Abstract
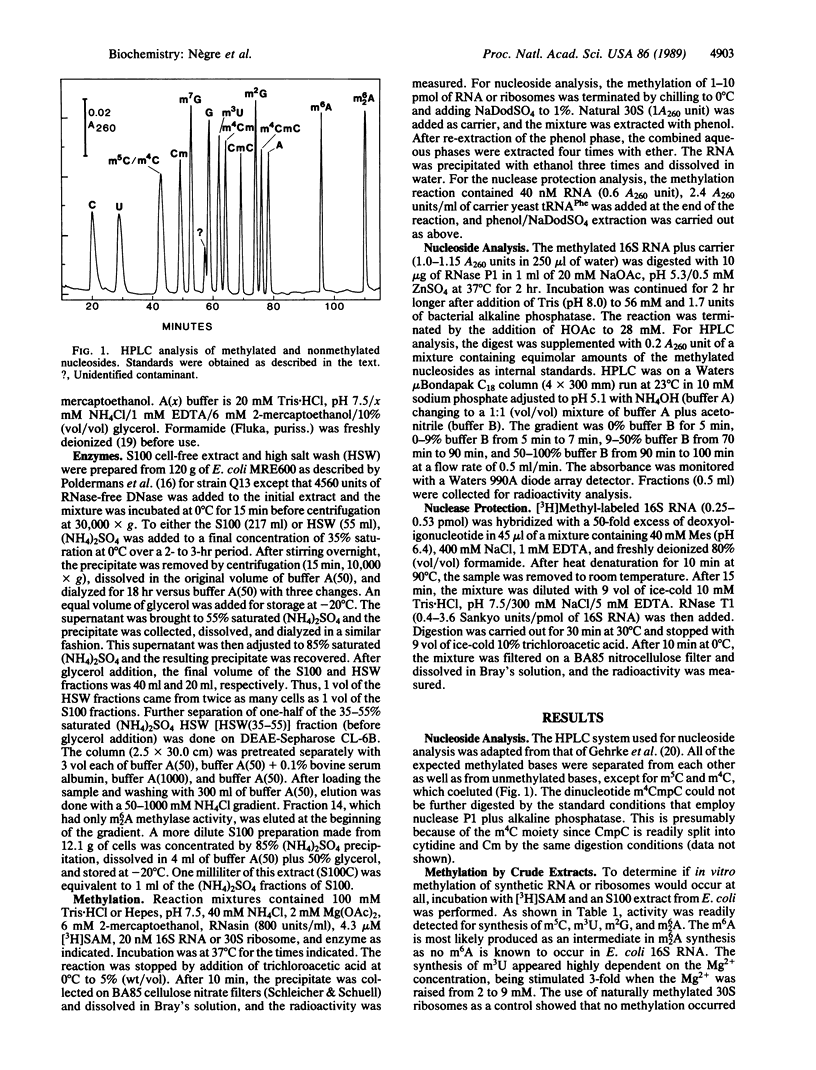
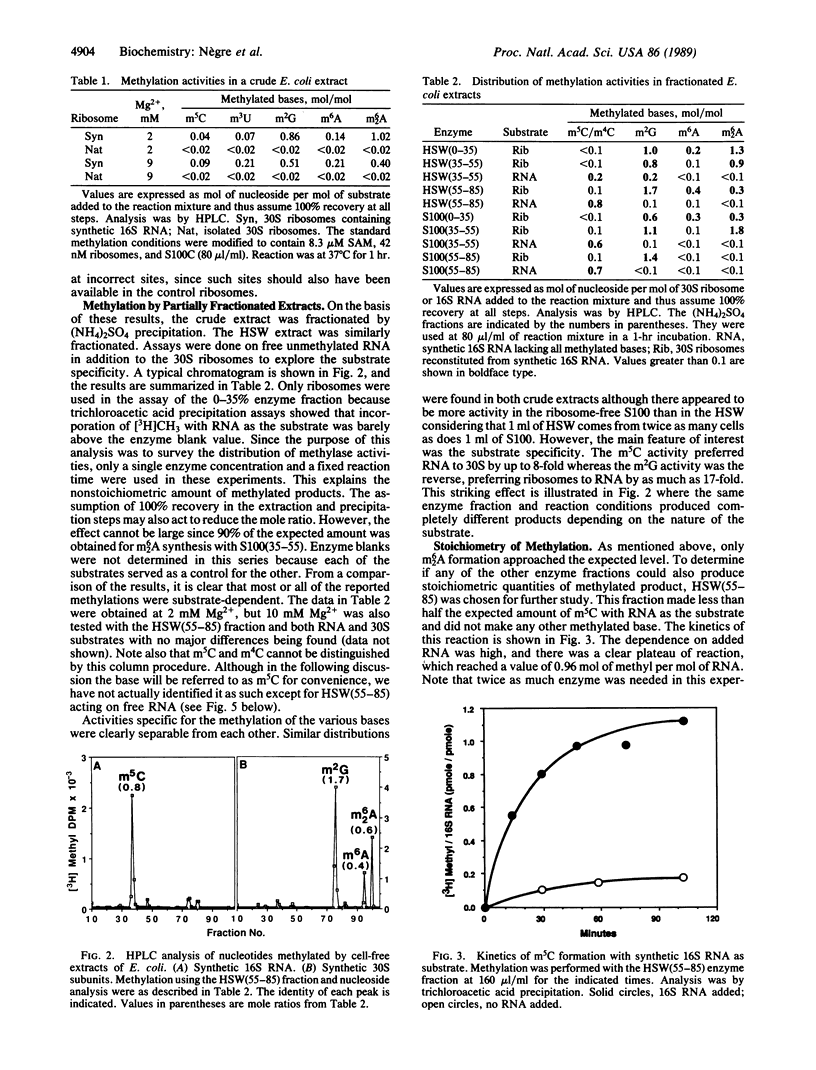
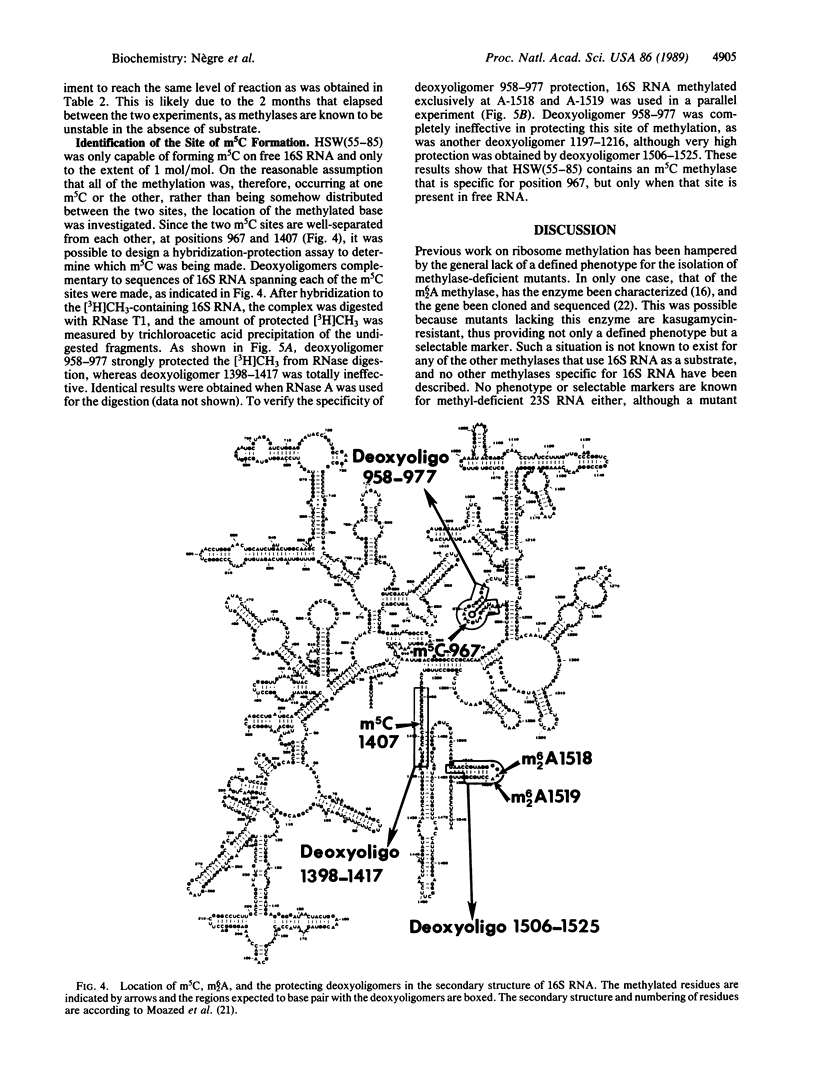
Treatment of synthetic 30S particles lacking all of the normally methylated nucleotides with S-adenosyl-[3H]methionine and either an S100 or ribosomal high salt wash extract resulted in ribosome-dependent incorporation of [3H]methyl groups into trichloroacetic acid-insoluble material. No incorporation was observed when naturally methylated isolated 30S particles were used, showing that methylation at unnatural sites did not occur. Enzymatic hydrolysis of the labeled RNA to nucleosides followed by HPLC analysis identified the [3H]methylated residues. Activities for the formation of N6-methyladenosine, N6-dimethyladenosine, 5-methylcytidine (m5C), 3-methyluridine, and N2-methylguanosine were found. Fractionation by ammonium sulfate partially resolved the different activities. All of the fractions with m5C activity were 6-8 times more active on synthetic unmethylated 16S RNA than on synthetic 30S ribosomes, whereas the N2-methylguanosine activity preferred 30S ribosomes to 16S RNA by a factor of more than 10. The N6-methyladenosine and N6-dimethyladenosine activities were 30S ribosome-specific. The m5C activity present in the 55-85% ammonium sulfate fraction of the high salt wash yielded a maximum of 1.0 mol of m5C per mol of 16S RNA, although two m5C residues, positions 967 and 1407, are found in vivo. RNase protection by hybridization with the appropriate oligodeoxynucleotide identified the methylated residue as C-967. Methylation of m5C-967 did not require prior methylation of G-966, and methylation of A-1518 and A-1519 was not dependent on prior methylation of G-1516.
Full text
PDF
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Björk G. R., Isaksson L. A. Isolation of mutants of Escherichia coli lac king 5-methyluracil in transfer ribonucleic acid or 1-methylguanine in ribosomal RNA. J Mol Biol. 1970 Jul 14;51(1):83–100. doi: 10.1016/0022-2836(70)90272-x. [DOI] [PubMed] [Google Scholar]
- Björk G. R., Kjellin-Stråby K. General screening procedure for RNA modificationless mutants: isolation of Escherichia coli strains with specific defects in RNA methylation. J Bacteriol. 1978 Feb;133(2):499–507. doi: 10.1128/jb.133.2.499-507.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelbi-Alix M. K., Expert-Bezançon A., Hayes F., Alix J. H., Branlant C. Properties of ribosomes and ribosomal RNAs synthesized by Escherichia coli grown in the presence of ethionine. Normal maturation of ribosomal RNA in the absence of methylation. Eur J Biochem. 1981 Apr;115(3):627–634. doi: 10.1111/j.1432-1033.1981.tb06248.x. [DOI] [PubMed] [Google Scholar]
- Cunningham P. R., Negre D., Weitzmann C., Denman R., Nurse K., Ofengand J. The role of 16S RNA in ribosome function: single base alterations and their effect on in vitro protein synthesis. Arch Biol Med Exp (Santiago) 1988 Dec;21(3-4):393–401. [PubMed] [Google Scholar]
- Denman R., Nègre D., Cunningham P. R., Nurse K., Colgan J., Weitzmann C., Ofengand J. Effect of point mutations in the decoding site (C1400) region of 16S ribosomal RNA on the ability of ribosomes to carry out individual steps of protein synthesis. Biochemistry. 1989 Feb 7;28(3):1012–1019. doi: 10.1021/bi00429a014. [DOI] [PubMed] [Google Scholar]
- Denman R., Weitzmann C., Cunningham P. R., Nègre D., Nurse K., Colgan J., Pan Y. C., Miedel M., Ofengand J. In vitro assembly of 30S and 70S bacterial ribosomes from 16S RNA containing single base substitutions, insertions, and deletions around the decoding site (C1400). Biochemistry. 1989 Feb 7;28(3):1002–1011. doi: 10.1021/bi00429a013. [DOI] [PubMed] [Google Scholar]
- Feunteun J., Rosset R., Ehresmann C., Stiegler P., Fellner P. Abnormal maturation of precursor 16S RNA in a ribosomal assembly defective mutant of E. coli. Nucleic Acids Res. 1974 Jan;1(1):141–147. doi: 10.1093/nar/1.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helser T. L., Davies J. E., Dahlberg J. E. Mechanism of kasugamycin resistance in Escherichia coli. Nat New Biol. 1972 Jan 5;235(53):6–9. doi: 10.1038/newbio235006a0. [DOI] [PubMed] [Google Scholar]
- Heus H. A., van Kimmenade J. M., van Knippenberg P. H., Haasnoot C. A., de Bruin S. H., Hilbers C. W. High-resolution proton magnetic resonance studies of the 3'-terminal colicin fragment of 16 S ribosomal RNA from Escherichia coli. Assignment of iminoproton resonances by nuclear Overhauser effect experiments and the influence of adenine dimethylation on the hairpin conformation. J Mol Biol. 1983 Nov 15;170(4):939–956. doi: 10.1016/s0022-2836(83)80197-1. [DOI] [PubMed] [Google Scholar]
- Hsuchen C. C., Dubin D. T. Methylation patterns of mycoplasma transfer and ribosomal ribonucleic acid. J Bacteriol. 1980 Dec;144(3):991–998. doi: 10.1128/jb.144.3.991-998.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaksson L. A. Partial purification of ribosomal RNA(m1G)- and rRNA(m2G)-methylases from Escherichia coli and demonstration of some proteins affecting their apparent activity. Biochim Biophys Acta. 1973 Jun 8;312(1):122–133. doi: 10.1016/0005-2787(73)90057-9. [DOI] [PubMed] [Google Scholar]
- Krzyzosiak W., Denman R., Nurse K., Hellmann W., Boublik M., Gehrke C. W., Agris P. F., Ofengand J. In vitro synthesis of 16S ribosomal RNA containing single base changes and assembly into a functional 30S ribosome. Biochemistry. 1987 Apr 21;26(8):2353–2364. doi: 10.1021/bi00382a042. [DOI] [PubMed] [Google Scholar]
- Melançon P., Gravel M., Boileau G., Brakier-Gingras L. Reassembly of active 30S ribosomal subunits with an unmethylated in vitro transcribed 16S rRNA. Biochem Cell Biol. 1987 Dec;65(12):1022–1030. doi: 10.1139/o87-134. [DOI] [PubMed] [Google Scholar]
- Moazed D., Stern S., Noller H. F. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J Mol Biol. 1986 Feb 5;187(3):399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- Narayan P., Rottman F. M. An in vitro system for accurate methylation of internal adenosine residues in messenger RNA. Science. 1988 Nov 25;242(4882):1159–1162. doi: 10.1126/science.3187541. [DOI] [PubMed] [Google Scholar]
- Noller H. F. Structure of ribosomal RNA. Annu Rev Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- Poldermans B., Bakker H., Van Knippenberg P. H. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3' end of 16S ribosomal RNA of Escherichia coli. IV. The effect of the methylgroups on ribosomal subunit interaction. Nucleic Acids Res. 1980 Jan 11;8(1):143–151. doi: 10.1093/nar/8.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldermans B., Roza L., Van Knippenberg P. H. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3' end of 16 S ribosomal RNA of Escherichia coli. III. Purification and properties of the methylating enzyme and methylase-30 S interactions. J Biol Chem. 1979 Sep 25;254(18):9094–9100. [PubMed] [Google Scholar]
- Poldermans B., Van Buul C. P., Van Knippenberg P. H. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3' end of 16 S ribosomal RNA of Escherichia coli. II. The effect of the absence of the methyl groups on initiation of protein biosynthesis. J Biol Chem. 1979 Sep 25;254(18):9090–9093. [PubMed] [Google Scholar]
- Shapiro S. K., Ehninger D. J. Methods for the analysis and preparation of adenosylmethionine and adenosylhomocysteine. Anal Biochem. 1966 May;15(2):323–333. doi: 10.1016/0003-2697(66)90038-8. [DOI] [PubMed] [Google Scholar]
- van Buul C. P., Visser W., van Knippenberg P. H. Increased translational fidelity caused by the antibiotic kasugamycin and ribosomal ambiguity in mutants harbouring the ksgA gene. FEBS Lett. 1984 Nov 5;177(1):119–124. doi: 10.1016/0014-5793(84)80994-1. [DOI] [PubMed] [Google Scholar]
- van Buul C. P., van Knippenberg P. H. Nucleotide sequence of the ksgA gene of Escherichia coli: comparison of methyltransferases effecting dimethylation of adenosine in ribosomal RNA. Gene. 1985;38(1-3):65–72. doi: 10.1016/0378-1119(85)90204-5. [DOI] [PubMed] [Google Scholar]


