Abstract
Dendritic spines are small actin-rich structures that receive the majority of excitatory synaptic input in the brain. The actin-based dynamics of spines are thought to mediate synaptic plasticity, which underlies cognitive processes, such as learning and memory. However, little is known about the molecular mechanisms that regulate actin dynamics in spines and synapses. In this study we show the multifunctional actin-binding protein vasodilator-stimulated phosphoprotein (VASP) regulates the density, size, and morphology of dendritic spines by inducing actin assembly in these structures. Knockdown of endogenous VASP by siRNA led to a significant decrease in the density of spines and synapses, whereas expression of siRNA-resistant VASP rescued this defect. The ability of VASP to modulate spine and synapse formation, maturation, and spine head enlargement is dependent on its actin binding Ena/VASP homology 2 (EVH2) domain and its EVH1 domain, which contributes to VASP localization to actin-rich structures. Moreover, VASP increases the amount of PSD-scaffolding proteins and the number of surface GluR1-containing α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptors (AMPARs) in spines. VASP knockdown results in a reduction in surface AMPAR density, suggesting a role for this protein in regulating synaptic strength. Consistent with this, VASP significantly enhances the retention of GluR1 in spines as determined by fluorescence recovery after photobleaching and increases AMPAR-mediated synaptic transmission. Collectively, our results suggest that actin polymerization and bundling by VASP are critical for spine formation, expansion, and modulating synaptic strength.
Keywords: Actin, Cytoskeleton, Neuron, Neurotransmitter Receptors, Synapses, Dendritic Spines, Synaptic Strength, VASP
Introduction
Neurons communicate via specialized structures called synapses that are composed of presynaptic and postsynaptic terminals. In excitatory synapses the majority of synaptic input takes place on dendritic spines, which are actin-rich structures composed of a bulbous head and a thin neck connected to dendritic shafts (1, 2). Dendritic spines play a central role in cognitive processes, and changes in their size, number, and morphology are associated with numerous neurological disorders (3). Emerging evidence indicates these processes are regulated by polymerization and reorganization of the actin cytoskeleton, pointing to the importance of actin dynamics in modulating synaptic function (4–6). Alterations in actin remodeling, in turn, are mediated by actin-binding proteins, but the role these proteins play in modulating the development, morphology, and function of spines and synapses is not well understood.
VASP is an actin-binding protein that regulates actin polymerization and bundling via direct interaction with both globular (G) and filamentous (F) actin (7–9). In non-neuronal cells, VASP2 localizes to dynamic actin structures, such as focal adhesions, filopodia, and the leading edge of lamellipodia, where it regulates actin-based cellular processes (10–12). In the nervous system, VASP family proteins are required for proper positioning of neurons and neuritogenesis in the neocortex as well as filopodia formation in cortical and hippocampal neurons (13–17). In addition, loss of VASP family proteins significantly impairs normal brain development, indicating an important role for VASP proteins in the central nervous system (15).
VASP contains three conserved domains, including an EVH1 domain, a proline-rich domain (PRD), and an EVH2 domain, that have different roles in VASP function (18). The EVH1 domain mediates VASP localization to actin-rich structures, possibly via association with proline-rich proteins (10, 18–21). The central PRD contains binding sites for WW- and SH3-domain-containing proteins as well as the G-actin binding protein profilin (18, 22–25). The C-terminal EVH2 domain consists of a G-actin binding motif, an F-actin binding domain, and a coiled-coil domain for VASP tetramerization (8, 9, 26). Like the EVH1 domain, the EVH2 domain contributes to VASP targeting to lamellipodia (12, 27). Moreover, it is involved in bundling F-actin, protecting barbed ends from capping, and mediating filopodia formation (9, 20, 27, 28).
Here, we show that VASP regulates actin polymerization in dendritic spines to modulate spine and synapse formation as well as spine head enlargement. In addition, VASP increases the amount and retention of GluR1-containing AMPARs in spines to regulate synaptic strength, which is thought to be the basis of learning and memory.
EXPERIMENTAL PROCEDURES
Reagents
SV2 monoclonal antibody was from Developmental Studies Hybridoma Bank (University of Iowa, Iowa City, IA). PSD95 antibodies were purchased from Chemicon (Temecula, CA) and NeuroMAB (Davis, CA). Homer 1b/c antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). Shank1 antibody was purchased from Abcam (Cambridge, MA). VASP antibody was kindly provided by Frank Gertler (MIT, Boston, MA). Mena antibody (clone 21) was purchased from BD Biosciences. Alexa Fluor® 546 phalloidin, β-actin AC-15 monoclonal antibody, unlabeled phalloidin, strychnine, and bicuculline methiodide were obtained from Sigma. GluR1 antibody was from Calbiochem. GFP antibody, Alexa Fluor® 568 G-actin, Alexa Fluor® 647 phalloidin, ProLong Gold antifade reagent, Alexa Fluor® 488, 555, and 647 anti-mouse, Alexa Fluor® 488 and 555 anti-rabbit, and Alexa Fluor® 680 anti-mouse were from Invitrogen. IRDye 800 anti-mouse was obtained from Rockland Immunochemicals (Gilbertsville, PA). Aqua Ply/Mount was from Polysciences (Warrington, PA). Tetrodotoxin was purchased from Tocris Bioscience (Ellisville, MO). ATP was from Fisher.
Plasmids
VASP cDNA was a generous gift from Jüergen Wehland (Technical University of Braunschweig, Braunschweig, Germany). Full-length human VASP tagged with enhanced green fluorescent protein was cloned into a neuronal expression vector, generously provided by Freda Miller, which contains a neuronal-specific α1-tubulin promoter (29). GluR1-GFP was kindly provided by Julius Zhu (University of Virginia, Charlottesville, VA). mCherry cDNA was generously provided by Roger Tsien (University of California, San Diego, CA) Deletion constructs were generated using PCR with the following primers: VASP ΔEVH1 (Δ1–117), forward (5′-GGTTCCAGATCTCCCCCTCCACCCCCAGCACTTCC-3′) and reverse (5′-GGCCTTCTCGAGTCAGGGAGAACCCCGCTTCC-3′); VASP ΔEVH2 (Δ226–380), forward (5′-GGTTCCGGATCCATGAGCGAGACGGTCATCTGTTCC-3′) and reverse (5′-GGCCTTCTCGAGGCCTGGGGCCCCAGCTCCCC-3′). Nested PCR methods were used to generate the ΔPRD (Δ118–225) construct, and the external primers were as follows: forward (5′-GGTTCCGGATCCATGAGCGAGACGGTCATCTGTTCC-3′) and reverse (5′-GGCCTTCTCGAGTCAGGGAGAACCCCGCTTCC-3′). Internal primers used were: forward (5′-GCGTTGGAAGGAGGTGGGCTGGCCGCAGCTATTGCTGG-3′) and reverse, (5′-CCAGCAATAGCTGCGGCCAGCCCACCTCCTTCCAACGC-3′). VASP siRNA constructs were generated by ligating 64-mer sense and antisense oligonucleotides into pSUPER vector as previously described (30). The VASP siRNA oligos contained the following 19-nucleotide target sequences: VASP #1, 5′-TGAAAGAGGAAATAATCGA, and VASP #2, 5′-TTGTGGAAGAGGTGCGGAA.
Cell Culture and Transfection
Low density hippocampal neurons were prepared and cultured as previously described (31). Briefly, neurons were plated at a density of 75,000 cells/mm2 and transfected by a modified calcium phosphate method at days 5–6 in culture unless otherwise specified (32). Human embryonic kidney 293T (HEK-293T) cells and rat 2 fibroblasts (R2Fs) (ATCC, Manassas, VA) were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum and penicillin/streptomycin. HEK-293T and R2Fs were transfected with Lipofectamine 2000 (Invitrogen) and Amaxa kits (Lonza Cologne), respectively, according to the manufacturer's instructions.
Microscopy
Neurons were imaged using a Retiga EXi CCD camera (QImaging) on an Olympus IX71 inverted microscope (Olympus, Melville, NY) with a PlanApo 60X OTIRFM objective (NA 1.45). Image acquisition was controlled with MetaMorph software (Molecular Devices, Sunnyvale, CA), which was interfaced with a Lambda 10–2 automated controller (Sutter Instruments). Alexa Fluor 488 and enhanced green fluorescent protein were imaged with an Endow GFP Bandpass filter cube (excitation HQ470/40, emission HQ525/50, Q495LP dichroic mirror) (Chroma, Brattleboro, VT). For Alexa Fluor 555 or 546, a TRITC/Cy3 cube (excitation HQ545/30, emission HQ610/75, Q570LP dichroic mirror) was used. Alexa Fluor 647 was imaged with a Cy5TM cube (excitation HQ620/60, emission HQ700/75, Q660LP dichroic mirror).
FRAP was performed on a Quorum WaveFX spinning disk confocal system with a Nikon Eclipse Ti microscope using a PlanApo 60X TIRF objective (NA 1.49). Four to six circular regions of interest (20 × 20 pixels) in spine heads were photobleached with a 405-nm diode laser for 700 ms with 100% laser power. Enhanced green fluorescent protein images were acquired at 20-s intervals using a Hamamatsu ImageEM-CCD camera and MetaMorph software. Enhanced green fluorescent protein and mCherry were excited with 491- and 561-nm laser lines, respectively. For FRAP analysis, the background subtracted fluorescent intensity at each time point t was corrected for the loss of fluorescence due to image acquisition. The corrected data were further normalized to the base-line fluorescence (Ipre), which is defined as 100% and graphed according to the equation FI (t) = (It × Inf, pre)/(Ipre × Inf,t) where nf represents a region that was not subjected to FRAP. A graph of the recovery traces were generated using the following equation: Fluorescence recovery (FR) = [FI (t) − FI (0)]/[FI(pre) − FI (0)]. The time constant was calculated according to the equation: FR = α − A1exp(−k1t) − A2exp(−k2t), where α is the mobile fraction, and Ax is the amplitude of the exponential process with rate constant kx (33).
Immunocytochemistry and Image Analysis
Neurons were fixed at days 11–12 in culture, permeabilized, and stained as previously described (34). To simultaneously stain for endogenous VASP and PSD95, neurons were fixed with cold 10% formalin for 15 min at room temperature, permeabilized, and stained as previously described (34). Neurons were stained for surface GluR1 as previously described (35, 36). Briefly, cells were incubated for 30–50 min at room temperature in an extracellular solution containing 150 mm NaCl, 5 mm KCl, 2 mm CaCl2, 10 mm HEPES, 30 mm glucose, 0.5 μm tetrodotoxin, 1 μm strychnine, and 20 μm bicuculline methiodide, pH 7.4. For live-cell GluR1 staining, neurons were incubated with GluR1 antibody for 20 min at 37 °C. These cells were subsequently fixed in 2% paraformaldehyde, 0.12 m sucrose and stained with secondary antibodies. To visualize presynaptic terminals in these neurons, cells were subsequently permeabilized with 0.2% Triton X-100 and immunostained for SV2.
The density of spines and synapses was quantified beginning within 5 μm of the soma, along primary and secondary dendrites as previously described (37). The average length of the dendrites analyzed was 60 μm. We define spines as dendritic protrusions that have a bulbous head with an average size of 0.5 μm2 that are in contact with presynaptic terminals. In our analyses, dendritic spines ranged in length from 1 to 4 μm. The spine/shaft ratio was calculated by measuring the background subtracted fluorescent intensity in individual spines and an equivalent area in the neighboring shaft. Statistical analyses were performed using Student's t test.
Actin Barbed End Staining
Barbed end staining was performed as previously described with minor modifications (38, 39). Briefly, neurons were permeabilized with 0.02% saponin in 20 mm HEPES, 138 mm NaCl, 4 mm MgCl2, 3 mm EGTA, 1% BSA, 1 mm ATP, and 3 μm unlabeled phalloidin, pH 7.4. After a brief wash, free barbed ends were stained with Alexa 568 G-actin in saponin-free solution. Cells were then fixed in 4% paraformaldehyde, 0.12 m sucrose and visualized in fluorescence.
Electrophysiology
Neurons were transfected with GFP-VASP at day 6 in culture, and whole-cell patch clamp recordings were obtained at day 14–16 in culture. Cells were placed in a recording chamber in an extracellular solution containing 140 mm NaCl, 3 mm KCl, 2 mm MgCl2, 2 mm CaCl2, 11 mm glucose, 25 mm HEPES, 0.5 μm tetrodotoxin, 20 μm bicuculline methiodide, and 1 μm strychnine, pH 7.4. Patch pipettes were filled with an intracellular solution composed of 115 mm cesium gluconate, 17.5 mm CsCl, 10 mm HEPES, 2 mm MgCl2, 10 mm EGTA, 4 mm K2ATP, 0.4 mm Na-GTP, pH 7.4, and cells were recorded at room temperature at a holding potential of −60 mV using a Multiclamp 700A amplifier (Molecular Devices). Recordings were pass-filtered at 2 kHz and sampled at 10 kHz. Membrane and access resistances were monitored continuously, and recording data were rejected if series access resistance varied more than 20%. The statistical significance was calculated using a paired t test.
RESULTS
VASP Is Concentrated in Dendritic Spines and Excitatory Synapses
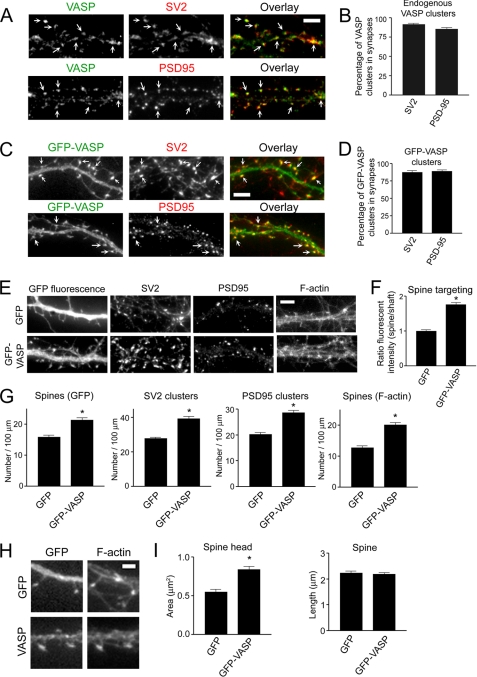
Ena/VASP proteins are highly expressed in the brain and in hippocampal pyramidal neurons, which mediate excitatory synaptic connections via dendritic spines (15, 40, 41), leading us to hypothesize that VASP plays a role in regulating spine and synapse development. To begin to test this hypothesis, we examined the localization of endogenous VASP with synaptic markers in low density cultures of hippocampal neurons. Endogenous VASP co-localized with the synaptic vesicle protein SV2 as well as the excitatory postsynaptic density protein PSD95 (Fig. 1A). Quantification showed that ∼85% of the VASP puncta colocalized with SV2 and PSD95 clusters (Fig. 1B), indicating VASP is enriched in spines and excitatory synapses. Like endogenous VASP, GFP-VASP is concentrated in spines and synapses with about 85% of GFP-VASP puncta co-localizing with SV2 and PSD95 clusters (Fig. 1, C and D). To confirm the enrichment of GFP-VASP to spines, we measured the ratio of the fluorescent intensity in spines to neighboring shafts from GFP-VASP-expressing neurons and normalized it to that observed in neurons expressing GFP. Indeed, the ratio of the fluorescent intensity was significantly enhanced in GFP-VASP-expressing neurons (Fig. 1F), indicating GFP-VASP is enriched in spines. Additionally, these results show that GFP-VASP localizes similarly to the endogenous protein and is a valid marker for examining VASP function in spines and synapses.
FIGURE 1.
VASP regulates spine and synapse formation and promotes spine head enlargement. A, hippocampal neurons were co-immunostained at day 12 in culture for endogenous VASP and the synaptic markers SV2 (upper panels) and PSD95 (lower panels). Endogenous VASP accumulated in puncta that co-localized with the synaptic markers (Overlays, right panels, arrows). Bar, 5 μm. B and D, shown is quantification of the percentage of endogenous VASP (B) and GFP-VASP (D) co-localizing with SV2 and PSD95. Error bars represent S.E. for at least 20 dendrites. C, neurons were transfected with GFP-VASP at day 5 in culture, fixed, and immunostained for SV2 (upper panels) and PSD95 (lower panels) at day 12. GFP-VASP localized in puncta with SV2 and PSD95 (Overlays, right panels, arrows). E, neurons were transfected with GFP or GFP-VASP and stained for SV2, PSD95, and F-actin (phalloidin). Bar, 5 μm. F, the ratio of the fluorescent intensity in spine heads to neighboring shafts for GFP-VASP-expressing neurons was normalized to that observed in control neurons expressing GFP. G, quantification of SV2 and PSD95 clusters and spine density as determined by GFP fluorescence and F-actin staining (phalloidin) is shown for GFP and GFP-VASP-expressing neurons. H, shown are higher magnification images of dendritic spines, visualized by GFP fluorescence and F-actin staining (phalloidin), from GFP and GFP-VASP-expressing neurons. Bar, 2 μm. I, quantification of spine length and area in GFP and GFP-VASP-expressing neurons is shown. Error bars represent S.E. for 40–50 dendrites (G) or 50–100 spines (F and I) from three separate experiments. Asterisks indicate p < 0.0001.
VASP Regulates the Formation of Spines and Synapses
To study the function of VASP in spines and synapses, GFP-VASP was expressed at relatively low levels, about 4-fold over endogenous (supplemental Fig. 1, A and B) in neurons. Expression of GFP-VASP resulted in a 35% increase in the number of spines compared with control neurons, as determined by GFP fluorescence (Fig. 1, E and G). Similar results were obtained when spine number was assessed by staining with phalloidin (Fig. 1, E and G), which binds to F-actin and can be used to visualize dendritic spines (4, 42). A 40% increase in synaptic density, as determined by staining for SV2 and PSD95, was observed in neurons expressing GFP-VASP compared with GFP expressing controls (Fig. 1, E and G). Even though VASP family proteins are reported to influence neurite outgrowth (14, 15), it is unlikely that the VASP effects we observe on spine and synapse formation are due to neuritogenesis. This process occurs predominately during days 1–4 in culture (31), and we did not transfect neurons with GFP-VASP until day 5 in culture. However, to address this concern, we transfected neurons with GFP-VASP at day 10 in culture, which is a time when neurites have already reached sufficient length to form synapses, and examined the effect of VASP on spines and synapses. Consistent with our previous results, expression of VASP led to 34.8 ± 3.8% (n = 20) increase in spine density. Collectively, these results suggest VASP is critical for the regulation of spine and synapse formation.
Because spines appeared to be larger in GFP-VASP-expressing neurons compared with controls (Fig. 1H), we quantified the spine head area and length of spines. Expression of GFP-VASP resulted in a 1.5-fold increase in spine head area but did not significantly affect spine length when compared with GFP expressing neurons (Fig. 1I), suggesting an important role for VASP in regulating spine head enlargement.
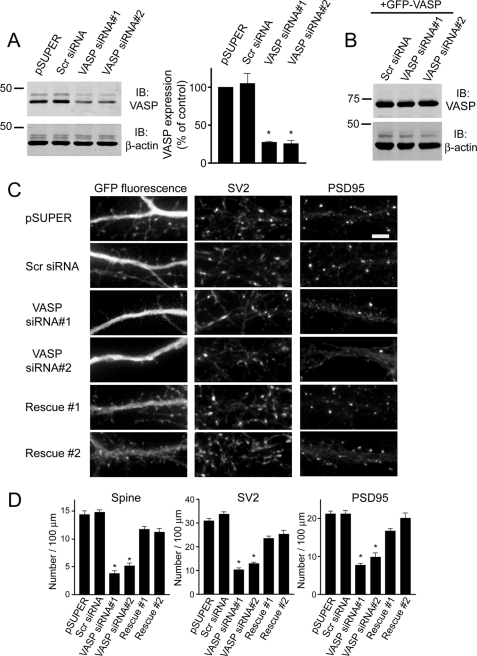
To further show that VASP regulates spine development, we generated two small interfering RNA (siRNA) constructs to knock down endogenous expression of VASP. The VASP siRNAs knocked down VASP expression by ∼70% in R2Fs compared with pSUPER empty vector or a scrambled siRNA (Fig. 2A). The siRNAs did not affect expression of Mena, another VASP family member, or other actin-binding proteins, such as N-WASP (supplemental Fig. 1, C and D), indicating their specificity for VASP. The VASP siRNAs were similarly effective in neurons, decreasing endogenous VASP expression by about 55% (supplemental Fig. 1, E and F). Transfection of neurons with VASP siRNA resulted in a significant decrease in the number of spines and synapses compared with control cells expressing pSUPER empty vector or scrambled siRNA (Fig. 2, C and D). Because our VASP siRNAs were specifically designed against the rat VASP sequence, it should not affect expression of human VASP due to several nucleotide mismatches. To confirm this, we expressed human GFP-VASP with VASP siRNA. As shown in Fig. 2B, VASP siRNA did not affect expression of GFP-VASP, which allowed us to perform “rescue” experiments in neurons. Expression of GFP-VASP in siRNA knockdown neurons rescued the siRNA-mediated defect on spines and synapses (Fig. 2, C and D). Thus, these results show the defect is due to loss of endogenous VASP and indicate an important role for VASP in regulating spine and synapse formation.
FIGURE 2.
Knockdown of endogenous VASP inhibits the formation of spines and synapses. A, cell lysates from R2Fs transfected with VASP siRNAs, pSUPER empty vector, or a non-silencing scrambled siRNA (Scr siRNA) were immunoblotted (IB) for VASP and β-actin (loading control). Quantification of blots from four separate experiments is shown (right panel). Error bars represent S.E. (*, p < 0.0001). B, cell lysates from R2Fs co-transfected with human GFP-VASP and scrambled siRNA (Scr siRNA) or VASP siRNAs were blotted for VASP and β-actin (loading control). C, neurons were co-transfected with GFP and either pSUPER empty vector, scrambled siRNA (Scr siRNA), or VASP siRNAs at day 6 in culture, fixed, and stained with synaptic markers at day 12. To show the siRNA-induced defect on spines and synapses was due to endogenous loss of VASP, neurons were co-transfected with human GFP-VASP and VASP siRNA#1 or VASP siRNA#2 (lower panels, Rescue #1 and Rescue #2). Bar, 5 μm. D, quantification of spine and synaptic density (SV2 and PSD95 clusters) in neurons transfected with the indicated constructs is shown. Error bars represent S.E. for 40–50 dendrites from at least three separate experiments (*, p < 0.0001). For panels A and D, asterisks denote a statistically significant difference compared with pSUPER-transfected cells.
The EVH Domains Are Necessary for VASP Recruitment and Function in Spines and Synapses
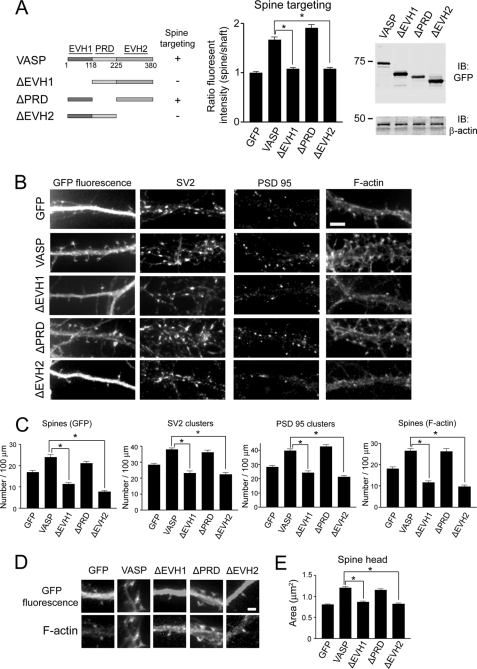
To identify the region of VASP that mediates its localization and function in spines and synapses, we generated various deletion constructs and expressed them as GFP fusion proteins (Fig. 3A). The relative expression level of all of the deletion constructs was similar to that observed with full-length GFP-VASP as determined by immunoblot analysis (Fig. 3A). When either the EVH1 or EVH2 domain was deleted (ΔEVH1 or ΔEVH2), VASP failed to localize to spines (Fig. 3A), whereas deletion of the PRD (ΔPRD) domain did not significantly affect VASP localization to spines (Fig. 3A). The central role of the EVH1 and EVH2 domains in targeting VASP to spines suggested these domains may also be important for VASP function in the development of spines and synapses. Indeed, deletion of either the EVH1 or EVH2 domain of VASP significantly impaired spine and synapse formation (Fig. 3, B and C). In neurons expressing ΔEVH1- or ΔEVH2-VASP, the number of spines was decreased by 54 and 68%, respectively, compared with full-length VASP (Fig. 3, B and C). Moreover, expression of ΔEVH1- or ΔEVH2-VASP resulted in a significant decrease in the number of SV2 and PSD95 clusters compared with full-length VASP (Fig. 3, B and C). In contrast, the number of spines and synapses in ΔPRD-VASP-expressing neurons was comparable with that observed in neurons expressing full-length VASP (Fig. 3, B and C).
FIGURE 3.
The EVH1 and EVH2 domains of VASP promote spine and synapse formation as well as spine head enlargement. A, domain structure of full-length VASP is shown. EVH1, PRD, and EVH2 domains are indicated. A schematic diagram of VASP deletion constructs is shown. The deletion mutants that localize to spines are indicated with a “+”. The ratio of the fluorescent intensity in spine heads to neighboring shafts was quantified in neurons expressing the indicated constructs and normalized to control neurons expressing GFP (middle panel). In the right panel, lysates from HEK-293T cells transfected with the indicated constructs were blotted for GFP and β-actin (loading control). IB, immunoblot. B, neurons were transfected with the indicated constructs and stained for SV2, PSD95, and F-actin (phalloidin). Bar, 5 μm. C, shown is quantification of synaptic density (SV2 and PSD95 clusters) and spine number by GFP fluorescence and F-actin staining from transfected neurons. D, shown are higher magnification images of dendritic spines from neurons expressing GFP, GFP-VASP, or the indicated VASP deletion mutants. Bar, 2 μm. E, shown is quantification of spine head area in neurons expressing the constructs from panel D. For panels A, C, and E, error bars represent S.E. for 40 dendrites (C) or 100 spines (A and E) from three separate experiments (*, p < 0.0001).
Because VASP also promotes spine head enlargement, we determined which domains of VASP are critical for this process. The spine head area in neurons expressing VASP mutants lacking either the EVH1 or EVH2 domain was reduced compared with full-length VASP (Fig. 3, D and E), indicating the importance of these domains in promoting spine head enlargement. In contrast, the PRD domain was not necessary for VASP-induced enlargement of spines heads (Fig. 3, D and E). Collectively, these results indicate the EVH1 and EVH2 domains of VASP regulate spine/synapse formation, targeting, and spine head enlargement.
VASP Regulates Actin Dynamics in Dendritic Spines
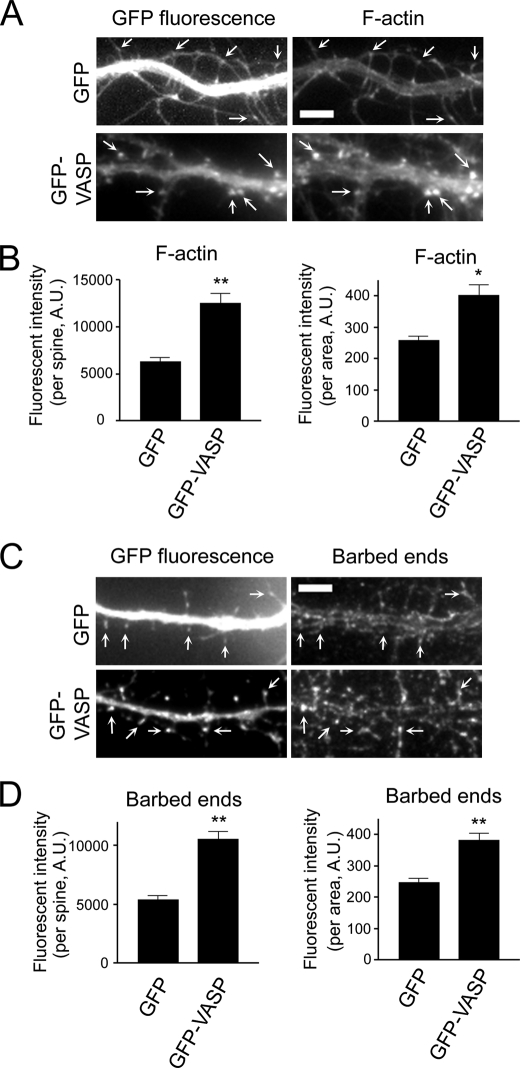
To further explore the effect of VASP on actin dynamics in spines, we stained GFP and GFP-VASP-expressing neurons with phalloidin to visualize F-actin. Quantification of the fluorescent intensity of phalloidin showed a 2.5-fold increase in F-actin staining in spines of GFP-VASP-expressing neurons compared with GFP controls (Fig. 4, A and B, and supplemental Fig. 2A). In addition, when we normalized the fluorescent intensity to the unit area, a significant increase in the normalized fluorescent intensity in spines was still observed in GFP-VASP-expressing neurons compared with controls (Fig. 4B), indicating VASP significantly enhances the amount of F-actin in spines.
FIGURE 4.
VASP promotes actin polymerization in spines. A, images of GFP and GFP-VASP-expressing neurons stained for F-actin are shown. Arrows indicate dendritic spines. Bar, 5 μm. B, quantification of the fluorescent intensity of F-actin staining in individual spines from GFP and GFP-VASP-expressing neurons is shown (left panel). The fluorescent intensity was normalized to the spine area (right panel). C, images of GFP and GFP-VASP-expressing neurons stained for free actin barbed ends are shown. Arrows indicate spines. Bar, 5 μm. D, quantification of the fluorescent intensity in individual spines from barbed end staining is shown (left panel). The fluorescent intensity was normalized to spine area (right panel). For panels B and D, error bars represent S.E. for 100 spines from three independent experiments (**, p < 0.0001; *, p < 0.01).
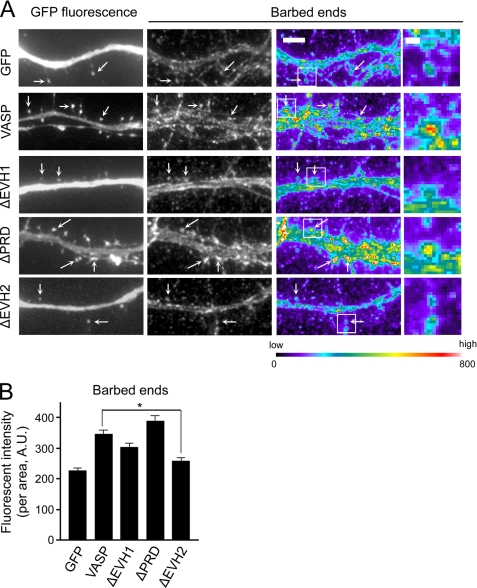
Does VASP promote actin polymerization in dendritic spines? In vitro, VASP induces actin polymerization in the presence of barbed end capping proteins, suggesting VASP protects F-actin from capping proteins to promote filament elongation (12, 28, 43). This led us to hypothesize that VASP stimulates actin polymerization in dendritic spines by protecting the barbed ends of actin filaments. To test this, we stained GFP and GFP-VASP-expressing neurons with fluorescent monomeric actin to label available barbed ends. Some barbed end staining of actin filaments was observed in spines of GFP expressing neurons (Fig. 4, C and D, and supplemental Fig. 2B), which is consistent with a previous study demonstrating free barbed ends in dendritic spines (44). Importantly, a 2-fold increase in the fluorescent intensity of barbed end staining in spines was observed in GFP-VASP-expressing neurons compared with controls (Fig. 4, C and D, and supplemental Fig. 2B). Similar results were obtained when the fluorescent intensity in spines was normalized to the unit area, indicating the enhanced staining was due to an increased number of available barbed ends and not to larger spine heads. When the actin binding EVH2 domain was deleted, VASP no longer protected barbed ends (Fig. 5, A and B). Interestingly, the number of barbed ends was still increased in ΔEVH1-VASP-expressing neurons compared with GFP controls, suggesting the EVH1 domain in not essential for this function of VASP. These results suggest VASP, through its EVH2 domain, promotes actin polymerization in spines by increasing the availability of barbed ends for further actin assembly.
FIGURE 5.
The EVH2 domain is essential for VASP to protect barbed ends. A, images from barbed end staining of neurons expressing the indicated constructs are shown in grayscale (middle panels) and pseudo-color coding (right panels). Pseudo-color coding indicates the range of fluorescent intensities to the assigned color. Bar, 5 μm. Arrows indicate spines. Higher magnification images of the boxed regions are shown in the far right panels. Bar, 1 μm. B, quantification of the fluorescent intensity in individual spines from neurons expressing GFP, GFP-VASP, or VASP deletion mutants and stained for free barbed ends is shown. The fluorescent intensity was normalized to the spine area. Error bars represent S.E. for at least 70 spines from three separate experiments (*, p < 0.0001).
VASP Modulates the Amount of GluR1-containing AMPARs and PSD-scaffolding Proteins in Spines
The spine head size, the PSD area, and the intact actin cytoskeleton control the anchoring of postsynaptic receptors, which can determine the efficacy of synaptic strength (42, 45–47). Because our results indicate VASP modulates spine head size and elongation of actin filaments, it could regulate the size of the PSD and synaptic strength. To test this, we examined the effect of VASP on the amount of several PSD-scaffolding proteins, including PSD95, Homer, and Shank, in spines because they have been shown to promote synapse maturation and modulate synaptic strength (48–50). Expression of GFP-VASP resulted in a significant increase in the intensity of PSD95 staining in spines (Fig. 6A and supplemental Fig. 3A). Quantification of the fluorescent intensity of PSD95 showed a 2-fold increase in the level of PSD95 in GFP-VASP-expressing neurons compared with GFP controls (Fig. 6B). Comparable results were obtained when the fluorescent intensity was normalized to the unit area (Fig. 6B). Moreover, VASP expression promoted a similar increase in the amount of Homer and Shank in spines (supplemental Fig. 3, B–E). These results suggest that VASP modulates the level of PSD-scaffolding proteins in spines and point to a role for VASP in regulating synaptic strength.
FIGURE 6.
VASP increases the amount of PSD95 and GluR1 in spines. A, images of GFP and GFP-VASP-expressing neurons stained for PSD95 are shown. Bar, 5 μm. Arrows indicate spines. B, quantification of the fluorescent intensity in individual spines of neurons stained for PSD95 is shown (left panel). The fluorescent intensity was normalized to spine area (right panel). C, GFP and GFP-VASP-expressing neurons were stained for surface GluR1 (sGluR1) under non-permeabilizing conditions. Neurons were then permeabilized and stained for SV2 to indicate synapses. The sGluR1 puncta at synapses are indicated (arrows). Bar, 5 μm. D, quantification of the fluorescent intensity of sGluR1 in spines from GFP and GFP-VASP-expressing neurons is shown (left panel). The fluorescent intensity was normalized to the spine area (right panel). Error bars represent S.E. for 100 spines (B and D) from three separate experiments (**, p < 0.0001; *, p < 0.03).
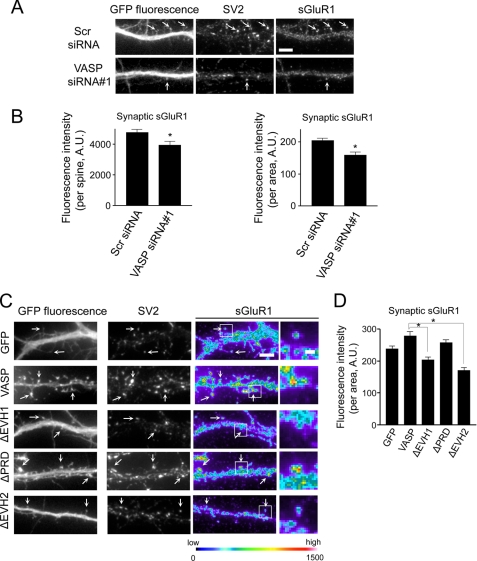
In excitatory synapses, synaptic strength is regulated by the release of glutamate neurotransmitter from presynaptic terminals and expression of glutamate receptors at the plasma membrane of postsynaptic terminals (51). Most rapid excitatory synaptic transmission takes place through AMPA-type glutamate receptors, which consists of GluR1–4 subunits (51). Because expression of AMPAR subunit GluR1 is critical for synaptic function (51), we examined the effect of VASP on surface GluR1 levels by staining with an antibody against the extracellular epitopes of this subunit in non-permeabilized cells. Neurons were subsequently permeabilized and immunostained for SV2 to show synapses. In neurons expressing GFP-VASP, the level of synaptic surface GluR1 (sGluR1) in spines was increased 1.5-fold compared with GFP controls (Fig. 6, C and D, and supplemental Fig. 3F). A significant increase in the amount of sGluR1 in spines was still observed when the fluorescent intensity of the GluR1 signal was normalized to the unit area (Fig. 6D). In contrast, expression of VASP siRNA#1 resulted in a significant decrease in the level of sGluR1 in spines (Fig. 7, A and B, and supplemental Fig. 3G). The effect of VASP on sGluR1 levels is mediated through its EVH1 and EVH2 domains as VASP failed to increase sGluR1 when these domains were deleted (Fig. 7, C and D).
FIGURE 7.
VASP regulates the level of sGluR1. A, neurons were co-transfected with GFP and scrambled siRNA (Scr siRNA) or VASP siRNA#1 at day 6 in culture, fixed, and stained for sGluR1 and SV2 at day 12. sGluR1 puncta at synapses are indicated (arrows). Bar, 5 μm. B, quantification of the fluorescent intensity of sGluR1 in individual spines from scrambled and VASP siRNA-expressing neurons is shown (left panel). The fluorescent intensity was normalized to the spine area (right panel). C, images from neurons expressing GFP, GFP-VASP, or VASP deletion mutants stained for SV2 and sGluR1 are shown. sGluR1 staining is shown in pseudo-color coding (right panels). Arrows indicate sGluR1 puncta that co-localize with SV2. Higher magnification images of the boxed regions are shown (far right panels). Βar, 1 μm. D, quantification of the fluorescent intensity of sGluR1 in individual spines from neurons expressing the indicated constructs is shown. The fluorescent intensity was normalized to spine area. Error bars represent S.E. for 75 spines (B and D) from three separate experiments (*, p < 0.0001).
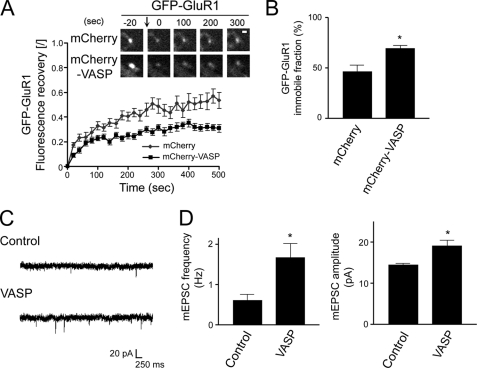
We next examined the effect of VASP on maintaining GluR1 in spines with FRAP in neurons expressing GFP-GluR1 and mCherry-VASP or mCherry as a control. As shown in Fig. 8A, GFP-GluR1 recovery was significantly slower in mCherry-VASP-expressing neurons compared with control mCherry-expressing neurons. The time constant of recovery, which is the inverse of the rate constant, for neurons expressing mCherry-VASP was 26.2 and 277.8 s for the fast and slow components, respectively, compared with 8.5 and 232.6 s for control mCherry-expressing neurons (Fig. 8A). In addition, the immobile fraction of GluR1 for mCherry-VASP and mCherry-expressing neurons was 69.3 and 46.5%, respectively, indicating a longer retention of GluR1 in spines of VASP-expressing neurons (Fig. 8B).
FIGURE 8.
VASP enhances the retention of GluR1 in spines to potentiate synaptic strength. A, neurons were transfected with GFP-GluR1 and either mCherry or mCherry-VASP at day 6 in culture and subjected to FRAP at day 10. Prebleach and subsequent recovery images of GFP-GluR1 are shown (upper panels). The bleached point is indicated (arrow). Bar, 1 μm. To calculate the fluorescent recovery, the normalized intensity was divided by the extent of bleaching (graph). B, quantification of the immobile fraction of GFP-GluR1 clusters in cells co-transfected with mCherry or mCherry-VASP is shown. Error bars represent S.E. for 19–31 spines. *, p < 0.005). C, shown are representative traces of mEPSCs recorded from a GFP-VASP expressing neuron and an untransfected neighboring cell (Control). D, quantification of the mEPSC frequency and amplitude in GFP-VASP expressing and control untransfected neurons is shown. Error bars represent S.E. for 15-paired neurons from 11 separate experiments (*, p < 0.01).
To further assess a role for VASP in regulating synaptic strength, we examined AMPAR-mediated synaptic transmission using whole-cell patch clamp recordings. The frequency and amplitude of miniature excitatory postsynaptic currents (mEPSCs) were measured in GFP-VASP-expressing neurons and neighboring untransfected cells. VASP expression led to a 2.7-fold increase in mEPSC frequency and a 1.3-fold increase in mEPSC amplitude compared with untransfected control neurons (Fig. 8, C and D). Taken together, these results indicate that VASP increases the surface level and retention of GluR1 in spines to potentiate synaptic strength.
DISCUSSION
Although emerging evidence points to the importance of the actin cytoskeleton in spine development and plasticity, the underlying molecular mechanisms that regulate actin dynamics in spines and synapses still remain largely unknown. Our results show that VASP promotes the formation of dendritic spines and synapses and induces enlargement of spine heads by regulating actin dynamics in these structures. The EVH2 domain, which binds G- and F-actin, is necessary for synaptic targeting of VASP as well as VASP regulation of spine head size and density. The EVH1 domain also contributes to VASP localization and function in spines and synapses, but the PRD region does not appear to be necessary in this regard. Additionally, VASP increases the amount of PSD-scaffolding proteins and GluR1 in spines and synapses, which results in an enhancement of synaptic transmission.
Knockdown of endogenous VASP using two siRNA constructs led to a significant reduction in the spine and synaptic density. The VASP siRNAs did not target another VASP family member, Mena, or other related actin-binding proteins, such as EVH1-containing N-WASP. This data strongly suggests that the siRNAs are specific for VASP and validates the use of this approach to alter expression of the endogenous protein. Moreover, the siRNAs specifically target the rat VASP sequence and do not affect expression of human VASP (Fig. 2B). Indeed, expression of human VASP in siRNA knockdown neurons rescued the spine and synapse defects, which further suggests the phenotypic changes in these structure is specifically due to the loss of endogenous VASP.
Our results raise the question as to how the EVH1 and EVH2 domains contribute to VASP targeting and function in spines. EVH1 domains are found in other postsynaptic proteins belonging to the Homer family where they are thought to serve as localization modules (4, 52). This domain is structurally similar to the pleckstrin homology (PH) domain and could potentially mimic the membrane-targeting function of the PH domain to recruit VASP to spines (52, 53). Alternatively, VASP may be recruited to spines by yet to be identified EVH1-binding proteins. The EVH2 domain mediates F-actin bundling through its ability to bind actin and promote tetramerization of VASP (8, 54). Deletion of the coiled-coil and F-actin binding regions, which are within the EVH2 domain, impairs spine development and spine head expansion, suggesting VASP may regulate these processes through its F-actin bundling activity (data not shown). Moreover, the EVH2 domain could promote spine and synapse development by protecting barbed ends of filaments from capping, thereby inducing actin assembly. The blockage of barbed ends by treatment with a low dose of cytochalasin D resulted in the displacement of VASP family proteins from actin-rich structures (12, 55), suggesting the interaction of VASP with barbed ends could target it to spines.
The PRD region of VASP binds to the actin polymerizing protein profilin II, which targets to spine heads upon neuronal activity where it is thought to stabilize spine morphology (56). It has been proposed that proline-rich motifs in proteins, such as VASP, serve to recruit profilin to the plasma membranes of spine heads (56), pointing to profilin as an effector of VASP in regulating the activities of spines and synapses. Surprisingly, we did not observe a requirement for profilin binding via the PRD region in VASP function in spines and synapses. It is possible that the interaction between VASP and profilin only contributes to spine function after stimulation when spines and synapses are actively remodeling and not during their development. However, it is more likely that VASP exhibits activities in spines and synapses that are mediated through other effectors and are not dependent on profilin.
In addition to its ability to modulate actin assembly, VASP is also involved in the organization of the actin network by mediating the cross-linking of actin filaments (8). Interestingly, the architecture of actin filaments in spines was recently shown to be organized into a branched, cross-linked network as demonstrated by platinum replica electron microscopy (57). This observation raises the possibility that VASP-promoted cross-linking of actin filaments may initiate and/or stabilize the enlargement of spine heads. Furthermore, F-actin cross-linking could potentiate synaptic strength by providing sites for enhancing the recruitment and interaction of signaling molecules, scaffolding proteins, and neurotransmitter receptors in spines.
VASP increases the amount of GluR1-containing AMPARs and PSD-scaffolding proteins in spines, suggesting that actin dynamics are critical for facilitating GluR1 delivery or persistence at synaptic membranes and in regulating the composition and stability of the PSD. Indeed, perturbation of actin polymerization has been shown to affect the distribution and stability of postsynaptic scaffolding proteins in synapses (42, 58). For example, several PSD proteins, including Shank, guanylate kinase-associated protein and Homer 1c are maintained in spines in an F-actin-dependent manner (58). Moreover, actin disassembly reduces the density of PSD95 and increases the non-synaptic distribution of this protein (42, 58, 59). Remodeling of the actin cytoskeleton has also been shown to play a role in regulating the localization of AMPARs to synaptic membranes (42, 60–62). Scaffolding proteins, such as PSD95, may contribute to this process by enhancing GluR1 synaptic clustering and by controlling the incorporation of AMPARs into synaptic membranes (48, 63, 64). The size of the PSD and anchoring of AMPARs to synaptic membranes determine the efficacy of synaptic strength, suggesting a pivotal function for the modulation of actin dynamics by VASP in regulating this process.
How does the actin cytoskeleton facilitate transport of AMPARs within dendritic spines? Growing evidence indicates that myosin motors, which travel along actin, are responsible for the insertion and removal of AMPARs, suggesting the integrity and organization of the actin cytoskeleton can affect receptor expression levels at synaptic membranes (35, 65–67). In support of this hypothesis, perturbation of actin dynamics has been reported to affect the internalization of GluR1-containing AMPARs (60). Thus, VASP-mediated actin reorganization may not only provide a stable platform for signaling molecules but could also actively modulate AMPAR movement into and out of synaptic membranes.
Our data are consistent with a working model in which VASP stimulates actin polymerization and bundling in spines. The growing actin filaments, induced by VASP, provide an underlying structure to support spine formation and enlargement. As spines develop, VASP-promoted actin remodeling stimulates recruitment of scaffolding proteins, such as PSD95, Homer, and Shank, to the postsynaptic density, which generates an expanded anchoring area for synaptic proteins. The dynamic actin filaments may also provide for the efficient delivery and retention of AMPARs in synapses.
Supplementary Material
Acknowledgments
We thank Frank Gertler, Freda Miller, Roger Tsien, Jüergen Wehland, and Julius Zhu for generously providing reagents. We are grateful to Lan Hu for technical assistance in preparing neuronal cultures. We thank Joe Roland, Andrew Benesh, Greg Mathews, and Rajalakshmi Nambiar for helpful suggestions and discussion.
This work was supported, in whole or in part, by National Institutes of Health Grant MH071674 and by National Center for Research Resources Grant S10RR025524 (to D. J. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- VASP
- vasodilator-stimulated phosphoprotein
- PRD
- proline-rich domain
- AMPAR
- α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor
- FRAP
- fluorescence recovery after photobleaching
- PSD95
- postsynaptic density protein 95
- R2F
- rat 2 fibroblast
- sGluR1
- surface GluR1
- mEPSC
- miniature excitatory postsynaptic current
- TRITC
- tetramethylrhodamine isothiocyanate
- A.U.
- arbitrary units.
REFERENCES
- 1.Gray E. G. (1959) J. Anat. 93, 420–433 [PMC free article] [PubMed] [Google Scholar]
- 2.Harris K. M., Kater S. B. (1994) Annu Rev. Neurosci. 17, 341–371 [DOI] [PubMed] [Google Scholar]
- 3.Fiala J. C., Spacek J., Harris K. M. (2002) Brain Res. Brain Res. Rev. 39, 29–54 [DOI] [PubMed] [Google Scholar]
- 4.Fischer M., Kaech S., Knutti D., Matus A. (1998) Neuron 20, 847–854 [DOI] [PubMed] [Google Scholar]
- 5.Matus A., Ackermann M., Pehling G., Byers H. R., Fujiwara K. (1982) Proc. Natl. Acad. Sci. U.S.A. 79, 7590–7594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zito K., Knott G., Shepherd G. M., Shenolikar S., Svoboda K. (2004) Neuron 44, 321–334 [DOI] [PubMed] [Google Scholar]
- 7.Reinhard M., Halbrügge M., Scheer U., Wiegand C., Jockusch B. M., Walter U. (1992) EMBO J. 11, 2063–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bachmann C., Fischer L., Walter U., Reinhard M. (1999) J. Biol. Chem. 274, 23549–23557 [DOI] [PubMed] [Google Scholar]
- 9.Walders-Harbeck B., Khaitlina S. Y., Hinssen H., Jockusch B. M., Illenberger S. (2002) FEBS Lett. 529, 275–280 [DOI] [PubMed] [Google Scholar]
- 10.Bear J. E., Loureiro J. J., Libova I., Fässler R., Wehland J., Gertler F. B. (2000) Cell 101, 717–728 [DOI] [PubMed] [Google Scholar]
- 11.Rottner K., Behrendt B., Small J. V., Wehland J. (1999) Nat. Cell Biol. 1, 321–322 [DOI] [PubMed] [Google Scholar]
- 12.Bear J. E., Svitkina T. M., Krause M., Schafer D. A., Loureiro J. J., Strasser G. A., Maly I. V., Chaga O. Y., Cooper J. A., Borisy G. G., Gertler F. B. (2002) Cell 109, 509–521 [DOI] [PubMed] [Google Scholar]
- 13.Lebrand C., Dent E. W., Strasser G. A., Lanier L. M., Krause M., Svitkina T. M., Borisy G. G., Gertler F. B. (2004) Neuron 42, 37–49 [DOI] [PubMed] [Google Scholar]
- 14.Dent E. W., Kwiatkowski A. V., Mebane L. M., Philippar U., Barzik M., Rubinson D. A., Gupton S., Van Veen J. E., Furman C., Zhang J., Alberts A. S., Mori S., Gertler F. B. (2007) Nat. Cell Biol. 9, 1347–1359 [DOI] [PubMed] [Google Scholar]
- 15.Kwiatkowski A. V., Rubinson D. A., Dent E. W., Edward van Veen J., Leslie J. D., Zhang J., Mebane L. M., Philippar U., Pinheiro E. M., Burds A. A., Bronson R. T., Mori S., Fässler R., Gertler F. B. (2007) Neuron 56, 441–455 [DOI] [PubMed] [Google Scholar]
- 16.Goh K. L., Cai L., Cepko C. L., Gertler F. B. (2002) Curr. Biol. 12, 565–569 [DOI] [PubMed] [Google Scholar]
- 17.Lin Y. L., Lei Y. T., Hong C. J., Hsueh Y. P. (2007) J. Cell Biol. 177, 829–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gertler F. B., Niebuhr K., Reinhard M., Wehland J., Soriano P. (1996) Cell 87, 227–239 [DOI] [PubMed] [Google Scholar]
- 19.Carl U. D., Pollmann M., Orr E., Gertlere F. B., Chakraborty T., Wehland J. (1999) Curr. Biol. 9, 715–718 [DOI] [PubMed] [Google Scholar]
- 20.Applewhite D. A., Barzik M., Kojima S., Svitkina T. M., Gertler F. B., Borisy G. G. (2007) Mol. Biol. Cell 18, 2579–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niebuhr K., Ebel F., Frank R., Reinhard M., Domann E., Carl U. D., Walter U., Gertler F. B., Wehland J., Chakraborty T. (1997) EMBO J. 16, 5433–5444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reinhard M., Giehl K., Abel K., Haffner C., Jarchau T., Hoppe V., Jockusch B. M., Walter U. (1995) EMBO J. 14, 1583–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang F., Laine R. O., Bubb M. R., Southwick F. S., Purich D. L. (1997) Biochemistry 36, 8384–8392 [DOI] [PubMed] [Google Scholar]
- 24.Ermekova K. S., Zambrano N., Linn H., Minopoli G., Gertler F., Russo T., Sudol M. (1997) J. Biol. Chem. 272, 32869–32877 [DOI] [PubMed] [Google Scholar]
- 25.Gertler F. B., Comer A. R., Juang J. L., Ahern S. M., Clark M. J., Liebl E. C., Hoffmann F. M. (1995) Genes Dev. 9, 521–533 [DOI] [PubMed] [Google Scholar]
- 26.Zimmermann J., Labudde D., Jarchau T., Walter U., Oschkinat H., Ball L. J. (2002) Biochemistry 41, 11143–11151 [DOI] [PubMed] [Google Scholar]
- 27.Loureiro J. J., Rubinson D. A., Bear J. E., Baltus G. A., Kwiatkowski A. V., Gertler F. B. (2002) Mol. Biol. Cell 13, 2533–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barzik M., Kotova T. I., Higgs H. N., Hazelwood L., Hanein D., Gertler F. B., Schafer D. A. (2005) J. Biol. Chem. 280, 28653–28662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gloster A., El-Bizri H., Bamji S. X., Rogers D., Miller F. D. (1999) J. Comp. Neurol. 405, 45–60 [DOI] [PubMed] [Google Scholar]
- 30.Zhang H., Macara I. G. (2008) Dev. Cell 14, 216–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goslin K., Asmussen H., Banker G. (1998) Rat Hippocampal Neurons in Low Density Culture, pp. 339–370, MIT Press, Cambridge, MA [Google Scholar]
- 32.Zhang H., Webb D. J., Asmussen H., Horwitz A. F. (2003) J. Cell Biol. 161, 131–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tyska M. J., Mooseker M. S. (2002) Biophys. J. 82, 1869–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wegner A. M., Nebhan C. A., Hu L., Majumdar D., Meier K. M., Weaver A. M., Webb D. J. (2008) J. Biol. Chem. 283, 15912–15920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lisé M. F., Wong T. P., Trinh A., Hines R. M., Liu L., Kang R., Hines D. J., Lu J., Goldenring J. R., Wang Y. T., El-Husseini A. (2006) J. Biol. Chem. 281, 3669–3678 [DOI] [PubMed] [Google Scholar]
- 36.Lu W., Man H., Ju W., Trimble W. S., MacDonald J. F., Wang Y. T. (2001) Neuron 29, 243–254 [DOI] [PubMed] [Google Scholar]
- 37.Zhang H., Webb D. J., Asmussen H., Niu S., Horwitz A. F. (2005) J. Neurosci. 25, 3379–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bryce N. S., Clark E. S., Leysath J. L., Currie J. D., Webb D. J., Weaver A. M. (2005) Curr. Biol. 15, 1276–1285 [DOI] [PubMed] [Google Scholar]
- 39.Chan A. Y., Raft S., Bailly M., Wyckoff J. B., Segall J. E., Condeelis J. S. (1998) J. Cell Sci. 111, 199–211 [DOI] [PubMed] [Google Scholar]
- 40.Lanier L. M., Gates M. A., Witke W., Menzies A. S., Wehman A. M., Macklis J. D., Kwiatkowski D., Soriano P., Gertler F. B. (1999) Neuron 22, 313–325 [DOI] [PubMed] [Google Scholar]
- 41.Aszódi A., Pfeifer A., Ahmad M., Glauner M., Zhou X. H., Ny L., Andersson K. E., Kehrel B., Offermanns S., Fässler R. (1999) EMBO J. 18, 37–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allison D. W., Gelfand V. I., Spector I., Craig A. M. (1998) J. Neurosci. 18, 2423–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Breitsprecher D., Kiesewetter A. K., Linkner J., Urbanke C., Resch G. P., Small J. V., Faix J. (2008) EMBO J. 27, 2943–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hotulainen P., Llano O., Smirnov S., Tanhuanpää K., Faix J., Rivera C., Lappalainen P. (2009) J. Cell Biol. 185, 323–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takumi Y., Ramírez-León V., Laake P., Rinvik E., Ottersen O. P. (1999) Nat. Neurosci. 2, 618–624 [DOI] [PubMed] [Google Scholar]
- 46.Matsuzaki M., Ellis-Davies G. C., Nemoto T., Miyashita Y., Iino M., Kasai H. (2001) Nat. Neurosci. 4, 1086–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim C. H., Lisman J. E. (1999) J. Neurosci. 19, 4314–4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.El-Husseini A. E., Schnell E., Chetkovich D. M., Nicoll R. A., Bredt D. S. (2000) Science 290, 1364–1368 [PubMed] [Google Scholar]
- 49.Hung A. Y., Futai K., Sala C., Valtschanoff J. G., Ryu J., Woodworth M. A., Kidd F. L., Sung C. C., Miyakawa T., Bear M. F., Weinberg R. J., Sheng M. (2008) J. Neurosci. 28, 1697–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sala C., Piëch V., Wilson N. R., Passafaro M., Liu G., Sheng M. (2001) Neuron 31, 115–130 [DOI] [PubMed] [Google Scholar]
- 51.Malinow R., Malenka R. C. (2002) Annu Rev. Neurosci. 25, 103–126 [DOI] [PubMed] [Google Scholar]
- 52.Prehoda K. E., Lee D. J., Lim W. A. (1999) Cell 97, 471–480 [DOI] [PubMed] [Google Scholar]
- 53.Fedorov A. A., Fedorov E., Gertler F., Almo S. C. (1999) Nat. Struct. Biol. 6, 661–665 [DOI] [PubMed] [Google Scholar]
- 54.Ahern-Djamali S. M., Comer A. R., Bachmann C., Kastenmeier A. S., Reddy S. K., Beckerle M. C., Walter U., Hoffmann F. M. (1998) Mol. Biol. Cell 9, 2157–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krause M., Leslie J. D., Stewart M., Lafuente E. M., Valderrama F., Jagannathan R., Strasser G. A., Rubinson D. A., Liu H., Way M., Yaffe M. B., Boussiotis V. A., Gertler F. B. (2004) Dev. Cell 7, 571–583 [DOI] [PubMed] [Google Scholar]
- 56.Ackermann M., Matus A. (2003) Nat. Neurosci. 6, 1194–1200 [DOI] [PubMed] [Google Scholar]
- 57.Korobova F., Svitkina T. (2010) Mol. Biol. Cell 21, 165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuriu T., Inoue A., Bito H., Sobue K., Okabe S. (2006) J. Neurosci. 26, 7693–7706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang W., Benson D. L. (2001) J. Neurosci. 21, 5169–5181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou Q., Xiao M., Nicoll R. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 1261–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shen L., Liang F., Walensky L. D., Huganir R. L. (2000) J. Neurosci. 20, 7932–7940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rocca D. L., Martin S., Jenkins E. L., Hanley J. G. (2008) Nat. Cell Biol. 10, 259–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ehrlich I., Malinow R. (2004) J. Neurosci. 24, 916–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schnell E., Sizemore M., Karimzadegan S., Chen L., Bredt D. S., Nicoll R. A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13902–13907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Z., Edwards J. G., Riley N., Provance D. W., Jr., Karcher R., Li X. D., Davison I. G., Ikebe M., Mercer J. A., Kauer J. A., Ehlers M. D. (2008) Cell 135, 535–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Osterweil E., Wells D. G., Mooseker M. S. (2005) J. Cell Biol. 168, 329–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Correia S. S., Bassani S., Brown T. C., Lisé M. F., Backos D. S., El-Husseini A., Passafaro M., Esteban J. A. (2008) Nat. Neurosci. 11, 457–466 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.