Abstract
We demonstrate here that the bioactive lipid sphingosine 1-phosphate (S1P) uses sphingosine 1-phosphate receptor 4 (S1P4) and human epidermal growth factor receptor 2 (HER2) to stimulate the extracellular signal regulated protein kinase 1/2 (ERK-1/2) pathway in MDA-MB-453 cells. This was based on several lines of evidence. First, the S1P stimulation of ERK-1/2 was abolished by JTE013, which we show here is an S1P2/4 antagonist and reduced by siRNA knockdown of S1P4. Second, the S1P-stimulated activation of ERK-1/2 was almost completely abolished by a HER2 inhibitor (ErbB2 inhibitor II) and reduced by siRNA knockdown of HER2 expression. Third, phyto-S1P, which is an S1P4 agonist, stimulated ERK-1/2 activation in an S1P4- and HER2-dependent manner. Fourth, FTY720 phosphate, which is an agonist at S1P1,3,4,5 but not S1P2 stimulated activation of ERK-1/2. Fifth, S1P stimulated the tyrosine phosphorylation of HER2, which was reduced by JTE013. HER2 which is an orphan receptor tyrosine kinase is the preferred dimerization partner of the EGF receptor. However, EGF-stimulated activation of ERK-1/2 was not affected by siRNA knockdown of HER2 or by ErbB2 (epidermal growth factor receptor 2 (or HER2)) inhibitor II in MDA-MB-453 cells. Moreover, S1P-stimulated activation of ERK-1/2 does not require an EGF receptor. Thus, S1P and EGF function in a mutually exclusive manner. In conclusion, the magnitude of the signaling gain on the ERK-1/2 pathway produced in response to S1P can be increased by HER2 in MDA-MB-453 cells. The linkage of S1P with an oncogene suggests that S1P and specifically S1P4 may have an important role in breast cancer progression.
Keywords: G Protein-coupled Receptors (GPCR), Growth Factors, Receptor Tyrosine Kinase, Signal Transduction, Sphingolipid
Introduction
There is increasing evidence that suggests a role for the bioactive lipid sphingosine 1-phosphate (S1P)2 in breast cancer. S1P binds to a family of five GPCR, termed S1Pn (where n = 1–5), that are differentially coupled to heterotrimeric G proteins (Gi, Gq, and G12/13) to regulate various effectors such as MAP kinases linked to diverse cellular processes such as proliferation, cell survival, and differentiation (1–13). The specificity of signaling in terms of which kinase modules are activated is dependent upon the receptor sub-type involved and is cell context-specific. S1P is produced by the enzyme sphingosine kinase (SK), which catalyzes the phosphorylation of sphingosine to produce S1P. SK (which exists as two isoforms termed SK1 and SK2) is activated by agonists such as antigen, platelet-derived growth factor, nerve growth factor, tumor necrosis factor α, and epidermal growth factor (EGF), resulting in an increase in intracellular S1P. SK1 activation and/or translocation is regulated by phosphorylation catalyzed by ERK-1/2 and by CIB1 (calcium and integrin-binding protein 1) (14, 15).
S1P has an important role in breast cancer cells in terms of regulating survival, proliferation, and migration (16–18). For instance, ectopic expression of SK1 increased S1P levels, estrogen-dependent tumorigenesis, and blocked apoptosis of MCF-7 cells induced by anticancer drugs, sphingosine, and tumor necrosis factor α (16). SK1 and S1P also are required for EGF-induced MCF-7 migration, proliferation, and cell survival (18). S1P also stimulates breast cancer cell growth through activation of the serum response element and indirectly by enhancing insulin-like growth factor (IGF) II synthesis and function (19).
The HER2/neu/c-erbB-2 gene encodes a 185-kDa transmembrane receptor tyrosine kinase, which is related to other members of the EGF receptor family (20). Moreover, the overexpression of HER2/neu is found in up to 30% of primary breast cancers and increased tumor invasion, poor prognosis, and therapeutic resistance is correlated with its expression (21). A soluble ligand for HER2 has not been identified, although HER2 operates as a shared receptor subunit of other ErbBs. In this regard, HER2 is a heterodimerization partner of the EGF receptor (22). HER2 delays EGF dissociation from its receptor, improves coupling of the EGF receptor and stimulation of the ERK-1/2 pathway, and impedes EGF receptor down-regulation. Thus, HER2 is a master regulator that drives epithelial cell proliferation. An example of this is evident from studies that demonstrate that the ectopic expression of HER2/neu in MCF-7 (estrogen receptor (ER)-positive/HER2-negative) cells stimulates the PI3K/Akt pathway and down-regulates p53 (23), which increases cell survival.
Breast cancer cell lines can be categorized into three major phenotypic groups that include the following: (i) luminal epithelial-like ER-positive/HER2-negative cells (e.g. MCF-7 cells, weakly invasive); (ii) weakly luminal epithelial-like HER2-positive cells (e.g. MDA-MB-453 cells); and (iii) stromal/mesenchymal phenotype (e.g. MDA-MB-231), which are characterized as ER-negative/HER2-negative cells. In this study, we have used MDA-MB-453 cells to investigate the role of S1P in regulating the ERK-1/2 pathway, which is well established as having a role in cancer metastasis.
EXPERIMENTAL PROCEDURES
Materials
All general biochemicals were from Sigma-Aldrich. High glucose Dulbecco's modified Eagle's medium and European fetal calf serum, penicillin-streptomycin were from Invitrogen. DharmaFECTTM 2 reagent was from Dharmacon (Dharmacon, Cromlington, UK). BioScriptTM was from Bioline (London, UK). HER2 and S1P2 and S1P4 siRNA and anti-phosphorylated ERK-1/2 antibody were from Santa Cruz Biotechnology. EGF receptor siRNA was a gift from V. Natarajan (University of Chicago). Anti-ERK2 and anti-HRP tyrosine phosphate antibodies were from BD Transduction Laboratories. Anti-HER2 antibody was from New England Biolabs Ltd. S1P and phyto-S1P were from Avanti Polar Lipids, EGF was from Sigma, JTE013 was from Tocris Biosciences (Bristol, UK), and CAY10444 and SEW2871 were from Cayman Chemicals (Tallinn, Estonia). Recombinant heregulin, ErbB2 inhibitor II, AG 879, and AG 1478 were from Merck Biosciences (Nottingham, UK). FTY720 and FTY720 phosphate were gifts from R. Bittman (The City University of New York).
Cell Culture
MDA-MB-231 and MDA-MB-453 breast cancer cell lines were obtained from the ATCC (Rockville, MD) and were grown in a monolayer culture in high glucose DMEM with 10% European FCS and 1% Pen-Strep (104 units/ml penicillin G sodium, 10 mg/ml streptomycin sulfate) at 37 °C with 5% CO2. HEK 293 cells were grown as above with the exception that minimum essential medium was used instead of DMEM. MCF-7 Neo and MCF-7 HER2 breast cancer cells (from R. Schiff, Baylor College) were grown in a monolayer culture in DMEM with 10% European FCS and 1% Pen-Strep, 0.4% geneticin, and 15 μg/ml insulin at 37 °C with 5% CO2. HTC4 cells stably expressing S1P2, S1P3, or S1P4 receptors were maintained in DMEM containing 10% fetal bovine serum and 500 μg/ml of geneticin.
siRNA Treatment
Knockdown of HER2 expression was achieved using sequence-specific HER2 siRNA (HER2 receptor siRNA (AAGGGGCUGGCUCCGAUGUAUUUdTdT and AAAUACAUCGGAGCCAGCCCCUUdTdT)) and scrambled siRNA as control (GCGUCGGAGUGGCAUCUUAAUGUdTdT and ACAUUAAGAUGCCACUCCGACGCdTdT). The HER2 receptor-targeted sequence is AAGGGGCUGGCUCCGAUGUAUUU.
siRNA transfection was performed according to the protocol detailed by Dharmacon. Briefly, cells grown on 24-well plates were transfected with 100–400 nm siRNA prepared in a mix with DharmaFECTTM 2 reagent and DMEM containing 10% European FCS. Cells were also treated with EGF receptor siRNA (100 nm, Dharmacon) in the same manner. The cells were cultured for 48 h before being serum-starved for 24 h prior to stimulation.
RNA Extraction and Real-time Quantitative RT-PCR of S1P Receptor mRNA
RNA was isolated from MDA-MB-453 cells using TRIzol (Invitrogen). cDNA synthesis was performed by using the SuperScript First Strand Synthesis kit (Invitrogen) as recommended by the manufacturer. Amplification was performed for 40 cycles at 94 °C for 30 s, 50 °C for 60 s, and 72 °C for 60 s with initial activation of enzyme at 94 °C for 1 min using the ABI Model 7300 Real Time PCR machine. The following primer pairs were used: GAPDH, (forward) CTGCACCACCAACTGCTTAG and (reverse) GGGCCATCCACAGTCTTCT; S1P1, (forward) GCTGGGTCATCTCCCTCAT and (reverse) GCAGTTCCAGCCCATGAT; S1P2, (forward) CCAACAAGGTCCAGGAACAC and (reverse) GCAACAGAGGATGACGATGA; S1P3, (forward) TCAGGGAGGGCAGTATGTTC and (reverse) CCAGTAAGCTGCAGGTGGA; S1P4, (forward) GAAGACGGTGCTGATGATCC and (reverse) CAGAGGTTGGAGCCAAAGAC; and S1P5, (forward) GGTGAGCGAGGTCATCGT and (reverse) CCAGGAGCAGGAACATGG.
Quantitative values were obtained from the threshold cycle value (Ct). GAPDH was quantified as an internal RNA control, and each sample was normalized on the basis of its GAPDH content. Samples were run in quadruplicate. Data are representative of three independent experiments.
Transfection
HEK 293 and MDA-MB-453 cells were transfected with HA-tagged S1P4 plasmid constructs using LipofectamineTM 2000 reagent according to the manufacturer's instructions. Transfection was performed for 24 h at 37 °C before serum starvation for a further 24 h prior to harvesting in Laemmli buffer and analysis by SDS-PAGE and Western blotting.
Western Blotting
Analysis of proteins by SDS-PAGE and Western blotting was performed as described previously (24) using anti-phosphorylated ERK1/2, anti-ERK2, anti-HER2, and anti-HRP phosphotyrosine antibodies.
Calcium Assays
Monitoring of S1P-evoked changes in intracellular Ca2+ was done in HTC4 cells stably expressing S1P1, S1P2, S1P3, or S1P4 receptors originally described by An et al. (10), which were generously provided by Dr. Edward Goetzl (University of California, San Francisco). The assay protocol was identical to that described in our previous publication (25). Wild type HTC4 cells do not respond to S1P with changes in [Ca2+]i. HTC4 cells stably expressing each receptor were plated onto black-wall clear-bottom 96-well plates (Corning Inc. Life Sciences, Acton, MA) at a density of 5 × 104 cells/well and cultured overnight. The following day, the culture medium was replaced with modified Krebs buffer (120 mm NaCl, 5 mm KCl, 0.62 mm MgSO4, 1.8 mm CaCl2, 10 mm HEPES, 6 mm glucose, pH 7.4), and the cells were serum-starved for 4 h. Subsequently, cells were loaded with Fura-2 AM (Invitrogen) for 30 min in modified Krebs buffer containing 2% (v/v) pluronic acid. After incubating the cells with Fura-2 AM, the cells were rinsed with Krebs buffer, and changes in the intracellular Ca2+ concentration were monitored by determining the ratio of emitted light intensities at 520 nm in response to excitation at 340 and 380 nm using FLEXstation II (Molecular Devices, Sunnyvale, CA). Each well was monitored for 80 s. To test antagonist activity of the inhibitors JTE013 and CAY10444, increasing concentrations of the inhibitors were mixed with a constant concentration of S1P and added automatically after 15 s of baseline measurement. Each test was performed in quadruplicate. Significant difference between two experimental groups was determined by the Student's t test at a p value of 0.05. Data were plotted and fitted a sigmoid function by using the nonlinear curve-fitting feature of KaleidaGraph (Synergy Software, Essex Junction, VT).
Immunoprecipitation
The medium was removed, and cells were lysed in ice-cold immunoprecipitation buffer (1 ml) containing 20 mm Tris/HCl, 137 mm NaCl, 2.7 mm KCl, 1 mm MgCl2, 1 mm CaCl2, 1% (v/v) Nonidet P-40 (Nonidet P-40), 10% (v/v) glycerol, 1 mg/ml bovine serum albumin, 0.5 mm sodium orthovanadate, 0.2 mm phenylmethylsulfonyl fluoride, leupeptin, and aprotinin (all protease inhibitors were at 10 μg/ml, pH 8) for 10 min at 4 °C. The material was harvested, centrifuged at 22,000 × g for 5 min at 4 °C, and 200–400 μl of cell lysate supernatant (equalized for protein, 0.5–1 mg/ml) combined with 20 μl of protein A or G-Sepharose and incubated for 20 min at 4°c. The samples were centrifuged at 22,000 × g, and the supernatant was taken for immunoprecipitation with anti-HER2 or anti-ERK-1/2 antibody (5 μg of anti-HER2 antibody or 3 μg of anti-ERK-1/2 antibody and 20–40 μl of one part immunoprecipitation buffer and one part protein A or G Sepharose CL4B, respectively). After agitation for 2 h at 4 °C, the immune complex was collected by centrifugation at 22,000 × g for 15 s at 4 °C. Immunoprecipitates were washed twice with buffer A containing 10 mm HEPES, pH 7, 100 mm NaCl, 0.2 mm phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 20 μg/ml aprotinin, and 0.5% (v/v) Nonidet P-40 and once in buffer A without Nonidet P-40. The immunoprecipitates were then combined with boiling sample buffer containing 62 mm Tris/HCl, pH 6.7, 1.25% (w/v) SDS, 10% (v/v) glycerol, 3.75% (v/v) mercaptoethanol, and 0.05% (w/v) bromphenol blue. The samples were then subjected to SDS-PAGE and Western blotting.
Myelin Basic Protein Kinase Assay
Immunoprecipitates were combined with a phosphorylation mixture containing 20 mm HEPES, pH 7, 5 mm-MgCl2, 25 μm ATP, and 5μCi [γ-32P]ATP, and 1 μg myelin basic protein (MBP) and incubated at 30 °C for 10 min. Incubations were terminated by addition of boiling sample buffer and subjected to SDS-PAGE.
Immunohistochemistry
The cell pellet slides were first dewaxed and rehydrated through a series of xylene and alcohol washes. Antigen retrieval was performed for S1P2 by microwaving the slides under pressure in a Tris EDTA buffer for 5 min (pH 8.0) and for S1P4 in 10 mm citrate buffer at 96 °C in for 20 min. Endogenous peroxidase was blocked using 3% hydrogen peroxide for 20 min, and nonspecific background staining was reduced by blocking with a 1:20 concentration of horse serum diluted in Tris-buffered saline for 30 min. The sections were incubated with the primary antibody for S1P2 and S1P4 (Exalpha, Shirley, MA). Each antibody was incubated at a dilution of 1:100 at 4 °C overnight. EnVision-HRP conjugate (DAKO, Cambridgeshire, UK) was used for signal amplification and positive staining was identified using 3,3′-diaminobenzidine chromagen (Vector Laboratories). The slides were then counterstained with hematoxylin and Scott's tap water substitute before dehydration and mounting (26, 27).
Densitometry
Densitometric quantification of Western blots was performed using the Molecular Analyst Software (Bio-Rad). Statistical analysis was performed with GraphPad Prism software, using one-way analysis of variance followed by a Newman-Keuls post hoc test.
RESULTS
Regulation of ERK-1/2 by S1P in MDA-MB-453 Cells
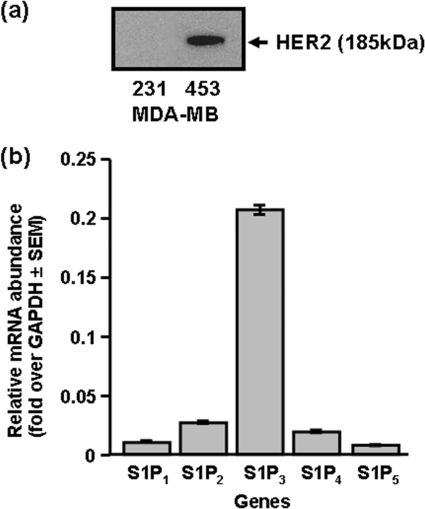
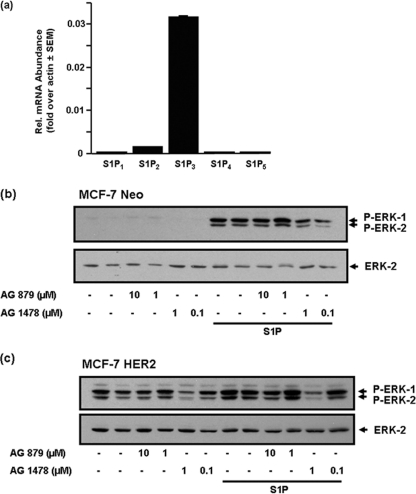
MDA-MB-453 cells are HER2+ as evidenced by immunodetection of HER2 (Mr = 185 kDa) with anti-HER2 antibody (Fig. 1a). In these experiments, MDA-MB-231 cells were used as a negative control for Western blotting, as these cells are HER2-null (Fig. 1a). Quantitative real-time PCR revealed that S1P3 was the most abundant S1P receptor mRNA transcript, with small quantities of S1P2/4, and very minor expression of S1P1/5 mRNA (Fig. 1b).
FIGURE 1.
S1P receptors and HER2 in MDA-MB-453 cells. a, Western blot of MDA-MB-453 and MDA-MB-231 cell lysates with anti-HER2 antibody. b, bar graph showing relative mRNA expression of S1P1–5 in MDA-MB-453 cells, determined using QRT-PCR.
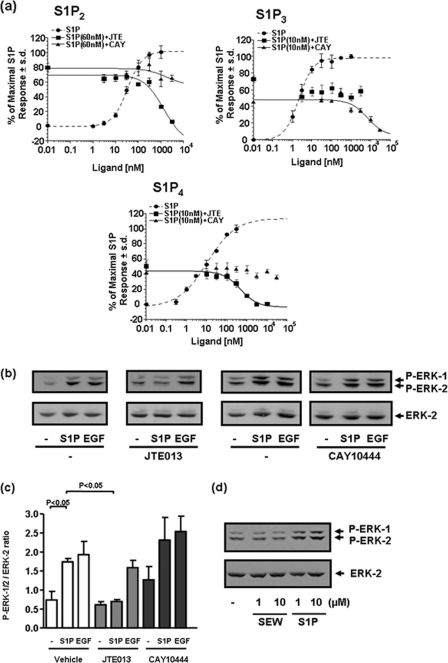
Treatment of MDA-MB-453 cells with S1P stimulated the phosphorylation/activation of ERK-1/2, which was reduced by pretreating these cells with pertussis toxin (PTX, which uncouples GPCR from Gi) (Fig. 2, a and b). These data suggest that S1P uses a heterotrimeric G protein-coupled receptor to regulate the ERK-1/2 pathway. PTX did not significantly reduce EGF-induced activation of ERK-1/2 (Fig. 2, a and b, p > 0.05 for EGF + PTX versus EGF). The identity of the S1P receptor involved in regulating the ERK-1/2 pathway was evaluated using pharmacological agents that demonstrate selectivity at S1P receptors. We used JTE013, which is reported to be an S1P2-selective antagonist (28), and CAY10444 (29), which is an S1P3 antagonist. To further characterize the pharmacological specificity of JTE013 and CAY10444, we used HTC4 cells in which S1P2,3,4 were separately stably expressed, and intracellular calcium mobilization was measured using the Fura-2 indicator dye. We found that JTE013 reduced S1P-stimulated calcium mobilization in cells overexpressing S1P2, whereas CAY10444 was very weakly effective at concentrations >1 μm (Fig. 3a). In contrast, JTE013 had no effect on calcium mobilization induced by S1P in cells overexpressing S1P3, whereas CAY10444 reduced this response (Fig. 3a), thereby confirming specificity of CAY10444 at S1P3. Surprisingly, we found that JTE013 can also function as an S1P4 antagonist, as evidenced by results showing that JTE013 potently reduced S1P-stimulated calcium mobilization (Ki = 236.9 nm) in cells overexpressing S1P4, whereas CAY10444 was without effect (Fig. 3a). As there are no selective S1P4 antagonists available, we used JTE013 as a tool to investigate the role of S1P2/4 in mediating the effect of S1P on the activation of ERK-1/2 in MDA-MB-453 cells. We found that the S1P-induced activation of ERK-1/2 was substantially reduced by JTE013 and was not affected by CAY10444 (Fig. 3, b and c). Confirmation that MDA-MB-453 cells lack functional S1P1 was evidenced by the finding that the S1P1-selective agonist SEW2871 was without effect on ERK-1/2 activation (Fig. 3d).
FIGURE 2.
S1P signaling in MDA-MB-453 cells. MDA-MB-453 cells were pretreated with PTX (0.1 μg/ml) for 20 h prior to stimulation with and without EGF (50 ng/ml) or S1P (5 μm) for 10 min. a, the Western blot shows the effect of PTX on S1P- and EGF-stimulated activation of ERK-1/2. Images are from the same Western blot. Phosphorylated ERK-1/2 was detected on Western blots probed with anti-phosphorylated ERK-1/2 antibody. Blots were also probed with anti-ERK-1/2 antibody to ensure equal protein loading. b, shown is a bar graph quantifying the effect of PTX on ERK-1/2 activation. Results are expressed as P-ERK-1/2:ERK-2 ratios (mean ± S.D.) for n = 3 experiments.
FIGURE 3.
S1P receptor specificity by JTE013 and CAY10444 and their effect in MDA-MB-453 cells. a, specificity of S1P antagonists: graphs showing the effect of JTE013 or CAY10444 on S1P-stimulated calcium mobilization in HTC4 cells overexpressing recombinant S1P2 or S1P3 or S1P4. b–d, MDA-MB-453 cells were pretreated with vehicle, JTE013 (10 μm), or CAY10444 (10 μm) for 15 min prior to stimulation with and without EGF (50 ng/ml) or S1P (5 μm or indicated concentration) or SEW 2871 (1 or 10 μm) for 10 min. b, the Western blot shows the effect of JTE013 or CAY10444 on S1P- and EGF-stimulated activation of ERK-1/2 in MDA-MB-453 cells. The images for control/JTE013 with and without S1P/EGF and control/CAY10444 with and without S1P/EGF experiments were taken from the same Western blots, respectively. c, shown is a bar graph showing the quantification of the effect of JTE013 and CAY10444 on ERK-1/2 activation; d, Western blot showing the lack of effect of SEW2781 on ERK-1/2 activation in MDA-MB-453 cells. In b and d, phosphorylated ERK-1/2 was detected on Western blots probed with anti-phosphorylated ERK-1/2 antibody. Blots were also probed with anti-ERK-1/2 antibody to ensure equal protein loading. In c, results are expressed as P-ERK-1/2:ERK-2 ratios (mean ± S.D.) for n = 3 experiments.
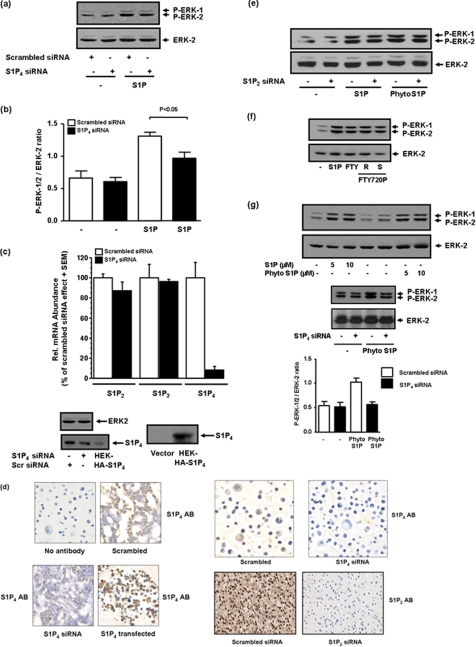
We also used specific siRNA approaches to knock down expression of the S1P4 receptor in MDA-MB-453 cells. In this regard, we found that siRNA knockdown of S1P4 reduced the S1P-stimulated activation of ERK-1/2 (Fig. 4, a and b). We confirmed by QRT-PCR that S1P4 siRNA reduced S1P4 mRNA transcript and had no effect on the S1P2 or S1P3 mRNA transcript (Fig. 4c). We also detected S1P4 protein (Mr = 42 kDa) in lysates of MDA-MB-453 cells on Western blots probed with anti-S1P4 antibody and found that S1P4 siRNA partially reduced expression (55 ± 12% reduction, n = 5) of the protein (Fig. 4c). In addition, immunohistochemical staining of MDA-MB-453 cells with anti-S1P4 antibody demonstrated that siRNA knockdown of S1P4 reduced immunostaining (Fig. 4d). We could also confirm increased immunostaining when MDA-MB-453 cells were transfected with a S1P4 plasmid construct (Fig. 4d). Collectively, these data confirm expression of S1P4 protein in MDA-MB-453 cells, specificity of the antibody, and effectiveness of S1P4 siRNA treatment.
FIGURE 4.
The effect of S1P, FTY720, FTY720 phosphate, and phyto-S1P on the ERK-1/2 pathway and role of S1P2/4 receptor in MDA-MB-453 cells. MDA-MB-453 cells were treated with scrambled siRNA or S1P4 siRNA (200 nm, 48 h) or S1P2 siRNA (100 nm, 48 h) prior to stimulation with and without S1P (1 μm). Cells were also stimulated with FTY720 or FTY720 phosphate or phyto-S1P (at the indicated concentration) for 10 min. Western blots are shown. a, the effect of siRNA knock down of S1P4 on S1P-stimulated activation of ERK-1/2. b, bar graph showing quantification of the effect of S1P4 siRNA on the S1P-induced activation of ERK-1/2 activation. c, quantification by QRT-PCR of S1P2,3,4 mRNA expression in cells treated with scrambled or S1P4 siRNA. Rel., relative. Also shown is a Western blot probed with anti-S1P4 antibody to show siRNA knock down of S1P4 in MDA-MB-453 cells (n = 5 cell samples). HEK-HA-S1P4 are samples from HA-S1P4 overexpressing HEK 293 cell. d, immunohistochemical staining showing the effect of S1P2 siRNA and S1P4 siRNA on the expression of S1P2 and S1P4 protein, respectively, in MDA-MB-453 cells. Cells were also co-stained with hematoxylin. AB denotes S1P2 or S1P4 antibody. Also shown is an S1P4 antibody immunostaining of MDA-MB-453 cells transfected with S1P4 plasmid construct. e, the lack of effect of S1P2 siRNA on S1P- or phyto-S1P (5 μm)-stimulated activation of ERK-1/2. f, the effect of FTY720 (5 μm), (R)- and (S)-FTY720 phosphate (each at 5 μm) on ERK-1/2 activation. g, the effect of siRNA knockdown of S1P4 on phyto-S1P (5 μm)-stimulated activation of ERK-1/2. Also included is a bar graph showing the quantification of the effect of S1P4 siRNA on phyto-S1P-induced activation of ERK-1/2. Results in b and g are expressed as P-ERK-1/2:ERK-2 ratios (mean ± S.D.) for n = 3 experiments. Results in a, e, f, and g are representative of three separate experiments. Phosphorylated ERK-1/2 was detected on Western blots probed with anti-phosphorylated ERK-1/2 antibody. ERK-2 was also detected with anti-ERK-2 antibody either on reprobes or in the same samples run on a separate SDS-PAGE to ensure comparable protein loading.
Evidence to exclude S1P2 was obtained using S1P2 siRNA, which did not reduce activation of ERK-1/2 by S1P (Fig. 4e). immunohistochemical staining of MDA-MB-453 cells with anti-S1P2 antibody demonstrated that siRNA knockdown of S1P2 eliminated immunostaining (Fig. 4d), thereby confirming expression of S1P2 protein in MDA-MB-453 cells, specificity of the antibody and successful knockdown of S1P2. Knockdown of S1P2 with specific siRNA was more effective than corresponding knockdown of S1P4 with its respective siRNA (Fig. 4d). The residual S1P4 expression after siRNA knockdown might explain the incomplete abolition of S1P-stimulated ERK-1/2 activation with S1P4 siRNA (Fig. 4a), whereas JTE013 via S1P4 antagonism is completely effective (Fig. 3b).
In addition, we used FTY720 (which is phosphorylated by SK2 to FTY720 phosphate) and FTY720 phosphate, which is an agonist at S1P1,3,4,5 but not S1P2 and which stimulated activation of ERK-1/2 in MDA-MB-453 cells (Fig. 4f). Finally, we used phyto-S1P, which is an agonist at S1P4 (30) and demonstrated that this agent also induced activation of ERK-1/2 (Fig. 4g), which was reduced by siRNA knockdown of S1P4 (Fig. 4g), but not S1P2 (Fig. 4e). S1P4 knockdown was more effective against phyto-S1P compared with S1P. This apparent anomaly might be explained by a model in which there is higher fractional receptor occupancy with phyto-S1P compared with S1P for ERK-1/2 activation and where efficacy for each phyto-S1P-bound S1P4 receptor is less than for each S1P-bound S1P4 receptor. We conclude that S1P4 mediates the effect of S1P on ERK-1/2 activation in MDA-MB-453 cells.
S1P Receptor Functional Interaction with HER2
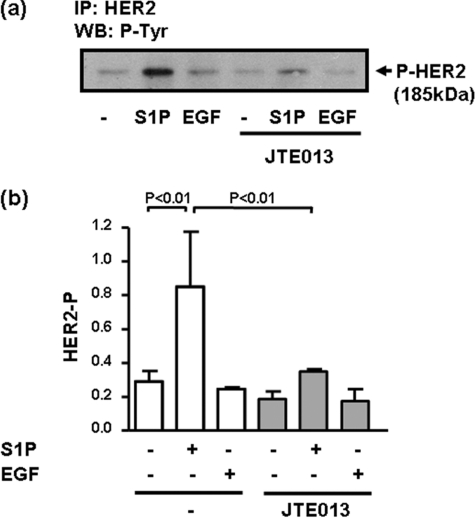
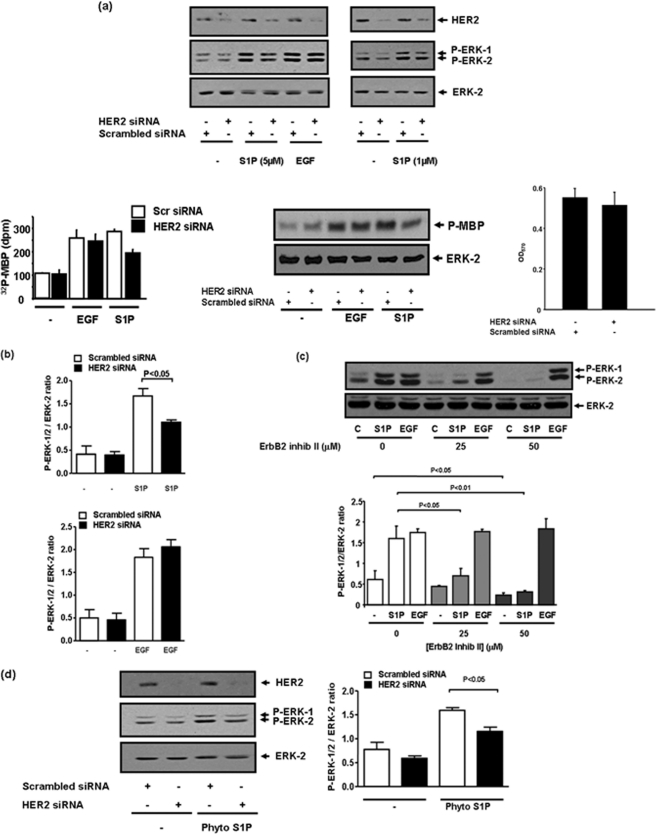
Evidence that a functional interaction occurs between S1P and HER2 was obtained by the finding that S1P but not EGF stimulated the tyrosine phosphorylation of HER2, and this was severely reduced by JTE013 (Fig. 5, a and b). Evidence for the involvement of HER2 in regulating S1P4 signaling to the ERK-1/2 pathway was demonstrated by results showing that siRNA knockdown HER2 expression reduced S1P-stimulation of ERK-1/2 by ∼50% compared with cells treated with scrambled siRNA (Fig. 6, a and b). The incomplete reduction in ERK-1/2 activation might be due to residual HER2 expression after siRNA treatment. To confirm data obtained by Western blotting with antiphospho-ERK-1/2 antibody, we immunoprecipitated ERK-1/2 with anti-ERK-1/2 antibody and measured ERK-1/2 activity against MBP in the immunoprecipitates. This assay confirmed that S1P or EGF stimulated ERK-1/2 activity (Fig. 6a). Moreover, siRNA knockdown of HER2 reduced the stimulation of ERK-1/2 activity by S1P and had no effect on the response to EGF (Fig. 6a). HER2 siRNA had no effect on cell integrity as assessed using an 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay (Fig. 6a). We also used the HER2 inhibitor, ErbB2 inhibitor II (4-(3-phenoxyphenyl)-5-cyano-2H-1,2,3-triazole), which is a cell-permeable HER2 ATP binding kinase inhibitor and reduces phosphorylation of HER2 in MDA-MB-453 cells but not that of an overexpressed EGF receptor in MDA-MB-468 cells, even at concentrations as high as 100 μm (31). The inhibitor was discovered from a computer-aided drug design approach and searched from molecule libraries. Modeling has also been used to demonstrate binding of the inhibitor in the HER2 ATP binding site (31).
FIGURE 5.
S1P stimulates tyrosine phosphorylation of HER2. MDA-MB-453 cells were pretreated with JTE013 (10 μm) for 15 min prior to stimulation with and without EGF (50 ng/ml) or S1P (5 μm) for 10 min. a, the Western blot (WB) is of anti-HER2 immunoprecipitates probed with HRP-anti-phosphotyrosine antibody and demonstrates that S1P but not EGF increases the tyrosine phosphorylation of HER2. b, bar graph showing quantification of the effect of S1P, EGF and JTE013 on HER2 tyrosine phosphorylation (mean ± S.D.) for n = 3 experiments.
FIGURE 6.
Role of HER2 in regulating S1P-induced activation of ERK-1/2 in MDA-MB-453 cells. MDA-MB-453 cells were pretreated with scrambled siRNA or a HER2 siRNA (100 or 400 nm, 48 h) to knock down expression of HER2 or ErbB2 inhibitor II (at indicated concentrations) for 10 min prior to stimulation with and without S1P (at the indicated concentrations) or EGF (50 ng/ml) or phyto-S1P (1 μm) or FTY720 (5 μm) for 10 min. a, Western blots showing the effect of siRNA (400 nm) knockdown of HER2 on S1P- (1 and 5 μm) and EGF-stimulated activation of ERK-1/2. In addition, cell lysates were immunoprecipitated with anti-ERK-1/2 antibody and analyzed for ERK-1/2 activity with MBP as the substrate. The autoradiograph shows the siRNA knockdown of HER2 reduces S1P- (5 μm, 10 min) but not EGF-stimulated activation of ERK-1/2 (upper panel). Total ERK-2 inputs are also shown in the lower panel. The bar graph demonstrates quantification (mean ± S.D.) of the kinase assay using Cherenkov counting (p < 0.01 for S1P-stimulated HER2 siRNA-treated cells versus S1P-stimulated scrambled siRNA-treated cells, n = 4). Also shown is the siRNA knockdown of HER2 (Mr = 185 kDa) detected on Western blots probed with anti-HER2 antibody. The bar graph shows that siRNA knockdown of HER2 had no effect on cell viability as assessed in the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. b, bar graph showing quantification of the effect of siRNA knockdown of HER2 on S1P (1 μm)- and EGF-stimulated ERK-1/2 activation (mean ± S.D.). c, Western blot showing the effect of ErbB2 inhibitor II on S1P (5 μm)-stimulated activation of ERK-1/2 and lack of effect on the response to EGF. Also shown is a bar graph of the effect of ErbB2 inhibitor (inhib) II on S1P- and EGF-stimulated ERK-1/2 activation. d, Western blot showing the effect of siRNA (100 nm) knockdown of HER2 on phyto-S1P- (1 μm) stimulated activation of ERK-1/2. Also included is a bar graph showing quantification of the effect of HER2 siRNA on the phyto-S1P-induced activation of ERK-1/2 activation. These are representative results from three separate experiments. Results in b–d are expressed as P-ERK-1/2:ERK-2 ratios (mean ± S.D.) for n = 3 experiments. Phosphorylated ERK-1/2 was detected on Western blots with anti-phosphorylated ERK-1/2 antibody. Blots were also probed with anti-ERK-1/2 antibody to ensure equal protein loading.
We found that the pretreatment of MDA-MB-453 cells with the ErbB2 inhibitor II reduced basal ERK-1/2 phosphorylation (Fig. 6c), suggesting a tonic influence of HER2 on this pathway. However, this does not explain the involvement of HER2 in S1P4 receptor signaling, as S1P-stimulated ERK-1/2 activation was almost completely abolished by ErbB2 inhibitor II, whereas the response to EGF was unaffected (Fig. 6c). The ability of ErbB2 inhibitor II to reduce basal ERK-1/2 activation differs from HER2 siRNA, which had no significant effect (Fig. 6, a and b). These findings suggest that the knockdown of HER2 with siRNA, which is incomplete, might not be sufficient to ablate basal ERK-1/2 activation but is able to reduce S1P signaling via S1P4. Thus, basal and S1P-stimulated ERK-1/2 activation may have different requirements for HER2, e.g. less HER2 is required to sustain the basal ERK-1/2 activation compared with the S1P-stimulated activation of ERK-1/2.
In addition, phyto-S1P-stimulated activation of ERK-1/2 was also reduced by siRNA knockdown of HER2 (Fig. 6d). We have therefore demonstrated that S1P binding to S1P4 engages HER2 to regulate the ERK-1/2 pathway in MDA-MB-453 cells.
EGF Regulation of ERK-1/2 in MDA-MB-453 and MCF-7 Cells
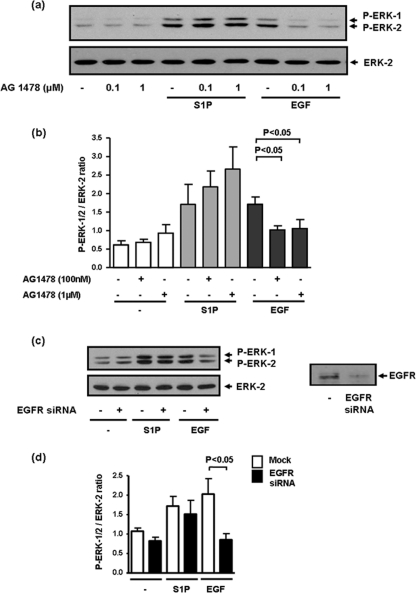
HER2 is an orphan receptor tyrosine kinase and is the preferred dimerization partner of the EGF receptor. However, we have demonstrated here that the EGF-induced activation of ERK-1/2 was not reduced by the siRNA knockdown of HER2 (Fig. 6, a and b). Thus, S1P and EGF function in a mutually exclusive manner. In addition, S1P does not use the EGF receptor tyrosine kinase to regulate ERK-1/2. Thus, the activation of ERK-1/2 by EGF was reduced by the EGF receptor tyrosine kinase inhibitor AG 1478 (Fig. 7, a and b) and by siRNA knockdown of EGF receptor expression (Fig. 7, c and d). However, the S1P-stimulated activation of ERK-1/2 was not modulated by AG 1478 (Fig. 7, a and b) or by siRNA knockdown of the EGF receptor (Fig. 7, c and d). Further evidence to support divergent signaling by S1P and EGF with respect to HER2 was the finding that unlike S1P, EGF did not induce the tyrosine phosphorylation of HER2 (Fig. 5, a and b).
FIGURE 7.
EGF-stimulated activation of ERK-1/2 occurs via a divergent signaling pathway compared with S1P in MDA-MB-453 cells. MDA-MB-453 cells were pretreated with AG 1478 (0.1 or 1 μm, 15 min) or siRNA EGF receptor (100 nm, 48 h) prior to stimulation with and without S1P (5 μm) or EGF (50 ng/ml) for 10 min. a and c, Western blot showing the effect of AG 1478 (a) and siRNA (c) knockdown of EGF receptor on the EGF- and S1P-stimulated activation of ERK-1/2. Also shown in c is the siRNA knockdown of EGF receptor (Mr = 170 kDa) detected on Western blots probed with anti-EGF receptor antibody. Bar graphs showing quantification of the effect of AG 1478 (b) and siRNA (d) knockdown of EGF receptors on EGF- and S1P-stimulated ERK-1/2 activation. Results in b and d are expressed as P-ERK-1/2:ERK-2 ratios (mean ± S.D.) for n = 3 experiments. In a and c, phosphorylated ERK-1/2 was detected on Western blots probed with anti-phosphorylated ERK-1/2 antibody. Blots were also probed with anti-ERK-1/2 antibody to ensure equal protein loading.
S1P3 has no functional role in terms of regulating ERK-1/2 signaling in response to S1P in MDA-MB-453 cells (Fig. 3b). On the contrary, S1P4 signaling appears to predominate. We therefore asked the question: what is the molecular organization of signaling from the S1P3 receptor in other breast cancer cell types that lack S1P4? For this purpose, we used a breast cancer cell line, ER+ MCF-7 cells, where S1P3 mRNA is abundantly expressed but where S1P4 mRNA is absent (Fig. 8a). Moreover, S1P-stimulated activation of ERK-1/2 is known to be mediated by the S1P3 receptor and involves transactivation of the EGF receptor (S1P increases tyrosine phosphorylation of the EGF receptor) in these cells (32). Indeed, we have confirmed that siRNA knockdown of S1P3 or use of the S1P3 antagonist, CAY10444, reduced the S1P-stimulated activation of ERK-1/2 in both MCF-7 Neo (expressing the neomycin resistance gene) and MCF-7 HER2 cells (HER218 cells, stably expressing HER2) by >90% (data not shown). We show here that the stimulation of ERK-1/2 by S1P in MCF-7 Neo and MCF-7 HER2 cells was reduced by pre-treating cells with AG 1478 (Fig. 8, b and c). The higher basal ERK-1/2 activation in MCF-7 HER2 cells compared with MCF-7 Neo cells might be due to the EGF receptor, as this was also reduced by AG 1478 (Fig. 8c). In conclusion, S1P/S1P3 stimulation of ERK-1/2 is characterized by a requirement for EGF receptor tyrosine kinase activity in MCF-7 cells. In addition, the S1P stimulation of ERK-1/2 in MCF-7 Neo (which lack HER2) and MCF-7 HER2 cells was unaffected by the HER2 kinase inhibitor, AG 879 (Fig. 8, b and c).
FIGURE 8.
The requirement for EGF receptor in the S1P stimulation of ERK-1/2 in MCF-7 cells. a, Bar graph showing relative (Rel.) expression of S1P1–5 mRNA transcript in MCF-7 cells, determined using QRT-PCR. MCF-7 Neo cells (b) or MCF-7 HER2 cells (c) were pre-treated with either vehicle (DMSO), AG 1478 or AG 879 at the indicated concentrations for 15 min and then treated with and without S1P (1 μm) for 5 min. The Western blots show inhibition of S1P-stimulated ERK-1/2 by AG 1478 but not AG 879. In b and c, phosphorylated ERK-1/2 was detected on Western blots probed with anti-phosphorylated ERK-1/2 antibody. Blots were also probed with anti-ERK-2 antibody to ensure equal protein loading. Results in b and c are representative from three separate experiments.
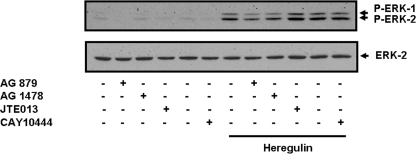
We also tested whether heregulin might participate in S1P signaling in MDA-MB-453 cells. Interestingly, heregulin-stimulated activation of ERK-1/2 was reduced by AG 879 and AG 1478 in these cells (Fig. 9). However, the stimulation of ERK-1/2 by heregulin was insensitive to the S1P2/4 antagonist JTE013 and the S1P3 antagonist CAY10444 (Fig. 9).
FIGURE 9.
Heregulin-stimulated activation of ERK-1/2 does not involve S1P4. MDA-MB-453 cells were pretreated with AG 1478 (100 nm) or AG 879 (1 μm) or CAY10444 (10 μm) or JTE013 (5 μm) for 15 min before treatment with heregulin (25 ng/ml) for 10 min. The Western blot shows the lack of effect of JTE013 and CAY10444 on the heregulin-stimulated activation of ERK-1/2. Phosphorylated ERK-1/2 was detected on Western blots probed with anti-phosphorylated ERK-1/2 antibody. Blots were also probed with anti-ERK-1/2 antibody to ensure equal protein loading. Results are representative of three separate experiments.
DISCUSSION
We demonstrate here that S1P stimulates the ERK-1/2 pathway via a mechanism that involves HER2 and S1P4 in MDA-MB-453 cells. This novel mechanism is based on several lines of evidence. First, we demonstrated that the S1P-induced activation of ERK-1/2 was reduced by JTE013, but not by CAY10444, an S1P3 selective antagonist. We have shown here for the first time, that in addition to being an S1P2 receptor antagonist, JTE013 is also a potent antagonist of S1P4 and can block S1P-induced mobilization of calcium in cells overexpressing this receptor. This is an important finding because JTE013 is used widely and is considered to have a high degree of specificity for S1P2. Second, siRNA knockdown of S1P4 reduced the activation of ERK-1/2 by S1P. Third, phyto-S1P, which is a selective S1P4 agonist, stimulated ERK-1/2 activation indicating that S1P4 specific ligands are able to activate this kinase pathway in MDA-MB-453 cells. Moreover, siRNA knockdown of S1P4 reduced the activation of ERK-1/2 by phyto-S1P. A role for S1P2 in the regulation of ERK-1/2 is excluded based on results showing the following: (i) siRNA knockdown of S1P2 had no effect on the stimulation of ERK-1/2 by either S1P or phyto-S1P; and (ii) FTY720 (the phosphorylated equivalent of which does not bind to S1P2) stimulated the activation of ERK-1/2. Three additional results provide evidence for a role of HER2 in regulating S1P4 signaling in MDA-MB-453 cells. First, the S1P-stimulated activation of ERK-1/2 was almost completely abolished by treatment of cells with ErbB2 inhibitor II. Second, the S1P- and phyto-S1P-stimulated activation of ERK-1/2 was reduced by the siRNA knockdown of HER2 expression. Third, S1P stimulated the tyrosine phosphorylation of HER2, which was reduced by JTE013.
We also found that S1P stimulation of ERK-1/2 via S1P4 in MDA-MB-453 cells does not involve participation of the EGF receptor, thereby excluding EGF release as a possible mechanism mediating the effects of S1P on this pathway. This contrasts with a role for EGF receptor downstream of S1P3 in MCF-7 cells (this article and Ref. 32). Regarding S1P4/ERK-1/2 signaling in MDA-MB-453 cells, the lack of participation of EGF receptor is based on the finding that the EGF receptor tyrosine kinase inhibitor AG 1478 and siRNA knockdown of EGF receptor expression reduced the EGF-dependent activation of ERK-1/2 but failed to modulate the response to S1P. Moreover, the EGF receptor does not require HER2 to activate the ERK-1/2 pathway based on results showing that EGF-stimulated activation of ERK-1/2 was not reduced by ErbB2 inhibitor 2 or siRNA knockdown of HER2. Furthermore, EGF failed to stimulate the tyrosine phosphorylation of HER2. Taken together, these findings demonstrate that S1P and EGF function in a mutually exclusive manner with S1P using S1P4 and HER2 to regulate ERK-1/2, whereas the EGF stimulation of this kinase pathway does not involve HER2.
Transactivation of the EGF receptor represents the paradigm for cross-talk between GPCRs and receptor tyrosine kinase (RTK) signaling pathways leading to regulation of ERK-1/2. Geschwind et al. (33) demonstrated, in a variety of squamous cell carcinoma cell lines of the head and neck, that treatment with the GPCR agonists lysophosphatidic acid, bradykinin, thrombin, and carbachol resulted in rapid tyrosine phosphorylation of the EGF receptor and concomitant activation of the ERK-1/2 pathway. In addition, these authors (33) reported that the activation of the ERK-1/2 pathway by lysophosphatidic acid was dependent both on metalloproteinase function and EGF receptor tyrosine kinase activity. Furthermore, Schaffer et al. (34) demonstrated that GPCR ligands induced tyrosine phosphorylation of EGF receptor as well as downstream signaling events such as recruitment of the adapter protein Shc and activation of ERK-1/2, JNK, and p38. Moreover, these authors reported that EGF receptor transactivation involves amphiregulin, heparin-binding EGF-like factor (Hb-EGF), and TGFα. These growth factors are released as a consequence of the action of metalloproteinases ADAM (a disintegrin and metalloproteinase) 10, 15, and 17 and are cell context-specific. Shida and colleagues (35) demonstrated that S1P induces rapid and transient phosphorylation of the EGF receptor via a Gi- and matrix metalloproteinase-dependent mechanism in MKN28 and MKN74 cells. S1P also induces rapid tyrosine phosphorylation of the hepatic growth factor receptor (c-Met) in these cells, although, in contrast with EGF receptor transactivation, this involves a matrix-metalloproteinase- and Gi-independent mechanism. JTE013 reduced the S1P-stimulated tyrosine phosphorylation of EGF receptor and c-Met. However, our findings concerning the specificity of JTE013, suggest that there is a need to evaluate whether the S1P4 receptor is involved in this process. The EGF receptor tyrosine kinase inhibitor AG 1478 was shown to have no effect on the S1P-stimulated tyrosine phosphorylation of c-Met, thereby excluding EGF receptor as an intermediate between S1P2 and c-Met. Shida and colleagues (36) also demonstrated that lysophosphatidic acid and S1P induce the tyrosine phosphorylation of HER2 in MKN28 and MKN74 cells and this was dependent upon metalloproteinase-dependent release of EGF receptor ligands.
With respect to the studies described above, our findings clearly describe a novel mechanism of HER2 transactivation and stimulation of the ERK-1/2 pathway in response to S1P in MDA-MB-453 cells. In this regard, we have previously demonstrated that GPCR and RTK can form functional signaling units that result in the RTK enhancing stimulation of the ERK-1/2 pathway by the respective GPCR ligand (24, 37, 38). For instance, we demonstrated that the S1P1 and PDGFβ receptor form a functional signaling unit, where the PDGFβ receptor tyrosine kinase enhances S1P stimulation of ERK-1/2 mediated by S1P1 (24, 37). It is therefore possible that S1P4 and HER2 might form similar signaling units to regulate the ERK-1/2 pathway in MDA-MB-453 cells. In this case, the tyrosine phosphorylation of HER2 in response to S1P might produce a signaling platform that enables recruitment of regulatory/adaptor proteins via their SH2 interaction with phosphotyrosines on HER2 and which may facilitate stronger activation of the ERK-1/2 pathway in response to S1P. This possibility requires formal testing.
We have also shown that heregulin does not use either S1P2/4 or S1P3 to regulate the ERK-1/2 pathway. In addition, the potential release of heregulin in response to S1P is unlikely because heregulin signaling is sensitive to AG 1478 and therefore requires EGF receptor, whereas S1P responses are insensitive to AG 1478.
In conclusion, we have demonstrated that the magnitude of the signaling gain on the ERK-1/2 pathway produced in response to S1P can be increased by an oncogene HER2 and in an ER− breast cancer cell line. This is unusual as S1P4 expression and function is largely restricted to lymphoid cells such as T-cells (39). Therefore, S1P4 expression and function may exhibit some promiscuity in cancer cells. In addition, the interaction between S1P4 and HER2 may provide new targets for drug intervention to reduce breast cancer progression. Further research is required to validate this possibility. In addition, JTE013 can be considered a potent antagonist of S1P4, providing both a useful tool for interrogating the function of this receptor in breast cancer and also as a prototype for further compound optimization and translational approaches to target S1P4 in ER−/HER2+ breast cancer.
This work was supported by Cancer Research UK Grant 23158/A7536 (to S. P. and N. J. P.), National Institutes of Health Grant CA 92160 from USPHS (to G. T.), the Royal College of Surgeons (Glasgow and Edinburgh branches), and a Glasgow Royal Infirmary endowment fund (to J. E.).
- S1P
- sphingosine 1-phosphate
- SK
- sphingosine kinase
- GPCR
- G protein-coupled receptor
- ER
- estrogen receptor
- QRT-PCR
- quantitative RT-PCR
- PTX
- pertussis toxin.
REFERENCES
- 1.Pyne S., Pyne N. J. (2000) Biochem. J. 349, 385–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee M. J., Van Brocklyn J. R., Thangada S., Liu C. H., Hand A. R., Menzeleev R., Spiegel S., Hla T. (1998) Science 279, 1552–1555 [DOI] [PubMed] [Google Scholar]
- 3.Hla T., Maciag T. (1990) J. Biol. Chem. 265, 9308–9313 [PubMed] [Google Scholar]
- 4.Okazaki H., Ishizaka N., Sakurai T., Kurokawa K., Goto K., Kumada M., Takuwa Y. (1993) Biochem. Biophys. Res. Comm. 190, 1104–1109 [DOI] [PubMed] [Google Scholar]
- 5.Gräler M. H., Bernhardt G., Lipp M. (1998) Genomics 53, 164–169 [DOI] [PubMed] [Google Scholar]
- 6.Glickman M., Malek R. L., Kwitek-Black A. E., Jacob H. J., Lee N. H. (1999) Mol. Cell. Neurosci. 14, 141–152 [DOI] [PubMed] [Google Scholar]
- 7.Yamazaki Y., Kon J., Sato K., Tomura H., Sato M., Yoneya T., Okazaki H., Okajima F., Ohta H. (2000) Biochem. Biophys. Res. Comm. 268, 583–589 [DOI] [PubMed] [Google Scholar]
- 8.Lee M. J., Evans M., Hla T. (1996) J. Biol. Chem. 271, 11272–11279 [DOI] [PubMed] [Google Scholar]
- 9.Kon J., Sato K., Watanabe T., Tomura H., Kuwabara A., Kimura T., Tamama K.., Ishizuka T., Murata N., Kanda T., Kobayashi I., Ohta H., Ui M., Okajima F. (1999) J. Biol. Chem. 274, 23940–23947 [DOI] [PubMed] [Google Scholar]
- 10.An S., Bleu T., Zheng Y. (1999) Mol. Pharmacol. 55, 787–794 [PubMed] [Google Scholar]
- 11.An S., Goetzl E. J., Lee H. (1998) J. Cell. Biochem. Suppl 30/31, 147–157 [PubMed] [Google Scholar]
- 12.Ancellin N., Hla T. (1999) J. Biol. Chem. 274, 18997–19002 [DOI] [PubMed] [Google Scholar]
- 13.Takuwa Y. (2002) Biochim. Biophys. Acta 1582, 112–120 [DOI] [PubMed] [Google Scholar]
- 14.Pitson S. M., Moretti P. A., Zebol J. R., Lynn H. E., Xia P., Vadas M. A., Wattenberg B. W. (2003) EMBO J. 22, 5491–5500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarman K. E., Moretti P. A., Zebol J. R., Pitson S. M. (2010) J. Biol. Chem. 285, 483–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nava V. E., Hobson J. P., Murthy S., Milstien S., Spiegel S. (2002) Exp. Cell Res. 281, 115–127 [DOI] [PubMed] [Google Scholar]
- 17.Wang F., Van Brocklyn J. R., Edsall L., Nava V. E., Spiegel S. (1999) Cancer Res. 59, 6185–6191 [PubMed] [Google Scholar]
- 18.Sarkar S., Maceyka M., Hait N. C., Paugh S. W., Sankala H., Milstien S., Spiegel S. (2005) FEBS Lett. 579, 5313–5317 [DOI] [PubMed] [Google Scholar]
- 19.Goetzl E. J., Dolezalova H., Kong Y., Zeng L. (1999) Cancer Res. 59, 4732–4737 [PubMed] [Google Scholar]
- 20.Schechter A. L., Hung M. C., Vaidyanathan L., Weinberg R. A., Yang-Feng T. L., Francke U., Ullrich A., Coussens L. (1985) Science 229, 976–978 [DOI] [PubMed] [Google Scholar]
- 21.Borg A., Tandon A. K., Sigurdsson H., Clark G. M., Fernö M., Fuqua S. A., Killander D., McGuire W. L. (1990) Cancer Res. 50, 4332–4337 [PubMed] [Google Scholar]
- 22.Graus-Porta D., Beerli R. R., Daly J. M., Hynes N. E. (1997) EMBO J. 16, 1647–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng L., Ren J. Q., Li H., Kong Z. L., Zhu H. G. (2004) Cell Res. 14, 497–506 [DOI] [PubMed] [Google Scholar]
- 24.Alderton F., Rakhit S., Kong K. C., Palmer T., Sambi B., Pyne S., Pyne N. J. (2001) J. Biol. Chem. 276, 28578–28585 [DOI] [PubMed] [Google Scholar]
- 25.Lu X., Sun C., Valentine W. J., Shuyu E., Liu J., Tigyi G., Bittman R. (2009) J. Org. Chem. 74, 3192–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGlynn L. M., Kirkegaard T., Edwards J., Tovey S., Cameron D., Twelves C., Bartlett J. M., Cooke T. G. (2009) Clin. Cancer Res. 15, 1487–1495 [DOI] [PubMed] [Google Scholar]
- 27.Tovey S. M., Witton C. J., Bartlett J. M., Stanton P. D., Reeves J. R., Cooke T. G. (2004) Breast Cancer Research 6, R246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohmori T., Yatomi Y., Osada M., Kazama F., Takafuta T., Ikeda H., Ozaki Y. (2003) Cardiovasc. Res. 58, 170–177 [DOI] [PubMed] [Google Scholar]
- 29.Koide Y., Hasegawa T., Takahashi A., Endo A., Mochizuki N., Nakagawa M., Nishida A. (2002) J. Med. Chem. 45, 4629–4638 [DOI] [PubMed] [Google Scholar]
- 30.Inagaki Y., Pham T. T., Fujiwara Y., Kohno T., Osborne D. A., Igarashi Y., Tigyi G., Parrill A. L. (2005) Biochem. J. 389, 187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng Z. Y., Li W. J., He F., Zhou J. M., Zhu X. F. (2007) Bioorg. Med. Chem. 15: 1533–1538 [DOI] [PubMed] [Google Scholar]
- 32.Sukocheva O., Wadham C., Holmes A., Albanese N., Verrier E., Feng F., Bernal A., Derian C. K., Ullrich A., Vadas M. A., Xia P. (2006) J. Cell Biol. 173, 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gschwind A., Prenzel N., Ullrich A. (2002) Cancer Res. 62, 6329–6336 [PubMed] [Google Scholar]
- 34.Schäfer B., Gschwind A., Ullrich A. (2004) Oncogene 23, 991–999 [DOI] [PubMed] [Google Scholar]
- 35.Shida D., Kitayama J., Yamaguchi H., Yamashita H., Mori K., Watanabe T., Yatomi Y., Nagawa H. (2004) FEBS Lett. 577, 333–338 [DOI] [PubMed] [Google Scholar]
- 36.Shida D., Kitayama J., Yamaguchi H., Yamashita H., Mori K., Watanabe T., Nagawa H. (2005) Biochem. Biophys. Res. Comm. 327, 907–914 [DOI] [PubMed] [Google Scholar]
- 37.Waters C. M., Connell M. C., Pyne S., Pyne N. J. (2005) Cell. Signal. 17, 263–277 [DOI] [PubMed] [Google Scholar]
- 38.Pyne N. J., Pyne S. (2008) Biochim. Biophys. Acta 1781, 467–476 [DOI] [PubMed] [Google Scholar]
- 39.Wang W., Graeler M. H., Goetzl E. J. (2005) FASEB J. 19, 1731–1733 [DOI] [PubMed] [Google Scholar]