Abstract
Hereditary cancer syndromes provide powerful insights into dysfunctional signaling pathways that lead to sporadic cancers. Beckwith-Wiedemann syndrome (BWS) is a hereditary human cancer stem cell syndrome currently linked to deregulated imprinting at chromosome 11p15 and uniparental disomy. However, causal molecular defects and genetic models have remained elusive to date in the majority of cases. The non-pleckstrin homology domain β-spectrin (β2SP) (the official name for human is Spectrin, beta, nonerythrocytic 1 (SPTBN1), isoform 2; the official name for mouse is Spectrin beta 2 (Spnb2), isoform 2), a scaffolding protein, functions as a potent TGF-β signaling member adaptor in tumor suppression and development. Yet, the role of the β2SP in human tumor syndromes remains unclear. Here, we report that β2SP+/− mice are born with many phenotypic characteristics observed in BWS patients, suggesting that β2SP mutant mice phenocopy BWS, and β2SP loss could be one of the mechanisms associated with BWS. Our results also suggest that epigenetic silencing of β2SP is a new potential causal factor in human BWS patients. Furthermore, β2SP+/− mice provide an important animal model for BWS, as well as sporadic cancers associated with it, including lethal gastrointestinal and pancreatic cancer. Thus, these studies could lead to further insight into defects generated by dysfunctional stem cells and identification of new treatment strategies and functional markers for the early detection of these lethal cancers that otherwise cannot be detected at an early stage.
Keywords: DNA Methylation, Epigenetics, Insulin-like Growth Factor (IGF), Stem Cell, Transforming Growth Factor β (TGF-β), Beckwith-Wiedemann Syndrome, SPTBN1, Gastrointestinal and Hepatocellular Cancer, Spectrin, ß2SP
Introduction
Hereditary cancer syndromes provide powerful insights into our understanding of somatic mutations present in sporadic cancers, as well as implicated cell signaling pathways (1–5). One clear example is the identification of germ line, inactivating mutations in the adenomatous polyposis coli gene, that encodes a 300-kDa wnt pathway adaptor protein (4). Although germ line mutations in adenomatous polyposis coli are responsible for familial adenomatous polyposis, a rare condition affecting about 1 in 7000 individuals in the United States, somatic mutations in the adenomatous polyposis coli gene are present in more than 70% of colonic adenomatous polyps and carcinomas (5). Beckwith-Wiedemann syndrome (BWS)2 is a hereditary stem cell cancer syndrome currently linked to deregulation of an imprinted cluster on human chromosome 11p15 (6–8). Yet, causal molecular defects and genetic models of this overgrowth syndrome have remained elusive to date in many cases. BWS is associated with an 800-fold increased risk of embryonal neoplasms of childhood and to a lesser extent, hepatocellular carcinoma and renal cell carcinomas. 85% of BWS cases are sporadic, whereas 15% are familial and exhibit an autosomal dominant pattern of inheritance with linkage to chromosome 11p15 (9). Tumor risk estimates vary between 4 and 21% in affected individuals (9). BWS has an incidence of 1/6,000–10,000 births in the United States and a prevalence of 0.07/1,000 births (10). Surprisingly, a 4–9-fold increase in incidence has been recently observed in offspring that result from in vitro fertilization (11, 12).
Molecular defects underlying BWS are only partially understood. Several lines of study suggest that different or sometimes overlapping molecular errors may play a causative role in this disease. Paternal uniparental disomy (UPD, where both homologs of a chromosome pair are inherited exclusively from one parent, resulting in either overexpression or absence of a parent-specific transcript) and loss of imprinting at the insulin-like growth factor 2 (IGF2) gene locus on chromosome 11 associated with overexpression of IGF2, occur in 20 and 10% of cases, respectively. Moreover, decreased expression from mutations of cyclin-dependent kinase inhibitor 1C (CDKN1C or p57Kip2) or from loss of maternal methylation of potassium voltage-gated channel (KCNQ1, previously known as KvLQT1) overlapping transcript 1 (KCNQ1OT1), a non-protein-coding antisense RNA that regulates CDKN1C imprinting has also been reported (8, 13, 14). Furthermore, germ line mutations (homozygous frameshift) of NLRP2 (a member of the Nucleotide-binding oligomerization domain, Leucine-rich Repeat and Pyrin domain family) are also associated with BWS (15). In addition, the transcriptional insulator CCCTC-binding factor, a highly conserved zinc finger protein, has been implicated in BWS and has diverse regulatory functions, including transcriptional activation/repression, insulation, imprinting, and X chromosome inactivation (16, 17). CCCTC-binding factor interacts with itself or chromodomain helicase DNA-binding protein 8, forming “active chromatin hubs” mediating long range chromatin interactions between multiple loci such as the IGF2/H19 gene locus which is associated with BWS (8, 18). Despite these advances in the field, no clear mouse model with cancer development has emerged for BWS to date.
The non-pleckstrin homology (PH) domain β-spectrin (β2SP) (official name for human is Spectrin, beta, nonerythrocytic 1 (SPTBN1), isoform 2; official name for mouse is Spectrin beta 2 (Spnb2), isoform 2; also known as embryonic liver fodrin isoform; human gene ID, 6711; mouse gene ID, 20742/OMIM ID 182790), a TGF-β/Smad3/4 adaptor protein, is a potent suppressor of tumorigenesis, but the role of the β2SP in human gastrointestinal tumor syndromes remains unclear (2, 19–21). In the present study, we report a serendipitous observation that β2SP+/− mice are born with many phenotypic characteristics observed in BWS patients. These include dramatic visceromegaly, macroglossia, abnormal ear folds, midface dysmorphology, followed in later months by the development of multiple cancers, including carcinomas of the gastrointestinal tract (liver, stomach, intestine, and pancreas), as well as renal and adrenal adenocarcinomas. Therefore, epigenetic regulation of β2SP expression in human BWS, and the β2SP+/− mouse phenotype could offer valuable models for further genetic studies of this disease, as well as sporadic cancers associated with this stem cell disorder.
EXPERIMENTAL PROCEDURES
Human BWS Tissues and Primary Human BWS Cell Lines
Seven BWS tissues from BWS patients were diagnosed based upon three major criteria (8) (supplemental Fig. 1). Seven human BWS cell lines were developed by Dr. Weksberg (Ontario, Canada) with institutional review board and research ethics approval. Cells were cultured in MEM-α with 10% FBS. The cell lines were named for the molecular abnormality identified (UPD, KvDMR-loss of methylation in KCNQ1OT1 region or CDKN1C-mutation; Plus (“+”) shows molecular defect, and minus (“−”) shows absence of defect). In addition, cell lines were given a tumor (T) designation if the patient had a tumor or a “no tumor” (NT) designation if no tumor has been detected. Cell lines used in this study were: CDKN1C+NT (referred to as BWSC-1); KvDMR-NT (referred to as BWSC-2); KvDMR+T, hepatoblastoma (referred as BWSC-3); UPD+NT (referred to as BWSC-4); UPD+T, hepatoblastoma (referred to as BWSC-5); two tongue tissue (UPD+) cell lines derived from the same case with biopsies at separate sites (8369F referred to as BWSC-6 and 8370F referred to as BWSC-7). BWSC-2 and BWSC-3 were derived from normal monozygotic twin (absence of KvDMR molecular defect but it had some clinical signs of BWS) and BWS monozygotic twin with KvDMR molecular defect, respectively. Normal human hepatocytes were from the Liver Tissue Cell Distribution System, University of Pittsburgh and University of Minnesota.
Plasmids and Antibodies
The cDNA sequence of β2SP was amplified by gene-specific primers and inserted into pcDNA3.1/V5-His-TOPO (Invitrogen) for transfection studies. Rabbit anti-β2SP-N is an epitope-specific antibody recognizing the β2SP N terminus (36 amino acids). The mouse growth hormone (GH) ELISA kit was from Millipore.
Bisulfite Sequencing and DNA Methylation Analysis
Genomic DNA was bisulfite-modified with an EpiTect Bisulfite kit (Qiagen) according to the manufacturer's protocols. Prediction of CpG islands in the β2SP promoter and primer design for methylation-specific PCR used urogene software. DNA methylation analysis was by a web tool from RIKEN. Primer pairs (Pms-1) used for BWS cell lines and BWS nontumor tissues methylation-specific PCR and bisulfite sequencing were methylated forward, 5′-CGG TGT TTT TAT AAA TTT TTT TTG CGT C-3′ and reverse, 5′-AAT TCC ATT ATA CCC GAC GTA ACG C-3′; and unmethylated forward, 5′-TTG GTG TTT TTA TAA ATT TTT TTT GTG TTG A-3′ and reverse, 5′-CAA TTC CAT TAT ACC CAA CAT AAC ACC C-3′. Primer Pairs (Pms-2) used for formaldehyde-fixed paraffin-embedded BWS tumor tissues were methylated forward, 5′-TAG TTT TGT TTG GGA AGG TAT TAT C-3′ and reverse, 5′-TAT AAT TTT ATC AAA AAC CAC TCG C −3′ and unmethylated forward, 5′-GTA GTT TTG TTT GGG AAG GTA TTA TTG-3′ and reverse, 5′-TTA TAA TTT TAT CAA AAA CCA CTC ACC-3′.
Generation of β2SP+/− Mice and Genotype Analysis
A total of 29 β2SP+/− mice were generated from β2SP+/− mice intercrossed with β2SP+/− mice. Genotypes were determined by Southern blotting or PCR. All animal procedures were approved by the Institutional Animal Care and Use Committee of Georgetown University Medical Center, Washington, D. C.
Statistics
Statistical analysis was performed by one-way analysis of variance (ANOVA) and unpaired Student's t test using the INSTAT 3.00 package (GraphPad, San Diego, CA).
RESULTS
Phenotype and Cancer Development in β2SP+/− Heterozygote Mice
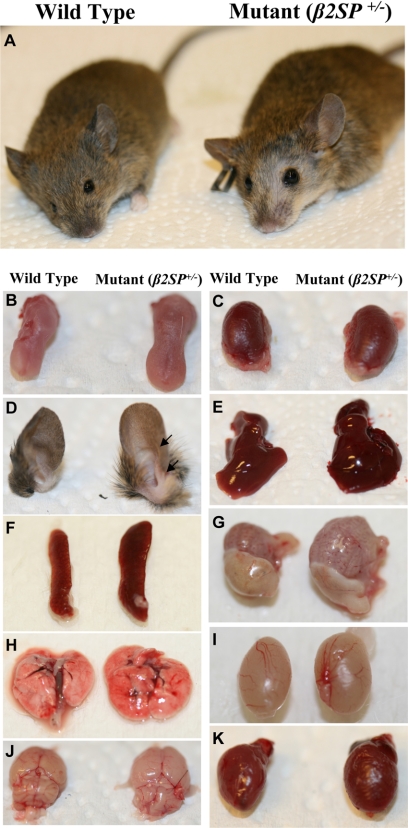
Mice with homozygous deletion of β2SP (β2SP−/−) die during midgestation (21). Analysis of 29 β2SP+/− heterozygous mice revealed a 25% increase (35.65 ± 5.72g versus 44.55 ± 8.30g; p < 0.01) in the average body size and mass compared with wild type mice. β2SP+/− heterozygous mice liver increased 23% (1.32 ± 0.19g versus 1.62 ± 0.29, p < 0.1). This was accompanied by macroglossia, hyperplasia, multiple ear folds, frontal balding, increased incidence of sudden death in the male mutant mice, visceromegaly with multilobed livers, cardiomegaly, renal hypertrophy, and testicular enlargement (Fig. 1). The phenotypic resemblance between the β2SP+/− heterozygote mice and BWS patients is considerable (Table 1). In BWS, postnatal gigantism (height more than 2 S.D. above normal) is observed in 45% of cases. Macroglossia occurs in 92% of patients with BWS. Comparing β2SP+/− heterozygote mice with other BWS mice models, Table 1 indicates that those mice (IGF2R−/−, IGF-II overexpression, p57−/−, and GPC3−/−) overlap with many of the clinical features of BWS, but a prominent BWS feature, tumor formation, was absent.
FIGURE 1.
Wild type versus β2SP+/− BWS-like phenotype. A–D, gross comparison of wild type (A, left) versus mutant β2SP+/− (A, right) mouse, wild type (B, left) versus β2SP+/− (B, right) tongue, wild type (C, left) versus β2SP+/− (C, right) kidney, wild type (D, left) versus β2SP+/− (D, right) ear. E, wild type (left) versus β2SP+/− (right) liver. F, wild type (left) versus β2SP+/− (right) spleen. G, wild type (left) versus β2SP+/− (right) stomach. H, wild type (left) versus β2SP+/− (right) lung. I, wild type (left) versus β2SP+/− (right) testis. J, wild type (left) versus β2SP+/− (right) brain. K, wild type (left) versus β2SP+/− (right) heart.
TABLE 1.
Comparison of mouse phenotypes with human Beckwith-Wiedemann syndrome
Data were extracted from the following original reports or reviews: (36, 37, 39, 40, 44–49). +, observed commonly; +/−, less commonly observed; −, not seen at all.
| Manifestation | BWS | β2SP+/− | IGF2R−/− | IGF-II overexpression | p57−/− | GPC3−/− |
|---|---|---|---|---|---|---|
| Overgrowth | + | + | + | + | − | + |
| Increased circulating IGF-II | ND | ND | + | − | ND | − |
| Perinatal death | +/− | +/− | + | + | + | + |
| Kidney dysplasia | +/− | +/− | − | − | + | + |
| Thymoma | +/− | + | − | − | − | − |
| Breast adenomas | +/− | + | − | − | − | + |
| Adrenocortical carcinomas | + | + | − | − | − | − |
| Renal cell carcinomas | +/− | + | − | − | − | − |
| Optic nerve gliomas | +/− | + | − | − | − | − |
| Hepatocellular carcinomas | +/− | + | − | − | − | − |
| Thyroid carcinomas | +/− | + | − | − | − | − |
| Pancreatic carcinomas | + | + | − | − | − | − |
| Adrenal cysts | + | +/− | − | − | − | − |
| Lymphomas | +/− | + | − | − | − | − |
a ND, not studied.
Abnormalities including those of the ear are present in greater than 50% of BWS cases. Visceromegaly due to cellular hyperplasia of livers, kidneys, and pancreas occurs in a majority of cases and is sometimes accompanied by cardiomegaly (Fig. 1K). Interestingly, hyperplasia of three or more organs is associated with an increased risk of tumors (9, 22). Examination of heterozygous β2SP+/− mice revealed a 37.9% (Table 2) to 40% (23) increase in incidence of tumors of the liver. New findings, in addition to the large size, macroglossia, and multiple ear folds, include adrenal, pancreatic, kidney, ovarian, and other tumors, not previously described (Table 2). These studies suggest that loss of β2SP in the whole animal leads to multiple tumors. Because β2SP has been earlier shown to be important for the maintenance of TGF-β signaling (21), these results suggest either a universal role for this tumor suppressor pathway at key regulatory steps or in a specific cell subtype such as a stem or progenitor cell (2, 3).
TABLE 2.
Classification of tumors in β2SP+/− mutant mice
Total β2SP+/− mice = 29; total β2SP+/− mice with tumors = 13; β2SP+/− mice with multiple tumors = 5. NA, not observed or not applicable.
| Categorizies of tumors | β2SP+/− |
|
|---|---|---|
| Site | Incidence | |
| Hepatocellular carcinoma | Liver | 37.9% (11/29) |
| Hepatoblastoma | NA | NA |
| Adenocarcinoma/ lymphoma | Small bowel | 6.9% (2/29) |
| Adenocarcinoma | Lung | 6.9% (2/29) |
| Sarcoma | Abdominal/mesencymal | 3.4% (1/29) |
| Clear cell carcinoma | Kidney | 10.3% (3/29) |
| Lymphoma | Spleen | 6.9% (2/29) |
| Adenoma | Breast | 3.4% (1/29) |
| Squamous cell carcinoma | NA | NA |
| Thymoma | NA | NA |
| Carcinoma | Testis | 3.4% (1/29) |
| Glioma | NA | NA |
| Epithelial tumor | Ovary | 3.4% (1/29) |
Loss of β2SP Is Observed in Human Beckwith-Wiedemann Nontumor Tissues and Cell Lines
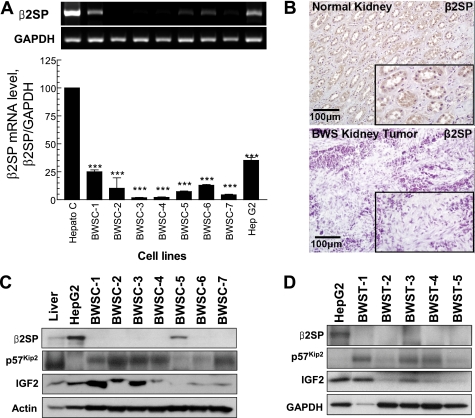
The surprising and dramatic phenotype of visceromegaly and multiple cancers led us to hypothesize that deregulation of the β2SP gene or protein may be an important effector of human BWS. We proceeded to analyze the role of β2SP as well as other molecules implicated in BWS: IGF2, IGF2R, and p57Kip2 from seven human BWS tissues and seven BWS cell lines. These human BWS cell lines displayed a significant decrease (p < 0.001) in β2SP mRNA with a 64–98% reduction compared with normal hepatocytes (Fig. 2A). Immunohistochemical labeling results revealed loss of β2SP expression in BWS kidney tumor compared with normal kidney (Fig. 2B). Western blot analysis of human BWS cell lines (BWSC) (Fig. 2C) and human BWS nontumor tissues (BWST) (Fig. 2D) shows complete loss of β2SP protein expression in tumor tissues compared with normal human liver and β2SP positive control (HepG2 cells), except for one, UPD+T, hepatoblastoma cells (BWSC-5), where β2SP protein expression is decreased. Our results indicated that a loss (or decrease) of β2SP expression occurs in nearly all BWS patients' tissues or cell lines examined, no matter what type molecular defects, even in unexplained cases of BWS, such as KvDMR-NT (BWSC-2) cell line, which is derived from a normal monozygotic twin (absence of KvDMR molecular defect, but it had some clinical signs of BWS) and a BWS monozygotic twin with KvDMR molecular defect (Fig. 2 and supplemental Fig. 1).
FIGURE 2.
Loss of β2SP protein expression in BWS. A, expression of β2SP RNA is decreased greater than ∼50% in all tested human BWS cells compared with HepG2 cells assessed by quantitative PCR analyses. B, immunohistochemical labeling of β2SP in normal and BWS kidney tumor revealed loss of β2SP expression in BWS kidney tumor compared with normal kidney. C, Western blot analysis human of BWSC demonstrates loss of β2SP expression. D, Western blot analysis of β2SP in human BWST is shown. Results shown in A reflect a mean ± S.E. from three independent experiments, performed in triplicate. ***, p < 0.001 compared with control values, determined by t test.
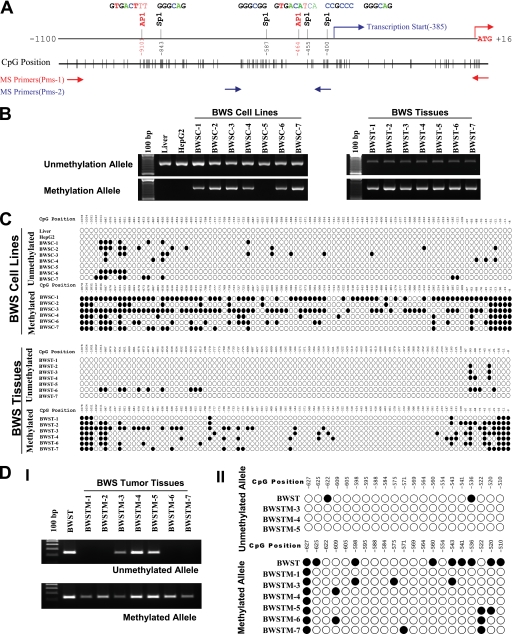
β2SP Is Silenced at Its Promoter by DNA Methylation in Human BWS Nontumor/Tumor Tissues and Cell Lines
DNA methylation patterns are often altered significantly in cancer cells including those from BWS patients. Growing evidence suggests that aberrant DNA methylation of CpG islands around promoter regions can have the same effect as coding region mutations, leading to the inactivation of tumor suppressor genes (24). Because the promoter region of β2SP contains four typical CpG islands (Fig. 3A), we examined their methylation state in genomic DNA isolated from eight cell lines (seven BWS cell lines and the β2SP-positive HepG2 cell line), seven human BWS tissues, and normal human liver tissue utilizing methylation-specific PCR (Fig. 3B) and bisulfite sequencing (Fig. 3C). These results showed a positive correlation between low levels or lack of β2SP expression and methylation in the vicinity of the β2SP promoter in human BWS cells (Fig. 2). In six human BWS cell lines we observed either loss or markedly decreased expression of β2SP (CDKN1c, KvDMR−, KvDMR+, UPD− Tongue-1(UPD) and Tongue-2(UPD)), cytosine residues of CpG dinucleotide in the β2SP promoter region (−1100 to −386) and 5′ UTR (−385 to −1) were almost completely methylated, whereas those cytosine residues in two cell lines (HepG2 and UPD+) and normal human liver, which express normal levels of β2SP, were entirely methylation-free (Fig. 3C). Thus, our results confirmed hypermethylation in this region of the β2SP promoter. To confirm epigenetic silencing of β2SP in individuals with BWS further, we analyzed BWS primary tissues (including nontumor tissues ranging from tongue to placenta with and without loss of imprinting at the IGF2 locus) and observed that loss of β2SP occurs irrespective of IGF2 loss of imprinting in all seven of the tissues examined (Fig. 3, B and C). Moreover, we also determined the methylation state of β2SP at the promoter region (−627 to −510) in genomic DNA isolated from seven formaldehyde-fixed paraffin-embedded BWS tumor tissues utilizing methylation-specific PCR (Fig. 3DI) and bisulfite sequencing (Fig. 3DII). The data demonstrate a comprehensive profile of β2SP silencing at its promoter by DNA methylation in human BWS cell lines, nontumor as well as tumor tissues.
FIGURE 3.
DNA methylation pattern of β2SP gene promoter in BWS. A, schematic representation of β2SP promoter and CpG islands. B, methylation status of the β2SP promoter in BWS cell lines and tissues detected by MS-PCR. C, DNA methylation pattern of the β2SP gene promoter in BWS cell lines and tissues identified by bisulfite sequencing. D, methylation status of the β2SP promoter in BWS tumor tissues (BWSTM) detected by MS-PCR (I) and bisulfite sequencing (II). Genomic DNA isolation from seven formaldehyde-fixed paraffin-embedded BWS tumor tissues is shown.
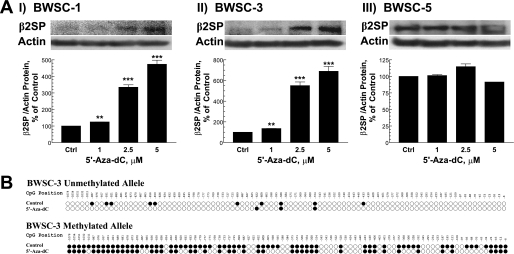
Reactivation of the β2SP Gene Expression by the DNA Methylation Inhibitor
5′-aza-2′-Deoxycytidine (5′-aza-dC), an inhibitor of DNA methylation, can reactivate gene expression when hypermethylation of CpG islands is the cause of reduced gene expression (25). To demonstrate regulation of β2SP expression by DNA methylation, three BWS cell lines (two β2SP-negative and one β2SP-positive) were treated with 5′-aza-dC for 6 days. As shown in Fig. 4A, 2.5 μm 5′-aza-dC reactivated β2SP expression in two β2SP-negative BWS cell lines. To confirm this result, we determined DNA methylation levels in β2SP-negative BWS cell line (BWS-3) with 5 μm 5′-aza-dC for 6 days. Results demonstrate significantly decreased methylation levels in both β2SP alleles compared with untreated cells (Fig. 4B). These studies further support the hypothesis that loss of β2SP is a causal event of human BWS.
FIGURE 4.
Reactivation of β2SP gene expression by DNA methylation inhibition. A, effect of 5-aza-dC on β2SP gene expression in BWS cell line by immunoblotting assay. B, DNA methylation pattern of the β2SP gene promoter in BWSC-3 treated with 5 μm 5′-aza-dC for 6 days identified by bisulfite sequencing. Results shown in A reflects a mean ± S.E. (error bars) from three independent experiments. **, p < 0.01 and ***, p < 0.001 compared with untreated (control) values determined by t test.
Increased IGF2 Expression in β2SP+/− Mice Is Similar to That Observed in Human BWS
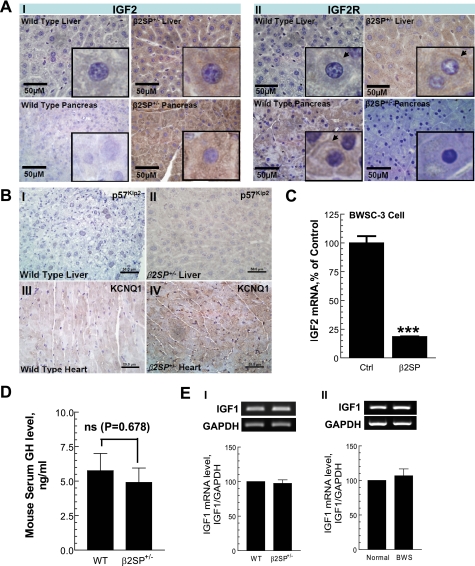
Increased IGF2 leading to BWS has been reported (8). We performed broad microarray and proteomic analyses on β2SP+/− liver tissues to determine alterations in pathways involved in BWS (supplemental Fig. 2). Microarray profiles of β2SP+/− liver tissues indicated a marked increase IGF2 expression with minor increased IGF2R or p57Kip2 levels (supplemental Fig. 2). Immunohistochemical analysis confirmed elevated expression of IGF2 in β2SP+/− liver and pancreatic tissues (Fig. 5AI). In contrast, IGF2 receptor is not activated in β2SP+/− pancreatic tissues, except for β2SP+/− liver tissue where a minor increase was noted (Fig. 5AII). p57Kip2 expression levels were slightly increased in β2SP+/− (Fig. 5B, I and II) tissues. However, KCNQ1 expression appears to be increased in β2SP+/− cardiac tissue (Fig. 5B, III and IV).
FIGURE 5.
Increased IGF2 expression in β2SP+/− mice is similar to that observed in human BWS. A, immunohistochemical analysis examining IGF2 (I) and IGF2R (II) in wild type and mutant β2SP liver and pancreas. Increased IGF2 expression in β2SP+/− tissues is shown (arrows). B, p57Kip2 expression in wild type (I) and β2SP+/− (II) mouse liver tissues assessed by immunohistochemistry. p57Kip2 expression increased in β2SP+/− liver tissues compared with wild type. Potassium voltage-gated channel (KCNQ1) expression in wild type (III) and β2SP+/− (IV) mouse heart tissues was assessed by immunohistochemistry. KCNQ1 expression increased in β2SP+/− liver tissues compared with wild type. C, BWSC-3 showing a high level of IGF2 RNA by quantitative PCR. IGF2 RNA levels decrease in cells transfected with full-length β2SP plasmid. D, GH protein level in β2SP+/− mice serum measured by ELISA. E, IGF1 mRNA level in β2SP+/− liver tissues (I) and BWS liver (II). Results shown in C reflect a mean ± S.E. (error bar) from three independent experiments. ***, p < 0.001 compared with untreated (control) values determined by t test.
We next investigated whether increased IGF2 levels in BWS cells could be secondary to loss of repression by β2SP. To determine this, β2SP was ectopically expressed in β2SP-negative BWS cell lines, and levels of IGF2 mRNA were determined. These studies revealed markedly reduced IGF2 mRNA levels (Fig. 5C) in β2SP-transfected BWS cells (a >5-fold reduction). These results indicate that β2SP is required for suppression of IGF2 signaling.
Considering that GH and IGF1 may alter in β2SP+/− mice and may be involved in overgrowth, we measured β2SP+/− the mouse serum GH protein level by ELISA (Fig. 5D) and IGF1 mRNA level by RT-PCR (Fig. 5EI). Results show that the levels of GH and IGF1 were not changed in β2SP+/− mice compared with wild type mice. We also measured the mRNA level in BWS patients' liver by RT-PCR (Fig. 5EII). Results indicated that there are no alterations between normal and BWS, which is consistent with the observation from β2SP+/− mice. These results suggest that loss of β2SP does not affect the GH and IGF1 axis.
DISCUSSION
BWS is considered to be linked to a cluster of imprinted genes at human chromosome 11p15.5, including increased IGF2, loss of H19, loss of p57Kip2, and loss of KCNQ1. The human syndrome is characterized by somatic overgrowth, macroglossia, abdominal wall defects, visceromegaly, and an increased susceptibility to a spectrum of tumors that include childhood tumors (26–32). The identification of these genes as well as others including Glypican 3 has led to the generation of multiple single/double gene mouse mutants (13, 33–37, 39–43). Yet to date, none of the mouse models develops the full panoply of symptoms and signs of BWS, particularly tumor formation (33). For instance, mutant mice with loss of KCNQ1 develop deafness, abnormalities in inner ear development, mucous neck cell hyperplasia resulting in a 3-fold increase in stomach weight, but no features of BWS, suggesting that KCNQ1 is not responsible for BWS (34). Mice with a maternally inherited deletion of H19 bear a 27% increase in birth weight, and paternal inheritance of the disruption has no effect (35). Deletion of the maternally inherited IGF2R (a maternally expressed imprinted gene in mouse but not man, partially functioning as a scavenger for IGF2, mediating its degradation) allele in mice results in increased free IGF2 available for growth factor signaling through the IGF1 receptor, with a 40% increase in birth weight, cardiac defects, and perinatal lethality (36). H19 and IGF2R double heterozygote mice overexpress 7- and 11-fold greater tissue and serum levels of IGF2 and develop a 2-fold increase in birth weight, embryonic lethality, omphalocele, visceromegaly, adrenal cysts, and cleft palate (37). Similar to H19 and IGF2R double heterozygote mice, IGF2-overexpression transgenic chimeric mice made with transgenic ES cells also leads to development of most of the BWS phenotypes, including prenatal overgrowth, polyhydramnios, fetal and neonatal lethality, disproportionate organ overgrowth such as tongue enlargement and skeletal abnormalities (13). Mice with loss-of-function mutations in p57KIP2 (a maternally expressed gene encoding a G1 cyclin-dependent kinase inhibitor) and loss of Igf2 imprinting develop placentomegaly, multitissue dysplasia, kidney dysplasia, macroglossia, cleft palate, omphalocele, and polydactyly, but not tumors (39–41). Targeted deletions of the murine homolog of glypican-3 gene (GPC3) encoding an extracellular proteoglycan believed to interact with IGF2 and/or other growth factors during development result in several features common to BWS and Simpson-Golabi-Behmel syndrome, namely overgrowth and cystic kidneys (42, 43). Although the data above clearly support the role for IGF2 as the effector for overgrowth, none of the genes appears to be the sole effector for BWS, particularly tumor formation, suggesting that other key proteins remain to be sought. Our surprising findings indicate that the β2SP mutant mice phenocopy BWS by both a genetic and biochemical basis.
Although genetic and biochemical data support the proposed β2SP protein function in human BWS, the situation in vivo is undoubtedly far more complex. Various types of imprinted gene abnormalities on the short arm of chromosome 11 are involved in the etiology of BWS, particularly in the IGF2 gene region (11p15.5). Our data point to an additional or perhaps related mechanism for deregulation of IGF2 involving β2SP. The resemblance of this mouse model to human BWS is substantial but not perfect. The tumors in human BWS are mostly of embryonal cell types, including nephroblastoma, hepatoblastoma, and pancreatoblastoma, whereas those in the mouse model are largely carcinomas or other “adult” type neoplasms. Neonatal hypoglycemia is observed in 13% of cases and has not been observed in the mutant mice. These distinctions may at least in part reflect species differences, or they may reflect the difficulties in discerning between embryonal and adult tumors in rodents, such as hepatoblastomas and hepatocellular carcinomas, and in part from the surprising discovery of the syndrome through tumors in surviving older mice in our colony.
Importantly, we report for the first time and provide strong evidence that β2SP is epigenetically silenced in individuals with BWS by examining affected nontumor tissues as well as cell lines. In addition, we observe silencing of β2SP in tissues from affected BWS patients where no loss of imprinting at the IGF2 locus is seen, further supporting the theory that loss of β2SP is one of the causal factors leading to the syndrome. Thus, mutations of β2SP in cancers could lead to new diagnostic, prognostic insights, and new therapeutic strategies into difficult to manage subsets of colorectal and hepatocellular cancers (19, 20). Furthermore, the unexpectedly high frequency of BWS in in vitro fertilization offspring is intriguing and highlights a previously undescribed role of the TGF-β pathway and new insight into epigenetic regulation of stem cells at crucial stages of differentiation and growth (11, 12, 38).
Genetic studies have revealed the importance of TGF-β signaling in mesoderm and endoderm development as well as in suppression of multiple cancers, not only gastrointestinal cancers (1, 2). It is possible that in BWS, as in sporadic hepatocellular cancer, derangement in TGF-β signaling in progenitor cells contributes to malignant transformation and eventual cancer development (2, 3). Future studies on genomic imprinting as an epigenetic mechanism controlling parental-origin-specific gene expression, and its perturbations should yield new therapeutics for cancers. Indeed insights into consequences of epigenotype switches at birth, and in BWS from β2SP as a non-pleckstrin homology domain β-spectrin and its scaffolding function may be of profound significance.
Supplementary Material
Acknowledgments
We thank Drs. Michael Zasloff, John J. Milburn Jessup, Ying Li, Young Woo Kim, Stephen Byers, Moin Ahmad, Aziza Shad, Bruce Beckwith, Said Sebti, and Anton Wellstein for critical review and helpful suggestions with the manuscript; Zhongxian Jiao, Ed Flores, Susette Mueller, and Geeta Upadhyay for excellent technical expertise.
This work was supported, in whole or in part, by National Institutes of Health Grants P01 CA130821, R01 CA106614A, and R01 CA042857 (to L. M.) and R01 DK58637 (to B. M.). This work was also supported by a Veterans Affairs merit award (to L. M.), the R. Robert and Sally D. Funderburg Research Scholar award (to L. M.), and the Benn Orr Scholar award (to L. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- BWS
- Beckwith-Wiedemann syndrome
- 5′-aza-dC
- 5′-aza-2′-deoxycytidine
- BWSC
- BWS cells
- BWST
- BWS nontumor tissue
- GH
- growth hormone
- IGF
- insulin-like growth factor
- KvDMR
- differentially methylated region (imprinting center) of KCNQ1OT1
- SPTBN1
- Spectrin beta, nonerythrocytic 1 (Homo sapiens, Gene ID: 6711, OMIM ID 182790)
- Spnb2
- Spectrin beta 2 (Mus musculus, Gene ID: 20742, MIM ID 182790)
- β2SP
- isoform 2 of SPTBN1 or Spnb2
- UPD
- uniparental disomy.
REFERENCES
- 1.Massagué J. (2008) Cell 134, 215–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mishra L., Derynck R., Mishra B. (2005) Science 310, 68–71 [DOI] [PubMed] [Google Scholar]
- 3.Tang Y., Kitisin K., Jogunoori W., Li C., Deng C. X., Mueller S. C., Ressom H. W., Rashid A., He A. R., Mendelson J. S., Jessup J. M., Shetty K., Zasloff M., Mishra B., Reddy E. P., Johnson L., Mishra L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2445–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neufeld K. L., Nix D. A., Bogerd H., Kang Y., Beckerle M. C., Cullen B. R., White R. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 12085–12090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Radtke F., Clevers H. (2005) Science 307, 1904–1909 [DOI] [PubMed] [Google Scholar]
- 6.Feinberg A. P., Ohlsson R., Henikoff S. (2006) Nat. Rev. Genet. 7, 21–33 [DOI] [PubMed] [Google Scholar]
- 7.Feinberg A. P. (2007) Nature 447, 433–440 [DOI] [PubMed] [Google Scholar]
- 8.Weksberg R., Shuman C., Smith A. C. (2005) Am. J. Med. Genet. C Semin. Med. Genet. 137C, 12–23 [DOI] [PubMed] [Google Scholar]
- 9.Lapunzina P. (2005) Am. J. Med. Genet. C Semin. Med. Genet. 137C, 53–71 [DOI] [PubMed] [Google Scholar]
- 10.Pettenati M. J., Haines J. L., Higgins R. R., Wappner R. S., Palmer C. G., Weaver D. D. (1986) Hum. Genet. 74, 143–154 [DOI] [PubMed] [Google Scholar]
- 11.Halliday J., Oke K., Breheny S., Algar E., Amor D. J. (2004) Am. J. Hum. Genet. 75, 526–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen C., Reardon W. (2005) Bjog 112, 1589–1594 [DOI] [PubMed] [Google Scholar]
- 13.Sun F. L., Dean W. L., Kelsey G., Allen N. D., Reik W. (1997) Nature 389, 809–815 [DOI] [PubMed] [Google Scholar]
- 14.Murrell A., Heeson S., Cooper W. N., Douglas E., Apostolidou S., Moore G. E., Maher E. R., Reik W. (2004) Hum. Mol. Genet. 13, 247–255 [DOI] [PubMed] [Google Scholar]
- 15.Meyer E., Lim D., Pasha S., Tee L. J., Rahman F., Yates J. R., Woods C. G., Reik W., Maher E. R. (2009) PLoS Genet. 5, e1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phillips J. E., Corces V. G. (2009) Cell 137, 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ling J. Q., Li T., Hu J. F., Vu T. H., Chen H. L., Qiu X. W., Cherry A. M., Hoffman A. R. (2006) Science 312, 269–272 [DOI] [PubMed] [Google Scholar]
- 18.Ishihara K., Oshimura M., Nakao M. (2006) Mol. Cell 23, 733–742 [DOI] [PubMed] [Google Scholar]
- 19.Tang Y., Katuri V., Srinivasan R., Fogt F., Redman R., Anand G., Said A., Fishbein T., Zasloff M., Reddy E. P., Mishra B., Mishra L. (2005) Cancer Res. 65, 4228–4237 [DOI] [PubMed] [Google Scholar]
- 20.Lin L., Amin R., Gallicano G. I., Glasgow E., Jogunoori W., Jessup J. M., Zasloff M., Marshall J. L., Shetty K., Johnson L., Mishra L., He A. R. (2009) Oncogene 28, 961–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang Y., Katuri V., Dillner A., Mishra B., Deng C. X., Mishra L. (2003) Science 299, 574–577 [DOI] [PubMed] [Google Scholar]
- 22.Enklaar T., Zabel B. U., Prawitt D. (2006) Expert Rev. Mol. Med. 8, 1–19 [DOI] [PubMed] [Google Scholar]
- 23.Kitisin K., Ganesan N., Tang Y., Jogunoori W., Volpe E. A., Kim S. S., Katuri V., Kallakury B., Pishvaian M., Albanese C., Mendelson J., Zasloff M., Rashid A., Fishbein T., Evans S. R., Sidawy A., Reddy E. P., Mishra B., Johnson L. B., Shetty K., Mishra L. (2007) Oncogene 26, 7103–7110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones P. A., Baylin S. B. (2007) Cell 128, 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cameron E. E., Bachman K. E., Myöhänen S., Herman J. G., Baylin S. B. (1999) Nat. Genet. 21, 103–107 [DOI] [PubMed] [Google Scholar]
- 26.Miles H. L., Hofman P. L., Cutfield W. S. (2005) Rev. Endocr. Metab. Disord. 6, 261–268 [DOI] [PubMed] [Google Scholar]
- 27.Spencer G. S., Schabel F., Frisch H. (1980) Arch. Dis. Child 55, 151–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiedemann H. R. (1964) J. Genet. Hum. 13, 223–232 [PubMed] [Google Scholar]
- 29.Combs J. T., Grunt J. A., Brandt I. K. (1966) N. Engl. J. Med. 275, 236–243 [DOI] [PubMed] [Google Scholar]
- 30.Fitzpatrick G. V., Soloway P. D., Higgins M. J. (2002) Nat. Genet. 32, 426–431 [DOI] [PubMed] [Google Scholar]
- 31.Luton D., Lepercq J., Sibony O., Barbet P., Le Bouc Y., Chavinié J., Lewin F. (1996) Fetal Diagn. Ther. 11, 154–158 [DOI] [PubMed] [Google Scholar]
- 32.John R. M., Ainscough J. F., Barton S. C., Surani M. A. (2001) Hum. Mol. Genet. 10, 1601–1609 [DOI] [PubMed] [Google Scholar]
- 33.Li M., Squire J. A., Weksberg R. (1998) Clin. Genet. 53, 165–170 [PubMed] [Google Scholar]
- 34.Lee M. P., Ravenel J. D., Hu R. J., Lustig L. R., Tomaselli G., Berger R. D., Brandenburg S. A., Litzi T. J., Bunton T. E., Limb C., Francis H., Gorelikow M., Gu H., Washington K., Argani P., Goldenring J. R, Coffey R. J., Feinberg A. P. (2000) J. Clin. Invest. 106, 1447–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leighton P. A., Ingram R. S., Eggenschwiler J., Efstratiadis A., Tilghman S. M. (1995) Nature 375, 34–39 [DOI] [PubMed] [Google Scholar]
- 36.Lau M. M., Stewart C. E., Liu Z., Bhatt H., Rotwein P., Stewart C. L. (1994) Genes Dev. 8, 2953–2963 [DOI] [PubMed] [Google Scholar]
- 37.Ludwig T., Eggenschwiler J., Fisher P., D'Ercole A. J., Davenport M. L., Efstratiadis A. (1996) Dev. Biol. 177, 517–535 [DOI] [PubMed] [Google Scholar]
- 38.Gosden R., Trasler J., Lucifero D., Faddy M. (2003) Lancet 361, 1975–1977 [DOI] [PubMed] [Google Scholar]
- 39.Yan Y., Frisén J., Lee M. H., Massagué J., Barbacid M. (1997) Genes Dev. 11, 973–983 [DOI] [PubMed] [Google Scholar]
- 40.Zhang P., Liégeois N. J., Wong C., Finegold M., Hou H., Thompson J. C., Silverman A., Harper J. W., DePinho R. A., Elledge S. J. (1997) Nature 387, 151–158 [DOI] [PubMed] [Google Scholar]
- 41.Caspary T., Cleary M. A., Perlman E. J., Zhang P., Elledge S. J., Tilghman S. M. (1999) Genes Dev. 13, 3115–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pilia G., Hughes-Benzie R. M., MacKenzie A., Baybayan P., Chen E. Y., Huber R., Neri G., Cao A., Forabosco A., Schlessinger D. (1996) Nat. Genet. 12, 241–247 [DOI] [PubMed] [Google Scholar]
- 43.Song H. H., Shi W., Filmus J. (1997) J. Biol. Chem. 272, 7574–7577 [DOI] [PubMed] [Google Scholar]
- 44.Eggenschwiler J., Ludwig T., Fisher P., Leighton P. A., Tilghman S. M., Efstratiadis A. (1997) Genes Dev. 11, 3128–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garganta C. L., Bodurtha J. N. (1992) Am. J. Med. Genet. 44, 129–135 [DOI] [PubMed] [Google Scholar]
- 46.Hughes-Benzie R. M., Pilia G., Xuan J. Y., Hunter A. G., Chen E., Golabi M., Hurst J. A., Kobori J., Marymee K., Pagon R. A., Punnett H. H., Schelley S., Tolmie J. L., Wohlferd M. M., Grossman T., Schlessinger D., MacKenzie A. E. (1996) Am. J. Med. Genet. 66, 227–234 [DOI] [PubMed] [Google Scholar]
- 47.Neri G., Gurrieri F., Zanni G., Lin A. (1998) Am. J. Med. Genet. 79, 279–283 [DOI] [PubMed] [Google Scholar]
- 48.Wang Z. Q., Fung M. R., Barlow D. P., Wagner E. F. (1994) Nature 372, 464–467 [DOI] [PubMed] [Google Scholar]
- 49.Weng E. Y., Moeschler J. B., Graham J. M., Jr. (1995) Am. J. Med. Genet. 56, 366–373 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.