected to a transwell migration assay. Three triplicate repeats were performed for each transfection and the experiment was repeated three times. *, p < 0.05.
Because TIMP-1 was shown to interfere with MMP-9 homodimerization, we then tested if the loss of homodimer formation of MMP-9 by TIMP-1 fails to enhance cell migration. By employing a transwell migration assay, we observed that co-expression of TIMP-1, but not TIMP-2, with pro-MMP-9 in COS-1 cells inhibited pro-MMP-9-enhanced cell migration, but had no apparent effect on pro-MMP9/PEXMMP-2/Myc-transfected cells (Fig. 3D). In addition, both TIMP-1 and TIMP-2 did not significantly inhibit MMP9/PEXMMP-2/Myc enhancement of COS-1 cell migration (Fig. 3D). These results suggest a unique role for TIMP-1 interference of both MMP-9 homodimerization and enhancement of cell migration.
Minimal Motif Required for MMP-9 Homodimer Formation
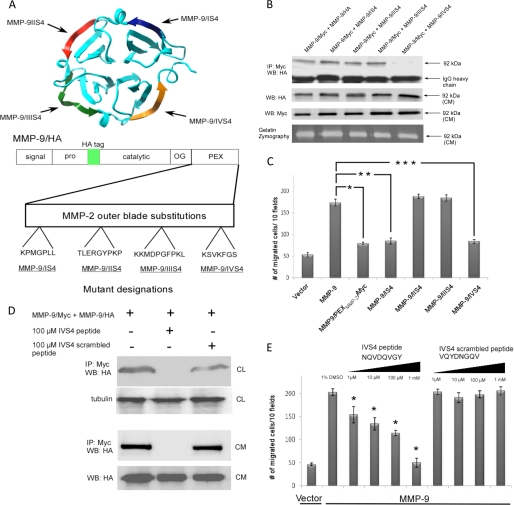
The PEX domain of MMPs exhibit similar structures composed of a disc-like shape, with the chain folded into a β-propeller structure that has a pseudo 4-fold symmetry. Each blade contains four anti-parallel β-strands with peptide loops linking one strand to the next as illustrated in Fig. 1B. Based on the crystal structure analysis of purified MMP-9 PEX domain from E. coli (6), homodimerization of MMP-9 occurs through an interaction of the outermost strand of the fourth blade of the PEX domains. However, the crystal structure of the MMP-9 PEX domain using the recombinant protein from E. coli has not been completely validated in a mammalian cell system. To test if the interaction interface of MMP-9 homodimer formation occurs through the outermost strand of the fourth blade of the PEX domains in mammalian cells, we employed a genetic approach to generate substituted mutations for the outermost β-strands of each blade within the MMP-9 PEX domain. The outermost β-strand of the fourth blade of the MMP-9/HA PEX domain (688NQVDQVGY695) was substituted by the corresponding region from MMP-2 PEX domain (586KSVKFGS592) to generate MMP-9/IVS4 chimera (Figs. 1A and 4A). As controls, three additional mutations for the outermost strands of blades I, II, and III were engineered by replacing with the corresponding sequence of MMP-2 (MMP-9/IS4, MMP-9/IIS4, and MMP-9/IIIS4) (Fig. 4A). Employing a co-immunoprecipitation assay for COS-1 cells co-transfected with different combinations of cDNAs, we observed that mutations of blades I, II, and III of the MMP-9 PEX domain had no effect on MMP-9 homodimer formation; whereas mutation of blade IV (MMP-9/IVS4) failed to dimerize (Fig. 4B). This defect was not due to the loss of the secretory ability of the mutant as evidenced by Western blot and gelatin zymography of the conditioned medium from transfected COS-1 cells (Fig. 4B). The four mutants and wild type MMP-9 were then evaluated for their ability to enhance cell migration using a transwell migration assay (Fig. 4C). Mutation of the fourth β-strand of the PEX domain of MMP-9 (MMP-9/IVS4) failed to enhance COS-1 cells migration, as compared with wild type MMP-9, whereas mutations of blades II and III induced cell migration as well as wild type MMP-9 (Fig. 4C). Unexpectedly, mutation of the first blade of the PEX domain of MMP-9 (MMP-9/IS4) also failed to induce cell migration. These data suggest that both blades I and IV of the MMP-9 PEX domain are required for the enhanced cell migration. Because the outermost β-strand of blade I is not involved in homodimer formation, another explanation for failure of migration of the MMP-9/IS4 mutant needs to be sought.
FIGURE 4.
Blade IV of the PEX domain of MMP-9 is required for homodimerization and cell migration. A, ribbon diagram of MMP-9 PEX domain (PDB 1ITV). Each outermost β-strand from the four blades was swapped with the corresponding MMP-2 PEX domain sequences (upper panel). Lower panel, schematic diagram of substitution mutations at the outermost β-strands of four blades of MMP-9 by corresponding MMP-2 sequences. B, requirement of blade IV for MMP-9 homodimerization: COS-1 cells transfected with MMP-9 chimeric cDNAs were examined by co-immunoprecipitation (IP) assay. Mutation at blade IV of the PEX domain of MMP-9 fails to co-precipitate with wild type MMP-9 (upper panel). The aliquot of condition medium (CM) was examined by Western blotting (WB) using anti-HA and anti-Myc antibody (middle panel) and by gelatin zymography (lower panel) for control of protein expression. C, mutations of the PEX domain or the outermost IS4 and IVS4 motifs of MMP-9 fail to enhance cell migration. COS-1 cells transfected with wild type and mutant MMP-9 cDNAs were examined by a transwell cell migration assay. Three triplicate repeats were performed for each transfection and the experiment was repeated three times. *, p < 0.05. D, design of an inhibitory peptide interfering with MMP-9 homodimer formation. A peptide mimicking MMP-9/IVS4 was chemically synthesized and incubated (100 μm) with COS-1 cell-transfected cDNAs as indicated. Scrambled peptide was used as a control. Both the cell lysates (CL) and conditioned medium were examined by a co-immunoprecipitation assay (upper and middle panel). An aliquot of the conditioned medium and cell lysate were examined by Western blotting using anti-HA and anti-α/β-tubulin as a control. E, dose-dependent inhibition (from 1 μm to 1 mm) of MMP-9-mediated cell migration by IVS4 peptides. COS-1 cells transfected with an empty vector or MMP-9 cDNAs were preincubated with 1% DMSO, the IVS4 peptide (NQVDQVGY), and IVS4 scrambled peptide (VQYDNGQV) for 30 min followed by a transwell chamber migration assay in the presence of different concentrations of peptides for 6 h. Each data point was performed in triplicate and the experiments were repeated three times (*, p < 0.05).
Blocking MMP-9-induced Cell Migration by Targeting the Homodimerization of MMP-9
Increased cell migration is involved in many pathological conditions including atherosclerosis and cancer; up-regulated MMP-9 has been implicated in these processes (4). One approach to blunt MMP-9-induced cell migration might be to target homodimer formation, which is a prerequisite for MMP-9-induced cell migration. Given the observation that the outermost β-strand of the fourth blade of the MMP-9 PEX domain is required for MMP-9 homodimerization formation, an 8-amino acid peptide (NQVDQVGY) was synthesized to mimic the outer β-strand of blade IV of the MMP-9 PEX domain (IVS4 peptide). As a control, a scrambled peptide with the same amino acids but rearranged in a different order (VQYDNGQV) was designed and synthesized. To test if the IVS4 peptide blocked MMP-9 homodimer formation, COS-1 cells transfected with MMP-9 cDNA were treated with 100 μm peptides for 24 h prior to a co-immunoprecipitation assay. As shown in Fig. 4D, the IVS4 peptide, but not scrambled peptide, efficiently blocked MMP-9 homodimer formation both in the conditioned medium and the cell lysate. To examine the potential therapeutic effect of this peptide, a transwell migration assay was performed. Interference with homodimerization by the peptide inhibited MMP-9-induced cell migration in a dose-dependent manner, whereas the scrambled peptide had no apparent effect (Fig. 4E). This IVS4 peptide had no effect on cell migration induced by MT1-MMP (supplemental Fig. S1F), an integral membrane matrix metalloproteinase capable of stimulating cell migration (27). These data suggest that the IVS4 peptide specifically inhibits MMP-9-induced cell migration through interference with MMP-9 homodimer formation.
Cross-talk between MMP-9 and CD44 Regulates Cell Migration
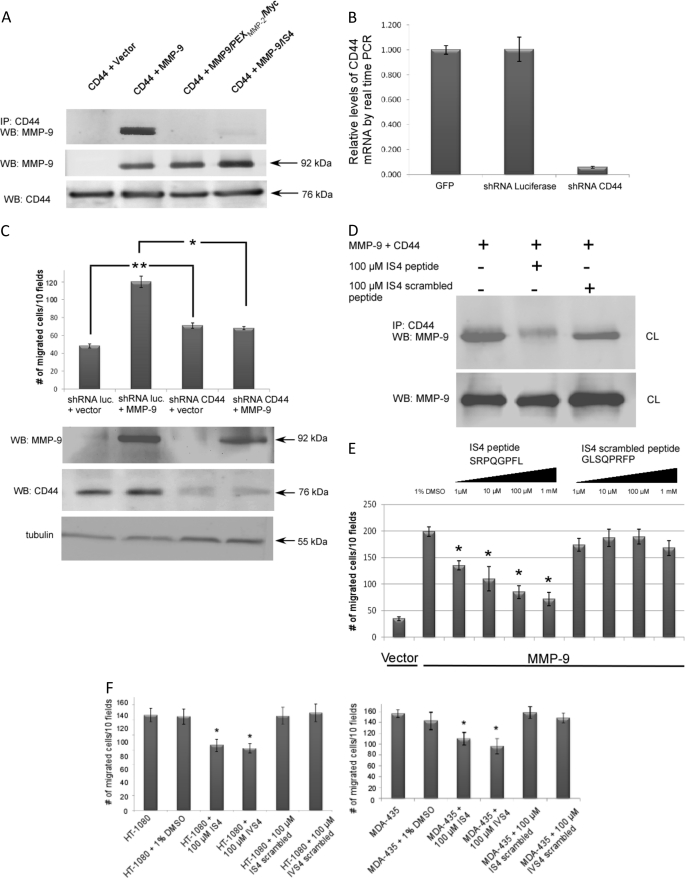
Although the outermost β-strand of blade I of MMP-9 is not required for MMP-9 homodimer formation (Fig. 4B), this motif is essential for MMP-9-induced cell migration (Fig. 4C). Given the evidence that MMP-9 interacts with CD44, a cell surface glycoprotein involved in cell-cell interactions, cell adhesion, and migration (7, 9, 28), we reasoned that MMP-9 may also form a heterodimer with CD44, which signals for cell migration. As examined by real time RT-PCR and immunofluorescent staining (Fig. 5B and supplemental Fig. S2B), COS-1 cells express endogenous CD44. Co-immunoprecipitation between CD44 and MMP-9 was assessed to determine the interaction between MMP-9 and CD44 in MMP-9-transfected COS-1 cells. Indeed, MMP-9 and CD44 co-precipitated in the transfected COS-1 cells (Fig. 5A), which confirms heterodimer formation of the two molecules. In contrast, swapping the PEX domain of MMP-9 by that of MMP-2 (MMP9/PEXMMP-2/Myc) resulted in failure to complex with CD44, suggesting that heterodimer formation between CD44 and MMP-9 is through the PEX domain of MMP-9. To further determine which motif is required for the heterodimer formation, CD44 was co-expressed with MMP-9 mutants (MMP-9/IS4, MMP-9/IIS4, MMP-9/IIIS4, and MMP-9/IVS4) in transfected COS-1 cells, followed by co-immunoprecipitation. MMP-9/IS4 failed to form a complex with CD44, indicating that the outer β-strand of blade I interacts with CD44 at the cell surface (Fig. 5A). Mutations of outer β-strand of blades II, III, and IV of the MMP-9 PEX domain did not interfere with CD44/MMP-9 heterodimerization (supplemental Fig. S2A).
FIGURE 5.
CD44 serves as a docking molecule for MMP-9 on the cell surface and facilitates MMP-9-mediated cell migration. Effect of specific peptides on cell migration. A, CD44 forms a complex with MMP-9 in co-transfected COS-1 cells examined by a co-immunoprecipitation (IP) assay. MMP-9 in the conditioned medium (CM) and CD44 in the cell lysate (CL) were used as control for protein expression. B, silencing of CD44 in COS-1 cells using a shRNA approach. COS-1 cells were infected with retrovirus containing shRNA against CD44 and luciferase control. Total RNAs were extracted followed by a real time RT-PCR analysis. The relative quantitative value of CD44 expression was normalized against housekeeping genes, HPRT1 and GAPDH. Each bar represents the mean ± S.E. C, MMP-9 enhancement of cell migration is dependent on CD44. CD44-silenced COS-1 cells were transfected with MMP-9 or vector control followed by a transwell migration assay (upper panel). Each data point was performed in triplicate and the experiment was repeated three times (*, p < 0.05; **, p < 0.01). Western blotting for MMP-9, CD44, and α/β-tubulin (lower panel) was performed to monitor protein expression in the transfected COS-1 cells. D, peptides mimicking the outermost b-stand of blade I interfere with MMP-9 heterodimer formation (upper panel). 20 μg of total cell lysates were examined by Western blotting using anti-MMP-9 antibody to determine equal expression of MMP-9 (lower panel). IS4 and IS4 scrambled peptides (100 μm) were incubated with the cell lysate for 24 h prior to a co-immunoprecipitation assay. E, dose-dependent inhibition (from 1 μm to 1 mm) of MMP-9-mediated cell migration by IS4 peptides. COS-1 cells transfected with MMP-9 cDNAs or vector control were preincubated with 1% DMSO, IS4 peptide (SRPQGPFL), or IS4 scrambled peptide (GLSQPRFP) at different doses for 30 min followed by a transwell chamber migration assay. Each data point was performed in triplicate and the experiment was repeated three times (*, p < 0.05). F, inhibition of cell migration by MMP-9-specific peptides in human cancer cells expressing endogenous MMP-9. Human fibrosarcoma HT-1080 cells and breast cancer MDA-MB-435 cells were preincubated with control and specific peptides (100 μm) for 30 min followed by a transwell chamber migration assay. Each data point was performed in triplicate and the experiment was repeated three times (*, p < 0.05).
To explore the role of CD44 in MMP-9-induced cell migration, we silenced endogenous CD44 expression in COS-1 cells using a short hairpin RNA (shRNA) approach. As determined by real time RT-PCR, mRNA of CD44 was suppressed more than 20-fold in CD44 shRNA expressing COS-1 cells (Fig. 5B). No detectable CD44 protein was found in COS-1 cells expressing CD44 shRNA examined by immunofluorescent staining and flow cytometry analysis (supplemental Fig. S2, B and C). By functional assay, the enhanced cell migration of MMP-9-transfected COS-1 cells was reduced significantly when CD44 was silenced, indicating that CD44 is a critical molecule in the MMP-9 cell migration signaling pathway (Fig. 5C). Interestingly, CD44-silenced COS-1 cells migrated to higher levels as compared with shRNA luciferase control and wild type COS-1 cells. This observation might be due to decreased cell-cell and cell-matrix interactions by silencing CD44 expression in COS-1 cells (29).
To further examine the importance of cross-talk between CD44 and MMP-9 in cell migration, an 8-amino acid peptide (SRPQGPFL) was synthesized to mimic the outermost β-strand of the first blade of the MMP-9 PEX domain. As a control, a scrambled peptide with the same amino acids but rearranged in a different order (GLSQPRFP) was synthesized. COS-1 cells transfected with CD44 and MMP-9 cDNAs were treated with IS4 and IS4 scrambled peptides for 24 h followed by monitoring complex formations between CD44 and MMP-9 using a co-immunoprecipitation assay. IS4 peptide interfered with the CD44/MMP-9 heterodimer, whereas the IS4 scrambled peptide had minimal effect (Fig. 5D). In addition, the IS4 peptide displayed dose-dependent inhibition of MMP-9-induced cell migration, but not the scrambled peptide (Fig. 5E). The IVS4 peptide also inhibited cell migration. Although the potency of IS4 and IVS4 peptides (IC50: 12 and 50 μm, respectively) are suboptimal from a therapeutic standpoint, generation of the mimicking inhibitory peptides provide a proof of concept for specific targeting of an important molecule in human disease. To further validate the efficiency and relevance of the IS4 and IVS4 peptide effect, HT-1080 and MDA-MB-435, human cancer cell lines, which produce high levels of MMP-9 (30, 31), were tested. Using the transwell migration assay, both IS4 and IVS4 peptides significantly inhibited migration of these two cell lines (Fig. 5F).
MMP-9-CD44-EGFR Axis in Cell Migration
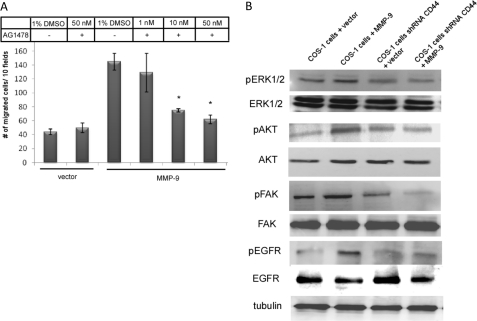
It has been reported that CD44 activates receptor tyrosine kinases initiating signaling for cell migration (11). To further investigate which receptor tyrosine kinases are involved in the MMP-9-CD44 signaling pathway for cell migration, eight small molecule inhibitors (AG1024, PD173074, PHA665752, Genistein, PP2, AG490, AG1296, and AG1478) targeting different receptor tyrosine kinase pathways were employed and tested in a transwell migration assay. Among the eight tested inhibitors, three inhibitors (AG1478, AG1024, and PD173074) significantly reduced cell migration in MMP-9-transfected COS-1 cells as compared with the DMSO control (supplemental Fig. S3A). The most efficient inhibitor, an EGFR inhibitor (AG1478), demonstrated a dose-dependent inhibition of cell migration (Fig. 6A), suggesting that EGFR may be activated in MMP-9-transfected COS-1 cells.
FIGURE 6.
CD44 activates EGFR to regulate MMP-9-enhanced cell migration. A, dose-dependent inhibition of MMP-9-mediated cell migration using an EGFR inhibitor (AG1478). COS-1 cells transfected with vector or MMP-9 cDNAs were treated with different concentrations of AG1478 for 30 min before being subjected to a transwell migration assay; p values reflect comparison with MMP-9-transfected cells: *, p < 0.05. B, activation of EGFR downstream effectors in COS-1 cells transfected with MMP-9 cDNAs, but not in CD44-silenced COS-1 cells. Cell lysates of transfected COS-1 cells were prepared and subjected to Western blot analysis using antibodies against pERK1/2, ERK1/2, pAKT, AKT, pFAK, FAK, pEGFR, EGFR, and α/β-tubulin antibodies.
To test if MMP-9-CD44 interactions results in activation of EGFR, COS-1 and CD44-silenced COS-1 cells transfected with MMP-9 cDNA as well as vector control were examined by Western blotting using a specific anti-phosphorylated EGFR antibody. As shown in Fig. 6B, expression of MMP-9 in wild type COS-1 cells resulted in increased phosphorylation of EGFR as compared with vector control. This activation of EGFR did not occur in CD44-silenced COS-1 cells transfected with MMP-9 cDNA. These data are consistent with a pathway by which a complex formation between MMP-9 and CD44 leads to activation of EGFR. To further dissect the MMP-9-CD44-EGFR pathway, we examined downstream effectors of activated EGFR, including pERK, pAKT, and pFAK, which have been implicated in EGFR-induced cell migration (11). An increase of pERK1/2, pAKT, pFAK (less visibly evident), and pEGFR was observed in MMP-9-transfected COS-1 cells, but not MMP-9-transfected CD44-silenced COS-1 cells or vector control, confirming the MMP-9-CD44-EGFR signaling axis in MMP-9-induced cell migration (Fig. 6B and supplemental Fig. S3B).
In another approach to identify signaling pathway(s) differentially activated in MMP-9 expressing COS-1 cells, we employed Kinexus Antibody Microarrays. These arrays simultaneously detected the presence and relative quantities of 500 pan-specific and 300 phospho-site antibodies in MMP-9-transfected COS-1 cells, as compared with vector control cells. We confirmed the elevated activity of EGFR, pFAK, pERK, and pAKT in MMP-9-transfected COS-1 cells (data not shown). This study demonstrates that MMP-9-enhanced protease-independent cell migration involves the coordination of PEX domain homodimerization and CD44 heterodimerization leading to EGFR activation.
DISCUSSION
The proteolytic activity of MMP-9 has been implicated in various physiologic and pathologic conditions. Inhibition of the catalytic domain has been a long term focus of MMP research. More recently, the PEX domain of MMPs was demonstrated to be critical for mediating protein-protein interactions and enhancing cell migration (7, 16, 32). We previously demonstrated that the proteolytic activity of MMP-9 is not required for MMP-9-induced cell migration (16). In the current report, we have further dissected the mechanism of MMP-9-induced cell migration in cells expressing recombinant or endogenous MMP-9. By swapping the MMP-9 PEX with that of MMP-2, we demonstrate the unique homo- and heterodimerization properties of the MMP-9 PEX domain in cell migration. Based on our mutagenesis analysis, specific peptides preventing MMP-9 dimerization have been developed and demonstrated to interfere with MMP-9-induced cell migration.
The proteolytic activity of secreted MMPs is inhibited by TIMP binding in a 1:1 ratio to the catalytic core domain. It has also been shown that the MMP-9 PEX domain binds specifically to TIMP-1, whereas the PEX domain of MMP-2 binds to TIMP-2 (24, 25). Based on a crystallography analysis, TIMP-2 forms a complex with pro-MMP-2 through interaction with blade IV of the PEX domain of MMP-2 (24). A precise interaction between the pro-MMP-9 PEX domain and TIMP-1, however, has not been solved by crystallography. Using a biochemical approach, we herein confirm that TIMP-1 interacts with pro-MMP-9. This observation raises the question: can TIMP-1 competitively interfere with pro-MMP-9 homodimerization and hence, inhibit MMP-9 biological properties? Using a biochemical assay, we show that TIMP-1 interfered with MMP-9 homodimerization either competitively, caused by overlapping contact areas, or allosterically, caused by conformational changes to PEX-9 that render it incompetent for homodimerization. This interaction reduced MMP-9-mediated cell migration.
TIMP-1 has been demonstrated to form a complex in a 1:1 ratio with MMP-9 (33). Roderfeld et al. (34) reported that TIMP-1:MMP-9 forms in the Golgi apparatus prior to secretion using a fluorescence resonance energy transfer (FRET) approach. A similar result was reported using pulse-chase analysis (26). However, given the evidence that most MMP-9 bound to the cell surface is TIMP free (35, 36) and specific peptides blocking MMP-9 dimerization inhibits MMP-9-enhanced cell migration (Fig. 5, E and F), we hypothesize that pro-MMP-9·TIMP-1 complexes dissociate before interacting with cell surface molecule(s), e.g. CD44. Docking of the TIMP-1-free MMP-9 monomer at the cell surface by CD44 leads to MMP-9 dimerization and cell migration. In support of our hypothesis, it has been demonstrated that cell surface-bound MMP-9 is less accessible for binding to TIMPs than soluble MMP-9, which is possibly due to the dimerization of MMP-9. A 21-fold higher IC50 in terms of inhibition of membrane-bound MMP-9 by TIMPs as compared with soluble MMP-9 has been reported (36). Due to the limitation of current technology, direct evidence for the effect of TIMP-1 on MMP-9 dimerization and migration remains to be fully understood.
Homodimer formation of MMP-9 features functions that are not present in monomeric PEX of MMP-9, such as electrostatic potential at the physical-chemical level (6). Moreover, an extended hydrophobic surface patch, accessible to solvent in the monomeric MMP-9 PEX domain, becomes buried upon dimerization. Finally, the dimeric complex brings all domains, including the catalytic domains of the two MMP-9 monomers, within a defined distance from each other. This distance restraint is likely to influence a multitude of functions, including accessibility for proteolytic activation (26), localization to the extracellular matrix (37), and substrate recognition and processing (38).
Employing an ectopic expression system followed by gelatin zymography, we demonstrated that ∼16% of MMP-9 is secreted as homodimer, whereas ∼84% of MMP-9 is secreted as the monomeric form (supplemental Fig. S4A). Because MMP-9 examined in this study was overexpressed in COS-1 cells, which might not be representative of pathophysiologic conditions in most cell types, HT-1080 human fibrosarcoma and MDA-MB-435 human breast cancer cell lines were utilized to further determine the ratio of dimer versus monomer of MMP-9. Both cell lines expressed comparable levels of MMP-9 dimers (18% in HT1080 cells and 9% in MDA-MB-435 cells) as examined by gelatin zymogram (supplemental Fig. S4B).
Because the SDS detergent used in gelatin zymography may dissociate MMP dimers into monomers, thereby affecting the MMP-9 dimers:monomers ratio, we employed a gel filtration technique to separate dimers from monomers prior to gelatin zymography (supplemental Fig. S4C). This approach confirmed the MMP-9 dimerization data and is in agreement with a previous report showing that MMP-9 dimers are SDS stable (26).
We, and others, have demonstrated that MMP-9 plays an important role in cell migration (3, 16, 31, 39). However, given technical limitations, the individual role of MMP-9 dimers versus monomers in cell migration has not been previously determined. By using mutagenesis and biochemical approaches, we demonstrated that MMP-9 dimers, but not monomers, are required for full enhancement of MMP-9-mediated cell migration. Based on diminished migration of cells transfected with MMP-9 mutants, which are unable to form dimers, we conclude that this minor population of MMP-9 (10–15% MMP-9 dimers; supplemental Fig. S4A) is responsible for functional activity. Furthermore, peptides interfering with MMP-9 dimerization abrogated MMP-9-enhanced cell migration in COS-1 cells transfected with MMP-9 cDNA and in cells expressing endogenous MMP-9 (HT-1080 and MDA-435) (Fig. 5F). These data further support our conclusion that dimerization of MMP-9 is required for MMP-9-enhanced cell migration.
Expression of MMP-9 in cells has been found to result in activation of MAPK and PI3K pathways; activation of these signaling molecules has been linked to the phosphorylation status of receptor tyrosine kinases (40, 41). However, it remains to be explained how MMP-9 initiates signaling transduction leading to enhanced cell migration. To pinpoint this signaling mechanism, we first screened the effect of inhibitors against different receptor tyrosine kinase pathways on MMP-9-mediated cell migration. We identified that phosphorylation of EGFR is involved in the MMP-9 signaling cascade. This observation was supported by four lines of evidence: 1) inhibition of EGFR activation by specific inhibitors abrogates MMP-9-mediated cell migration; 2) activation of EGFR in MMP-9-transfected cells was identified by antibody array analysis; 3) enhanced phosphorylation of EGFR in MMP-9-transfected cells was confirmed by Western blotting; and 4) downstream effectors of active EGFR, including pERK1/2, pAKT, and pFAK in MMP-9 expressing cells were up-regulated or phosphorylated, as examined by antibody array and immunoblotting assays.
It has been reported that CD44 and EGFR interactions promote cell migration involving activation of Akt, FAK, and MMP-2 (11). In a separate report, Yu and Stamenkovic (9) demonstrated that CD44 serves as a cell surface docking molecule for MMP-9. We now present data showing that MMP-9 transduces signals to activate EGFR through cross-talk with CD44 to initiate a cell migration signaling cascade. This conclusion is based on the fact that silencing of CD44 in MMP-9 expressing cells abrogated activation of EGFR. In addition, mutations of blades II, III, and IV did not interfere with the binding of CD44, whereas blade I of the MMP-9 PEX domain mediates MMP-9 and CD44 interactions, increases phosphorylation of EGFR which, in turn, activates FAK, AKT, and ERK leading to cell migration. Collectively, these data suggest that homodimerization of MMP-9 is not a prerequisite for binding CD44 as evidenced by co-immunoprecipitation between MMP-9/Blade IV (only observed as a monomeric form) and CD44 (Fig. 4B and supplemental Fig. S2A).
Docking of pro-MMP-9 at the cell surface by CD44 was reported to lead to activation of pro-MMP-9 in osteoclasts and results in enhanced cell migration (42). In contrast, we demonstrate that proteolytic activity of MMP-9 is not a prerequisite for MMP-9-mediated cell migration. Four lines of evidence support our previous conclusion that MMP-9-mediated COS-1 cell migration is independent of enzymatic activity (16): 1) constitutively inactive MMP-9 (MMP-9E-A mutation) induced cell migration in COS-1 cells as well as wild type MMP-9; 2) synthetic MMP inhibitors did not interfere with MMP-9-mediated COS-1 cell migration; 3) TIMP-1, but not TIMP-2 abrogates MMP-9-mediated cell migration; and 4) no activated MMP-9 was detected by the fluorogenic peptide assay, gelatin zymography, and Western blotting in our transfected cells. This discrepancy could be due to cell type differences, but the exact mechanism remains to be examined.
Targeting the PEX domain of MMP-9 has been our goal in recent years because most MMP inhibitors blocking enzymatic activities failed to prove useful in several clinical trials (43, 44). Three approaches have been used to target the PEX domain of MMP-9: 1) anti-functional antibody: an anti-functional antibody targeting the MMP-9 PEX domain blocked Schwann cell migration (45); 2) recombinant protein of the PEX domain of MMP-9: Ezhilarasan et al. (46) reported that excess recombinant MMP-9 PEX protein retarded endothelial cell migration; and 3) inhibitory peptides: Bjorklund et al. (47) reported that random peptides isolated from a phage display peptide library inhibit cell migration and tumor formation by targeting either the MMP-9 catalytic domain or PEX domain. These studies emphasized the importance of the PEX domain of MMP-9 in cell migration and targeting this domain is a viable approach to abrogate pathologic cell migration. To date, no small molecular compounds have been developed for targeting the PEX domain of MMP-9. Our strategy for the development of synthetic compounds targeting the functional MMP-9 PEX domain is to identify targeting motifs within the PEX domain based on a mutagenesis approach followed by chemical synthesis. Our peptides mimicking the outermost β-strand of blades I and IV of the MMP-9 PEX domain inhibited MMP-9-mediated cell migration in a dose-dependent manner. The core structure of these peptides is being evaluated by NMR and chemical synthesis will be followed.
In summary, our studies demonstrate a novel axis of MMP-9-CD44-EGFR in which MMP-9 initiates cross-talk between CD44 and EGFR, which in turn activates downstream effectors for cell migration. We further pinpoint two critical motifs in the PEX domain of MMP-9 required for cell migration. The synthetic inhibitory peptides mimicking these motifs demonstrate proof of principle that developing pharmaceutical compounds targeting these regions is a useful approach to impair cell migration during pathological processes.
Supplementary Material
Acknowledgments
Special thanks to Jennifer DeLeon and Kevin Zarrabi for critical reading and A. J. Campbell for the help with Figs. 1A and 4A.
This work was supported, in whole or in part, by National Institutes of Health Grant 5RO1CA11355301A1 and a Walk-for-Beauty Foundation grant (to J. C.), NYSTAR Faculty Development Program Grant C040076 (to N. S. S.), a Veterans Affairs Merit Review grant, and the Carol Baldwin Breast Cancer Foundation grant (to S. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- MMP
- matrix metalloproteinase
- PEX
- hemopexin domain
- TIMP
- tissue inhibitor of metalloproteinase
- EGFR
- epidermal growth factor receptor
- DMSO
- dimethyl sulfoxide.
REFERENCES
- 1.Morrison C. J., Butler G. S., Rodríguez D., Overall C. M. (2009) Curr. Opin. Cell. Biol. 21, 645–653 [DOI] [PubMed] [Google Scholar]
- 2.Nagase H., Woessner J. F., Jr. (1999) J. Biol. Chem. 274, 21491–21494 [DOI] [PubMed] [Google Scholar]
- 3.Björklund M., Koivunen E. (2005) Biochim. Biophys. Acta 1755, 37–69 [DOI] [PubMed] [Google Scholar]
- 4.Sternlicht M. D., Werb Z. (2001) Annu. Rev. Cell. Dev. Biol. 17, 463–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zucker S., Cao J., Chen W. T. (2000) Oncogene 19, 6642–6650 [DOI] [PubMed] [Google Scholar]
- 6.Cha H., Kopetzki E., Huber R., Lanzendörfer M., Brandstetter H. (2002) J. Mol. Biol. 320, 1065–1079 [DOI] [PubMed] [Google Scholar]
- 7.Redondo-Muñoz J., Ugarte-Berzal E., García-Marco J. A., del Cerro M. H., Van den Steen P. E., Opdenakker G., Terol M. J., García-Pardo A. (2008) Blood 112, 169–178 [DOI] [PubMed] [Google Scholar]
- 8.Van den Steen P. E., Van Aelst I., Hvidberg V., Piccard H., Fiten P., Jacobsen C., Moestrup S. K., Fry S., Royle L., Wormald M. R., Wallis R., Rudd P. M., Dwek R. A., Opdenakker G. (2006) J. Biol. Chem. 281, 18626–18637 [DOI] [PubMed] [Google Scholar]
- 9.Yu Q., Stamenkovic I. (1999) Genes Dev. 13, 35–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monferran S., Paupert J., Dauvillier S., Salles B., Muller C. (2004) EMBO J. 23, 3758–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim Y., Lee Y. S., Choe J., Lee H., Kim Y. M., Jeoung D. (2008) J. Biol. Chem. 283, 22513–22528 [DOI] [PubMed] [Google Scholar]
- 12.Tsukita S., Oishi K., Sato N., Sagara J., Kawai A., Tsukita S. (1994) J. Cell. Biol. 126, 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bourguignon L. Y., Gilad E., Brightman A., Diedrich F., Singleton P. (2006) J. Biol. Chem. 281, 14026–14040 [DOI] [PubMed] [Google Scholar]
- 14.Murai T., Miyauchi T., Yanagida T., Sako Y. (2006) Biochem. J. 395, 65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Massova I., Kotra L. P., Fridman R., Mobashery S. (1998) FASEB J. 12, 1075–1095 [PubMed] [Google Scholar]
- 16.Dufour A., Sampson N. S., Zucker S., Cao J. (2008) J. Cell. Physiol. 217, 643–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zucker S., Lysik R. M., Zarrabi M. H., Moll U. (1993) Cancer Res. 53, 140–146 [PubMed] [Google Scholar]
- 18.Cao J., Drews M., Lee H. M., Conner C., Bahou W. F., Zucker S. (1998) J. Biol. Chem. 273, 34745–34752 [DOI] [PubMed] [Google Scholar]
- 19.Cao J., Chiarelli C., Richman O., Zarrabi K., Kozarekar P., Zucker S. (2008) J. Biol. Chem. 283, 6232–6240 [DOI] [PubMed] [Google Scholar]
- 20.Zucker S., Wieman J. M., Lysik R. M., Wilkie D., Ramamurthy N. S., Golub L. M., Lane B. (1987) Cancer Res. 47, 1608–1614 [PubMed] [Google Scholar]
- 21.Imai K., Okada Y. (2008) Nat. Protoc. 3, 1111–1124 [DOI] [PubMed] [Google Scholar]
- 22.Cao J., Rehemtulla A., Bahou W., Zucker S. (1996) J. Biol. Chem. 271, 30174–30180 [DOI] [PubMed] [Google Scholar]
- 23.Davis B. G. (2004) Science 303, 480–482 [DOI] [PubMed] [Google Scholar]
- 24.Morgunova E., Tuuttila A., Bergmann U., Tryggvason K. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7414–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldberg G. I., Strongin A., Collier I. E., Genrich L. T., Marmer B. L. (1992) J. Biol. Chem. 267, 4583–4591 [PubMed] [Google Scholar]
- 26.Olson M. W., Bernardo M. M., Pietila M., Gervasi D. C., Toth M., Kotra L. P., Massova I., Mobashery S., Fridman R. (2000) J. Biol. Chem. 275, 2661–2668 [DOI] [PubMed] [Google Scholar]
- 27.Cao J., Kozarekar P., Pavlaki M., Chiarelli C., Bahou W. F., Zucker S. (2004) J. Biol. Chem. 279, 14129–14139 [DOI] [PubMed] [Google Scholar]
- 28.Yu Q., Stamenkovic I. (2000) Genes Dev. 14, 163–176 [PMC free article] [PubMed] [Google Scholar]
- 29.Acharya P. S., Majumdar S., Jacob M., Hayden J., Mrass P., Weninger W., Assoian R. K., Puré E. (2008) J. Cell. Sci. 121, 1393–1402 [DOI] [PubMed] [Google Scholar]
- 30.Partridge J. J., Madsen M. A., Ardi V. C., Papagiannakopoulos T., Kupriyanova T. A., Quigley J. P., Deryugina E. I. (2007) J. Biol. Chem. 282, 35964–35977 [DOI] [PubMed] [Google Scholar]
- 31.Rolli M., Fransvea E., Pilch J., Saven A., Felding-Habermann B. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9482–9487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stefanidakis M., Karjalainen K., Jaalouk D. E., Gahmberg C. G., O'Brien S., Pasqualini R., Arap W., Koivunen E. (2009) Blood 114, 3008–3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogata Y., Itoh Y., Nagase H. (1995) J. Biol. Chem. 270, 18506–18511 [DOI] [PubMed] [Google Scholar]
- 34.Roderfeld M., Graf J., Giese B., Salguero-Palacios R., Tschuschner A., Müller-Newen G., Roeb E. (2007) Biol. Chem. 388, 1227–1234 [DOI] [PubMed] [Google Scholar]
- 35.Toth M., Gervasi D. C., Fridman R. (1997) Cancer Res. 57, 3159–3167 [PubMed] [Google Scholar]
- 36.Owen C. A., Hu Z., Barrick B., Shapiro S. D. (2003) Am. J. Respir. Cell Mol. Biol. 29, 283–294 [DOI] [PubMed] [Google Scholar]
- 37.Olson M. W., Toth M., Gervasi D. C., Sado Y., Ninomiya Y., Fridman R. (1998) J. Biol. Chem. 273, 10672–10681 [DOI] [PubMed] [Google Scholar]
- 38.Collier I. E., Saffarian S., Marmer B. L., Elson E. L., Goldberg G. (2001) Biophys. J. 81, 2370–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burg-Roderfeld M., Roderfeld M., Wagner S., Henkel C., Grötzinger J., Roeb E. (2007) Int. J. Oncol. 30, 985–992 [DOI] [PubMed] [Google Scholar]
- 40.Ellerbroek S. M., Halbleib J. M., Benavidez M., Warmka J. K., Wattenberg E. V., Stack M. S., Hudson L. G. (2001) Cancer Res. 61, 1855–1861 [PubMed] [Google Scholar]
- 41.Yao J. S., Chen Y., Zhai W., Xu K., Young W. L., Yang G. Y. (2004) Circ. Res. 95, 364–371 [DOI] [PubMed] [Google Scholar]
- 42.Samanna V., Ma T., Mak T. W., Rogers M., Chellaiah M. A. (2007) J. Cell. Physiol. 213, 710–720 [DOI] [PubMed] [Google Scholar]
- 43.Coussens L. M., Fingleton B., Matrisian L. M. (2002) Science 295, 2387–2392 [DOI] [PubMed] [Google Scholar]
- 44.Pavlaki M., Zucker S. (2003) Cancer Metastasis Rev. 22, 177–203 [DOI] [PubMed] [Google Scholar]
- 45.Mantuano E., Inoue G., Li X., Takahashi K., Gaultier A., Gonias S. L., Campana W. M. (2008) J. Neurosci. 28, 11571–11582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ezhilarasan R., Jadhav U., Mohanam I., Rao J. S., Gujrati M., Mohanam S. (2009) Int. J. Cancer. 124, 306–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Björklund M., Heikkilä P., Koivunen E. (2004) J. Biol. Chem. 279, 29589–29597 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.