Abstract
The insulin-like family of factors are involved in the regulation of a variety of physiological processes, but the function of the family member termed insulin-like 6 (Insl6) in skeletal muscle has not been reported. We show that Insl6 is a myokine that is up-regulated in skeletal muscle downstream of Akt signaling and in regenerating muscle in response to cardiotoxin (CTX)-induced injury. In the CTX injury model, myofiber regeneration was improved by the intramuscular or systemic delivery of an adenovirus expressing Insl6. Skeletal muscle-specific Insl6 transgenic mice exhibited normal muscle mass under basal conditions but elevated satellite cell activation and enhanced muscle regeneration in response to CTX injury. The Insl6-mediated regenerative response was associated with reductions in muscle cell apoptosis and reduced serum levels of creatine kinase M. Overexpression of Insl6 stimulated proliferation and reduced apoptosis in cultured myogenic cells. Conversely, knockdown of Insl6 reduced proliferation and increased apoptosis. These data indicate that Insl6 is an injury-regulated myokine that functions as a myogenic regenerative factor.
Keywords: Aging, Akt PKB, Insulin-like Growth Factor (IGF), Protein Secretion, Stem Cell, Cardiotoxin, Myokine, Regeneration
Introduction
Accumulating evidence suggests that skeletal muscle secretes proteins that impact the properties of other cell types. It is well-established that skeletal muscle can produce extracellular signaling factors that are required for coordinated blood vessel recruitment as myofibers undergo hypertrophy or ischemic repair (1–3). Contracting muscle can also produce and release IL-6, IL-8, and IL-15, which can affect systemic metabolism (4), and it is the source of other metabolic regulatory factors, including FGF21 and Visfatin (5–7). These muscle-produced cytokines, which exert their effects in an autocrine, paracrine, or endocrine manner, have been termed “myokines” (8).
Skeletal muscle is a highly regenerative tissue. This regenerative capacity is due to the action of satellite cells that function as muscle progenitors. Adult satellite cells are located beneath the basal lamina, where they are normally quiescent. In response to muscle injury, satellite cells reenter the cell cycle, proliferate and eventually differentiate into myofibers (reviewed in Refs. 9, 10). A number of secreted factors, including insulin-like growth factor (IGF),3 TGF-β, HGF, and IL-6, play a role in satellite cell activation and differentiation (reviewed in Ref. 11). It is thought that a well orchestrated balance between the growth and differentiation of satellite cells is critical for muscle regeneration in response to injury.
The cardiotoxin (CTX) injury model is widely used to examine skeletal muscle regeneration. The injection of CTX induces into rodent muscle induces inflammation and myofiber degeneration that is followed by a regenerative response (10, 11). In this model, the stages of regeneration include the activation of satellite cells within 2 h of injury, a satellite cell proliferative stage that peaks at 2–3 days followed by the differentiation of myofibers between 5 and 7 days (12). The restoration of the cellular architecture is typically complete by 2–3 weeks after injury. Impaired muscle regeneration in the CTX model is a hallmark of the aging process. This decline of muscle regeneration with age is thought to be due primarily to defects in satellite cell function (reviewed in Refs. 13–15).
The phosphatidylinositol 3-kinase (PI(3)K)/Akt signaling axis is critical in the control of muscle mass (1, 16, 17). Numerous studies have also shown that activation IGF-PI(3)K-Akt signaling accelerates muscle regeneration in models of muscle atrophy and trauma (reviewed in Refs. 11, 18, 19). These regenerative effects are associated with enhanced satellite cell activation and protection from cell death (19, 20). We generated skeletal muscle-specific, inducible Akt1 transgenic mice and demonstrated that transgene expression led to a marked increase in myofiber hypertrophy with corresponding increase in muscle strength (21). In this model, transgene activation in mature myofibers, via the muscle creatine kinase (MCK) promoter, is associated with marked satellite cell activation (19). Thus, we postulate that the activation of Akt signaling in myofibers results in production and release of myokines, which act on the satellite cell population in a paracrine manner. In previous studies, we have employed this transgenic system to isolate myokine candidates that regulate metabolism and cardiovascular function (3, 5). The purpose of this study was to identify candidate Akt-regulated myokines that activate satellite cells and consequently stimulate muscle regeneration in response to injury. In this report, we demonstrate that insulin-like 6 (Insl6) is a muscle-derived factor that is up-regulated by injury or the induction of Akt signaling. Overexpression of Insl6 stimulates satellite cell activation and accelerates muscle regeneration in the CTX injury model. Cell culture experiments show that Insl6 promotes muscle progenitor cell proliferation and survival.
EXPERIMENTAL PROCEDURES
Materials
Cardiotoxin was obtained from EMD Chemicals (San Diego, CA). AdEAsyTM system was purchased from Q-Biogene (Carlsbad, CA). Dulbecco's modified Eagle's medium (DMEM), trypsin, fetal bovine serum (FBS), penicillin-streptomycin mixture, and LipofectamineTM RNAiMAX were obtained from Invitrogen (Carlsbad, CA). Phospho Akt (Ser-473) and HA antibodies were purchased from Cell Signaling Technology (Danvers, MA). BrdU was purchased from BD Bioscience (San Diego, CA). Anti-BrdU antibody was purchased from Roche Diagnostics (Indianapolis, IN). Protease inhibitor mixture and RIPA buffer were purchased from Sigma. The pre-designed Insl6 siRNA and negative control siRNA were obtained from Qiagen (Valencia, CA).
Mice
Skeletal muscle-specific inducible Akt1-Tg mice (double transgenic mice, DTG) were generated by crossing 1256 [3Emut] MCK-rtTA TG mice with TRE-myrAkt1 TG mice as described (21). For Akt1 transgene expression, DTG mice were treated with doxycycline (0.5 mg/ml) in drinking water and DTG mice treated with normal water were used as controls. C57Bl/6 male mice (Charles River Laboratory, Wilmington, MA) were 2 months old and weighed 20–25 g. 18 months old C57Bl/6 male mice were purchased from NIA (Bethesda, MD). Mice were housed at 24 °C on a fixed 12-h light/dark cycle and fed a normal chow diet (Teklad global 18% protein rodent diet, 2018, Harlan). Study protocols were approved by the Institutional Animal Care and Use Committee at Boston University.
Generation of Insl6 Transgenic Mice
The mouse Insl6 cDNA sequence was amplified by PCR and subcloned downstream of the mouse 4.8 kbp murine MCK sequence in the MCK-pBS2 SK(+) vector at the EcoRV cloning site (22). This plasmid was incubated with endonucleases KpnI and XbaI, and the restriction digestion products were separated by agarose gel electrophoresis. The DNA fragment corresponding to the MCK-Insl6 fragment was excised from the gel and subject to further purification. The purified MCK-Insl6 fragment was used for pronuclear injection for fertilized C57Bl/6 oocytes according to established techniques.
Insl6 Antibody Preparation
Rabbit anti-mouse Insl6, affinity-purified antibody was prepared by Bethyl Laboratories, Inc (Montgomery, TX). Peptide Cys-MAVASLPFVDF was used as the immunogen. Hyperimmune sera from rabbits were processed over an immunosorbent to produce affinity-purified antibody.
Adenoviral Vector Construction
Replication-defective adenovirus construct expressing Insl6 (Adeno-Insl6) was made with AdEAsyTM system from Q-Biogene (Carlsbad, CA) according to the manufacturer's protocol. In brief, the mouse Insl6 cDNA vector was purchased from American Type Culture Collection. Full-length Insl6 cDNA was amplified by PCR and subcloned into the CMV p-shuttle vector. This vector was recombined with the Ad5ΔE1/ΔE3 vector, and the resulting vector construct containing mouse Insl6 gene was transfected into HEK293 cells. Virus particles were amplified in HEK293 cells and purified by ultracentrifugation with a cesium chloride gradient solution. Viral titers were determined and expressed as plaque-forming units (pfu).
Muscle Injury
A 10 μm solution of CTX or equal volume of PBS was injected into to either tibialis anterior (TA) or gastrocnemius (GA) muscles at 2 μl/g body weight using an insulin syringe (12, 23). The needle was inserted parallel to the muscle fiber longitude until reaching the tendon near the knee and then slowly withdrawn while simultaneously injecting the CTX solution in its path.
Adenovirus Delivery
For intramuscular delivery, 5 × 109 pfu of Adeno-βgal, as a control, or Adeno-Insl6 was injected in muscle 48 h after the CTX injury. For systemic delivery, 1010 pfu of either Adeno-βgal or Adeno-Insl6 in 100 μl of saline was injected into the jugular vein at the time of CTX injection into muscle.
Western Blotting
Skeletal muscle tissues were homogenized in radioimmune precipitation assay buffer, which was supplemented with protease inhibitor mixture. Equal amount of proteins were loaded on NuPAGE Gels (Invitrogen) and run as instructed by the manufacturer. The transfer to PVDF membranes was done under wet conditions. Membranes were blocked in 5% milk or BSA solution, and then incubated with primary antibodies at 4 °C overnight. Membranes were washed several times in TBST while agitating, and then incubated with HRP-conjugated secondary antibodies at room temperature for 1–2 h. Chemiluminescent antigen expression was captured on film by ECL using a Western blotting detection kit (GE Healthcare).
Histological Analysis
Skeletal muscle tissues were embedded in OCT compound from Fisher scientific (Pittsburgh, PA) and snap-frozen in liquid nitrogen. Serial cryostat sections (8 μm) were fixed for 10 min in 4% paraformaldehyde, and were stained with H&E for histological analysis. For quantification of new myotube formation, sections containing the largest lesion areas were selected for analysis. The core of the lesion, indicated by the region that regenerated the least, was visualized using a ×20 objective, and the image was captured to a computer screen. All the centrally nucleated fibers within this field were counted. For some analyses, serial cryostat sections (8 μm) were stained with anti-HA, Insl6, Ki67, BrdU, and Ncam1 primary antibodies. Fluorescent dye conjugated secondary antibodies were used to visualize the antigen expression in situ. DAPI was used to counterstain the cell nuclei. Fifteen randomly chosen microscopic fields from three different sections in each tissue block were examined for the presence of antigen-expressing cells.
Microarray Analysis
Total RNA from whole gastrocnemius muscle was used to generate cDNA using the poly(dT) primer in the Superscript Choice system (Invitrogen) according to the manufacturer's protocol (Affymetrix) (5, 24, 25). The resulting cDNA was used to generate biotin-labeled cRNA by incorporating biotinylated CTP and UTP using the ENZO Bioarray High Yield transcript labeling kit (Affymetrix). cRNA samples were hybridized to Affymetrix GeneChip Mouse Expression Set 430 microarrays A and B for 16 h at 45 °C, and bound sequences were quantified by staining and scanning according to Affymetrix protocols. The arrays were then scanned at 488 nm using a G25000A gene array scanner (Agilent, Palo Alto, CA). Scanned images were quantified using Microarray Suite 5.0 software (MAS 5.0) (Affymetrix). Transcript expression levels were then scaled to an average intensity of 500 units on each chip.
Quantitative Real-time PCR
Total RNA from skeletal muscle was prepared by Qiagen RNeasy kit (Valencia, CA) using protocols provided by the manufacturer. cDNA was produced using ThermoScript RT-PCR Systems (Invitrogen, Carlsbad, CA). The primers sequences for qRT-PCR are described in supplemental Fig. S1. Transcript levels were determined relative to the signal from 36B4, and normalized to the mean value of samples from control mice. Primers were purchased from Integrated DNA technology (Coralville, IA), and primer sequences are available upon request.
TUNEL Staining in Tissue
Skeletal muscle tissues cryostat sections (8 μm) were fixed for 10 min in 4% paraformaldehyde at room temperature. The Apo-BrdU In Situ DNA Fragmentation Assay kit from MBL Inc. (Woburn, MA) was used to label DNA breaks, thereby detecting apoptotic cells, according to the manufacturer's instructions.
Creatine Kinase M
Creatine Kinase M levels in mouse serum was measured in Charles River Laboratories Research Animal Diagnostic Services (Wilmington, MA) (assay code: CP-SCH-20).
Cell Culture
Mouse C2C12 cells obtained from the American Type Culture Collection were maintained in growth medium (DMEM supplemented with 20% FBS). Human quadriceps muscle primary cells were isolated from quadriceps muscle biopsy as previously described (26). The cells were maintained in DMEM supplemented with 20% FBS.
Cell Transduction in Vitro
For adenoviral infection in vitro, C2C12 or human primary skeletal muscle cells were typically incubated with adenovirus at a multiplicity of infection (MOI) of 250–500 in culture medium for 16 h. The virus was removed when the medium was replaced with fresh medium. Cell lysates were collected 48 h after virus infection and subject to Western blot analysis. The siRNAs targeting mouse Insl6 and unrelated siRNAs were transfected using LipofectamineTM RNAiMAX reagent (Invitrogen) according to the manufacturer's instructions.
Cell Proliferation Assays
Cell proliferation was assessed by direct cell counting using a hemocytometer at 48 h after treatment with siRNA or adenoviral vector. DNA synthesis was measured as [3H]thymidine (GE Healthcare) incorporation. C2C12 or human primary skeletal muscle cells were plated in 6-well plates and cultured in growth medium. When the cells are 50–60% confluent, culture medium was switched to DMEM containing 2% FBS. Cells were transfected with adenovirus vectors at an MOI of 250–500 or siRNA reagents for overnight. At 48 h post-transfection, cells were incubated with [3H]thymidine (1.0 μCi/well) for 4 h. Cells were then washed cells twice with ice-cold PBS and incubated with 10% trichloroacetic acid for 30 min at 4 °C. The lysed cell extract was neutralized with a 0.25 n NaOH solution, and the incorporation of radioactivity was determined by scintillation counting.
Cell Death Assays
C2C12 myoblast cultures were transduced with adenovirus or siRNA reagents overnight in DMEM medium containing 5% FBS, and CTX was added a final concentration of 100 nm on the second day. Apoptosis assays were carried out 48 h after the virus transfection. Cell death detection ELISAPLUS was purchased from Roche Diagnostics (Indianapolis, IN). Cytoplasmic histone-associated-DNA-fragments were quantified according to the manufacturer's protocol. Caspase 3/7 activity of the cell lysate was measured by Apo-ONE caspase-3/7 assay kit from Promega (Madison, WI) per the manufacturer's protocol.
Statistical Analysis
All values are presented as mean ± S.E. Student's t tests were performed to assess the statistical significance of 2-way analyses. For multiple comparisons, ANOVA was employed. p values of less than 0.05 were considered statistically significant.
RESULTS
Insl6 Is Up-regulated by Akt Activation in the Skeletal Muscle
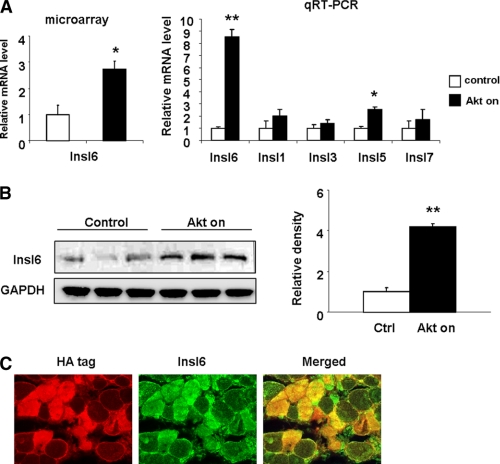
Skeletal muscle-specific, conditional Akt1 transgenic mice (21) were used for this study as to identify candidate paracrine factors that contribute to satellite cell activation. Male mice at 8 weeks of age were put on drinking water containing doxycycline for 2 weeks to activate myogenic Akt signaling. Age and gender matched transgenic mice without doxycycline treatment were used as control. Microarray expression analysis was performed on total RNA isolated from the GA muscles to identify transcripts that were differentially regulated by Akt activation using Affymetrix GeneChip Mouse Genome 430 2.0 arrays (24, 25). Approximately 400 pre-identified transcripts were differentially regulated following 2 weeks of Akt transgene activation. This set of 400 transcripts was further queried for transcripts encoding secreted proteins by Signal IP and SMART software. Of the differentially regulated genes predicted to encode secreted proteins, Insl6 was of interest because it is a member of the IGF/insulin/relaxin superfamily with unknown biological function(s). Insl6 was up-regulated 2.5-fold in microarray analysis (Fig. 1A), but transcripts encoding IGF, Insl1 (relaxin) or other members of the insulin-like family did not change (data not shown). The results of the microarray analysis were corroborated by qRT-PCR. The Insl6 transcript level was induced ∼8.5-fold in Akt-activated GA muscle, whereas transcripts encoding other members of the insulin-like family including Insl1 (relaxin), Insl3, Insl5, and Insl7 showed little or no change.
FIGURE 1.
Up-regulation of Insl6 in Akt-activated skeletal muscle. Skeletal muscle-specific, Akt1 transgenic mice were treated with either doxycycline or vehicle for 2 weeks (A). Total RNA was isolated from GA muscles and subject to cDNA synthesis and microarray hybridization (left panel). Relative transcript of Insl6 (n = 3 in each group) from the microarray analysis. Relative transcript expression of Insl1, 3, 5, 6, and 7 in total RNA isolated from GA muscle of control and Akt-induced transgenic mice as measured by qRT-PCR (right panel). B, GA muscle lysate from control and Akt-induced transgenic mice were analyzed by Western blotting for Insl6 and GAPDH as a loading control. C, representative image of GA muscle sections from Akt-induced transgenic immunoblotted with Insl6 antibody (green) and hemagglutinin antibody (HA) that is fused to the Akt transgene (red). Sections from transgenic mice were double stained with anti-HA primary antibody with rhodamine-conjugated secondary antibody (red) and anti-Insl6 primary antibody with fluorescein-conjugated secondary antibody (green). A representative image is shown. Results are presented by mean ± S.E. *, p < 0.05; **, p < 0.01.
A polyclonal antibody was raised against a peptide sequence derived from what is predicted to be the “A-chain” of Insl6 (27). The specificity of the antibody was demonstrated by performing Western blot analysis on C2C12 cells transduced with adenoviral vectors expressing full-length murine Insl6 or a control vector (supplemental Fig. S2A). Western immunoblot analysis using this antibody on a GA muscle revealed that Insl6 protein was up-regulated more than 4-fold following 2 weeks of Akt1 transgene induction (Fig. 1B). Immunohistochemical analysis revealed that Insl6 induction was predominantly localized to the myofibers in GA muscle that were positive for the HA-tagged Akt1 transgene (Fig. 1C).
Insl6 Is Up-regulated in Skeletal Muscle upon CTX Injury
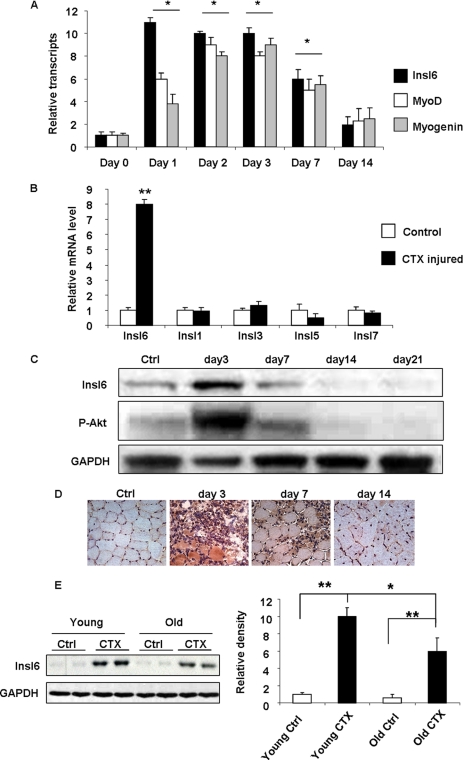
To examine Insl6 regulation in a model of skeletal muscle injury and regeneration, a solution CTX was injected in to TA muscle. Tissues were analyzed at day 1, 2, 3, 7, and 14 (n = 3–5 at each time point). As seen in Fig. 2a, qRT-PCR analysis revealed that the maximal induction of Insl6 transcript occurred at the acute phase of injury (1–3 days) after cardiotoxin delivery which corresponds to the peak satellite cell proliferation in this model. Transcript expression of the myogenic nuclear factors MyoD and myogenin, that serve as satellite cell markers in this model (9, 12), closely paralleled the expression of Insl6. Insl6 expression returned to baseline by 14 days post-injury. In contrast, the other insulin-like family members, Insl1 (relaxin), Insl3, Insl5, and Insl7, did not display regulation in the CTX model when examined at the 3 day time point (Fig. 2B).
FIGURE 2.
Insl6 expression in the cardiotoxin model of skeletal muscle injury. A cardiotoxin solution (10 μm) or an equal volume of PBS (control) was injected into TA muscle at 2 μl/g body weight. A, relative mRNA transcripts of Insl6 (black), MyoD (white), and Myogenin (gray) in CTX or PBS-injected TA muscle were assessed at the indicated time points by qRT-PCR (n = 4–5). Statistical comparisons were made between the indicated day and control (day 0) for each transcript. B, relative transcript expression of Insl1, -3, -5, -6, and -7 in TA muscle at 3 days post CTX (black) or PBS (white) injection were measured by qRT-PCR (n = 5). C, time course of Insl6 protein expression and the level of Akt phosphorylation at serine residue 473 in CTX-injured TA muscle relative to control (Ctrl). A representative immunoblot is shown. D, representative images of TA muscle sections stained with anti-Insl6 antibody at different time points after injury by CTX injection. E, aging skeletal muscle exhibits decreased Insl6 induction by injury. Western blot analysis of Insl6 in TA muscle samples obtained from young (2 months old) and aged (18 months old) C57Bl/6 mice 3 days after the CTX or PBS injection. The results are presented as the mean ± S.E., n = 5–6, **, p < 0.01; *, p < 0.05.
Western blot analysis confirmed the dynamic regulation of Insl6 protein expression during regeneration in TA muscle, with high levels of expression occurring at the 3 day time point (Fig. 2C). The time course of Insl6 protein expression in CTX-injured muscle was similar to the transient increase in the activating phosphorylation of Akt at Ser-473. Immunohistochemical analysis of Insl6 expression in sections of TA muscle was also performed (Fig. 2D). The strongest signal for Insl6 occurred in myofibers at the 3 day time point. At 7 days post-injury, Insl6 protein could also be detected within nascent myofibers that are characterized by centralized nuclei. By 21 days post-injury, muscle architecture had largely been restored, and little or no expression of Insl6 protein was detected.
The induction of Insl6 in response to CTX injury was also compared in muscle of old versus young mice (Fig. 2E). The TA muscles of young (2 months) and old (18 months) mice were treated with CTX solution. Quantitative Western blot analysis of Insl6 performed on the protein samples harvested before and 3 days after the injury revealed that injury-induced Insl6 expression was reduced by a factor of approximately two in old mice relative to young mice.
Insl6 Accelerates Muscle Regeneration in CTX Injury Model
Because the induction of Insl6 expression correlated with a muscle wound healing response, we conducted adenovirus-mediated gain-of-function studies to examine the role of Insl6 in injury-induced myogenesis. An adenoviral vector expressing the mouse full-length Insl6 cDNA (Adeno-Insl6) was injected into uninjured GA muscle. An equivalent amount of adenoviral vector expressing β-galactosidase (Adeno-β-gal) was used as control. The Adeno-Insl6 treatment led to an appreciable increase (∼16-fold) in Insl6 transcript expression in GA muscles (supplemental Fig. S2B), that was comparable to levels of Insl6 expression achieved by CTX-induced injury (Fig. 2A). Immunohistochemical analysis revealed increased Insl6 protein expression in myofibers of Adeno-Insl6-treated muscle compared with control, and the analysis of β-galactosidase activity revealed uniform transgene expression in GA muscle (supplemental Fig. S2C). However, the delivery of Insl6 to uninjured TA muscle did not affect myofiber cross-sectional area or the frequencies of BrdU- and MyoD-positive cells in tissue sections (data not shown).
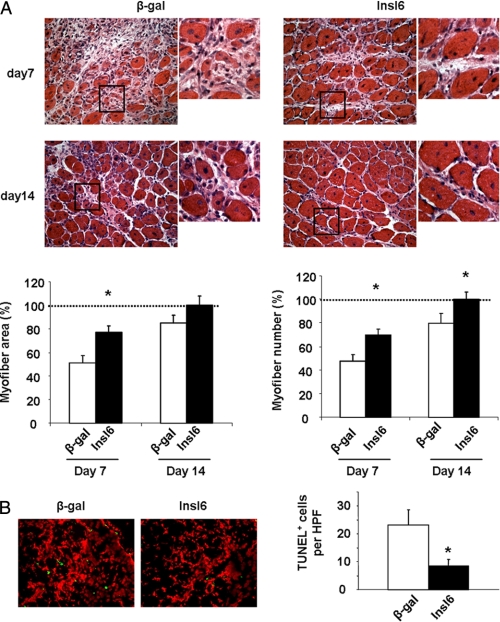
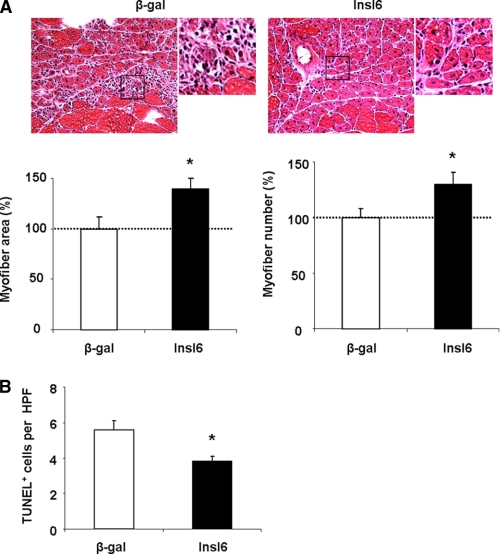
To examine the consequences of Insl6 overexpression on muscle regeneration, adenoviral vectors were injected intramuscularly at 48 h post-CTX delivery, after the edema had subsided. Muscle was harvested at 7, 14, and 21 days post-injury for analyses. The density of myofibers with centrally located nuclei and myofiber cross-sectional area was assessed in H&E-stained sections to evaluate the extents of muscle regeneration. Based upon these parameters, Adeno-Insl6-treated muscles contained significantly more regenerating myofibers than control muscles at day 7 post-injury. Similar trends were also observed 14 days following injury, but the differences between test and control groups was less notable (Fig. 3A). Terminal deoxynucleotide transferase TdT-mediated dUTP-digoxigenin nick-end (TUNEL) staining at day 7 also revealed less apoptotic cells present in Adeno-Insl6-treated muscle than in the control group (Fig. 3B). Comparison between the muscles injected with Adeno-β-gal and Adeno-Insl6 revealed no difference after 21 days. At this time point both experimental groups of muscle appeared histologically normal, and no significant scar formation was observed (data not shown).
FIGURE 3.
Intramuscular Adeno-Insl6 treatment facilitates muscle regeneration in the cardiotoxin model of skeletal muscle injury. Cardiotoxin solution was injected into TA muscle, and 5 × 109 pfu of Adeno-βgal or Adeno-Insl6 was injected into the same muscle 48 h after the injury. A, representative image of H&E-stained TA muscle sections at day 7 and day 14 after the cardiotoxin injection. Histological analysis of muscle regeneration was performed by counting nascent myofiber number and measuring the total cross-section at day 7 post injury. Data are expressed as a percentage relative to the mean value of the Insl6-treated group at day 14 post-injury (n = 6–8 in each group). B, representative image of a TA muscle section stained for TUNEL for each experimental group at 7 days post-injury. Quantitative analysis of TUNEL-positive cells per high power microscopic field (HPF) was performed. The results are presented as the mean ± S.E. *, p < 0.05.
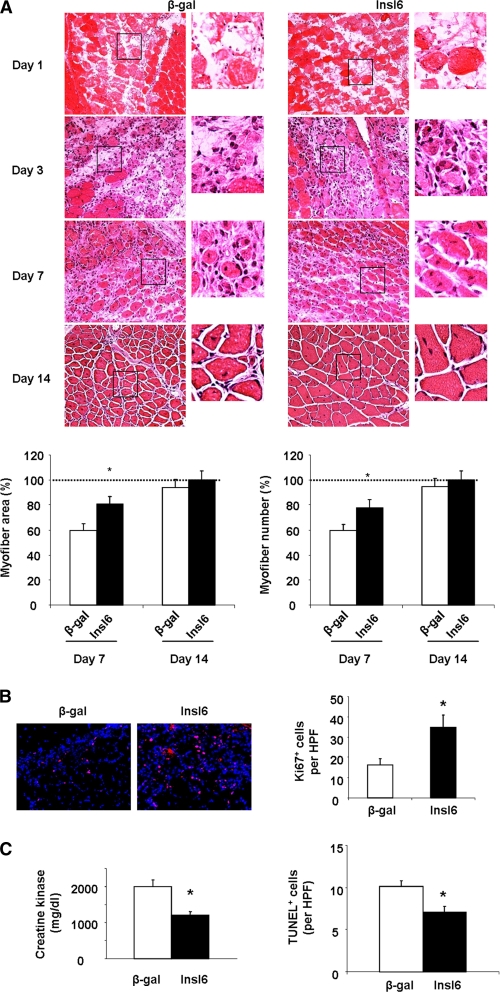
Because Insl6 is a secreted protein (27), the effects of intravenous Insl6 delivery on muscle regeneration were assessed. This mode of delivery allowed for the simultaneous delivery of adenoviral delivery and the injection of CTX in TA muscle because the transgene is expressed predominantly from the liver (28). The transduction of the liver was confirmed by qRT-PCR, which showed an 8-fold increase in Insl6 transcript in the livers of mice treated with Adeno-Insl6 (supplemental Fig. S2D). The systemic delivery of Adeno-Insl6 or control vectors allowed measurements at earlier time points than with the intramuscular delivery experiments because it was not necessary to wait 48 h for edema to subside for adenovirus delivery. Consistent with a prior report (29), CTX injection led to pronounced muscle necrosis and the accumulation of inflammatory cells at 24 h, and no myofiber regeneration was observable at this time point in the H&E-stained muscle sections (Fig. 4A). Although both Adeno-Insl6- and Adeno-β-gal-treated groups displayed very few nascent myofibers with centralized nuclei at 3 days post-injury, Ki67 staining revealed markedly more proliferating cells in the Adeno-Insl6 group (Fig. 4, A and B). By day 7, a conspicuously larger percentage of nascent myofibers were observed in muscles of Insl6-treated mice (Fig. 4A). At this time point the improvement in myofiber regeneration in the Insl6-treated group is associated with reduced serum creatine kinase M (CKM) level and a reduction in TUNEL-positive cells in the CTX-treated muscle (Fig. 4C). At 14 days after post-injury, a near complete restoration of muscle cellular architecture was observed in both Adeno-β-gal- and Adeno-Insl6- treated groups (Fig. 4A).
FIGURE 4.
Systemic delivery of Adeno-Insl6 accelerates muscle repair. Cardiotoxin solution was injected into TA muscle, and 1 × 1010 pfu of either Adeno-βgal or Adeno-Insl6 was simultaneously injected into the jugular vein. A, representative image of H&E stained TA muscle cross-sections at day 1, 3, 7, and 14 postoperation. Muscle regeneration was quantified by counting nascent myofibers and myofiber cross-section area measurements. Data are expressed as a percentage relative to the mean value of the Insl6-treated group at day 14 post-injury (n = 4–5 in each group). B, representative image of Ki67 immunohistochemical staining of CTX-treated TA muscle at 3 days post-injury. Quantitative analysis of Ki67+ cells per high power microscopic field was performed (red, Ki67, blue, DAPI). C, creatine kinase levels were measured in serum at the different experimental groups at 7 days post-injury (left panel). TUNEL staining was performed on histological sections from CTX-treated TA muscle at 7 days post-injury (right panel). Data are presented as the mean ± S.E. *, p < 0.05.
Insl6 Overexpression in Aged Skeletal Muscle Stimulates a Regenerative Response
Aged muscles recover poorly following damage, and this regenerative defect may be due to changes in the function of satellite cells as they age (30–32). Because injury-induced Insl6 expression is reduced by aging (Fig. 2E), we tested whether the delivery of Insl6 could promote regeneration in the injured muscle of aged mice. GA muscles of 18 months old C57 mice were pretreated with CTX solution, Adeno-Insl6 or Adeno-β-gal were intramuscularly delivered 48 h later, and muscle regeneration was evaluated at day 7 post-injury. As seen in Fig. 5A, Insl6 treatment led to increases in nascent myofiber number and an increase in myofiber cross-sectional area. Correlating with the enhanced myogenesis, less TUNEL-positive cells were observed in Insl6-treated muscles (Fig. 5B).
FIGURE 5.
Insl6 overexpression promotes muscle regeneration in old mice. Approximately 10 μl of CTX solution (10 μm) was injected in 4 different positions on GA muscle. At 48 h after the injury, 5 × 109 total pfu of Adeno-βgal or Adeno-Insl6 was also injected into 4 regions of GA muscle. A, representative image of H&E-stained GA muscle cross-sections at day 7 after the cardiotoxin injection (n = 4–6 in each group). Histological analysis of muscle regeneration was assessed by counting nascent myofibers and measuring myofiber cross-section area at day 7 post-injury. Data are expressed as a percentage relative to the mean value for the Adeno-βgal-treated group. B, in vivo TUNEL staining was performed, and positive cells are quantified in histological sections. Data are presented as the mean ± S.E. *, p < 0.05.
Transgenic Mice Expressing Insl6 Mice Exhibit Enhanced Satellite Cell Activation and Muscle Repair following Injury
To examine the consequences of chronic Insl6 overexpression in muscle, transgenic mice were constructed that express murine Insl6 from the MCK promoter (supplemental Fig. S3A). Western blot analysis revealed appreciable Insl6 over-expression in the extensor digitorum longus, soleus, gastrocnemius, and tibialis anterior muscles of the transgenic mice (supplemental Fig. S3B). A low level of Insl6 transgene expression could also be detected in heart, but not in liver. Analysis of mice at 3 months of age did not detect changes in body weight, gastrocnemius muscle weight or cross-sectional area between transgenic and wild-type mice (supplemental Fig. S4A). Similarly, no differences were detected in tibialis anterior muscle weight or histological appearance between wild-type and transgenic mice (Fig. S4B). These data were corroborated by analyses with a second Insl6 transgenic mouse line (data not shown).
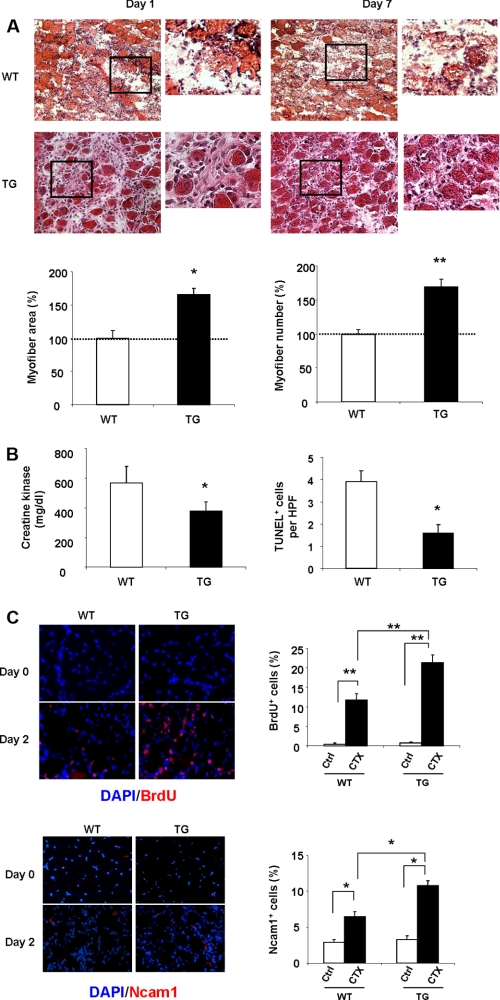
CTX solution was delivered to the TA muscles of Insl6 transgenic and their wild-type littermate controls. Histological analyses revealed an improved regenerative response in transgenic mice compared with wild-type based upon quantitation of myofiber area and number at the 7 day time point (Fig. 6A). No differences in muscle histology could be detected at the 1 day time point, with more than 95% of the myofibers destroyed in the affected muscle of both strains. CTX-treated Insl6 transgenic mice displayed reductions in serum CKM levels and a decrease in the frequency of TUNEL-positive cells in the treated muscle compared with CTX-treated wild-type mice at the 7 day time point (Fig. 6B).
FIGURE 6.
Insl6 transgenic mice exhibit improved muscle regeneration and enhanced satellite cell activation following CTX injury. A, representative images of H&E-stained TA muscle sections at day 1 and day 7 post-injury. Muscle regeneration was quantified in histological sections by counting nascent myofibers and measuring the cross-sectional area of myofibers at day 7. Data are expressed as a percentage relative to the mean value of WT mouse at 7 days post-injury. B, creatine kinase levels were measured in serum at day 7. TUNEL staining was performed on histological sections from the 7 day time point. C, representative images of TA muscle sections stained with BrdU or Ncam1 (red), with a DAPI counter stain (blue) at 2 days post injury (n = 6). Quantitative data are presented as the percentage of BrdU- or Ncam1-positive cells relative to the total number of DAPI-positive cells. The results are presented as the mean ± S.E. **, p < 0.01; *, p < 0.05.
Because the transgenic mouse reagent eliminated the need for Insl6 delivery via an adenoviral vector that could confound cellular analyses, additional experiments were performed to assess the status of proliferating satellite cells in the two strains of mice using BrdU incorporation and NCAM1 staining (10, 33). Uninjured muscle displayed very low levels of BrdU incorporation regardless of genotype (Fig. 6C). In contrast, CTX delivery led to a marked increase in BrdU incorporation that was significantly greater in the Insl6 transgenic than wild-type mice. Similarly, there was no significant difference between the percentage of NCAM1-positive cells in uninjured muscle between wild-type and transgenic mice. The frequency of NCAM1-positive cells increased 2 days following CTX delivery, and the magnitude of the increase was greater in the Insl6 transgene-positive muscle (Fig. 6C). Collectively, these data suggest that Insl6 promotes regeneration, at least in part, by promoting the activation of satellite cells.
Insl6 Is a Regulator of Myogenic Cell Proliferation
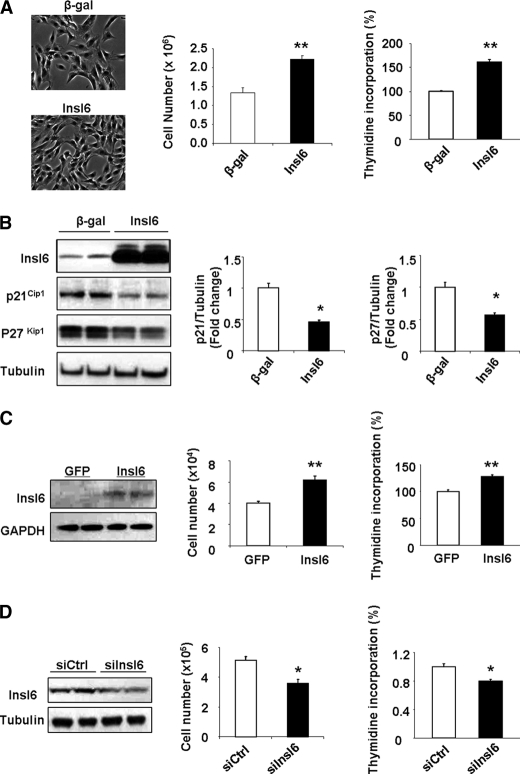
To investigate the effects of Insl6 on myogenic cells in vitro, C2C12 myoblasts were transduced with Adeno-Insl6 or Adeno-β-gal in proliferation media and pulse-labeled with [3H]thymidine. The overexpression of Insl6 was associated with a significant increase in [3H]thymidine into DNA and an increase in cell number (Fig. 7A). Consistent with these data, transduction with Adeno-Insl6 led to down-regulation of the cell cycle inhibitors p21Cip1 and p27Kip1 in proliferating C2C12 myoblasts (Fig. 7B). The mitogenic action of Insl6 on myogenic cells was corroborated with cultured human skeletal muscle primary cells isolated from biopsy of human quadriceps muscle (26). As shown in Fig. 7C, transduction with Adeno-Insl6 stimulates skeletal muscle primary cell proliferation as assessed by the [3H]thymidine incorporation assay and measurements of cell number.
FIGURE 7.
Insl6 is an endogenous regulator of muscle cell proliferation. A, C2C12 cells were transduced with adenovirus or siRNA reagent. [3H]thymidine pulse labeling was performed 48 h after the transfection. A representative image of C2C12 myoblasts transduced with Adeno-Insl6 or Adeno-β-gal is shown (left panel). Total cell number was assessed 48 h following adenovirus transduction. Data are expressed as a percentage relative to the mean of Adeno-βgal experimental group. B, Insl6 down-regulates cyclin-dependent kinase inhibitors p21Cip1 and p27Kip1. Cells were treated as in A and Insl6, p21Cip1 and p27Kip1 levels were determined by Western blot analysis. A representative blot is shown. The levels of p21Cip1 and p27Kip1 levels were quantified relative to the expression of tubulin that was used as a loading control. Data are expressed relative to the mean of the ratio of p21Cip1 or p27Kip1 relative to tubulin in the Adeno-βgal-treated cells (n = 6). C, human skeletal muscle primary cells were transduced as described above with Adeno-Insl6 or Adeno-GFP as control. A representative Western blot is shown to indicate Insl6 expression. The incorporation of [3H]thymidine into DNA and cell number was assessed at 48 h after transduction with adenovirus. D, Insl6 deficiency leads to cell growth retardation. C2C12 myoblasts were transfected with an siRNA targeting Insl6 or a negative control siRNA. A representative Western immunoblot is shown to indicate the degree of Insl6 ablation. The incorporation of [3H]thymidine and total cell number was assessed 48 h after treatment with siRNA. The values are presented by mean ± S.E. **, p < 0.01; *, p < 0.05.
To further elucidate the physiological role of Insl6 in myoblast proliferation, siRNA was used to suppress endogenous Insl6 expression in C2C12 cells. As shown Fig. 7D, a 50% ablation of endogenous Insl6 resulted in a reduction in [3H]thymidine incorporation into DNA, suggesting that Insl6 is an endogenous regulator of myogenic cell proliferation.
Insl6 Reduces CTX-induced Cell Apoptosis
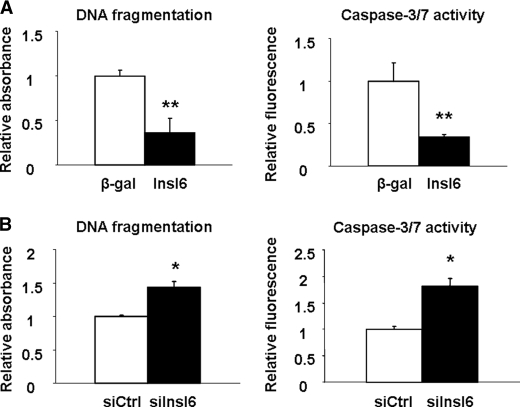
CTX is reported to induce apoptosis in skeletal muscle and neuroblastoma cells (34, 35). Thus C2C12 myoblasts were transduced with Adeno-Insl6 or Adeno-β-gal and incubated with CTX for 18 h. The degree of CTX-induced apoptosis was markedly reduced in Adeno-Insl6-transduced cells as determined by measuring the formation of histone-associated DNA fragments (Fig. 8A). Consistent with these observations, cells treated in this manner also displayed a reduction in caspase 3 and 7 activity. Conversely, the knock-down of Insl6 by siRNA increased the frequency of CTX-induced apoptosis (Fig. 8B). Whereas cells treated with siRNA targeting Insl6 displayed an increase in histone-associated DNA fragmentation and caspase 3 and 7 activity in the presence or absence of CTX, transfection with Insl6 siRNA had no effect on the low level of cell death observed in the absence of CTX (data not shown).
FIGURE 8.
Insl6 promotes C2C12 cell survival under conditions of CTX-induced stress. A, C2C12 cells transduced with Adeno-βgal or Adeno-Insl6 for 16 h followed by 24 h of incubation with 100 nm CTX. Cellular apoptosis was quantified by measuring cytoplasm histone-associated-DNA-fragments and homogenous caspase 3/7 activity. B, C2C12 cells were transduced with Insl6 or non-related siRNA for 16 h followed by 24 h of incubation with 100 nm of CTX. Cellular apoptosis was quantified as described above. Values are mean ± S.E. Data are expressed as a proportion relative to the mean value of the control group. **, p < 0.01; *, p < 0.05. n = 6 per group.
DISCUSSION
In this report, we show that resting skeletal muscle expresses low levels of Insl6, but this factor is markedly induced by muscle injury. We also show that the overexpression of Insl6 accelerates muscle regeneration in the CTX injury model. Insl6-mediated improvement in skeletal muscle was documented by an acceleration in the appearance of nascent myofibers containing centrally located nuclei and increases in myofiber cross-sectional area. The increase in muscle regenerative response could be documented either by the acute delivery of an adenoviral vector expressing Insl6 and in transgenic strains of mice that overexpress Insl6 from a muscle-specific promoter. The improvement in regeneration was accompanied by an increase in the frequency of proliferative cells and by a decrease in the frequency of apoptotic cells in the damaged muscle. The increase in Insl6 expression also led to an increase in the frequency of cells positive for Ncam1, a satellite cell marker, and a reduction in circulating creatine kinase M levels, a marker of necrosis. Finally, Insl6 induction by injury was reduced in 18 month old mice, but overexpression of Insl6 in aged muscle stimulated the regenerative response. In the light of these data, we propose that Insl6 functions as a “myokine” that is involved in the muscle regenerative process. Its pro-regenerative properties appear to rely, at least in part, on its mitogenic effect on muscle progenitor cells and by its ability to promote cell survival.
It has been proposed that skeletal muscle secretes factors, referred to as “myokines,” that influence the behavior of neighboring or remote cells (4, 8). Insl6 has been previously identified as a secreted protein (27), and several lines of evidence suggest that Insl6 can be designated as an Akt-regulated myokine. The up-regulation of Insl6 was initially observed in a mouse model of inducible, muscle-specific Akt induction that is associated with marked satellite cell activation (19, 21). In the CTX-injury model, the induction of Insl6 paralleled the time course of the Akt phosphorylation increase. Other myokines regulated by Akt signaling include the metabolic factor FGF-21 (5), and the cardiovascular-protective factor Fstl1 (3). Furthermore, myogenic Akt signaling leads to the secretion of VEGF by muscle, and this mechanism is posited to maintain tissue perfusion as myofibers undergo Akt-mediated hypertrophic growth (1).
It has been previously reported that Insl6 is expressed in male germ cells (36, 37), and that Insl6 is essential for spermatogenesis (38). Insl6 is a member of the insulin-like/relaxin family of proteins that have diverse roles in reproductive processes (39–41). With regard to muscle regeneration, it has previously been shown that Insl1, also referred to as relaxin, will promote muscle repair in a laceration model through its ability to diminish fibrosis (42, 43). Thus, it is possible that multiple members of the insulin-like family display muscle regenerative properties. However, only Insl6, but not other member of the relaxin family, was consistently up-regulated during the CTX-induced muscle injury time course. Furthermore, Akt induction in muscle leads to marked induction of Insl6, but has little or no effect on transcript levels of other relaxin family members.
Skeletal muscle is a highly regenerative tissue, and muscle injury leads to the release of various secreted factors that are thought to have roles at different stages of muscle regeneration (11, 44). Some of the factors released during the early phase of muscle injury, such as IL-6, bFGF, and HGF, will stimulate myoblast proliferation in vitro. However, these factors are either ineffective or detrimental when applied to injured muscles in vivo (45–47). It is also reported that transcripts of myostatin accumulate throughout the regeneration process (48), but this factor inhibits muscle regeneration through the suppression of satellite cell proliferation (49, 50). In contrast, IGF-1 and -2 appear to be major stimulatory factors in skeletal muscle regeneration. Both IGF-1 and -2 are induced by muscle injury (51), and these factors can stimulate the proliferation and differentiation of myoblasts (11, 52). The administration of IGF-1 will promote the regeneration of injured muscle (53–55). IGF-1 will also promote myofiber hypertrophy, and transgenic mice that overexpress this growth factor exhibit increased muscle mass (56–58).
In contrast to the well described anabolic functions of the IGF isoforms, Insl6 overexpression did not stimulate myogenesis in normal muscle in either the adenovirus or transgenic models. Consistent with these observations, Insl6 does not induce C2C12 cell hypertrophy nor promote myogenic differentiation in culture. In contrast, Insl6 stimulated the proliferation and survival of myogenic cells in culture, consistent with a role as a satellite cell activator in injured muscle. Although insulin-like family proteins are related to insulin and IGF based on the protein sequence and structural homologies, the receptor-mediated signaling pathways triggered by these factors are remarkably different. Whereas IGF and insulin activate tyrosine kinase receptors, most insulin-like family proteins signal through G-protein coupled receptors to control cAMP-mediated signaling events (59). However, the receptor for Insl6 has yet to be identified, and nothing is known about the downstream signaling molecules that respond to this ligand.
A potential limitation of our study is that Insl6 function was primarily assessed by gain-of-function approaches. However, the level of overexpression by intramuscular adenovirus-mediated Insl6 delivery (16-fold) is comparable to the induction of Insl6 expression by CTX injury (8–10-fold). Furthermore, the gain-of-function studies in vivo were corroborated by loss-of-function studies in vitro. In this regard, we found that siRNA knockdown of Insl6 inhibits myoblast proliferation and promotes apoptosis, whereas the overexpression of Insl6 has the opposite action. In conclusion, our study shows that Insl6 is an injury-induced myokine that plays a role in facilitating a muscle-regenerative response. Strategies to enhance Insl6-mediated signaling could be a useful treatment for skeletal muscle myopathies or trauma-induced muscle injuries.
Supplementary Material

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- IGF
- insulin-like growth factor
- CTX
- cardiotoxin
- GA
- gastrocnemius
- TA
- tibialis anterior
- pfu
- plaque-forming units
- Insl
- insulin-like
- CKM
- creatine kinase M.
REFERENCES
- 1.Takahashi A., Kureishi Y., Yang J., Luo Z., Guo K., Mukhopadhyay D., Ivashchenko Y., Branellec D., Walsh K. (2002) Mol. Cell. Biol. 22, 4803–4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arany Z., Foo S. Y., Ma Y., Ruas J. L., Bommi-Reddy A., Girnun G., Cooper M., Laznik D., Chinsomboon J., Rangwala S. M., Baek K. H., Rosenzweig A., Spiegelman B. M. (2008) Nature 451, 1008–1012 [DOI] [PubMed] [Google Scholar]
- 3.Ouchi N., Oshima Y., Ohashi K., Higuchi A., Ikegami C., Izumiya Y., Walsh K. (2008) J. Biol. Chem. 283, 32802–32811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pedersen B. K., Febbraio M. (2005) Brain Behav. Immun 19, 371–376 [DOI] [PubMed] [Google Scholar]
- 5.Izumiya Y., Bina H. A., Ouchi N., Akasaki Y., Kharitonenkov A., Walsh K. (2008) FEBS Lett. 582, 3805–3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hojman P., Pedersen M., Nielsen A. R., Krogh-Madsen R., Yfanti C., Akerstrom T., Nielsen S., Pedersen B. K. (2009) Diabetes 58, 2797–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krzysik-Walker S. M., Ocón-Grove O. M., Maddineni S. R., Hendricks G. L., 3rd, Ramachandran R. (2008) Endocrinology 149, 1543–1550 [DOI] [PubMed] [Google Scholar]
- 8.Pedersen B. K., Akerström T. C., Nielsen A. R., Fischer C. P. (2007) J. Appl. Physiol. 103, 1093–1098 [DOI] [PubMed] [Google Scholar]
- 9.Collins C. A. (2006) Curr. Opin. Pharmacol. 6, 301–306 [DOI] [PubMed] [Google Scholar]
- 10.Shi X., Garry D. J. (2006) Genes Dev. 20, 1692–1708 [DOI] [PubMed] [Google Scholar]
- 11.Chargé S. B., Rudnicki M. A. (2004) Physiol. Rev. 84, 209–238 [DOI] [PubMed] [Google Scholar]
- 12.Yan Z., Choi S., Liu X., Zhang M., Schageman J. J., Lee S. Y., Hart R., Lin L., Thurmond F. A., Williams R. S. (2003) J. Biol. Chem. 278, 8826–8836 [DOI] [PubMed] [Google Scholar]
- 13.Brack A. S., Rando T. A. (2007) Stem Cell Rev 3, 226–237 [DOI] [PubMed] [Google Scholar]
- 14.Gopinath S. D., Rando T. A. (2008) Aging Cell 7, 590–598 [DOI] [PubMed] [Google Scholar]
- 15.Machida S., Narusawa M. (2006) Ann. N.Y. Acad. Sci. 1067, 349–353 [DOI] [PubMed] [Google Scholar]
- 16.Bodine S. C., Stitt T. N., Gonzalez M., Kline W. O., Stover G. L., Bauerlein R., Zlotchenko E., Scrimgeour A., Lawrence J. C., Glass D. J., Yancopoulos G. D. (2001) Nat. Cell Biol. 3, 1014–1019 [DOI] [PubMed] [Google Scholar]
- 17.Rommel C., Bodine S. C., Clarke B. A., Rossman R., Nunez L., Stitt T. N., Yancopoulos G. D., Glass D. J. (2001) Nat. Cell Biol. 3, 1009–1013 [DOI] [PubMed] [Google Scholar]
- 18.MacGregor J., Parkhouse W. S. (1996) Can J. Appl. Physiol. 21, 236–250 [DOI] [PubMed] [Google Scholar]
- 19.Peter A. K., Ko C. Y., Kim M. H., Hsu N., Ouchi N., Rhie S., Izumiya Y., Zeng L., Walsh K., Crosbie R. H. (2009) Hum Mol. Genet. 18, 318–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujio Y., Guo K., Mano T., Mitsuuchi Y., Testa J. R., Walsh K. (1999) Mol. Cell. Biol. 19, 5073–5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izumiya Y., Hopkins T., Morris C., Sato K., Zeng L., Viereck J., Hamilton J. A., Ouchi N., LeBrasseur N. K., Walsh K. (2008) Cell Metab. 7, 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sternberg E. A., Spizz G., Perry W. M., Vizard D., Weil T., Olson E. N. (1988) Mol. Cell. Biol. 8, 2896–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garry D. J., Meeson A., Elterman J., Zhao Y., Yang P., Bassel-Duby R., Williams R. S. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5416–5421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schiekofer S., Belisle K., Galasso G., Schneider J. G., Boehm B. O., Burster T., Schmitz G., Walsh K. (2008) Angiogenesis 11, 289–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schiekofer S., Shiojima I., Sato K., Galasso G., Oshima Y., Walsh K. (2006) Physiol. Genomics 27, 156–170 [DOI] [PubMed] [Google Scholar]
- 26.Ozden S., Huerre M., Riviere J. P., Coffey L. L., Afonso P. V., Mouly V., de Monredon J., Roger J. C., El Amrani M., Yvin J. L., Jaffar M. C., Frenkiel M. P., Sourisseau M., Schwartz O., Butler-Browne G., Desprès P., Gessain A., Ceccaldi P. E. (2007) PLoS ONE 2, e527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu C., Walker W. H., Sun J., Weisz O. A., Gibbs R. B., Witchel S. F., Sperling M. A., Menon R. K. (2006) Endocrinology 147, 5611–5623 [DOI] [PubMed] [Google Scholar]
- 28.Shibata R., Sato K., Kumada M., Izumiya Y., Sonoda M., Kihara S., Ouchi N., Walsh K. (2007) Cardiovasc. Res. 74, 471–479 [DOI] [PubMed] [Google Scholar]
- 29.Goetsch S. C., Hawke T. J., Gallardo T. D., Richardson J. A., Garry D. J. (2003) Physiol. Genomics 14, 261–271 [DOI] [PubMed] [Google Scholar]
- 30.McArdle A., Broome C. S., Kayani A. C., Tully M. D., Close G. L., Vasilaki A., Jackson M. J. (2006) Exp. Gerontol. 41, 497–500 [DOI] [PubMed] [Google Scholar]
- 31.Conboy I. M., Conboy M. J., Smythe G. M., Rando T. A. (2003) Science 302, 1575–1577 [DOI] [PubMed] [Google Scholar]
- 32.Miller J. B., Emerson C. P., Jr. (2003) Sci. Aging Knowledge Environ. pe34. [DOI] [PubMed] [Google Scholar]
- 33.Mackey A. L., Kjaer M., Charifi N., Henriksson J., Bojsen-Moller J., Holm L., Kadi F. (2009) Muscle Nerve 40, 455–465 [DOI] [PubMed] [Google Scholar]
- 34.Chen K. C., Chiou Y. L., Kao P. H., Lin S. R., Chang L. S. (2008) Toxicon 51, 624–634 [DOI] [PubMed] [Google Scholar]
- 35.Sinha-Hikim I., Braga M., Shen R., Sinha Hikim A. P. (2007) Apoptosis 12, 1965–1978 [DOI] [PubMed] [Google Scholar]
- 36.Brailoiu G. C., Dun S. L., Yin D., Yang J., Chang J. K., Dun N. J. (2005) Brain Res. 1040, 187–190 [DOI] [PubMed] [Google Scholar]
- 37.Lok S., Johnston D. S., Conklin D., Lofton-Day C. E., Adams R. L., Jelmberg A. C., Whitmore T. E., Schrader S., Griswold M. D., Jaspers S. R. (2000) Biol. Reprod. 62, 1593–1599 [DOI] [PubMed] [Google Scholar]
- 38.Burnicka-Turek O., Shirneshan K., Paprotta I., Grzmil P., Meinhardt A., Engel W., Adham I. M. (2009) Endocrinology 150, 4348–4357 [DOI] [PubMed] [Google Scholar]
- 39.Ivell R., Grutzner F. (2009) Endocrinology 150, 3986–3990 [DOI] [PubMed] [Google Scholar]
- 40.Hsu S. Y., Semyonov J., Park J. I., Chang C. L. (2005) Ann. N.Y. Acad. Sci. 1041, 520–529 [DOI] [PubMed] [Google Scholar]
- 41.Wilkinson T. N., Speed T. P., Tregear G. W., Bathgate R. A. (2005) BMC Evol. Biol. 5, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y., Negishi S., Sakamoto M., Usas A., Huard J. (2005) Ann. N.Y. Acad. Sci. 1041, 395–397 [DOI] [PubMed] [Google Scholar]
- 43.Negishi S., Li Y., Usas A., Fu F. H., Huard J. (2005) Am. J. Sports Med. 33, 1816–1824 [DOI] [PubMed] [Google Scholar]
- 44.Ten Broek R. W., Grefte S., Von den Hoff J. W. (2010) J. Cell. Physiol. 224, 7–16 [DOI] [PubMed] [Google Scholar]
- 45.Miller K. J., Thaloor D., Matteson S., Pavlath G. K. (2000) Am. J. Physiol. Cell Physiol. 278, C174–C181 [DOI] [PubMed] [Google Scholar]
- 46.Mitchell C. A., McGeachie J. K., Grounds M. D. (1996) Growth Factors 13, 37–55 [DOI] [PubMed] [Google Scholar]
- 47.Li Y., Foster W., Deasy B. M., Chan Y., Prisk V., Tang Y., Cummins J., Huard J. (2004) Am. J. Pathol. 164, 1007–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Armand A. S., Della Gaspera B., Launay T., Charbonnier F., Gallien C. L., Chanoine C. (2003) Dev. Dyn. 227, 256–265 [DOI] [PubMed] [Google Scholar]
- 49.Patel K., Amthor H. (2005) Neuromuscul. Disord. 15, 117–126 [DOI] [PubMed] [Google Scholar]
- 50.Gonzalez-Cadavid N. F., Bhasin S. (2004) Curr. Opin. Clin. Nutr. Metab. Care. 7, 451–457 [DOI] [PubMed] [Google Scholar]
- 51.Hayashi S., Aso H., Watanabe K., Nara H., Rose M. T., Ohwada S., Yamaguchi T. (2004) Histochem. Cell Biol. 122, 427–434 [DOI] [PubMed] [Google Scholar]
- 52.Allen R. E., Boxhorn L. K. (1989) J. Cell. Physiol. 138, 311–315 [DOI] [PubMed] [Google Scholar]
- 53.Kasemkijwattana C., Menetrey J., Somogyl G., Moreland M. S., Fu F. H., Buranapanitkit B., Watkins S. C., Huard J. (1998) Cell Transplant 7, 585–598 [DOI] [PubMed] [Google Scholar]
- 54.Menetrey J., Kasemkijwattana C., Day C. S., Bosch P., Vogt M., Fu F. H., Moreland M. S., Huard J. (2000) J. Bone Joint Surg. Br. 82, 131–137 [DOI] [PubMed] [Google Scholar]
- 55.Sato K., Li Y., Foster W., Fukushima K., Badlani N., Adachi N., Usas A., Fu F. H., Huard J. (2003) Muscle Nerve 28, 365–372 [DOI] [PubMed] [Google Scholar]
- 56.Musarò A., McCullagh K., Paul A., Houghton L., Dobrowolny G., Molinaro M., Barton E. R., Sweeney H. L., Rosenthal N. (2001) Nat. Genet. 27, 195–200 [DOI] [PubMed] [Google Scholar]
- 57.Coleman M. E., DeMayo F., Yin K. C., Lee H. M., Geske R., Montgomery C., Schwartz R. J. (1995) J. Biol. Chem. 270, 12109–12116 [DOI] [PubMed] [Google Scholar]
- 58.Adams G. R., McCue S. A. (1998) J. Appl. Physiol. 84, 1716–1722 [DOI] [PubMed] [Google Scholar]
- 59.Bathgate R. A., Ivell R., Sanborn B. M., Sherwood O. D., Summers R. J. (2005) Ann. N.Y. Acad. Sci. 1041, 61–76 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.