Abstract
The type VI secretion system (T6SS) contributes to the virulence of Burkholderia cenocepacia, an opportunistic pathogen causing serious chronic infections in patients with cystic fibrosis. BcsKC is a highly conserved protein among the T6SSs in Gram-negative bacteria. Here, we show that BcsKC is required for Hcp secretion and cytoskeletal redistribution in macrophages upon bacterial infection. These two phenotypes are associated with a functional T6SS in B. cenocepacia. Experiments employing a bacterial two-hybrid system and pulldown assays demonstrated that BcsKC interacts with BcsLB, another conserved T6SS component. Internal deletions within BcsKC revealed that its N-terminal domain is necessary and sufficient for interaction with BcsLB. Fractionation experiments showed that BcsKC can be in the cytosol or tightly associated with the outer membrane and that BcsKC and BcsLB form a high molecular weight complex anchored to the outer membrane that requires BcsFH (a ClpV homolog) to be assembled. Together, our data show that BcsKC/BcsLB interaction is essential for the T6SS activity in B. cenocepacia.
Keywords: Bacterial Genetics, Membrane Proteins, Protein-Protein Interactions, Secretases, Subcellular Fractionation
Introduction
The Burkholderia cepacia complex comprises closely related species of Gram-negative bacteria commonly found in the environment (1, 2). B. cepacia complex bacteria are opportunistic human pathogens that establish chronic lung infections in immunocompromised patients with chronic granulomatous disease or, most commonly, cystic fibrosis (3). In most cystic fibrosis centers worldwide and more particularly in Canada, Burkholderia cenocepacia is one of the most common B. cepacia complex species recovered from patients (4, 5) and is frequently associated with the most severe infections (6).
Novel factors for B. cenocepacia survival in vivo were discovered in our laboratory by signature-tagged mutagenesis using a rat model of chronic lung infection (7), and these included a type 6 secretion system (T6SS)3 (7, 8). The T6SS is a newly recognized secretion system widespread among Gram-negative pathogens and symbionts that are in close interaction with eukaryotic cells (9–11). Several proteins specifically secreted by the T6SS into culture supernatants have been identified. These proteins usually lack any conventional N-terminal hydrophobic signal peptide and include the hemolysin coregulated-like protein (Hcp) (12), the valine-glycine repeat family proteins (VgrG) (13), and in some cases non-Hcp, non-VgrG substrates (12, 14, 15). Hcp is secreted by most of the T6SSs studied to date, and its detection in culture supernatants is considered the hallmark of a functional T6SS (8, 16–19). Hcp and VgrG proteins might also be part of the T6SS structural scaffold displayed at the bacterial surface and might be released into culture supernatants by shearing forces (20, 21).
The T6SS contributes to the pathogenicity of many bacteria (12, 16, 17, 22, 23). In Vibrio cholerae, bacteria-host contact and endocytosis are required to trigger the translocation of VgrG-1 into the host cell cytosol. Cytosolic VgrG-1 mediates actin cross-linking impairing the phagocytic cell functions (24). Although in some bacteria the T6SS has evolved to target eukaryotic cells, the Pseudomonas aeruginosa T6SS has been recently shown to deliver toxins into other bacteria. This novel T6SS property would confer a fitness advantage to P. aeruginosa growing in bacterial communities (25).
Typically, the T6SS is poorly expressed under in vitro laboratory conditions, but it is quorum sensing regulated (19, 26) and induced in vivo during infection (9, 11, 27). We have recently identified AtsR, a sensor kinase-response regulator hybrid that negatively regulates the T6SS expression in B. cenocepacia K56-2 (8). Deletion of atsR causes increased T6SS activity, as exemplified by hypersecretion of Hcp into culture supernatant (8). Infection of macrophages with the atsR deletion strain induces cytoskeletal rearrangements, resulting in macrophages displaying actin-rich cellular projections (8). This phenotype relies on T6SS activity and has only been described for B. cenocepacia, but the B. cenocepacia effector molecules and the eukaryotic targets involved in this process are unknown and currently under investigation in our laboratory.
Similarity between bacteriophage proteins and the T6S-associated Hcp and VgrG proteins suggests that the T6SS is structurally and evolutionarily related to bacteriophages (21, 28). Several components highly conserved among T6SSs have been investigated, and some light has been shed on their localization, function, and interaction with other T6SS components. These include: (i) the integral inner membrane IcmF-like protein (ImpLM) from Agrobacterium tumefaciens (29); (ii) the outer membrane lipoprotein SciN (30); (iii) the polytopic inner membrane protein SciZ anchoring the T6SS to the cell wall (31) in enteroaggregative Escherichia coli; (iv) the proteins VipA in V. cholerae (32) and its homolog IglA in Francisella novicida, which interact with VipB and IglB (11), respectively; and (v) the energizing component ClpV, which disintegrates cytosolic tubule like structures made of VipA and VipB (32).
This study focuses on BcsKC, a T6SS component from B. cenocepacia homologous to IglB. We show here that BcsKC is critical for the T6SS activity of B. cenocepacia K56-2. Experiments employing the bacterial two-hybrid system confirmed that BcsKC also interacts with BcsLB, a protein homologous to IglA, and support the notion that interactions between BcsLB- and BcsKC-like proteins are a common feature of T6SSs. Internal deletions of BcsKC allowed us to characterize the region required for BcsLB binding and demonstrated that the N terminus of BcsKC is essential and sufficient for interaction with BcsLB. Furthermore, cell fractionation experiments revealed that BcsKC can be tightly associated with the outer membrane and that BcsKC, in association with BcsLB, forms a membrane associated high molecular weight complex whose assembly requires BcsFH (ClpV homolog). Together, this work establishes BcsKC as a critical component of the T6S apparatus.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Culture Media
Bacterial strains and plasmids used in this study are listed in supplemental Table S1. Bacteria were routinely cultured in LB (Difco) at 37 °C. E. coli cultures were supplemented, as required, with the following antibiotics (final concentrations): 100 μg/ml ampicillin, 30 μg/ml tetracycline, 30 μg/ml kanamycin, 30 μg/ml chloramphenicol, 50 μg/ml trimethoprim, and 50 μg/ml gentamicin. B. cenocepacia cultures were supplemented, as required, with 100 μg/ml trimethoprim, 100 μg/ml tetracycline, and 50 μg/ml gentamicin. We used the unified nomenclature proposed by Cascales (33) and assigned the alphabetical character designated by Shalom et al. (34) to the T6SS genes from B. cenocepacia K56-2.
General Molecular Techniques
DNA manipulations were performed as described previously (35). T4 polynucleotide kinase and T4 DNA ligase (Roche Applied Science) were used as recommended by the manufacturers. Transformation of E. coli DH5α, E. coli SY327, and E. coli KEM5 was done by the calcium chloride protocol (36). Mobilization of complementing plasmids and mutagenesis plasmids into B. cenocepacia K56-2 was performed by triparental mating using E. coli DH5α carrying the helper plasmid pRK2013 (37, 38). DNA amplification by PCR was performed using a PTC-221 DNA engine (MJ Research, Incline Village, Nevada) with Taq, Proof Start, or HotStar HiFidelity DNA polymerases (Qiagen). DNA sequences of all of the primers used in this study are described in supplemental Table S2. DNA sequencing was performed at the DNA sequencing facility of York University (Toronto, Canada). The computer program BLAST was used to analyze the sequenced genome of B. cenocepacia strain J2315.
Mutagenesis of B. cenocepacia K56-2 and Complementing Plasmids
Unmarked and nonpolar deletions were performed as described previously (39). Details about the construction of the deletion plasmids are available in the supplemental materials. Plasmid pDelatsR was used to delete atsR (BCAM0379) in B. cenocepacia K56-2. Deletions were subsequently performed in the atsR-deleted mutant, B. cenocepacia K56-2 ΔatsR. Plasmids pDelT6SS, pDelTU1, pDelTU2, and pDelTU3 were used to delete the entire T6SS gene cluster: T6SS transcriptional units 1, 2, and 3, respectively. Each gene within transcriptional unit 2 was individually deleted. Plasmids pDelbcsM, pDelbcsLB, pDelbcsKC, pDA45, pDelbcsIE, pDelbcsHF, pDelbcsGG, pDelbcsFH, and pDelbcsEA were used to delete BCAL0340, 0341, 0342, 0343, 0344, 0345, 0346, 0347, and BCAL0348, respectively. Gene deletions were first analyzed by PCR and also confirmed by Southern blot hybridization.
The bcsKC gene was PCR-amplified using primer pair 3401–3393. Amplicons were digested with the restriction enzymes KpnI-SacI and cloned into a similarly digested pME6000 plasmid, giving rise to pBcsKC. We took advantage of two SphI restriction sites within the bcsKC gene sequence to generate an in-frame deletion of 0.45 kb. The bcsKC gene was PCR-amplified as described above and digested with KpnI, SacI, and SphI, generating three fragments (0.13, 0.45, and 0.93 kb). Only the 0.13- and 0.93-kb fragments were gel-purified using the QiaQuick gel extraction kit (Qiagen) and cloned as above into the KpnI-SacI-digested pME6000 plasmid, giving rise to pBcsKCΔ40–189.
A FLAG epitope was fused to the N termini of BcsKC and BcsKCΔ40–189 as follows. The bcsKC and bcsKCΔ40–189 genes were PCR-amplified using primer pair 3700–3701. Amplicons were digested with the restriction enzymes HindIII-BamHI and cloned into a similarly digested pEL-1 plasmid giving rise to pBcsKC-FLAG and pBcsKCΔ40–189-FLAG.
Preparation of Culture Supernatant Proteins
Culture supernatant proteins were precipitated with trichloroacetic acid as described previously (8). Ten μg of protein were loaded on a 16% SDS-PAGE. Detection was performed with a Brilliant Blue G colloidal staining according to the manufacturer's recommendations (Sigma).
Pulldown Assay and MS
Protein G-agarose beads (Roche Applied Science; 20-μl packed volume/sample) were washed three times with 20 volumes of TBS. Five μg of FLAG M2 monoclonal antibody (Sigma) per sample were incubated with the bead suspensions with constant agitation for 2 h at 4 °C using a Barnstead Thermolyne LABQUAKE (Barnstead International, Dubuque, IA). The beads were washed three times with 20 volumes of TBS. Overnight cultures of B. cenocepacia K56-2 mutants ΔatsR ΔbcsKC (pEL-1), ΔatsR ΔbcsKC (pBcsKC-FLAG), and ΔatsR ΔbcsKC (pBcsKCΔ40–189-FLAG) were 10-fold diluted in 50 ml of prewarmed LB. After 3 h of incubation at 37 °C, the cultures were centrifuged for 10 min at 10,000 × g at 4 °C. Bacterial pellets were washed twice with 10 ml of TBS and resuspended in 4 ml of TBS containing a protease inhibitor mixture (Roche Applied Science). The cells were lysed by three passages in a French press (Thermo Scientific) using a miniature French pressure cell at 600 p.s.i. The cell lysates were centrifuged first at 5000 × g at 4 °C for 15 min, and the supernatants were transferred in fresh tubes, centrifuged at 21,000 × g for 15 min, and collected. The bead suspensions conjugated to the FLAG antibody were added to 1 ml of each supernatant and incubated for 3 h at 4 °C with constant rotation. The beads were washed three times with 10 volumes of TBS and resuspended in 100 μl of TBS. Twenty μl of bead suspension were boiled and loaded in duplicate on a 16% SDS-PAGE gel. After electrophoresis, detection was performed on half of the gel by silver staining adapted from Blum et al. (40). The other half of the gel was transferred to a nitrocellulose membrane for immunoblot analysis (see below).
Protein bands were excised from a silver-stained one-dimensional gel using an Ettan Spot-Picker (Amersham Biosciences). In-gel trypsin digestion was performed in the Functional Proteomics Facility of the University of Western Ontario using the Waters MassPREP automated station. Five μl of submitted sample was injected for LC/MS/MS analysis as described elsewhere (8). The data were processed using the Proteinlynx Global Server software, and a database search was performed using MASCOT search engine (Matrix Science, London, UK). This search also provides a score estimating the probability that the observed match between experimental data and each protein in the database is a random event. The higher the protein score, the higher the confidence that the protein in the sample corresponds to the match.
Bacterial Two-hybrid System, β-Galactosidase Assay, and Plasmid Constructions
In vivo interaction between BcsKC and BcsLB was investigated using a bacterial adenylate cyclase two-hybrid system kit (Euromedex). This system is based on the interaction-mediated reconstitution of the adenylate cyclase activity in an adenylate cyclase-deficient E. coli reporter strain (41). The catalytic domain of adenylate cyclase (CyaA) from Bordetella pertussis consists of two complementary fragments, T25 and T18, that are not active when physically separated. When these two fragments are fused to interacting polypeptides and coexpressed in E. coli, heterodimerization of the hybrid proteins restores the activity CyaA, leading to cyclic AMP synthesis and transcriptional activation of the lac operon. Therefore, interaction between two hybrid proteins in E. coli will generate high levels of β-galactosidase activity, which can be readily scored on X-gal-containing medium and quantified.
BcsLB and BcsKC were fused to T25 and T18, respectively (details are available in the supplemental materials). Plasmid pT25BcsLB encodes a hybrid protein made of BcsLB fused to the C terminus of the T25 fragment (T25-BcsLB). Plasmids pT18BcsKC and pT18BcsKCΔ40–189 encode hybrid proteins made of BcsKC or a truncated variant fused to the C-terminal end of T18. Plasmids pBcsKCT18 and pBcsKCΔ40–189T18 encode hybrid proteins made of BcsKC or a truncated variant fused to the N-terminal end of T18.
Sequential 25-amino acid deletions were performed within BcsKC (details are available in the supplemental materials). Deletion of amino acids 38–62, 63–87, 88–112, 113–137, 138–162, 163–187, 188–212, 213–237, and 213–496 were performed, giving rise to plasmids pBcsKCΔ38–62T18, pBcsKCΔ63–87T18, pBcsKCΔ88–112T18, pBcsKCΔ113–137T18, pBcsKCΔ138–162T18, pBcsKCΔ163–187T18, pBcsKCΔ188–212T18, pBcsKCΔ213–237T18, and pBcsKCΔ213–496T18, respectively, expressing truncated BcsKC fused to the N terminus of T18.
The E. coli KEM5 reporter strain (lacking any endogenous adenylate cyclase activity) harboring pT25BcsLB was cotransformed with the recombinant plasmids expressing hybrid proteins fused to the T18 domain. Bacterial cultures were grown for 16 h in LB at 30 °C with antibiotics as required and isopropyl β-d-1-thiogalactopyranoside (0.5 mm). Samples were taken to measure the bacterial growth (A600 nm) and the β-galactosidase activity (A420 nm and A550 nm) upon lysis and incubation with O-nitrophenyl-β-d-galactoside according to Miller (42). Enzyme activities (Miller units) were calculated using the equation [(A420 nm − 1.75 A550 nm) × 1000]/[reaction time (min) × culture volume (ml) × A600 nm]. Each sample was analyzed in triplicate during three independent experiments. Four μl of each culture were also spotted on a LB agar plate containing X-gal (40 μg/ml), isopropyl β-d-1-thiogalactopyranoside (0.5 mm), and antibiotics as required. Incubation was performed for 24 and 48 h at 30 and 37 °C.
Cell Fractionation, Sucrose Density Gradient, and Western Blot Analysis
Overnight cultures were diluted in prewarmed LB. After 3 h of incubation at 37 °C, the cultures were centrifuged for 10 min at 10,000 × g at 4 °C. Bacterial pellets were washed with 50 mm Tris-HCl, pH 7.4, and resuspended in 5 ml of the same buffer containing a protease inhibitor mixture, PMSF (1 mm), DNase (50 μg/ml), and RNase (50 μg/ml). The cells were lysed by three passages in a French press at 600 p.s.i. Unbroken cells were removed by centrifugation twice at 20,000 × g for 15 min at 4 °C. An aliquot of the supernatant corresponding to the total cell lysate was stored at −20 °C, and the remainder was processed to separate the soluble fraction from the total membrane fraction as follows. The membranes were pelleted by ultracentrifugation at 150,000 × g for 1 h at 4 °C in a Beckman 70Ti rotor. An aliquot of the supernatant corresponding to the soluble fraction was stored at −20 °C. To increase the purity of the membrane preparation, the pellet was resuspended in 2 ml of 50 mm Tris-HCl, pH 7.4, containing 10% (w/w) sucrose, a protease inhibitor mixture, and 1 mm PMSF and loaded on top of a discontinuous sucrose gradient consisting of 4 ml of 15% (w/w) sucrose placed on top of 4 ml of 55% (w/w) sucrose made in 50 mm Tris-HCl, pH 7.4. Ultracentrifugation was performed at 37,000 rpm for 1 h at 4 °C in a Beckman SW41. The membranes were collected using a syringe by puncturing the side of the tube slightly below the membrane ring. An aliquot corresponding to the total membrane fraction was stored at −20 °C.
The protein concentration of each fraction was determined by Bradford assay (Bio-Rad), and 10 μg of protein were loaded on 12% SDS-PAGE gels. Unless indicated, the samples were not boiled prior to loading. After electrophoresis, the gels were transferred to nitrocellulose membranes for immunoblot analysis. The membranes were incubated with the 4RA2 monoclonal antibody (Neoclone) cross-reacting with the B. cenocepacia RNA polymerase subunit 1 mm (cytosolic control) and the FLAG M2 monoclonal antibody (Sigma). The Alexa Fluor 680 goat anti-mouse IgG (Molecular Probes) was used as a secondary antibody. Detection was performed using the Odyssey Infrared Imager (LI-COR Biosciences).
Protein solubilization experiments were performed based on the work of Ma et al. (29). Briefly, the membrane fractions were incubated with either 50 mm Tris-HCl, pH 7.4 (control), 1 m NaCl, 6 m urea, 3% Triton X-100, 2% N-lauroylsarcosine, or 1% SDS in ice for 1 h and centrifuged at 150,000 × g for 1 h at 4 °C in a Beckman 100Ti rotor to separate soluble and insoluble fractions. The pellet was resuspended in 50 mm Tris-HCl, pH 7.4, in the same volume as the soluble fraction and analyzed by Western blot.
To increase the separation of the inner and outer membranes, the cells were incubated at room temperature for 15 min in 20% (w/w) sucrose made in 50 mm Tris-HCl, pH 7.4, with protease inhibitor. DNase (50 μg/ml), RNase (50 μg/ml), EDTA (0.5 μm), and lysozyme (0.1 mg/ml) were added prior to cell lysis. The soluble lysate was loaded on top of a discontinuous sucrose gradient consisting of 4 ml of 25% (w/w) sucrose placed on top of 2 ml of 60% (w/w) sucrose made in 50 mm Tris-HCl, pH 7.4, and centrifuged as indicated previously. The sucrose concentration of the purified total membranes was lowered to 30% (w/w) sucrose and loaded on top of a second discontinuous sucrose gradient consisting of 0.6 ml of 60% and 1.16-ml layers of 56, 53, 50, 47, 44, 41, 38, 35, and 32% (w/w) sucrose. The membrane fractionation was performed by ultracentrifugation at 37,000 rpm for 40 h at 4 °C in a Beckman SW41 rotor. The gradient was collected from top to bottom using a syringe by puncturing holes in the tube every 0.3 cm. Protein content was assessed by measuring the absorption at 280 nm, and the quality of the fractionation was assessed by the detection of the outer membrane porin BCAM1931, by measuring the enzymatic activity of the NADH oxidase (inner membrane marker) (43) and the refractive index of each fraction. Ten-μl aliquots of the collected fractions were loaded on a 12% SDS-PAGE gel. After electrophoresis, the gel was transferred to a nitrocellulose membrane for immunoblot analysis or silver-stained. The samples need to be boiled to detect BCAM1931.
Electroelution
Purified total membranes from B. cenocepacia K56-2 ΔatsR ΔbcsKC (pBcsKC-FLAG) and B. cenocepacia K56-2 ΔatsR ΔT6SS (pBcsKC-FLAG) were prepared as above. One mg of total membrane proteins was loaded in several wells and separated by electrophoresis on 12% SDS-PAGE gels. After electrophoresis, a 4-mm-wide band from the top of the separating gel (containing the high molecular weight proteins) was excised from each gel and minced. The proteins were electroeluted in volatile buffer (50 mm NH4HCO3, 0.1% SDS) using a model 422 Electro-Eluter (Bio-Rad) and Green Membrane caps (molecular mass cutoff of 3,500 Da) for 6 h at 20 mA. After the elution the volatile buffer was removed using a spin vacuum, leaving concentrated proteins. The pellets were resuspended with 80 μl of volatile buffer, and protein concentration was estimated by measuring absorbance at 280 nm using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). Seventy μg of protein were boiled for 10 min and loaded on a 16% SDS-PAGE gel. Protein was detected by silver staining, and selected protein bands were excised and analyzed by mass spectrometry as indicated above.
Macrophage Infection Assay
The C57BL/6 murine bone marrow-derived macrophage cell line ANA-1 (44) was maintained in DMEM with 10% (v/v) FBS and grown at 37 °C in a humidified atmosphere of 95 and 5% CO2. Bacteria were added to ANA-1 cells at a multiplicity of infection of 50:1 as described previously (8) and incubated for 4 h at 37 °C. The images were digitally processed using the Northern Eclipse version 6.0 imaging analysis software (Empix Imaging, Mississauga, Canada).
RESULTS
BcsKC Is Essential for T6SS Activity
The bcsKC gene (BCAL0342) encodes a 496-amino acid protein (∼55 kDa). BLASTP analysis revealed that BcsKC contains a domain of unknown function, DUF877, and belongs to a family of uncharacterized bacterial proteins that are associated with the T6S locus of many Gram-negative bacteria including Burkholderia mallei (TssB), P. aeruginosa (PA0084), V. cholerae (VipB), Salmonella enterica serovar Typhimurium (SciI), Edwardsiella tarda (EvpB), and F. novicida (IglB).
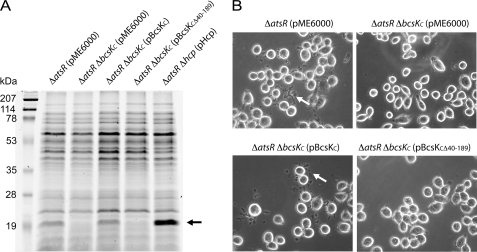
We have previously demonstrated that inactivation of atsR in B. cenocepacia K56-2 leads to increased T6SS activity, as exemplified by (i) hypersecretion of an Hcp-like protein and (ii) cytoskeletal reorganization with the formation of cellular projections upon infection of ANA-1 murine macrophages (8). Following these two phenotypes we investigated the importance of BcsKC in the T6SS activity of B. cenocepacia K56-2. Analysis of culture supernatants (Fig. 1A) revealed that in contrast to K56-2 ΔatsR, the K56-2 ΔatsR ΔbcsKC double mutant was unable to release Hcp, consistent with a secretion defect. Hcp secretion was restored upon introduction of pBcsKC, which constitutively expresses BcsKC into K56-2 ΔatsR ΔbcsKC (Fig. 1A). Furthermore, more than 60% of ANA-1 macrophages displayed cellular projections upon infection with K56-2 ΔatsR (Fig. 1B, upper left panel), whereas this phenotype was absent in macrophages infected with K56-2 ΔatsR ΔbcsKC. Introduction of pBcsKC into K56-2 ΔatsR ΔbcsKC (Fig. 1B, lower left panel) restored the ability of these bacteria to mediate cytoskeletal rearrangements in macrophages. Identical results were obtained with pBcsKC-FLAG (supplemental Fig. S1), indicating that fusion of a FLAG epitope to the N terminus of BcsKC does not alter the protein function. Together, these results demonstrate that BcsKC is essential for T6SS function in B. cenocepacia K56-2.
FIGURE 1.
BcsKC is required for T6SS activity. A, secretion assay. SDS-polyacrylamide gel electrophoresis analysis of concentrated culture supernatants recovered from B. cenocepacia K56-2 ΔatsR containing the control plasmid pME6000 (ΔatsR pME6000), K56-2 ΔatsR ΔbcsKC (pME6000), K56-2 ΔatsR ΔbcsKC (pBcsKC), K56-2 ΔatsR ΔbcsKC (pBcsKCΔ40–189), and K56-2 ΔatsR Δhcp (pHcp). Shown at left is the molecular mass marker with the indicated sizes (kDa). The black arrow highlights the position of secreted Hcp. B, phase contrast microscopy of infected ANA-1 macrophages. The infections were performed at a multiplicity of infection of 50:1 for 4 h with the following strains: B. cenocepacia K56-2 ΔatsR (pME6000), K56-2 ΔatsR ΔbcsKC (pME6000), K56-2 ΔatsR ΔbcsKC (pBcsKC), and K56-2 ΔatsR ΔbcsKC (pBcsKCΔ40–189). The white arrows indicate cellular projections.
The N Terminus of BcsKC Is Required for T6SS Activity
We took advantage of two conveniently located SphI restriction sites in bcsKC to generate a 0.45-kb truncation of the gene, causing an in frame deletion of 150 amino acids within the BcsKC N terminus (BcsKCΔ40–189). Plasmid pBcsKCΔ40–189 could not restore Hcp secretion in K56-2 ΔatsR ΔbcsKC (Fig. 1A). Moreover, ANA-1 macrophages infected with K56-2 ΔatsR ΔbcsKC (pBcsKCΔ40–189) or K56-2 ΔatsR ΔbcsKC (pBcsKCΔ40–189-FLAG) did not display the cellular projections typical of T6SS activity (Fig. 1B, lower right panel, and supplemental Fig. S1). Western blot analysis of bacterial cell lysates prepared from K56-2 ΔatsR ΔbcsKC (pBcsKC-FLAG) and K56-2 ΔatsR ΔbcsKC (pBcsKCΔ40–189-FLAG) demonstrated that BcsKC-FLAG and BcsKCΔ40–189-FLAG were similarly expressed (see below). Thus, a lack of complementation by pBcsKCΔ40–189-FLAG was not due to a defect in protein expression. Together, these data indicate that the N-terminal portion of BcsKC (amino acids 40–189) is critical for the T6SS activity, suggesting that this region may be required for protein-protein interaction essential for T6SS assembly/stability, correct localization of the protein within the cell, or the intrinsic function of the protein.
BcsKC Interacts with BcsLB, and the N Terminus of BcsKC Is Required for the Interaction
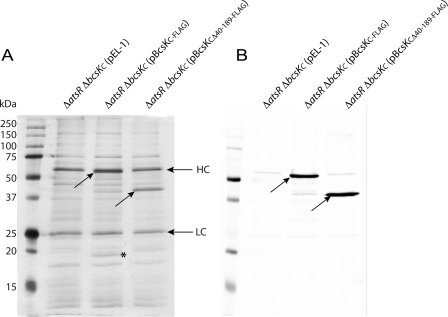
Studies in V. cholerae and F. novicida show that BcsKC homologs interact with a small T6SS protein homologous to BcsLB from B. cenocepacia (32, 45). To investigate whether BcsKC also interacts with BcsLB or with other proteins, we performed pulldown assays using cell lysates from B. cenocepacia K56-2 ΔatsR ΔbcsKC (pEL-1), K56-2 ΔatsR ΔbcsKC (pBcsKC-FLAG), and K56-2 ΔatsR ΔbcsKC (pBcsKCΔ40–189-FLAG). As shown in Fig. 2A, an additional protein with an apparent mass of ∼20 kDa was copurified with BcsKC-FLAG but not with BcsKCΔ40–189-FLAG, suggesting that the N-terminal portion of BcsKC is required to bind the 20-kDa protein. Mass spectrometry was performed on the 20-kDa polypeptide and on the corresponding region in lane K56-2 ΔatsR ΔbcsKC (pBcsKCΔ40–189-FLAG) as control. Mass spectra were searched against the NCBI database with the MASCOT search engine. Identified peptides from the 20-kDa protein matched with good confidence BcsLB (BCAL0341; 19.2 kDa) with a total protein score of 467 and were absent in the control lane K56-2 ΔatsR ΔbcsKC (pBcsKCΔ40–189-FLAG).
FIGURE 2.
BcsKC interacts with BcsLB. Shown is an immunoprecipitation assay using anti-FLAG coupled protein G-agarose beads and cell lysates from B. cenocepacia K56-2 ΔatsR ΔbcsKC harboring pEL-1 (control), pBcsKC-FLAG, or pBcsKCΔ40–189-FLAG. A, silver staining. B, anti-FLAG Western blot analysis. The diagonal arrows highlight the positions of BcsKC-FLAG and BcsKCΔ40–189-FLAG, whereas the asterisk indicates the position of the copurified 20-kDa protein, BcsLB. Protein bands of ∼25 and 53 kDa (indicated LC and HC, respectively) in all samples correspond to the light and heavy chains of the anti-FLAG antibody.
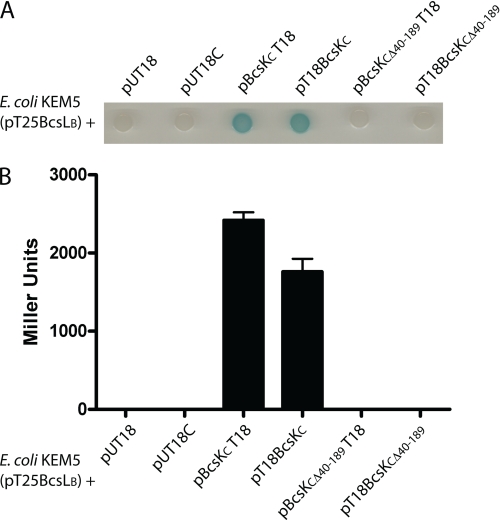
Bacterial two-hybrid experiments confirmed the BcsKC/BcsLB interaction. BcsLB was used as bait and fused to the T25 domain of the adenylate cyclase from B. pertussis (pT25BcsLB). BcsKC and BcsKCΔ40–189 were used as prey and fused either to the N terminus of the adenylate cyclase T18 domain using the pUT18 plasmid (pBcsKCT18 and pBcsKCΔ40–189T18) or to the C terminus of the T18 domain using the pUT18C plasmid (pT18BcsKC and pT18BcsKCΔ40–189). E. coli KEM5 (adenylate cyclase-deficient) harboring pT25BcsLB was cotransformed with the recombinant plasmids expressing BcsKC hybrid proteins. Bacterial cultures were spotted on a X-gal containing agar plate, and transcriptional activation of the lac operon indicative of protein interaction was visualized by the formation of blue colonies (Fig. 3A). The extent of interaction was also estimated by measuring the β-galactosidase activity (Fig. 3B). E. coli KEM5 colonies harboring pT25BcsLB together with the vectors pUT18 or pUT18C remained white, indicating that BcsLB fused to the T25 domain does not interact with the T18 domain alone expressed from pUT18 or pUT18C plasmids. In contrast, E. coli KEM5 (pT25BcsLB + pBcsKCT18) and E. coli KEM5 (pT25BcsLB + pT18BcsKC) formed blue colonies, denoting interacting hybrid proteins and thus confirming that BcsKC interacts with BcsLB. However, E. coli KEM5 (pT25BcsLB + pBcsKCΔ40–189T18) and E. coli KEM5 (pT25BcsLB + pT18BcsKCΔ40–189) formed white colonies, clearly demonstrating that BcsKCΔ40–189 does not interact with BcsLB and indicating that the N terminus of BcsKC is critical for interaction with BcsLB. These results were validated with β-galactosidase assays, thus confirming the interaction between full-length BcsKC and BcsLB and the critical role of the BcsKC N terminus in the interaction (Fig. 3B).
FIGURE 3.
The N terminus of BcsKC is required for interaction with BcsLB. A, protein interaction using the bacterial two-hybrid system. E. coli harboring pT25BcsLB was cotransformed with pUT18 and pUT18C control vectors or with the recombinant plasmids expressing BcsKC or BcsKCΔ40–189 hybrid proteins fused to the T18 domain in pUT18 (pT18BcsKC and pT18BcsKCΔ40–189) and in pUT18C (pBcsKCT18 and pBcsKCΔ40–189T18). Bacterial cultures were spotted on a X-gal containing agar plate and transcriptional activation of the lac operon indicative of protein interaction was visualized by the formation of blue colonies. B, quantification of the β-galactosidase activity of E. coli harboring pT25BcsLB cotransformed with pUT18, pUT18C, pBcsKCT18, pT18BcsKC, pBcsKCΔ40–189T18, and pT18BcsKCΔ40–189. The enzyme activities are expressed in Miller units. The data shown are the means of three independent experiments done in triplicate. The error bars represent the standard deviation.
To determine the minimal amino acid sequence of BcsKC required for BcsLB binding, we constructed a panel of sequential deletions of bcsKC (Fig. 4). Truncated BcsKC proteins were fused to the adenylate cyclase T18 domain and tested for interaction with T25BcsLB. Physical interactions between the hybrid proteins were detected on X-gal-containing plates. Among the deletions tested, only BcsKCΔ38–62T18, BcsKCΔ163–187T18, BcsKCΔ188–212T18, BcsKCΔ213–237T18, and BcsKCΔ213–496T18 could bind T25BcsLB and thus activate the lacZ reporter gene (Fig. 4). Interactions between BcsKCT18, BcsKCΔ38–62T18, BcsKCΔ213–237T18, and BcsKCΔ213–496T18 and T25BcsLB were detected on the plate after only 24 h of incubation at 30 °C (data not shown). By contrast, interactions between either BcsKCΔ163–187T18-T25BcsLB or BcsKCΔ188–212T18-T25BcsLB required 48 h of incubation, indicating that BcsKCΔ163–187T18 and BcsKCΔ188–212T18 interact weakly with T25BcsLB. After 1 week of incubation, BcsKCΔ113–137T18 and BcsKCΔ138–162T18 eventually turned blue, whereas BcsKCΔ63–87T18 and BcsKCΔ88–112T18 remained unable to activate lacZ (data not shown). Together, these results suggest that the N terminus of BcsKC (amino acids 2–212) is sufficient for the interaction with BcsLB. Within the BcsKC N-terminal domain, amino acids 88–162 are important for BcsKC/BcsLB interaction, and more precisely the region encompassing amino acid 63–112 is essential for interaction with BcsLB.
FIGURE 4.
Identification of BcsKC domains required for interaction with BcsLB. Protein interaction using the bacterial two-hybrid system. E. coli harboring pT25BcsLB was cotransformed with recombinant plasmids expressing sequentially truncated BcsKC hybrid proteins fused to the T18 domain in pUT18. Bacterial cultures were spotted on a X-gal containing agar plate, and transcriptional activation of the lac operon indicative of protein interaction was visualized by the formation of blue colonies.
BcsKC Is Tightly Associated with the Outer Membrane
BcsKC lacks a signal peptide sequence as predicted by the SignalP 3.0 program (54, 55), and in silico analysis of the BcsKC protein using different transmembrane helices prediction programs available at the ExPASY Proteomics and PSIPRED servers (46, 47) was not conclusive. BcsKC is predicted to have either no transmembrane domain (DAS, SOSUI, and TMHMM) or one transmembrane domain (HMMTOP, TMpred, TopPred, MEMSAT) whose position and orientation differ depending on the algorithm used (supplemental Fig. S2).
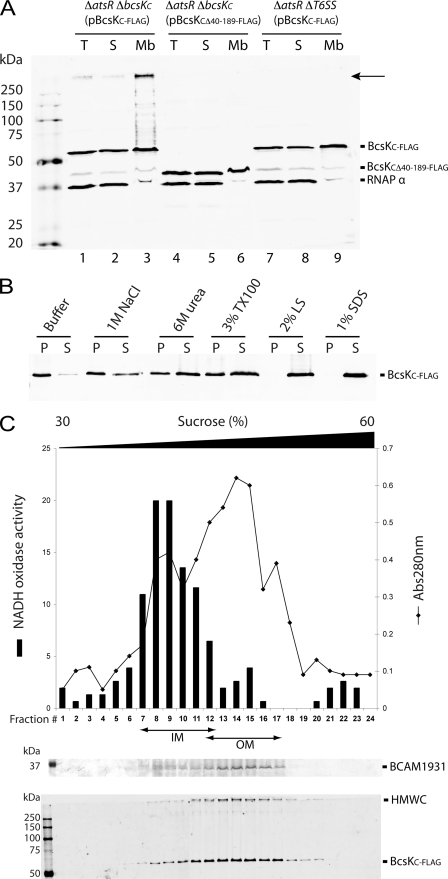
Cell fractionations were performed to determine the subcellular localization of BcsKC-FLAG and its truncated derivative BcsKCΔ40–189-FLAG. As shown in Fig. 5A, BcsKC-FLAG and BcsKCΔ40–189-FLAG proteins localized both in the soluble fraction and the membrane fraction, suggesting that deletion of the N terminus of BcsKC does not affect the protein localization. The membrane fraction containing BcsKC-FLAG was incubated with various reagents (Fig. 5B). BcsKC-FLAG was not released with high salt (1 m NaCl) but was partially solubilized by 6 m urea and nonionic detergent (3% Triton X-100). BcsKC-FLAG was completely solubilized by strong ionic detergents (1% SDS) and 2% N-lauroylsarcosine, which solubilizes both inner and outer membrane under the conditions tested (supplemental Fig. S3). Together, these data suggest that BcsKC is a tightly associated to the bacterial membrane. The membrane localization of BcsKC was further investigated by using sucrose density gradients to separate the inner from the outer membrane. As shown in Fig. 5C, BcsKC-FLAG did not cofractionate with the NADH oxidase activity but with BCAM1931, an outer membrane porin from B. cenocepacia, indicating that BcsKC-FLAG is associated with the outer membrane.
FIGURE 5.
BcsKC is tightly associated with the outer membrane. A, Western blot analysis of total cell lysate (T), soluble (S), and total membrane (Mb) fractions recovered from B. cenocepacia K56-2 ΔatsR ΔbcsKC (pBcsKC-FLAG), K56-2 ΔatsR ΔbcsKC (pBcsKCΔ40–189-FLAG), and B. cenocepacia K56-2 ΔatsR ΔT6SS (pBcsKC-FLAG) using anti-FLAG and anti-RNAP α subunit antibodies (cytosolic control). Ten μg of protein were loaded on a 12% SDS-PAGE gel. Molecular mass marker sizes (kDa) are shown on the left. The arrow highlights the position of the BcsKC-FLAG-containing complex. The faint bands seen in the membrane fractions (lines 3, 6, and 9) at the RNAP α subunit location (∼37 kDa) correspond to an unspecific protein cross-reacting with the anti-FLAG antibody (see Fig. 6B). B, protein solubilization. Total membranes were incubated with 50 mm Tris-HCl, pH 7.4 (Buffer), 1 m NaCl, 3% Triton X-100 (TX100), 2% N-lauroylsarcosine (LS), or 1% SDS. Insoluble (pellet, P) and soluble (supernatant, S) fractions were loaded on a 12% SDS-PAGE gel and analyzed by Western blot using an anti-FLAG antibody. C, sucrose density gradient. Inner and outer membranes were separated on a discontinuous sucrose gradient. The protein content was calculated by measuring the A280 nm, and the quality of the fractionation was assessed by measuring the enzymatic activity of the NADH oxidase (inner membrane marker) and by detecting the porin BCAM1931 (outer membrane marker). Fractions 1–24 were loaded on 12% SDS-PAGE gels and silver-stained or analyzed using an anti-FLAG antibody. The position of BCAM1931, BcsKC-FLAG and the high molecular mass complex (HMWC) are indicated. The positions of the inner membrane-containing (IM) and outer membrane-containing (OM) fractions are indicated.
BcsKC Forms a High Molecular Weight Complex Containing BcsLB and Also Requiring BcsFH for Its Formation
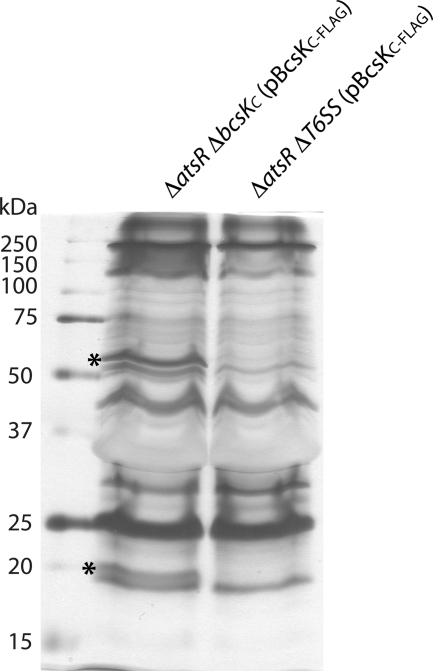
Expression of BcsKC-FLAG in B. cenocepacia K56-2 ΔatsR ΔbcsKC leads to the formation of a high molecular weight complex detected with the anti-FLAG antibody at the top of the separating gel (Fig. 5A, lane 3), suggesting multimerization of BcsKC-FLAG. This complex localizes almost exclusively in the membrane fraction (Fig. 5A, lanes 2 and 3) and is anchored to the outer membrane (Fig. 5C). The complex was absent when BcsKCΔ40–189-FLAG was expressed in K56-2 ΔatsR ΔbcsKC, suggesting that the N terminus of BcsKC is required for the complex formation (Fig. 5A, lanes 3 and 6). Moreover, although a complex is formed when BcsKC-FLAG is expressed in B. cenocepacia K56-2 ΔatsR ΔbcsKC, expression of BcsKC-FLAG in a strain where all the T6SS encoding genes have been deleted (K56-2 ΔatsR ΔT6SS) did not allow the complex formation, indicating that other T6SS components are required (Fig. 5A, lanes 3, 6, and 9).
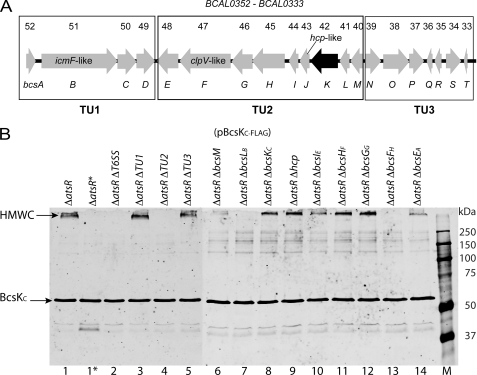
We investigated which T6SS components were required for the formation of the BcsKC-FLAG-containing complex. In B. cenocepacia K56-2 the T6SS gene cluster is organized in three putative transcriptional units (Fig. 6A). To identify the components involved in the BcsKC-containing complex formation, each of the three transcriptional units were individually deleted in the K56-2 ΔatsR (K56-2 ΔatsR ΔTU1, K56-2 ΔatsR ΔTU2, and K56-2 ΔatsR ΔTU3). The cell lysates were prepared from the different mutant strains expressing BcsKC-FLAG and analyzed by Western blot. As shown Fig. 6B, strains K56-2 ΔatsR ΔTU1 (pBcsKC-FLAG) (lane 3) and K56-2 ΔatsR ΔTU3 (pBcsKC-FLAG) (lane 5) but not K56-2 ΔatsR ΔTU2 (pBcsKC-FLAG) (lane 4) could produce the BcsKC-containing complex, suggesting that the TU2 contains the critical components for complex formation. Each gene of the TU2 was systematically deleted in K56-2 ΔatsR (Fig. 6A). Cell lysate analysis of individual gene mutants within the TU2 revealed that BcsLB and BcsFH were required for the formation of the BcsKC-FLAG-containing complex (Fig. 6B, lanes 7 and 13).
FIGURE 6.
BcsLB and BcsFH are required for the formation of the BcsKC-containing high molecular mass complex. A, genetic map of the T6SS gene cluster. This cluster has been designated bcs cluster for B. cenocepacia survival. The arrows represent the location and direction of gene transcription. BCAL gene designations according to the annotation of the B. cenocepacia J2315 genome (53) are shown above, and the bcs annotation of the genes is shown below. The genes icmF-like (BCAL0351, bcsBM), clpV-like (BCAL0347, bcsFH), and hcpBc (BCAL0343, bcsJD), which are hallmarks of T6SSs in other bacteria, are shown. The black arrow represents bcsKC. The putative transcriptional units TU1, TU2, and TU3 are boxed. B, anti-FLAG Western blot analysis of total cell lysates recovered from B. cenocepacia K56-2 mutant strains harboring pBcsKC-FLAG. Lane 1, K56-2 ΔatsR; lane 2, K56-2 ΔatsR ΔT6SS; lane 3, K56-2 ΔatsR ΔTU1; lane 4, K56-2 ΔatsR ΔTU2; lane 5, K56-2 ΔatsR ΔTU3; lane 6, K56-2 ΔatsR ΔbcsM; lane 7, K56-2 ΔatsR ΔbcsLB; lane 8, K56-2 ΔatsR ΔbcsKC; lane 9, K56-2 ΔatsR Δhcp; lane 10, K56-2 ΔatsR ΔbcsIE; lane 11, K56-2 ΔatsR ΔbcsHF; lane 12, K56-2 ΔatsR ΔbcsGG; lane 13, K56-2 ΔatsR ΔbcsFH; lane 14, K56-2 ΔatsR ΔbcsEA; lane M, molecular mass markers with the indicated sizes (kDa). The asterisk indicates that the sample has been boiled prior to loading. Black arrows indicate BcsKC or the high molecular mass complex (HMWC).
The complex is heat-sensitive because it was not detectable in samples that were heated prior to loading (Fig. 6B, lane 1*). To investigate the composition of the complex unheated membrane, samples prepared from K56-2 ΔatsR ΔbcsKC (pBcsKC-FLAG) and K56-2 ΔatsR ΔT6SS (pBcsKC-FLAG) were individually run on a SDS-PAGE gel. A slice from the top of the separating gels containing high molecular weight proteins (and the BcsKC-containing complex in the case of K56-2 ΔatsR ΔbcsKC (pBcsKC-FLAG)) was excised. The proteins were electroeluted and concentrated, and the samples were heated prior to loading on a SDS-PAGE gel to disassociate the components of the BcsKC-containing complex. Comparison of the protein profiles of K56-2 ΔatsR ΔbcsKC (pBcsKC-FLAG) and K56-2 ΔatsR ΔT6SS (pBcsKC-FLAG) revealed two extra bands in the electroeluted sample from K56-2 ΔatsR ΔbcsKC (pBcsKC-FLAG) with apparent masses of ∼55 and ∼20 kDa (Fig. 7). Mass spectrometry was performed on those two proteins. Identified peptides matched with a score of 507 and 643 to BCAL0342 (BcsKC) and BCAL0341 (BcsLB), respectively, indicating that BcsLB is also part of the complex containing BcsKC and again confirms the interaction between BcsKC and BcsLB.
FIGURE 7.
The high molecular mass complex contains BcsKC and BcsLB. Seventy μg of electroeluted high molecular mass proteins recovered from B. cenocepacia K56-2 ΔatsR ΔbcsKC (pBcsKC-FLAG) (left lane) and K56-2 ΔatsR ΔT6SS (pBcsKC-FLAG) (right lane) were boiled and loaded on a 16% SDS-PAGE gel. The asterisks highlight the positions of the 55- and 20-kDa proteins subjected to mass spectrometry analysis and corresponding to BcsKC and BcsLB, respectively. Shown at left are the molecular weight markers with the indicated sizes (kDa).
DISCUSSION
In this study, we show that deletion of bcsKC abolishes T6SS-associated phenotypes in B. cenocepacia (Hcp secretion and induction of the cytoskeletal rearrangements in macrophages after infection), demonstrating that BcsKC is a key component of the B. cenocepacia T6SS. Most T6SSs identified to date encode a BcsKC homolog, and the importance of BcsKC-like proteins in T6SS activity has already been underlined in other Gram-negative bacteria such as enteroaggregative E. coli, E. tarda, and F. novicida (11, 12, 14). In B. mallei, the BcsKC homolog TssB (BMAA0743) is not required for Hcp secretion, but it is necessary for virulence in hamsters (22). B. mallei contains three paralogs sharing more than 30% homology with TssB amino acid sequence. Redundancy might explain why Hcp is still secreted but does not explain the lack of virulence when TssB is mutated. Schell et al. (22) speculated that TssB is a secreted effector instead of a component of the T6S apparatus, but this remains to be demonstrated.
We also demonstrated that BcsKC could interact with BcsLB as shown by our immunoprecipitation and bacterial two-hybrid assays. This was an expected result because interactions between proteins homologous to BcsKC and BcsLB have already been reported in other T6S systems (11, 32, 45). For example, in F. novicida, the central region of IglA (BcsLB homolog) is required for interaction with IglB (BcsKC homolog) (45). Using a similar approach, our study reveals that the region required to bind BcsLB is limited to the N terminus of BcsKC, in particular to a segment encompassing amino acids 63–112. The N terminus of BcsKC, likely because of its ability to bind BcsLB, is essential for T6SS activity. PSIPRED analysis predicted this region to form two α helices preceding a β strand. Ab initio structural modeling of BcsKC, VipB, IglB, and PA0084 using the I-Tasser server (48) revealed a similar fold in the regions corresponding to amino acids 63–112 of BcsKC, which consists of paired α-helices (supplemental Fig. S4). Therefore, we propose that this is a conserved domain among BcsKC-like proteins for the interaction with BcsLB-like proteins.
Using a purified protein fusion containing the N terminus of ClpV and soluble cell lysate from V. cholerae Bönemann et al. (32) demonstrated that the N terminus of ClpV interacts with the VipA/VipB complex (BcsLB/BcsKC homolog). Interaction between BcsFH (ClpV homolog) and BcsKC/BcsLB has not been detected in our pulldown assays or our electroelution/resolution of the BcsKC-containing complex under the conditions tested. In V. cholerae ClpV is eight times less abundant than VipA and VipB (32). Thus, if the same ratio is true for B. cenocepacia, ClpV and BcsLB/BcsKC interactions might be more difficult to detect when BcsKC is used as bait in immunoprecipitation assays. Moreover, because the energizing component ClpV has been shown in V. cholerae to degrade BcsLB/BcsKC-like cytosolic complexes, it is possible that ClpV interacts with BcsLB/BcsKC transiently. Interaction between IglA/IglB in F. novicida has been shown to be required for the stability of each protein (45). In our experimental conditions BcsKC and derivatives are constitutively expressed on a multi-copy plasmid at higher levels than it would be in the wild type strain. Constitutive expression of BcsKCΔ40–189-FLAG (unable to bind BcsLB) in B. cenocepacia K56-2 ΔatsR or of BcsKC-FLAG in a B. cenocepacia K56-2 ΔatsR ΔbcsLB background likely bypassed the protein stability issue because no reduction of the protein expression levels or degradation products were noticed in the different cell lysates.
BcsLB and BcsKC homologs have been proposed to have different cellular localizations. In V. cholerae, the BcsLB and BcsKC homologs VipA and VipB have been described as nonsecreted cytosolic proteins (32). In F. novicida, the BcsLB and BcsKC homologs IglA and IglB have been described as strictly cytosolic proteins (11) or as surface-exposed proteins (49, 50). In E. tarda, EvpA (BcsLB) interacts with the periplasmic domain of IcmF (12), suggesting that BcsLB homologs might have a periplasmic location. By using a different approach and constitutively expressing BcsKC on a plasmid in B. cenocepacia K56-2 ΔatsR (overexpressing the other T6SS components), we show that BcsKC can be found in the cytosol but also in strong association with the outer membrane. In our fractionation analysis we noticed that BcsKC forms a high molecular weight complex anchored to the outer membrane. The formation of this complex depends on two other T6SS encoded proteins, BcsLB and BcsFH (ClpV homolog). Purification and analysis of the complex composition showed that BcsLB is also part of the complex (Fig. 7). The high molecular weight complex containing BcsKC and BcsLB may be indicative of the Type VI secretion structure; however, no other proteins could be detected as part of the complex under the conditions tested. If any other proteins are part of the complex, their amount is likely below the level of detection of our technique. In V. cholerae, BcsLB and BcsKC homologs VipA and VipB form cytosolic tubules disassembled by ClpV (32). However, in contrast to VipA/VipB tubules, the B. cenocepacia BcsLB/BcsKC complex was consistently detected in the membrane fraction, but not in the soluble fraction containing the cytosolic proteins. Furthermore, the BcsLB/BcsKC complex requires ClpV for its formation, whereas in V. cholerae the VipA/VipB tubule-like structures are severed by ClpV (32). Together, these data suggest that even though the BcsLB/BcsKC complex and the VipA/VipB tubules are made of BcsKC/BcsLB-like proteins, they have different localizations and likely reflect different stages in the biogenesis of the T6S apparatus. Leiman et al. (21) found that the characteristics of this VipA/B structure resemble those of the contracted T4 phage tail sheath (51) and speculated that VipA/B may provide energy to the T6SS-mediated secretion and membrane insertion process through conformational changes that mimic those that occur during phage tail contraction. Because of the similarities between the architectures of the VipA/VipB tubules and the bacteriophage T4 tail sheath, Bönemann et al. (52) proposed recently that VipA/VipB tubules might represent structural core components of the T6SS and that biogenesis of T6SSs might require ClpV to export VipA and VipB subunits into the periplasm, followed by VipA/VipB tubule reformation. We propose that the BcsLB/BcsKC complex that we describe in this study reflects precisely the tubule formation in the periplasmic space, which would be anchored to the outer membrane. Further characterization of BcsKC/BcsLB-like complexes is needed to understand their role in the functioning of the T6SS.
Supplementary Material
Acknowledgments
We thank Dr. M. Soledad Saldías and Slade Loutet for critical review of the manuscript, and Danuta Radzioch (McGill University, Department of Human Genetics, Montreal General Hospital Research Institute, Montreal, Canada) for the gift of the ANA-1 cell line.
This work was supported by a grant from the Canadian Cystic Fibrosis Foundation (to M. A. V.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Tables S1 and S2, and Figs. S1–S4.
- T6SS
- type VI secretion system.
REFERENCES
- 1.Vanlaere E., Baldwin A., Gevers D., Henry D., De Brandt E., LiPuma J. J., Mahenthiralingam E., Speert D. P., Dowson C., Vandamme P. (2009) Int. J. Syst. Evol. Microbiol. 59, 102–111 [DOI] [PubMed] [Google Scholar]
- 2.Vanlaere E., Lipuma J. J., Baldwin A., Henry D., De Brandt E., Mahenthiralingam E., Speert D., Dowson C., Vandamme P. (2008) Int. J. Syst. Evol. Microbiol. 58, 1580–1590 [DOI] [PubMed] [Google Scholar]
- 3.Govan J. R., Deretic V. (1996) Microbiol. Rev. 60, 539–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reik R., Spilker T., Lipuma J. J. (2005) J. Clin. Microbiol. 43, 2926–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Speert D. P., Henry D., Vandamme P., Corey M., Mahenthiralingam E. (2002) Emerg. Infect. Dis. 8, 181–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahenthiralingam E., Vandamme P., Campbell M. E., Henry D. A., Gravelle A. M., Wong L. T., Davidson A. G., Wilcox P. G., Nakielna B., Speert D. P. (2001) Clin. Infect. Dis. 33, 1469–1475 [DOI] [PubMed] [Google Scholar]
- 7.Hunt T. A., Kooi C., Sokol P. A., Valvano M. A. (2004) Infect. Immun. 72, 4010–4022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aubert D. F., Flannagan R. S., Valvano M. A. (2008) Infect. Immun. 76, 1979–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das S., Chaudhuri K. (2003) In Silico Biol. 3, 287–300 [PubMed] [Google Scholar]
- 10.Rao P. S., Yamada Y., Tan Y. P., Leung K. Y. (2004) Mol. Microbiol. 53, 573–586 [DOI] [PubMed] [Google Scholar]
- 11.de Bruin O. M., Ludu J. S., Nano F. E. (2007) BMC Microbiol. 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng J., Leung K. Y. (2007) Mol. Microbiol. 66, 1192–1206 [DOI] [PubMed] [Google Scholar]
- 13.Pukatzki S., Ma A. T., Revel A. T., Sturtevant D., Mekalanos J. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15508–15513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dudley E. G., Thomson N. R., Parkhill J., Morin N. P., Nataro J. P. (2006) Mol. Microbiol. 61, 1267–1282 [DOI] [PubMed] [Google Scholar]
- 15.Bladergroen M. R., Badelt K., Spaink H. P. (2003) Mol. Plant Microbe Interact. 16, 53–64 [DOI] [PubMed] [Google Scholar]
- 16.Pukatzki S., Ma A. T., Sturtevant D., Krastins B., Sarracino D., Nelson W. C., Heidelberg J. F., Mekalanos J. J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1528–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mougous J. D., Cuff M. E., Raunser S., Shen A., Zhou M., Gifford C. A., Goodman A. L., Joachimiak G., Ordoñez C. L., Lory S., Walz T., Joachimiak A., Mekalanos J. J. (2006) Science 312, 1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu H. Y., Chung P. C., Shih H. W., Wen S. R., Lai E. M. (2008) J. Bacteriol. 190, 2841–2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishikawa T., Rompikuntal P. K., Lindmark B., Milton D. L., Wai S. N. (2009) PLoS One 4, e6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Filloux A., Hachani A., Bleves S. (2008) Microbiology 154, 1570–1583 [DOI] [PubMed] [Google Scholar]
- 21.Leiman P. G., Basler M., Ramagopal U. A., Bonanno J. B., Sauder J. M., Pukatzki S., Burley S. K., Almo S. C., Mekalanos J. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4154–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schell M. A., Ulrich R. L., Ribot W. J., Brueggemann E. E., Hines H. B., Chen D., Lipscomb L., Kim H. S., Mrázek J., Nierman W. C., Deshazer D. (2007) Mol. Microbiol. 64, 1466–1485 [DOI] [PubMed] [Google Scholar]
- 23.Suarez G., Sierra J. C., Sha J., Wang S., Erova T. E., Fadl A. A., Foltz S. M., Horneman A. J., Chopra A. K. (2008) Microb. Pathog. (2008)44, 344–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma A. T., McAuley S., Pukatzki S., Mekalanos J. J. (2009) Cell Host Microbe 5, 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hood R. D., Singh P., Hsu F., Güvener T., Carl M. A., Trinidad R. R., Silverman J. M., Ohlson B. B., Hicks K. G., Plemel R. L., Li M., Schwarz S., Wang W. Y., Merz A. J., Goodlett D. R., Mougous J. D. (2010) Cell Host Microbe 7, 25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khajanchi B. K., Sha J., Kozlova E. V., Erova T. E., Suarez G., Sierra J. C., Popov V. L., Horneman A. J., Chopra A. K. (2009) Microbiology 155, 3518–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parsons D. A., Heffron F. (2005) Infect. Immun. 73, 4338–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pell L. G., Kanelis V., Donaldson L. W., Howell P. L., Davidson A. R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4160–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma L. S., Lin J. S., Lai E. M. (2009) J. Bacteriol. 191, 4316–4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aschtgen M. S., Bernard C. S., De Bentzmann S., Lloubès R., Cascales E. (2008) J. Bacteriol. 190, 7523–7531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aschtgen M. S., Gavioli M., Dessen A., Lloubes R., Cascales E. (2001) Mol. Microbiol. 75, 886–899 [DOI] [PubMed] [Google Scholar]
- 32.Bönemann G., Pietrosiuk A., Diemand A., Zentgraf H., Mogk A. (2009) EMBO J. 28, 315–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cascales E. (2008) EMBO Rep. 9, 735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shalom G., Shaw J. G., Thomas M. S. (2007) Microbiology 153, 2689–2699 [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 36.Cohen S. N., Chang A. C., Hsu L. (1972) Proc. Natl. Acad. Sci. U.S.A. 69, 2110–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Figurski D. H., Helinski D. R. (1979) Proc. Natl. Acad. Sci. U.S.A. 76, 1648–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Craig F. F., Coote J. G., Parton R., Freer J. H., Gilmour N. J. (1989) J. Gen. Microbiol. 135, 2885–2890 [DOI] [PubMed] [Google Scholar]
- 39.Flannagan R. S., Linn T., Valvano M. A. (2008) Environ. Microbiol. 10, 1652–1660 [DOI] [PubMed] [Google Scholar]
- 40.Blum H., Beier H., Gross H. J. (1987) Electrophoresis 8, 93–99 [Google Scholar]
- 41.Karimova G., Pidoux J., Ullmann A., Ladant D. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Putnam S. L., Koch A. L. (1975) Anal. Biochem. 63, 350–360 [DOI] [PubMed] [Google Scholar]
- 43.Osborn M. J., Gander J. E., Parisi E., Carson J. (1972) J. Biol. Chem. 247, 3962–3972 [PubMed] [Google Scholar]
- 44.Cox G. W., Mathieson B. J., Gandino L., Blasi E., Radzioch D., Varesio L. (1989) J. Natl. Cancer Inst. 81, 1492–1496 [DOI] [PubMed] [Google Scholar]
- 45.Bröms J. E., Lavander M., Sjöstedt A. (2009) J. Bacteriol. 191, 2431–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones D. T. (1999) J. Mol. Biol. 292, 195–202 [DOI] [PubMed] [Google Scholar]
- 47.McGuffin L. J., Bryson K., Jones D. T. (2000) Bioinformatics 16, 404–405 [DOI] [PubMed] [Google Scholar]
- 48.Zhang Y. (2008) BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Melillo A., Sledjeski D. D., Lipski S., Wooten R. M., Basrur V., Lafontaine E. R. (2006) FEMS Microbiol. Lett. 263, 102–108 [DOI] [PubMed] [Google Scholar]
- 50.Ludu J. S., de Bruin O. M., Duplantis B. N., Schmerk C. L., Chou A. Y., Elkins K. L., Nano F. E. (2008) J. Bacteriol. 190, 4584–4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moody M. F. (1967) J. Mol. Biol. 25, 167–200 [DOI] [PubMed] [Google Scholar]
- 52.Bönemann G., Pietrosiuk A., Mogk A. (2010) Mol Microbiol 76, 815–821 [DOI] [PubMed] [Google Scholar]
- 53.Holden M. T., Seth-Smith H. M., Crossman L. C., Sebaihia M., Bentley S. D., Cerdeño-Tárraga A. M., Thomson N. R., Bason N., Quail M. A., Sharp S., Cherevach I., Churcher C., Goodhead I., Hauser H., Holroyd N., Mungall K., Scott P., Walker D., White B., Rose H., Iversen P., Mil-Homens D., Rocha E. P., Fialho A. M., Baldwin A., Dowson C., Barrell B. G., Govan J. R., Vandamme P., Hart C. A., Mahenthiralingam E., Parkhill J. (2009) J. Bacteriol. 191, 261–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nielsen H., Engelbrecht J., Brunak S., von Heijne G. (1997) Protein Eng. 10, 1–6 [DOI] [PubMed] [Google Scholar]
- 55.Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004) J. Mol. Biol. 340, 783–795 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.