Abstract
Inwardly rectifying potassium channel Kir4.1 is critical for glial function, control of neuronal excitability, and systemic K+ homeostasis. Novel mutations in Kir4.1 have been associated with EAST/SeSAME syndrome, characterized by mental retardation, ataxia, seizures, hearing loss, and renal salt waste. Patients are homozygous for R65P, G77R, C140R or T164I; or compound heterozygous for A167V/R297C or R65P/R199Stop, a deletion of the C-terminal half of the protein. We investigated the functional significance of these mutations by radiotracer efflux and inside-out membrane patch clamping in COSm6 cells expressing homomeric Kir4.1 or heteromeric Kir4.1/Kir5.1 channels. All of the mutations compromised channel function, but the underlying mechanisms were different. R65P, T164I, and R297C caused an alkaline shift in pH sensitivity, indicating that these positions are crucial for pH sensing and pore gating. In R297C, this was due to disruption of intersubunit salt bridge Glu288–Arg297. C140R breaks the Cys108–Cys140 disulfide bond essential for protein folding and function. A167V did not affect channel properties but may contribute to decreased surface expression in A167V/R297C. In G77R, introduction of a positive charge within the bilayer may affect channel structure or gating. R199Stop led to a dramatic decrease in surface expression, but channel activity was restored by co-expression with intact subunits, suggesting remarkable tolerance for truncation of the cytoplasmic domain. These results provide an explanation for the molecular defects that underlie the EAST/SeSAME syndrome.
Keywords: Brain, Genetic Diseases, Ion Channels, Kidney, Potassium Channels, EAST/SeSAME Syndrome, Inward Rectifying Potassium Channels, KCNJ10, Kir4.1, Kir4.1/Kir5.1
Introduction
Kir4.1 (encoded by gene KCNJ10), a member of the inwardly rectifying potassium (Kir) channel family, is essential for the control of glial function and neuronal excitability, systemic K+ homeostasis, and renal salt exchange (1, 2). Kir4.1 subunits form homotetrameric channels (see Fig. 1A) or co-assemble with Kir5.1 (KCNJ16) in heterotetramers with distinct physiological properties (3–6). In renal tubular epithelia, these channels are responsible for maintenance of a negative membrane potential that drives ion pumps and exchangers (7, 8). In astrocytic glia, Kir4.1-containing channels account for the spatial buffering of K+ released by neurons during action potential propagation (1, 9–12). Kir4.1 is predominant in Müller glial cells, where it contributes to normal retinal physiology (13–18), and in satellite glial cells of sensory ganglia (19). Kir4.1 channels in oligodendrocytes are critical for myelination (20, 21), and in various cell types of the inner ear they play a role in K+ regulation and generation of the endocochlear potential, essential for normal development of the cochlea and audition (22–25).
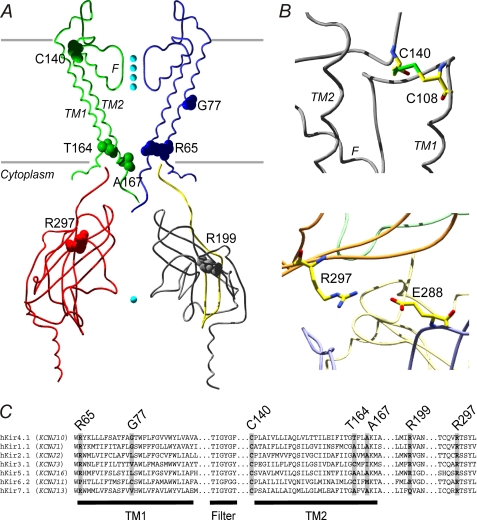
FIGURE 1.
Molecular basis of the EAST/SeSAME syndrome. Patients are homozygous for Kir4.1 mutation R65P, G77R, C140R or T164I; or compound heterozygous for R65P and R199Stop (a deletion of the C-terminal half of the protein) or A167V and R297C. A, predicted location of modified residues in the Kir4.1 channel, modeled on the crystal structure of chicken Kir2.2 (42) (Swiss-PdbViewer 4.0.1). Kir4.1 subunits consist of two membrane-spanning domains (TM1 and TM2) that flank a signature K+-selective pore (F), and functional channels are formed by the assembly of four subunits. Viewed in the plane of the membrane, only alternate transmembrane or cytoplasmic regions (uniquely colored) are displayed for clarity. Cyan spheres represent K+ ions. The area missing in R199Stop is shown in gray. B, close-up views showing predicted intrasubunit disulfide bond between Cys108 and Cys140 (top) and predicted salt bridge between Glu288 and Arg297 from adjacent subunits (bottom). The side chains are shown as sticks and colored according to atom type: oxygen, red; carbon, yellow; nitrogen, blue; sulfur, green. C, sequence alignment of human Kir4.1 (SwissProt accession number P78508.1) with hKir1.1 (ROMK, P48048.1), hKir2.1 (P63252.1), hKir3.1 (P48549.1), hKir5.1 (Q9NPI9.1), hKir6.2 (Q14654.2), and hKir7.1 (O60928.1) reveals different degrees of conservation of the positions mutated in EAST/SeSAME syndrome throughout the Kir channel family.
Polymorphisms in the KCNJ10 gene have been associated with decreased astrocytic Kir channel currents (26), seizure susceptibility, and epilepsy (27–30), but biophysical characterization of these variants failed to show effects on Kir4.1 channel properties (31). More recently, two independent studies described a novel syndrome, termed EAST or SeSAME, that presents with a unique set of symptoms including epilepsy, ataxia, mental retardation, hearing loss, and electrolyte imbalance related to renal salt loss (32, 33). Genetic screening revealed that EAST/SeSAME patients are homozygous or compound heterozygous for novel mutations in KCNJ10 that result in amino acid substitutions in Kir4.1 (R65P, G77R, C140R, T164I, A167V, or R297C) or a deletion of the C-terminal half of the protein (R199Stop; see Fig. 1, A and C). Loss of Kir4.1 function associated with these mutations has been reported (34, 35). Here, we used radiotracer efflux and electrophysiological measurements to investigate the functional significance of EAST/SeSAME mutations expressed heterologously in COSm6 cells. Our data show that each of these mutations compromises the function of both homomeric Kir4.1 and heteromeric Kir4.1/Kir5.1 channels and can account for the clinical phenotype. Kir4.1 function is reduced by distinct mechanisms, and collectively our findings provide novel insights into the critical role played by these residues in Kir4.1 channel function.
EXPERIMENTAL PROCEDURES
Expression of Wild-type and Mutant Kir4.1 Channels in COSm6 Cells
pEGFP-rKir4.1, in which rat Kir4.1 cDNA is fused to a N-terminal EGFP2 tag (36), was used as template into which EAST/SeSAME mutations were introduced, by means of the QuikChange II site-directed mutagenesis kit (Stratagene). The integrity of all of the constructs was verified by sequencing. COSm6 cells were plated in 6- or 12-well dishes and cultured in DMEM (Invitrogen) supplemented with 10% FBS, 105 units/liter penicillin, and 100 mg/liter streptomycin. At 80% confluency, the cells were transfected with the different plasmids using FuGENE 6 transfection reagent (Roche Applied Science). Typically, 1 μg of DNA was mixed with 3 μl of FuGENE 6, and the cells were incubated in the presence of the transfection mixture for 18–24 h. Heterozygous states were attained by transfecting cells with equal amounts of the relevant DNAs, while keeping the total amount of DNA constant at 0.5 μg/well in 12-well plates or 1 μg/well in 6-well plates. For experiments with heteromeric Kir4.1/Kir5.1 channels, pEGFP-rKir5.1 was added to the transfection reaction at a 1:1 Kir4.1 to Kir5.1 ratio, e.g. 0.5 μg of pEGFP-rKir5.1/well in 12-well plates. The biophysical properties of EGFP-tagged wild-type channels (pH dependence and rectification; see Figs. 4 and 5) were identical to those reported for nontagged channels in native cells (37) and heterologous expression systems (5, 6). However, we cannot discount the possibility that the fluorescent tag might affect localization or trafficking. Cells transfected with empty pEGFP vector (mock transfected) served as controls. Multiple transfections were done in parallel.
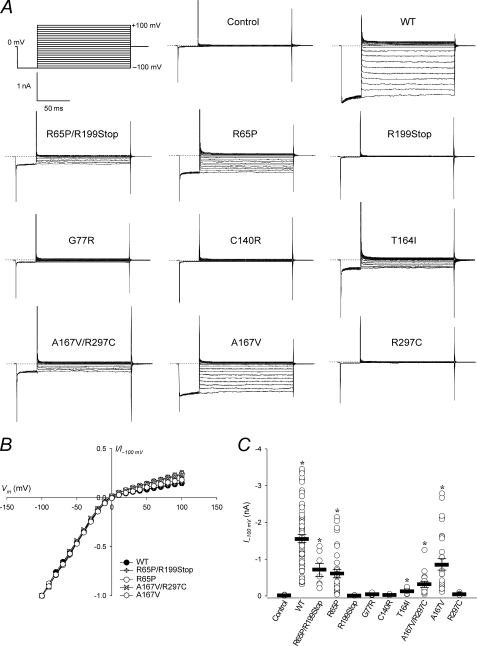
FIGURE 4.
Kir4.1 currents are decreased in intact cells expressing mutant channels. A, representative on-cell currents measured in mock transfected control cells and in cells expressing WT or mutant Kir4.1. The pulse protocol is shown in the top left panel. Pipette and external solution were KINT buffer at pH 7.5. The dotted lines indicate zero current. B, steady-state current-voltage relationships from experiments as in A, normalized to the value at −100 mV (I−100 mV). C, I−100 mV as a measure of current density. Circles represent data from individual patches (n = 10–70); bars indicate the means ± S.E. of all experiments. *, p < 0.05 as compared with the control (one-way analysis of variance and Kruskal-Wallis test).
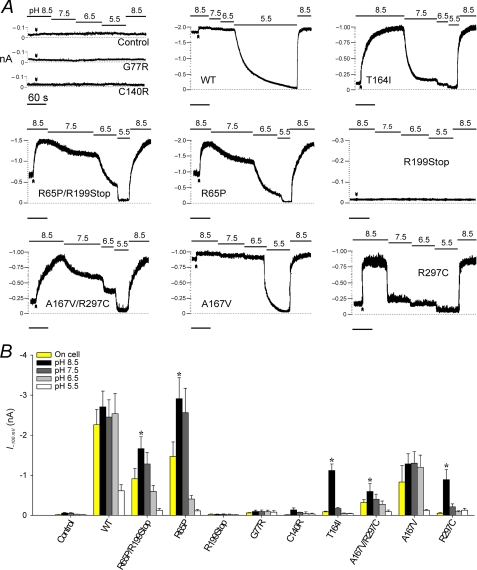
FIGURE 5.
R65P, R297C, and T164I form channels with increased pH sensitivity. A, representative current traces. Membrane potential was held at −100 mV, and inward currents were recorded continuously on cell and in inside-out excised patches exposed to KINT at pH 8.5–5.5; arrowheads mark the point of excision. The base-line current was determined at pH 5.5. The time constant of channel reactivation after acidic inhibition τpH was obtained by fitting the time course of recovery at pH 8.5 with a single exponential (Table 1). B, currents measured on-cell (yellow bars) and after exposure to pH 8.5–5.5 from experiments as in A. An asterisk denotes a significant difference (p < 0.05) between the currents measured on-cell and after excision into pH 8.5, as determined by paired Student's t test. The results are the means ± S.E. from 6-10 patches.
86Rb+ Efflux Assays
Cells in 12-well plates were incubated for 18 h in culture medium with 1 μCi/ml 86RbCl (PerkinElmer Life Sciences) and subsequently washed with Ringer's solution containing 118 mm NaCl, 2.5 mm CaCl2, 1.2 mm KH2PO4, 4.7 mm KCl, 25 mm NaHCO3, 1.2 mm MgSO4, and 10 mm Hepes (pH 7.4). The experiment was initiated by adding fresh Ringer's to the cells, and at selected time points the medium was collected for analysis and replaced with fresh solution. Upon completion of the assay, the cells were lysed with 2% SDS and collected. Radioactivity in the samples was then measured by liquid scintillation. The raw data are shown as 86Rb+ efflux relative to total counts, including all time points and the cell lysate for each construct. The rate constant of Kir4.1- or Kir4.1/Kir5.1-specific 86Rb+ efflux k2, proportional to Kir4.1- or Kir4.1/Kir5.1-dependent K+ conductance, was estimated by fitting the data to the following single exponential equation,
where the apparent rate constant for nonspecific efflux k1 was obtained from control cells. SigmaPlot 10.0 (Systat Software) was used for curve fitting.
Electrophysiology
One day after transfection, the cells in six-well plates were trypsinized and plated overnight onto sterile glass coverslips. Cells expressing the different EGFP-Kir4.1 fusion proteins fluoresced under ultraviolet light and were selected for patch-clamp analysis. The experiments were performed at room temperature in an oil gate chamber that allowed the solution bathing the exposed surface of the isolated patch to be changed rapidly (38). Micropipettes were prepared from nonheparinized hematocrit glass (Kimble-Chase) on a horizontal puller (Sutter Instrument Co.) and filled with a solution containing 140 mm KCl, 1 mm K-EGTA, and 10 mm Hepes (pH 7.5) to a typical electrode resistance of 1 mΩ. The membrane patches were voltage-clamped using an Axopatch one-dimensional amplifier (Molecular Devices), and the currents were recorded at −100 mV (pipette voltage, +100 mV) in on-cell and inside-out excised patch configuration. In other experiments, a pulse protocol was applied in which membrane potential (Vm) was held at 0 mV and stepped first to −100 mV for 30 ms and then to a test value for 140 ms before returning to the holding potential for an additional 30 ms. The test potential varied from −100 to +100 mV in 10-mV increments. Steady-state currents were measured at the end of 140 ms. The data were filtered at 2 kHz, and the signals were digitized at 5 kHz with a Digidata 1322A (Molecular Devices). pClamp and Axoscope software (Molecular Devices) was used for pulse protocol application and data acquisition. Bath solutions (KINT) contained 140 mm KCl, 1 mm K-EGTA, 1 mm K-EDTA to prevent channel rundown (39) and 10 mm Hepes (pH 8.5 or 7.5) or 10 mm Mes (pH 6.5, 6, or 5.5).
Cell Surface Biotinylation and Protein Fractioning
The cells plated in 100-mm dishes were transfected at 60% confluency with 4 μg of wild-type or mutant Kir4.1, as described above. After 48 h, cell surface protein biotinylation and extraction of cytosolic and surface membrane fractions were performed, using the Pierce cell surface protein isolation kit (Thermo Scientific). Briefly, the cells were washed in ice-cold PBS and incubated in Sulfo-NHS-SS-Biotin solution (0.25 mg/ml) at 4 °C for 30 min. At this time, the reaction was quenched, and the cells were collected and incubated for 30 min in lysis buffer; during the incubation, the reaction was sonicated to help disrupt the cells. The lysate was centrifuged, and the supernatant was added to columns containing immobilized neutravidin gel for biotin binding and incubated for 1 h at room temperature. The columns were centrifuged (1000 × g, 1 min), and the flow-through was collected as the cytosolic protein fraction and mixed with SDS-PAGE sample buffer (62.5 mm Tris/HCl, pH 6.8, 1% SDS, 10% glycerol) containing 5% mercaptoethanol. Subsequent to washing with a buffer containing 10% protease inhibitor mixture (Sigma), 400 μl of SDS-PAGE buffer with 50 mm DTT was added to the columns. After 1 h of incubation at room temperature, the columns were centrifuged, and the flow-through was recovered as the cell surface membrane protein fraction. Trace amounts of bromphenol blue were added to the membrane and cytoplasmic fractions before analysis by Western blot.
Western Blotting
Cytosolic and surface membrane protein fractions were separated by SDS-PAGE on 8% gels and transferred to PVDF membranes. These membranes were stained with India ink and analyzed by densitometry to determine the total protein (40). The various EGFP-Kir4.1 fusion proteins were detected with a mouse monoclonal anti-GFP antibody and an HRP-conjugated goat anti-mouse secondary antibody (Santa Cruz Biotechnology), using the SuperSignal® West Dura extended duration chemiluminescent substrate (Pierce). The intensity of the signal was normalized to protein density. Densitometry and fluorescence detection were performed using a gel documentation system (Versa Doc model 1000; Bio-Rad).
Data Analysis
The data were analyzed using Clampfit 10.1 (Molecular Devices) and Excel (Microsoft). SigmaPlot 10.0 and Prism 5.03 (GraphPad Software) were used for statistics and figure preparation. The results are presented as the means ± S.E. (standard error of the mean). Where indicated, one-way analysis of variance and Kruskal-Wallis test or paired Student's t test were applied to evaluate statistical differences between groups.
RESULTS
EAST/SeSAME Mutations Reduce Kir4.1-mediated K+ Conductance
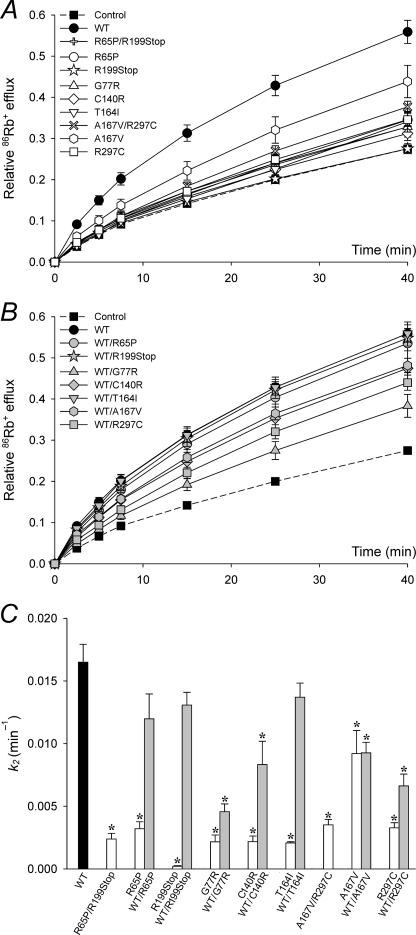
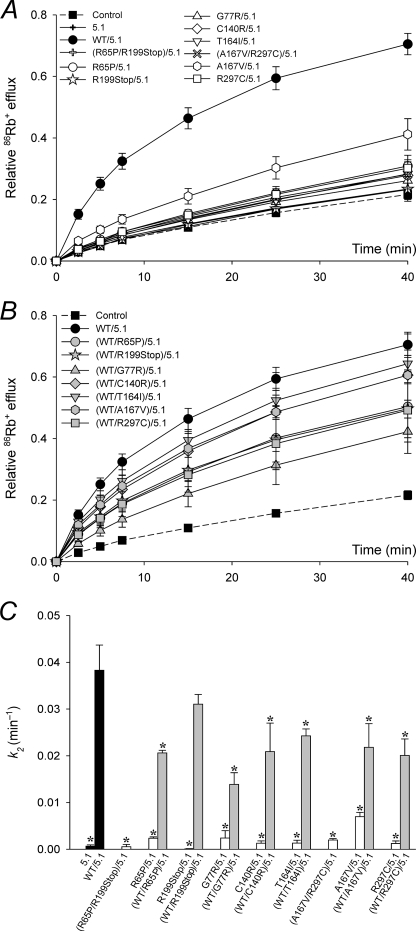
It has been suggested that EAST/SeSAME mutations (Fig. 1) compromise the activity of the Kir4.1 channel and contribute to the phenotypic manifestation of the syndrome (32, 33). To test this hypothesis, we first investigated the effect of these mutations on macroscopic K+ conductance, by means of radiolabeled Rb+ efflux assays in cells expressing homomeric Kir4.1 channels (Fig. 2). The efflux of 86Rb+ over time in mock transfected control cells and in cells transfected with WT or mutant Kir4.1 channels, independently and combined, is presented in Fig. 2 (A and B). The data were fitted to Equation 1 to obtain nonspecific and Kir4.1-mediated rate constants k1 and k2, respectively (Fig. 2C). k2 decreased significantly in cells expressing mutant channels (Fig. 2, A and C, white symbols and bars). We simulated the genetic composition of asymptomatic carriers of EAST/SeSAME mutations by co-expressing WT and mutant alleles (Fig. 2, B and C, gray symbols and bars). k2 increased to almost WT levels in WT/R65P, WT/R199Stop, and WT/T164I and to a lesser extent in WT/G77R, WT/C140R, and WT/R297C. Intriguingly, A167V reduced k2 by ∼40% in both homozygous and heterozygous conditions (Fig. 2C). This mutation thus appears benign on its own when in homomeric Kir4.1 channels but is found in heterozygous combination with R297C in the patient, where it could contribute to the phenotype.
FIGURE 2.
K+ conductance is reduced in EAST/SeSAME mutants. A and B, time course of relative 86Rb+ efflux in mock transfected control cells and in cells expressing WT or mutant Kir4.1 channels, in homozygous or compound heterozygous configuration (white symbols, A) or co-transfected with WT (gray symbols, B). Control and WT data are shown in both A and B to facilitate comparison. C, flux data were fitted to Equation 1 to obtain the rate constants for Kir4.1-dependent 86Rb+ efflux k2. Nonspecific k1 was estimated in control cells at 0.009 ± 0.0003 min−1 (n = 12), assuming k2 = 0. The data represent the means ± S.E. of 4–12 experiments, each in triplicate. *, p < 0.05 as compared with the WT (one-way analysis of variance and Kruskal-Wallis test).
We then determined the effect of EAST/SeSAME mutations in heteromeric Kir4.1/Kir5.1 channels (Fig. 3). In cells co-transfected with WT Kir4.1 and Kir5.1, K+ conductance was notably more sensitive to acidification than in cells expressing homomeric Kir4.1 channels (supplemental Fig. S1), consistent with the recognized properties of heteromeric Kir4.1/Kir5.1 channels (5, 6). As shown in Fig. 3, the function of Kir4.1/Kir5.1 channels was also compromised by EAST/SeSAME mutations (Fig. 3, A and C, white symbols and bars) to a similar extent to that seen in Kir4.1 homomers (Fig. 2) and was partially restored by co-transfection with WT Kir4.1 subunits (Fig. 3, B and C, gray symbols and bars). Collectively, the data show that all EAST/SeSAME Kir4.1 mutations decrease K+ conductance in both Kir4.1 homomers and Kir4.1/Kir5.1 heteromers and suggest that G77R, C140R, and R297C act as dominant negative subunits in the tetrameric complex when co-expressed with WT, resulting in decreased heterologous channel function. However, the absence of clinically relevant symptoms in the carrier parents and siblings of EAST/SeSAME patients indicates that the residual activity of the heterozygous channels is sufficient to avoid a clinical phenotype.
FIGURE 3.
Impaired Kir4.1 function is not rescued by Kir5.1 in heteromeric channels. A and B, time course of relative 86Rb+ efflux in control cells, in cells expressing Kir5.1 (5.1), and in cells co-expressing Kir5.1 and WT or mutant Kir4.1 channels, in homozygous or compound heterozygous configuration (white symbols, A) or co-transfected with WT Kir4.1 (gray symbols, B). Control and WT data are shown in both A and B to facilitate comparison. C, flux data were fitted to Equation 1 to obtain the rate constants for Kir4.1/Kir5.1-dependent 86Rb+ efflux k2. Nonspecific k1 was estimated in control cells at 0.007 ± 0.001 min−1 (n = 8), assuming k2 = 0. k2 was negligible in cells transfected with only Kir5.1, consistent with the inability of Kir5.1 subunits to assemble in functional homotetrameric channels (6). The data represent the means ± S.E. of 3–12 experiments, each in triplicate. *, p < 0.05 as compared with the WT (one-way analysis of variance and Kruskal-Wallis test).
Next, we used patch-clamp analysis to evaluate the impact of EAST/SeSAME mutations on the properties of Kir4.1 currents (Figs. 4 and 5). Characteristic inward rectification of Kir4.1 channels was observed in cells expressing WT and mutant channels (Fig. 4, A and B). On average, on-cell WT I−100 mV was 1.6 ± 0.1 nA; individual measurements varied from 0.35 to 3.5 nA but were at least 20-fold higher than in mock transfected control cells (0.020 ± 0.003 nA; Fig. 4C). R65P/R199Stop, R65P, T164I, A167V/R297C, and A167V showed reduced current density, and currents were essentially undetectable in cells expressing R199Stop, G77R, C140R, and R297C (Fig. 4, A and C).
R65P, T164I, and R297C Confer Increased pH Sensitivity
WT Kir4.1 channels are sensitive to acidic pH, i.e. conduction is maximal at pH 7.5 but inhibited at pH 6 (36). To assess pH sensing in EAST/SeSAME mutants, we tested the effect of intracellular pH in inside-out membrane patches from each group (Fig. 5). Typically, the cells were bathed in KINT at pH 8.5, and Vm was held at −100 mV. Data acquisition was initiated on cell, and currents were recorded continuously as the patch was excised and exposed to buffers of varying pH, each time until a steady-state was reached. In WT (Fig. 5A, top middle panel), currents remained unchanged at pH 7.5 and 6.5 but dropped to zero at pH 5.5. When WT patches were exposed to pH 8.5 following acidic inhibition, currents recovered quickly (τpH = ∼3 s; Table 1) and completely. In A167V, currents were smaller than in WT, but pH sensitivity was normal (Fig. 5 and Table 1). On the other hand, an alkaline shift in pH sensitivity was apparent for channels formed by R65P, T164I, and R297C subunits. In these, on-cell currents were greatly reduced (R65P, R65P/R199Stop, and A167V/R297C) or even insignificant (T164I and R297C) but rapidly increased to near WT levels upon excision into pH 8.5. Each of these channels was markedly inhibited at physiological pH, and recovery from acidic pH was much slower than in WT (Fig. 5 and Table 1). Consistent with a failure to generate functional channels, currents in G77R, C140R, or R199Stop were indistinguishable from those in mock transfected control cells at all pHs (Fig. 5).
TABLE 1.
pH sensitivity of WT and mutant Kir4.1 channels
Currents at pH 7.5 and 6.5 from Fig. 5B are shown relative to the current at pH 8.5. The time course of current recovery at pH 8.5 following exposure to pH 5.5 as in Fig. 5A was best fitted to a single exponential relationship to obtain the time constant of channel reactivation τpH. The results are the means ± S.E. from n patches.
| I7.5/I8.5 | I6.5/I8.5 | τpH | n | |
|---|---|---|---|---|
| (s) | ||||
| WT | 0.98 ± 0.01 | 0.92 ± 0.05 | 3.0 ± 0.4 | 10 |
| A167V | 0.97 ± 0.01 | 0.93 ± 0.02 | 5.6 ± 0.7 | 7 |
| R65P/R199Stop | 0.82 ± 0.01 | 0.41 ± 0.06 | 17.6 ± 5.7 | 8 |
| R65P | 0.69 ± 0.04 | 0.17 ± 0.05 | 17.3 ± 1.5 | 6 |
| A167V/R297C | 0.69 ± 0.06 | 0.46 ± 0.06 | 9.7 ± 0.6 | 10 |
| R297C | 0.35 ± 0.07 | 0.19 ± 0.09 | 1.1 ± 0.2 | 8 |
| T164I | 0.15 ± 0.01 | 0.04 ± 0.01 | 17.3 ± 4.6 | 6 |
Decreased Surface Expression Contributes to Loss of Function in R199Stop, A167V, and R297C
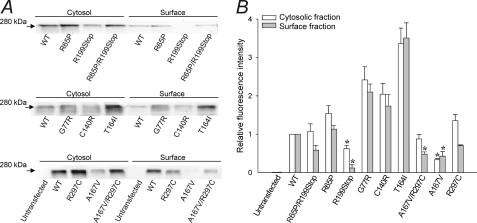
Finally, we explored the possibility that the observed effect of EAST/SeSAME mutants in Kir4.1 channel function may result from or be complicated by impaired surface expression. To this effect, we performed quantitative Western blot analysis of purified cytosolic and surface membrane protein fractions from untransfected cells and from cells expressing WT or mutant Kir4.1 (Fig. 6). Representative experiments are presented in Fig. 6A. In transfected cells, both fractions yielded a single band at ∼280 kDa, which corresponds to the predicted mass of EGFP-Kir4.1 tetramers; no signal was detected in untransfected cells. Fig. 6B shows the fluorescence intensity (proportional to Kir4.1 protein density) in internal (white bars) and surface (gray bars) membrane fractions of cells expressing mutant channels, relative to WT. Intracellular and cell surface fluorescence were normal in R65P, 2-fold increased in G77R and C140R, and over 3-fold increased in T164I. On the other hand, the signals decreased 40% in the cytosol, over 90% in the membrane of R199Stop, and 70% in both fractions of A167V. Relative surface expression was 60% in R65P/R199Stop, 50% in A167V/R297C, and 70% in R297C, whereas cytosolic levels were normal. These results indicate that, at least in our expression system, the reduced K+ conductance observed in channels containing R199Stop, A167V, and R297C subunits may partly result from defects in protein synthesis, stability, or expression at the surface. The relative absence of R199Stop in the surface membrane fraction provides validation for the biotinylation technique and indicates that there is relatively little, if any, contamination by the cytoplasmic fraction.
FIGURE 6.
Differential expression of EAST/SeSAME mutants in cytosolic and surface membrane fractions. A, representative Western blots in untransfected cells and in cells expressing WT or mutant Kir4.1 channels, following cell surface biotinylation. The EGFP-Kir4.1 fusion proteins were detected at 280 kDa with antibody against GFP. B, fluorescence intensity in cytosolic (white bars) and surface membrane (gray bars) fractions. The data were normalized to WT and are represented as the means ± S.E. of 3–8 experiments. An asterisk denotes a significant decrease with respect to WT (p < 0.05; one-way analysis of variance and Kruskal-Wallis test).
DISCUSSION
Loss of Kir4.1 Function Is Responsible for the EAST/SeSAME Phenotype
Kir4.1 channels are essential for brain, ear, and kidney physiology (1, 2, 22). Missense and nonsense mutations in KCNJ10, the gene encoding Kir4.1, have been identified in EAST/SeSAME syndrome patients (32, 33). Individuals affected by this disorder present with an array of neurological and renal symptoms, including seizures, ataxia, mental retardation, sensorineural deafness, and electrolyte imbalance. Identified patients are homozygous for R65P, G77R, C140R, or T164I or compound heterozygous for R65P/R199Stop or A167V/R297C (Fig. 1). Here, we show that all of these mutations disrupt channel function in Kir4.1 homomers and Kir4.1/Kir5.1 heteromers, in agreement with two other recent studies (34, 35). In addition, we investigate the molecular mechanisms responsible for the loss of Kir4.1 function in each case.
In COSm6 cells transfected with mutant channels, Kir4.1-mediated K+ conductance was significantly reduced, but in most cases not completely abolished (Figs. 2–5). Cells expressing the allele combinations found in patients retained up to 21% of the function as indicated by the rate of homomeric and heteromeric Kir4.1-dependent 86Rb+ efflux k2 (Figs. 2C and 3C), but seemingly this residual activity is insufficient to prevent the disease. On the other hand, 30–50% activity remained in cells co-expressing WT and C140R, R297C, or G777 (Fig. 2, B and C, and 3, B and C), and this corresponds to the expected function in heterozygous relatives of EAST/SeSAME patients who are reportedly asymptomatic. Taken together, these findings suggest a threshold of residual activity below which individuals will manifest the disorder and above which they will remain asymptomatic; in our expression system, this corresponds with a cut-off between 20 and 30% of residual activity. In turn, this may help predict the outcome of as yet unreported genotypes. For example, homozygotes for R199Stop or R297C should be symptomatic, because k2 was only up to ∼1 and ∼20% of WT in cells expressing these mutants in homozygous configuration (Figs. 2C and 3C). On the other hand, k2 was ∼60% of WT in cells expressing A167V (Fig. 2C) but only ∼18% in cells expressing A167V/Kir5.1 (Fig. 3C). This raises the possibility that homozygotes for A167V present with a milder or intermediate form of the syndrome as a result of compromised heteromeric Kir4.1/5.1 channel activity but not homomeric Kir4.1 activity.
Arg65, Thr164, and Arg297 Are Implicated in pH Sensing
Although they all led to the same clinical phenotype, each mutation affected channel properties in distinct ways, and this may help in understanding the role played by the relevant residues in Kir4.1 function. Gly77 is located halfway through TM1 (Fig. 1A), and in other Kir channels this position is conserved or occupied by uncharged residues such as Cys, Val, or Leu (Fig. 1C). In G77R, the positively charged side chain of Arg in the middle of the bilayer may significantly affect pore structure or gating. On the other hand, K+ conductance was decreased in cells expressing channels formed by R65P, T164I, A167V, and R297C, but rectification properties were normal (Figs. 2–4). This is in agreement with none of these residues being equivalent or in close proximity to those responsible for rectification in other Kir channels (41, 42) and suggests no significant effects on ion channel permeability properties. The moderate loss of function observed in A167V was largely a consequence of decreased surface expression (Fig. 6).
The disease phenotype in A167V/R297C and R65P/R199Stop likely arises from the increased pH sensitivity conferred by R297C and R65P, respectively (Fig. 5 and Table 1). Arg297 is conserved in Kir channels and is predicted to form a salt bridge with Glu288 in the adjacent subunit (Fig. 1B, bottom panel) that controls pH gating (43, 44). Consistent with this, we found that k2 was 7% of WT in cells expressing E288C and 17% in cells expressing the double mutant E288C/R297C in homomeric Kir4.1 channels (supplemental Fig. S2, A and C, white bars), and only 4 and 2% of WT in heteromeric Kir4.1/Kir5.1 channels (supplemental Fig. S2, B and C, gray bars). Neutralization of the equivalent residues in Kir1.1 and Kir2.1 causes Bartter's and Andersen's syndrome, respectively, because of increased pH sensitivity (45–47). Arg65 is relatively conserved throughout the Kir family, but intriguingly it corresponds to a proline at the equivalent residues in ATP-sensitive Kir6.1 and Kir6.2 subunits (Fig. 1C), and KATP channels are less sensitive to intracellular pH (48). Mutations that neutralize Arg82, the equivalent residue in Kir2.1 (KCNJ2), result in a complete loss of function (45) through unknown mechanisms. Here we provide evidence that this arginine at the base of TM1 may be part of the pH sensing and gating machinery in Kir channels. Accordingly, an alkaline shift in pH sensitivity has been observed in HEK293 cells and Xenopus oocytes expressing R65P (35).
T164I channels were inhibited at physiological intracellular pH (Figs. 2–5), and this was a consequence of a sharp alkaline shift in pH sensitivity (Fig. 5 and Table 1) that can entirely account for the clinical phenotype. Thr164 is located at the bundle crossing in TM2 (Fig. 1A) and has been proposed to form a hydrogen bond with Lys67 in TM1, equivalent to the Lys80–Ala177 pair in Kir1.1 that stabilizes the closed state at low pH (43, 49). Our data highlight the importance of the Lys67–Thr164 intrasubunit interaction in Kir4.1 channel pH sensing and gating. Accordingly, neutralizing mutations in Lys80 reduce the pH sensitivity in Kir1.1 channels, whereas A177T results in increased pH sensitivity and is associated with Bartter's syndrome (43, 50).
The Disulfide Bond between Cys108 and Cys140 Is Indispensable for Kir4.1 Channel Function
Kir4.1-mediated K+ conductance was severely compromised in C140R (Figs. 2–5), and this was not due to decreased surface expression (Fig. 6). Cys140 forms an intrasubunit disulfide bridge with Cys108 (Fig. 1B, top panel) that is absolutely conserved throughout the eukaryotic Kir channel family. Biochemical and structural data in Kir2 channels indicate that the disulfide bond between the equivalent residues (Cys123–Cys155) is essential for proper folding and function (51–53). In additional experiments, we observed that k2 was only 2% of WT in cells expressing C108E, 5% of WT in cells expressing the double mutant C108E/C140R in Kir4.1 homomers (supplemental Fig. S3, A and C, white bars), and only 3 and 2% of WT in Kir4.1/Kir5.1 heteromers (supplemental Fig. S3, B and C, gray bars). Our results suggest that this disulfide bond is critical to Kir4.1 function and cannot be effectively replaced by less stable interactions such as a salt bridge.
The C-terminal Domain of Kir4.1 Is Essential for Surface Expression, but Is It for Channel Activity?
R199Stop introduces a premature termination codon that deletes the C-terminal half of the protein, i.e. most of its cytoplasmic domain (Fig. 1). Complete loss of function in R199Stop corresponded with a dramatic drop in surface expression (Fig. 6), which may arise from defective protein synthesis, targeting, or insertion. In accordance with our findings, PDZ-binding and di-hydrophobic sequence motifs have been identified in the C terminus of Kir4.1 that are essential for cell surface retention and basolateral localization in renal tubular cells (54, 55). R199Stop is not dominant; k2 was 80% of WT in cells expressing WT/R199Stop and 80% of R65P in cells expressing R65P/R199Stop (Fig. 2C). This may be a consequence of higher expression levels of intact WT or R65P subunits under our experimental conditions. Intriguingly, however, it is also consistent with tetrameric channels containing up to two R199Stop subunits being active (based on binomial distribution). Further study may help elucidate whether truncated R199Stop subunits not only may be stabilized in the membrane by their intact counterparts but may contribute to functional channels.
In summary, Kir4.1 mutations R65P, G77R, C140R, T164I, R65P/R199Stop, and A167V/R297C led to a loss of function in homomeric Kir4.1 and heteromeric Kir4.1/Kir5.1 channels and contributed to the EAST/SeSAME phenotype. The mechanisms involved in each case are different, and our findings provide feed-forward genetic insights into the structural basis of pH sensing, gating, surface expression, and channel stability.
Supplementary Material
Acknowledgments
We thank Paola López-Pieraldi, Natalia Skatchkova, and Valentina Ghisays-Haydar for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants HL54171 (to C. G. N.), S11-NS48201 (to M. J. E.), U54-NS039408 (to S. N. S.), and G12-RR03035.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- EGFP
- enhanced GFP
- Mes
- 4-morpholineethanesulfonic acid.
REFERENCES
- 1.Butt A. M., Kalsi A. (2006) J. Cell. Mol. Med. 10, 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wagner C. A. (2010) J. Nephrol. 23, 5–8 [PubMed] [Google Scholar]
- 3.Hibino H., Fujita A., Iwai K., Yamada M., Kurachi Y. (2004) J. Biol. Chem. 279, 44065–44073 [DOI] [PubMed] [Google Scholar]
- 4.Ishii M., Fujita A., Iwai K., Kusaka S., Higashi K., Inanobe A., Hibino H., Kurachi Y. (2003) Am. J. Physiol. Cell. Physiol. 285, C260–C267 [DOI] [PubMed] [Google Scholar]
- 5.Tanemoto M., Kittaka N., Inanobe A., Kurachi Y. (2000) J. Physiol. 525, 587–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tucker S. J., Imbrici P., Salvatore L., D'Adamo M. C., Pessia M. (2000) J. Biol. Chem. 275, 16404–16407 [DOI] [PubMed] [Google Scholar]
- 7.Lachheb S., Cluzeaud F., Bens M., Genete M., Hibino H., Lourdel S., Kurachi Y., Vandewalle A., Teulon J., Paulais M. (2008) Am. J. Physiol. Renal Physiol. 294, F1398–F1407 [DOI] [PubMed] [Google Scholar]
- 8.Lourdel S., Paulais M., Cluzeaud F., Bens M., Tanemoto M., Kurachi Y., Vandewalle A., Teulon J. (2002) J. Physiol. 538, 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Djukic B., Casper K. B., Philpot B. D., Chin L. S., McCarthy K. D. (2007) J. Neurosci. 27, 11354–11365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kucheryavykh Y. V., Kucheryavykh L. Y., Nichols C. G., Maldonado H. M., Baksi K., Reichenbach A., Skatchkov S. N., Eaton M. J. (2007) Glia 55, 274–281 [DOI] [PubMed] [Google Scholar]
- 11.Neusch C., Papadopoulos N., Müller M., Maletzki I., Winter S. M., Hirrlinger J., Handschuh M., Bähr M., Richter D. W., Kirchhoff F., Hülsmann S. (2006) J. Neurophysiol. 95, 1843–1852 [DOI] [PubMed] [Google Scholar]
- 12.Olsen M. L., Campbell S. L., Sontheimer H. (2007) J. Neurophysiol. 98, 786–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bringmann A., Pannicke T., Grosche J., Francke M., Wiedemann P., Skatchkov S. N., Osborne N. N., Reichenbach A. (2006) Prog. Retin. Eye. Res. 25, 397–424 [DOI] [PubMed] [Google Scholar]
- 14.Kofuji P., Biedermann B., Siddharthan V., Raap M., Iandiev I., Milenkovic I., Thomzig A., Veh R. W., Bringmann A., Reichenbach A. (2002) Glia 39, 292–303 [DOI] [PubMed] [Google Scholar]
- 15.Raap M., Biedermann B., Braun P., Milenkovic I., Skatchkov S. N., Bringmann A., Reichenbach A. (2002) Neuroreport 13, 1037–1040 [DOI] [PubMed] [Google Scholar]
- 16.Satz J. S., Philp A. R., Nguyen H., Kusano H., Lee J., Turk R., Riker M. J., Hernández J., Weiss R. M., Anderson M. G., Mullins R. F., Moore S. A., Stone E. M., Campbell K. P. (2009) J. Neurosci. 29, 13136–13146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skatchkov S. N., Thomzig A., Eaton M. J., Biedermann B., Eulitz D., Bringmann A., Pannicke T., Veh R. W., Reichenbach A. (2001) Neuroreport 12, 1437–1441 [DOI] [PubMed] [Google Scholar]
- 18.Wu J., Marmorstein A. D., Kofuji P., Peachey N. S. (2004) Mol. Vis. 10, 650–654 [PMC free article] [PubMed] [Google Scholar]
- 19.Tang X., Schmidt T. M., Perez-Leighton C. E., Kofuji P. (2010) Neuroscience 166, 397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolton S., Butt A. M. (2006) Exp. Neurol. 202, 36–43 [DOI] [PubMed] [Google Scholar]
- 21.Neusch C., Rozengurt N., Jacobs R. E., Lester H. A., Kofuji P. (2001) J. Neurosci. 21, 5429–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hibino H., Nin F., Tsuzuki C., Kurachi Y. (2010) Pflugers Arch. 459, 521–533 [DOI] [PubMed] [Google Scholar]
- 23.Jagger D. J., Nevill G., Forge A. (2010) J. Assoc. Res. Otolaryngol. 11, 435–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rozengurt N., Lopez I., Chiu C. S., Kofuji P., Lester H. A., Neusch C. (2003) Hear. Res. 177, 71–80 [DOI] [PubMed] [Google Scholar]
- 25.Yang T., Gurrola J. G., 2nd, Wu H., Chiu S. M., Wangemann P., Snyder P. M., Smith R. J. (2009) Am. J. Hum. Genet. 84, 651–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inyushin M., Kucheryavykh L. Y., Kucheryavykh Y. V., Nichols C. G., Buono R. J., Ferraro T. N., Skatchkov S. N., Eaton M. J. (2010) Epilepsia 51, 1707–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buono R. J., Lohoff F. W., Sander T., Sperling M. R., O'Connor M. J., Dlugos D. J., Ryan S. G., Golden G. T., Zhao H., Scattergood T. M., Berrettini W. H., Ferraro T. N. (2004) Epilepsy Res. 58, 175–183 [DOI] [PubMed] [Google Scholar]
- 28.Ferraro T. N., Golden G. T., Smith G. G., Martin J. F., Lohoff F. W., Gieringer T. A., Zamboni D., Schwebel C. L., Press D. M., Kratzer S. O., Zhao H., Berrettini W. H., Buono R. J. (2004) Mamm. Genome 15, 239–251 [DOI] [PubMed] [Google Scholar]
- 29.Heuser K., Nagelhus E. A., Taubøll E., Indahl U., Berg P. R., Lien S., Nakken S., Gjerstad L., Ottersen O. P. (2010) Epilepsy Res. 88, 55–64 [DOI] [PubMed] [Google Scholar]
- 30.Lenzen K. P., Heils A., Lorenz S., Hempelmann A., Höfels S., Lohoff F. W., Schmitz B., Sander T. (2005) Epilepsy Res. 63, 113–118 [DOI] [PubMed] [Google Scholar]
- 31.Shang L., Lucchese C. J., Haider S., Tucker S. J. (2005) Brain Res. Mol. Brain Res. 139, 178–183 [DOI] [PubMed] [Google Scholar]
- 32.Bockenhauer D., Feather S., Stanescu H. C., Bandulik S., Zdebik A. A., Reichold M., Tobin J., Lieberer E., Sterner C., Landoure G., Arora R., Sirimanna T., Thompson D., Cross J. H., van't Hoff W., Al Masri O., Tullus K., Yeung S., Anikster Y., Klootwijk E., Hubank M., Dillon M. J., Heitzmann D., Arcos-Burgos M., Knepper M. A., Dobbie A., Gahl W. A., Warth R., Sheridan E., Kleta R. (2009) N. Engl. J. Med. 360, 1960–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scholl U. I., Choi M., Liu T., Ramaekers V. T., Häusler M. G., Grimmer J., Tobe S. W., Farhi A., Nelson-Williams C., Lifton R. P. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 5842–5847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang X., Hang D., Sand A., Kofuji P. (2010) Biochem. Biophys. Res. Commun. 399, 537–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reichold M., Zdebik A. A., Lieberer E., Rapedius M., Schmidt K., Bandulik S., Sterner C., Tegtmeier I., Penton D., Baukrowitz T., Hulton S. A., Witzgall R., Ben-Zeev B., Howie A. J., Kleta R., Bockenhauer D., Warth R. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 14490–14495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kucheryavykh Y. V., Pearson W. L., Kurata H. T., Eaton M. J., Skatchkov S. N., Nichols C. G. (2007) Channels 1, 172–178 [DOI] [PubMed] [Google Scholar]
- 37.Benfenati V., Caprini M., Nobile M., Rapisarda C., Ferroni S. (2006) J. Neurochem. 98, 430–445 [DOI] [PubMed] [Google Scholar]
- 38.Lederer W. J., Nichols C. G. (1989) J. Physiol. 419, 193–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin Y. W., Jia T., Weinsoft A. M., Shyng S. L. (2003) J. Gen. Physiol. 122, 225–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kucheryavykh L. Y., Kucheryavykh Y. V., Inyushin M., Shuba Y. M., Sanabria P., Cubano L. A., Skatchkov S. N., Eaton M. J. (2009) Open Neurosci. J. 3, 40–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurata H. T., Marton L. J., Nichols C. G. (2006) J. Gen. Physiol. 127, 467–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nichols C. G., Lopatin A. N. (1997) Annu. Rev. Physiol. 59, 171–191 [DOI] [PubMed] [Google Scholar]
- 43.Rapedius M., Fowler P. W., Shang L., Sansom M. S., Tucker S. J., Baukrowitz T. (2007) Neuron 55, 602–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rapedius M., Haider S., Browne K. F., Shang L., Sansom M. S., Baukrowitz T., Tucker S. J. (2006) EMBO Rep. 7, 611–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopes C. M., Zhang H., Rohacs T., Jin T., Yang J., Logothetis D. E. (2002) Neuron 34, 933–944 [DOI] [PubMed] [Google Scholar]
- 46.Schulte U., Hahn H., Konrad M., Jeck N., Derst C., Wild K., Weidemann S., Ruppersberg J. P., Fakler B., Ludwig J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 15298–15303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schulte U., Hahn H., Wiesinger H., Ruppersberg J. P., Fakler B. (1998) J. Biol. Chem. 273, 34575–34579 [DOI] [PubMed] [Google Scholar]
- 48.Schulte U., Fakler B. (2000) Eur. J. Biochem. 267, 5837–5841 [DOI] [PubMed] [Google Scholar]
- 49.Rapedius M., Paynter J. J., Fowler P. W., Shang L., Sansom M. S., Tucker S. J., Baukrowitz T. (2007) Channels 1, 327–330 [DOI] [PubMed] [Google Scholar]
- 50.Peters M., Ermert S., Jeck N., Derst C., Pechmann U., Weber S., Schlingmann K. P., Seyberth H. W., Waldegger S., Konrad M. (2003) Kidney Int. 64, 923–932 [DOI] [PubMed] [Google Scholar]
- 51.Cho H. C., Tsushima R. G., Nguyen T. T., Guy H. R., Backx P. H. (2000) Biochemistry 39, 4649–4657 [DOI] [PubMed] [Google Scholar]
- 52.Leyland M. L., Dart C., Spencer P. J., Sutcliffe M. J., Stanfield P. R. (1999) Pflugers Arch. 438, 778–781 [DOI] [PubMed] [Google Scholar]
- 53.Tao X., Avalos J. L., Chen J., MacKinnon R. (2009) Science 326, 1668–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanemoto M., Abe T., Onogawa T., Ito S. (2004) Am. J. Physiol. Renal Physiol. 287, F1148–F1153 [DOI] [PubMed] [Google Scholar]
- 55.Tanemoto M., Abe T., Ito S. (2005) J. Am. Soc. Nephrol. 16, 2608–2614 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.