Abstract
The multiple functions of the oncofetal protein survivin are dependent on its selective expression patterns within immunochemically distinct subcellular pools. The mechanism by which survivin localizes to these compartments, however, is only partly understood. Here we show that nuclear accumulation of survivin is promoted by CREB-binding protein (CBP)-dependent acetylation on lysine 129 (129K, Lys-129). We demonstrate a mechanism by which survivin acetylation at this position results in its homodimerization, while deacetylation promotes the formation of survivin monomers that heterodimerize with CRM1 and facilitate its nuclear export. Using proteomic analysis, we identified the oncogenic transcription factor STAT3 as a binding partner of nuclear survivin. We show that acetylated survivin binds to the N-terminal transcriptional activation domain of the STAT3 dimer and represses STAT3 transactivation of target gene promoters. Using multiplex PCR and DNA sequencing, we identified a single-nucleotide polymorphism (A → G) at Lys-129 that exists as a homozygous mutation in a neuroblastoma cell line and corresponds with a defect in survivin nuclear localization. Our results demonstrate that the dynamic equilibrium between survivin acetylation and deacetylation at amino acid 129 determines its interaction with CRM1, its subsequent subcellular localization, and its ability to inhibit STAT3 transactivation, providing a potential route for therapeutic intervention in STAT3-dependent tumors.
Keywords: Apoptosis, Nuclear Translocation, Post-translational Modification, STAT Transcription Factor, Transcription Repressor, Acetylation, Survivin
Introduction
Originally cloned as a member of the inhibitor of apoptosis (IAP)3 family (1) then shown to also associate with chromosomal passenger proteins (2), survivin plays a pivotal role in normal embryogenesis (2) and in the maintenance of both normal progenitor (3, 4) and cancer cells (5). These dual functions depend on protein existence within immunochemically distinct pools located within the mitochondria, cytoplasm, and nucleus (5, 6). In the mitochondria and cytoplasm (7), survivin inhibits caspase-dependent cell death (8). In the nucleus, survivin associates with the mitotic apparatus to regulate chromosomal migration and cytokinesis (9). In clinical studies, nuclear or cytoplasmic expression of survivin in tumor cells differentially correlates with patient outcome (10), indicating that regulation of nuclear-cytoplasmic shuttling may be an important factor in disease and/or in response to therapy. The export of survivin from the nucleus is dependent on its binding to the CRM1 export receptor that binds nuclear export signal (NES)-containing proteins (11). Structural analyses showed the CRM1 NES to be located within the survivin homodimerization domain, which is only accessible to CRM1 when survivin is in its monomeric state (12). To further elucidate its movement between these compartments, we determined the biochemical requirements of survivin for binding to CRM1 as a monomer.
An abundance of lysines at the C-terminal end of the survivin protein that create a basic pocket for potential protein-protein interactions (13, 14) suggested to us that survivin may undergo reversible post-translational modification at these residues to regulate its movement within the cell and contribute to its diverse functional repertoire. Given its basic overall structural similarity to another oncogenic protein, signal transducer and activator of transcription 3 (STAT3) (15), and the known mechanism of lysine acetylation to facilitate STAT3 nuclear-cytoplasmic shuttling (16), we hypothesized that acetylation of one or more of the survivin lysine residues might similarly aid in its nuclear transport. Here, we report that survivin is acetylated on multiple lysine residues by the histone acetyltransferase (HAT) CREB-binding protein (CBP) and that a single lysine residue (Lys-129) directs its localization to the nucleus by enhancing survivin homodimerization and thereby inhibiting CRM1-mediated nuclear export. Liquid chromatography-tandem mass spectrometry identified STAT3 as a binding partner of nuclear survivin and transcriptional assays showed that the STAT3/survivin complex inhibits STAT3 transactivation. Nuclear survivin negatively regulates STAT3 transcriptional activity in cancer cells and may represent a novel route for targeting STAT3-activated tumors.
EXPERIMENTAL PROCEDURES
Cells and Culture
HEK293T, HeLa and MCF7 cells were purchased from the American Type Culture Collection (ATCC). Coriell lymphoblasts were obtained from the Coriell Insitute for Medical Research. The NB7, NB8 and NB10 neuroblastoma cell lines were a kind gift from St. Jude Children's Research Hospital. All cell lines were grown in complete Dulbecco's Modified Eagle Medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum, penicillin and streptomycin in a 37 °C incubator maintained at 5% CO2. Human IL-6 and TSA were purchased from Sigma. For the IL-6 experiments, cells were starved for 24 h then treated with IL-6 at 100 ng/ml for 1 h. For the TSA experiments, cells were starved overnight, then treated with TSA at 5 μm for 6 h.
Plasmids and Transfections
6× MYC-survivin (17) was a gift from the laboratory of H. Cheung. HA-P300, HA-CBP, IRF-1, 6× MYC-Stat3, Flag-Stat3, Flag-Stat3 1–320, Flag-Stat3 1–465, Flag-Stat3 1–585, Flag-Stat3 1–688, Flag-Stat3 130–770, Flag-Stat3 320–770, Flag-Stat3 465–770, MYC-Stat3 685K, MYC-Stat3 705Y (16), Flag-survivin, eGFP-survivin constructs were generated as described previously (18). HEK293T cells were transfected with Lipofectamine 2000, according to the manufacturer's protocol (Invitrogen).
Site-directed Mutagenesis
Site-directed mutagenesis was performed with the Stratagene Quickchange II kit (Agilent Technologies) following the manufacturer's instructions.
Mass Spectrometry
To identify acetylated survivin residues, we cotransfected cDNAs encoding MYC-survivin and HA-CBP into HEK293T cells. Immunoprecipitated survivin protein from the transfectants was resolved by 12% SDS-PAGE and visualized by Coomassie Blue stain. The gel slice containing acetylated survivin was subjected to mass spectrometry analysis.
Survivin Interactomes
To identify survivin interactomes, tumor cell lysates were immunoprecipitated with a survivin or IgG-control antibody. Proteins were resolved by 12% SDS-PAGE and bands visualized by silver stain. Bands of interest were isolated and protein complexes analyzed by tandem mass spectrometry (MS/MS) using in-gel tryptic digestions of immunopurifications of the specific bait as compared with vector controls (19, 20). Tryptic peptides were resolved by RP liquid chromatography and nanoelectrospray MS/MS was performed on an ion trap mass spectrometer (LCQ DecaXP, ThermoElectron Corporation). The MS/MS-acquired data were searched against amino acid sequences in the International Protein Index human protein data base using SEQUEST sequence matching algorithm.
Immunoprecipitation and Immunoblotting
Lysates were precleared with the appropriate control IgG and protein G-agarose (EMD Chemicals) prior to incubation with primary antibodies and protein G Plus-agarose. Immunocomplexes were resolved by SDS-PAGE. For immunoblotting, samples were transferred to PVDF membranes that were then probed with the indicated antibodies. For the oligonucleotide agarose binding assay, whole cell lysates were incubated with agarose (Santa Cruz Biotechnology) and complexes analyzed by immunoblotting. For siRNA knockdown, cells were transiently transfected with siRNA specific for CBP or control siRNA (Santa Cruz Biotechnology) using Lipofectamine. Whole cell extracts were prepared at 48 h after transfection for immunoprecipitation and Western blot analysis. Immunoprecipitations were performed with mouse monoclonal anti-MYC (Santa Cruz Biotechnology) and anti-FLAG M2 antibody (Agilent Technologies). PVDF membranes were probed with rabbit anti-STAT3, mouse anti-survivin, mouse anti-MYC (Santa Cruz Biotechnology), anti-FLAG M2 antibody (Agilent Technologies), rabbit anti-GFP (Molecular Probes), and rabbit anti-acetylated lysine (Cell Signaling Technologies).
Gene Analysis
For PCR (qRT-PCR), HEK293T cells were transfected with cDNAs encoding STAT3 and CBP, with or without survivin 129K. Coriell lymphoblasts were starved overnight then treated with IL-6 for 6 h. Total RNA was extracted using the RNeasy Mini Kit (Qiagen Inc) 48 h after transfection. Two micrograms of total RNA was reverse transcribed into cDNA using the Omniscript RT kit (Qiagen). Gene expression was analyzed by RT-PCR with Power SYBR green Master Mix (Applied Biosystems) chemistry using the ABI HT7900 Sequence Detection System. Relative quantification was performed using the ΔΔCt method.
Tetra-primer PCR
Genotyping of the single nucleotide polymorphism at amino acid 129 of the canonical survivin gene product was carried out by tetra-primer ARMS PCR, performed as previously described (21). Briefly, the Takara Taq polymerase kit (Takara Bio) was used as indicated by the manufacturer's instructions with a total MgCl2 concentration of 3.5 mm. 0.1 μm of each outer primer and 1 μm of each inner primer (Table 1) were used with 3 ng/μl of genomic DNA prepared with the Qiagen Puregene Core Kit A (Qiagen). Touchdown PCR was performed for 30 s starting at an annealing temperature of 73 °C, decreasing by 1 degree for 10 cycles, followed by 25 cycles at 63 °C. 20 μl of each product was run on a 10% nondenaturing polyacrylamide gel.
Reporter Assays
HEK293T cells were transfected with a luciferase reporter construct containing 2× STAT3-binding SIE fragments derived from the promoter region of the mouse IRF1 gene, together with the indicated cDNAs, using Lipofectamine 2000. After 48 h, equal volumes of PBS and Dual-GloTM Luciferase Reagent (Promega) were added and absorbance of firefly luminescence measured. Then, Dual-GloTM Stop & Glo Reagent was added, and absorbance of Renilla luminescence measured. The experiments were performed in triplicate, for a minimum of three independent times. Two-tailed Student's t tests were performed to assess significance.
Immunofluorescence Microscopy
HeLa cells were grown to ∼50% confluence in an 8 chambered glass slide. Cells were fixed in 3.7% formaldehyde, 0.2% Triton X-100/PBS for 15 min at room temperature. Blocking was done in 1% BSA, 5% NGS/PBS for 1 h. Primary antibodies were rabbit-survivin (Santa Cruz Biotechnology), anti-STAT3 (Abnova), anti-Flag M2 (Agilent Technologies), and mouse monoclonal c-MYC (Santa Cruz Biotechnology). Secondary antibodies were anti-rabbit IgG conjugated to Dylight 488 (ThermoFisher Scientific) and goat-anti-mouse IgG conjugated to FITC (Santa Cruz Biotechnology). Slides were mounted with Prolong anti-fade reagent with DAPI (Invitrogen). Images were captured with a Nikon C1si confocal microscope.
FRET
HeLa cells seeded on a chambered slide were cotransfected with cDNAs encoding either MYC-survivin129E and Flag-survivin129E or MYC-survivin129K and Flag-survivin129K Cells were fixed and immunostained with rabbit anti-MYC and mouse anti-Flag as primary antibodies, and anti-mouse Dylight 594 and anti-rabbit Dylight 649 (Thermo Scientific) as secondary antibodies. Acceptor photobleaching was performed with a Nikon C1si confocal microscope. Green color was assigned to the 594 donor fluorescence, and red to the 649 acceptor. The donor fluorescence before and after photobleaching of the acceptor was measured, and percent efficiency of FRET was measured as E = 1 − (fluorescence before/fluorescence after).
RESULTS
Survivin Is Acetylated on Multiple Lysine Residues by CBP
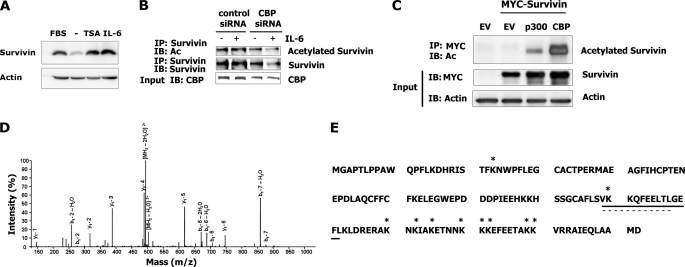
To determine whether survivin is endogenously acetylated, we treated MCF-7 or HeLa cells with the histone deacetylase (HDAC) inhibitor trichostatin A (TSA). Treatment with TSA led to an increase in survivin expression, similar to that observed after IL-6 treatment (Fig. 1A), suggesting that acetylation plays a role in survivin expression. The HAT proteins p300 and CBP acetylate numerous cancer-associated proteins, including p53, BRCA1, STAT3, and E2F/RB, to direct their function (22, 23). To determine whether CBP is responsible for survivin acetylation, we knocked down CBP with siRNA in MCF7 or HeLa cells. Down-regulation of CBP led to an inhibition of survivin acetylation (Fig. 1B), confirming CBP as an endogenous survivin acetyl-transferase. Consistently, ectopically expressed survivin in HEK293T cells was acetylated, as shown by cotransfection with CBP or p300 (Fig. 1C).
FIGURE 1.
The HAT protein CBP acetylates survivin. A, survivin expression in response to TSA in HeLa cells compared with serum alone (FBS), serum-free media (−), or following IL-6 treatment. B, survivin expression and acetylation in HeLa cells with or without CBP depletion, using a survivin or a pan-acetylation (Ac) antibody on survivin immunoprecipitants. IP, immunoprecipitation; IB, immunoblot. C, lysates from HEK293T cells transfected with cDNAs encoding a MYC-tagged survivin, p300 or CBP. Anti-MYC immunoprecipitants were analyzed with a pan-acetylation (Ac) antibody. EV, empty vector. D, 129-acetylated peptide of the survivin protein as identified by mass spectrometry. E, summary of the survivin-acetylated residues (asterisks) as identified by mass spectrometry. The solid underline denotes the location of the homodimer interface; the dashed line indicates the location of the nuclear export sequence.
To map the acetylated sites of survivin, CBP acetylated survivin proteins were immunopurified from HEK293T cell transfectants and submitted for mass spectrometric analysis. The acetylated survivin proteins were assigned discrete mass values by matching the measured masses with calculated mass values, as demonstrated for the lysine residue acetylated at amino acid 129 (Fig. 1D). The analysis revealed a total of 10 acetylated lysine residues, with the C-terminal region being the most heavily acetylated (Fig. 1E). Structural components within the C-terminal domain of the protein relevant to survivin function include a dimer interface (residues 89–102) required for survivin homodimerization (13), a nuclear export signal (residues 89–98) shown to bind the nuclear export protein CRM1 (11), and a microtubule interaction site (residues 99–142) (5). Thus, survivin is heavily acetylated in regions that are structurally accessible to protein dimerization and contain key domains required for survivin function.
dbSNP Reveals a Polymorphism at the Site Encoding Lysine at Amino Acid 129
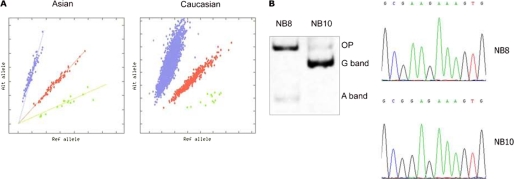
The unique distribution of the acetylated lysines within the survivin protein suggested to us that any one of these amino acid sites might be a prime target for mutational events leading to alterations in survivin activity within tumor cells. To identify genetic polymorphisms within survivin coding regions that could result in structural changes in the protein, we consulted the NCBI Single Nucleotide Polymorphism database (dbSNP). This search revealed rs2071214, an A → G polymorphism that alters a lysine (Lys-129) to glutamate (Glu-129). Among different ethnic populations represented in the genome databases (Perlegen Sciences and Genetic Association Information Network), from 4% to 42% of individuals were identified as heterozygous for glutamate (K/E) at this locus, while 2–17% were homozygous (E/E) (Fig. 2A). To identify tumor cell types that might express these variant proteins, we screened DNA sequences at this site using Tetra-Primer ARMS PCR followed by DNA sequencing and identified a neuroblastoma cell line (NB10) that was homozygous for the sequence encoding the derived allele (G/G), while other such cell lines (NB7 and NB8) were homozygous for the sequence encoding the ancestral allele (A/A) (Fig. 2B). Using indirect immunofluorescence staining of the endogenous proteins, we then analyzed the subcellular distribution of the proteins encoded by these alleles in the different neuroblastoma cell lines. While the protein encoded by the A/A alleles localized primarily within the nucleus in asynchronous proliferating cells, the protein encoded by G/G was ∼50% decreased in the nuclear compartment (not shown), suggesting that a variant at this locus might regulate survivin nuclear transport.
FIGURE 2.
A homozygous mutation in a neuroblastoma cell line is located at a survivin SNP site encoding lysine at position 129. A, population distributions of the ancestral (A) and derived (G) alleles encoding amino acid 129, as determined from dbSNP. Blue, homozygous AA; red, heterozygous AG; green, homozygous GG. B, tetra-primer PCR and DNA sequence analysis showing homozygosity of the A → G polymorphism in the NB10 neuroblastoma cell line but not in the NB8 cell line. OP, outer primer.
Lys-129 Acetylation Regulates Survivin Nuclear Localization
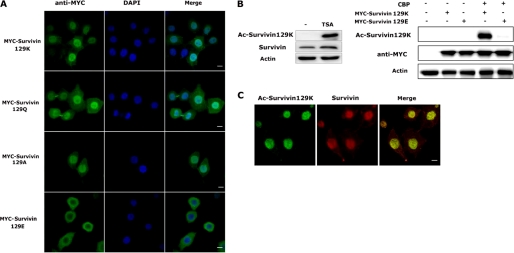
To further explore the function(s) of acetylation at Lys-129, we used site-directed mutagenesis to create a series of MYC-tagged mutant constructs that either could not be acetylated or that biochemically mimic acetylation (24, 25) at this locus. To this end, we mutated the Lys-129 residue to the neutral amino acid arginine (R); to the negatively charged amino acid glutamate (E); and to glutamine (Q) or alanine (A) (both mimic acetylation). We then transfected HeLa cells with the MYC-tagged survivin constructs encoding each of these mutant proteins and compared their subcellular localization to that of the 129K protein. Indirect immunofluorescence staining revealed that the 129K protein localized primarily within the nucleus (Fig. 3A). However, the 129 mutants had an altered subcellular localization, depending on whether they could be acetylated. Specifically, the Arg-129 and Glu-129 (129E) (unacetylatable) mutants localized primarily within the cytoplasm, while the Gln-129 (129Q) and Ala-129 (129A) (mimic acetylation) mutants localized primarily within the nucleus (Fig. 3A and data not shown). These data strongly suggest that acetylation at amino acid site 129 can direct survivin nuclear localization.
FIGURE 3.
Survivin acetylation at lysine 129 directs its nuclear localization. A, HeLa cells were transfected with cDNAs encoding MYC-tagged survivin129K, -129Q, -129A, or -129E then fixed and immunostained with an antibody to MYC and stained with DAPI. Images were taken with a Nikon confocal microscope. Scale bars, 20 μm. B, left panel, Western blot of lysates from HeLa cells treated with TSA and immunoblotted with a novel antibody to acetylated survivin 129K or anti-full-length survivin. right panel, Western blot of lysates from HeLa cells cotransfected with cDNAs encoding MYC-tagged survivin129K or survivin129E proteins and CBP and immunoblotted with a novel antibody to acetylated survivin129K, anti-MYC, or actin. C, immunofluorescence of fixed HeLa cells immunostained with the acetylated survivin129K antibody and the full-length survivin antibody. Images were taken with a Nikon Confocal microscope. Scale bars, 20 μm.
To generate an antibody that specifically detects acetylation at Lys-129, we immunized rabbits with a peptide acetylated at both Lys-129 and Lys-130, then immunodepleted the antisera with two peptides (C-FEETAKKVRRAIEQ-amide and C-FEETAE[K-acetyl]VRRAIEQ-amide) to eliminate binding to acetylated 130K. Expression of the endogenous acetylated 129K protein, as detected by this novel antibody, was markedly enhanced following treatment of HeLa cells with TSA (Fig. 3B). In addition, when HeLa cells were transfected with cDNAs encoding MYC-survivin129K or MYC-survivin129E proteins, the acetylated antibody specifically detected the 129K protein that was acetylated by CBP and not the 129E protein (Fig. 3B). Indirect immunofluorescence of HeLa cells immunostained with the acetylated antibody also showed preferential staining within cell nuclei (Fig. 3C), confirming the role for Lys-129 acetylation in nuclear localization.
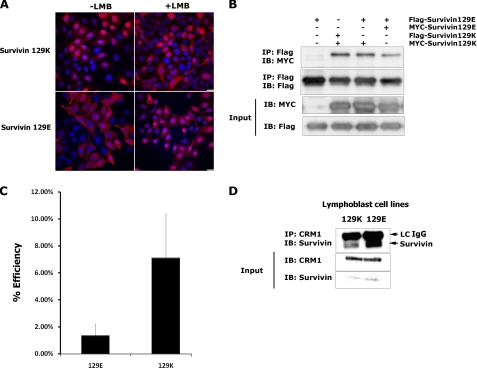
Deacetylated Survivin Preferentially Binds to CRM1 for Nuclear Export
Because an active nuclear import mechanism for survivin has not been demonstrated, active export by CRM1 is the only known means for regulating its nuclear concentration (11). To determine whether this export mechanism might be responsible for the absence of the acetylation-defective survivin 129 mutants from the nucleus, we treated HeLa cells expressing either the wild-type or acetylation-defective proteins with the CRM1 inhibitor leptomycin B (LMB) and visualized the cells by indirect immunofluorescence. The results in Fig. 4A demonstrate localization of the 129E protein to the nucleus following LMB, supporting a mechanism by which this variant protein undergoes active nuclear export by binding to CRM1. To determine whether the variant protein preferentially exists as a monomer and therefore explain its preferential binding to CRM1, we cotransfected HeLa cells with cDNAs encoding MYC-and Flag-tagged constructs of survivin129E and survivin129K and performed immunoprecipitation analyses using anti-MYC and anti-Flag antibodies. Fig. 4B shows that survivin129K forms more stable homodimers than survivin129E. To confirm this finding, we performed acceptor photobleaching fluorescence resonance energy transfer (FRET) on HeLa cells cotransfected with either MYC- and Flag-tagged survivin129E or MYC and Flag-tagged survivin129K. The results from this analysis show that the 129E protein has less FRET activity than the 129K protein (Fig. 4C), validating the observation that 129E is more likely to exist as a monomer. To demonstrate which survivin form binds to CRM1, we used Coriell lymphoblast cell lines that are homozygous for either A/A or G/G at the 129 locus, encoding survivin129K and -129E, respectively. Lysates prepared from these cells were immunoprecipitated with an antibody to CRM1 then immunoblotted with anti-survivin. Results show that CRM1 binds stronger to the 129E form, compared with the 129K form (Fig. 4D).
FIGURE 4.
Acetylated survivin forms homodimers while deacetylated survivin forms monomers and binds to CRM1. A, HeLa cells expressing MYC-tagged survivin129K or -129E were treated with or without the CRM1 inhibitor, leptomycin (LMB). Cells were fixed and immunostained with a MYC antibody and stained with DAPI. Images were taken using a Nikon Confocal microscope. Scale bars, 20 μm. B, lysates from HEK293T cells transfected with cDNAs encoding MYC- or Flag-tagged survivin 129K or 129E were immunoprecipitated with Flag antibody then immunoblotted with MYC or Flag antibody. C, acceptor photobleaching FRET analysis performed on HeLa cells cotransfected with cDNAs encoding MYC-survivin129E and Flag-survivin129E or MYC-survivin129K and Flag-survivin129K. D, lysates from Coriell lymphoblast cell lines homozygous for the A/A (129K) or G/G (129E) alleles at the 129 position were immunoprecipitated with CRM1 antibody then immunoblotted with survivin antibody. LC IgG, light chain immunoglobulin.
Nuclear Survivin Binds STAT3
As the only known nuclear role of survivin is as a chromosomal passenger protein during mitosis, we sought to determine potential additional functions for this protein within the nucleus. Thus, we performed electrospray liquid chromatography-tandem mass spectrometry, using protein from a tumor cell line immunoprecipitated with an antibody to survivin. Proteomic analysis of the individual silver-stained bands revealed STAT3 as a survivin-interacting protein.
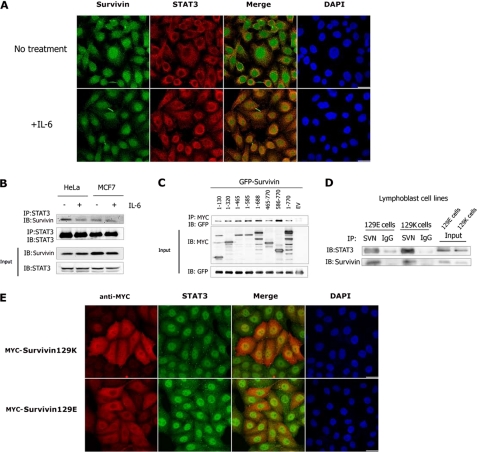
To confirm the nuclear survivin/STAT3 complex formation, we treated HeLa or MCF7 cells with IL-6 (26). Upon treatment, STAT3 translocated from the cytoplasm into the nucleus, where it strongly colocalized with survivin (Fig. 5A). As expected, we recovered survivin in STAT3 immunoprecipitates, obtained with endogenous pull-down assays and an antibody to STAT3, followed by Western blotting with anti-survivin (Fig. 5B). Interestingly, the survivin/STAT3 interaction was reduced in the presence of IL-6, suggesting that STAT3 forms a constitutive complex with survivin and that IL-6 may release STAT3 from the survivin binding complex. To identify the structural requirements for this interaction, we cotransfected HEK293T cells with STAT3 variants and survivin. Binding of survivin was strongest with the STAT3-(586–770) fragment (Fig. 5C), containing the STAT3 transactivation domain (TAD) (16), suggesting that survivin binds to STAT3 within the TAD.
FIGURE 5.
Nuclear survivin129K binds to the transactivation domain of STAT3. A, HeLa cells treated with or without IL-6, were fixed and immunostained with an antibody to survivin or STAT3, and stained with DAPI. Images were taken with a Nikon Confocal microscope. Bars, 20 μm. B, lysates from HeLa or MCF7 cells treated with or without IL-6 were immunoprecipitated with an antibody to STAT3 then immunoblotted with an antibody to survivin or STAT3. IP, immunoprecipitation; IB, immunoblot. C, HEK293T cells were cotransfected with cDNAs encoding MYC-tagged deletion mutants of STAT3 together with GFP-tagged survivin. Lysates were immunoprecipitated with a MYC antibody then immunoblotted with a GFP antibody. D, lysates from Coriell lymphoblast cell lines homozygous for the A/A (129K) or G/G (129E) alleles at the 129 position were immunoprecipitated with survivin or an IgG control then immunoblotted with anti-STAT3 or anti-survivin. E, HeLa cells expressing MYC-survivin 129K or 129E were fixed and immunostained with anti-MYC and anti-STAT3. Images were taken with a Nikon Confocal microscope. Scale bars, 20 μm.
To evaluate potential differences in STAT3 binding to the variant survivin proteins, we again used Coriell lymphoblasts endogenously expressing either survivin129K or survivin129E and performed immunoprecipitation analyses using anti-survivin and anti-STAT3 antibodies. Fig. 5D shows that STAT3 can bind to both forms of survivin, with an increase in binding observed with the 129K protein. In the MYC-transfected HeLa cells, the survivin129K/STAT3 complex was mainly found in the nucleus while the survivin129E/STAT3 complex was restricted to the cytoplasm (Fig. 5E), suggesting that survivin129K may be a component of the STAT3 enhanceosome (27).
Nuclear Survivin Inhibits STAT3 Transactivation
To delineate the potential role of survivin129K in gene regulation by STAT3, we examined the survivin/STAT3 complex for DNA binding activity by using the STAT3 consensus binding element (SIE) as the probe. Survivin129K was consistently recovered from the STAT3/DNA complex (Fig. 6A), confirming that it is indeed part of the STAT3 enhanceosome. Moreover, the DNA binding activity of STAT3 in the presence of survivin129K paralleled that seen with CBP alone, suggesting that survivin can facilitate STAT3 DNA binding in the absence of cytokine- (or CBP) mediated STAT3 activation. By forming a complex with STAT3, survivin did not disturb STAT3-DNA binding activity, rather STAT3/survivin forms a complex with DNA, suggesting an immediate involvement of survivin in STAT3 enhanceosome activity in promoter regulation.
FIGURE 6.
Acetylated survivin 129K inhibits STAT3 enhanceosome activity. A, HEK293T cells were cotransfected with cDNAs encoding MYC-STAT3 and the three survivin variants, in the presence or absence of CBP, as indicated. STAT3/Survivin complexes were affinity-purified using agarose beads conjugated with the SIE oligonucleotide and blotted with anti-MYC. B, HEK293T cells were cotransfected with survivin acetylation mutants, STAT3, CBP, and a STAT3-dependent luciferase activity reporter. The data are means ± S.E. for triplicate experiments (*, p < 0.005, two-tailed t test). EV, empty vector. C, HEK293T cells were cotransfected with cDNAs encoding STAT3 and CBP, with or without survivin, as indicated. RNA was purified, reverse transcribed into cDNA, and Q-RTPCR performed with primers to human BCLXL and MCL. ΔΔCts were calculated. The data are means ± S.E. for triplicate experiments (*, p < 0.005 in each comparison). D, HEK293T cells were transfected with EV or with cDNAs encoding MYC-STAT3, with or without the addition of IL-6 or CBP. For comparison, cells were cotransfected with cDNAs encoding MYC-survivin and either wild-type MYC-STAT3 or the phosphorylation-defective (Y705F), or acetylation-defective (K685R) MYC-STAT3 mutants. Lysates were affinity purified using agarose beads conjugated with SIE oligonucleotides then immunoblotted with anti-MYC, anti-STAT3-pY705, or a pan-acetylation (Ac) antibody. EV, empty vector; IB, immunoblot.
To determine if the STAT3/survivin complex alters transcription from the SIE, we cotransfected HEK293T cells with cDNAs encoding either survivin 129K or 1 of 10 survivin acetylation mutants together with STAT3, CBP, and a luciferase reporter construct driven by the SIE. Survivin129K repressed transcription from the STAT3 SIE, as demonstrated by a 5-fold decrease in reporter activity in the presence of this protein (Fig. 6B). Strikingly, when survivin acetylation at Lys-129 was inhibited by mutation to either glutamate or arginine, or when any of the other 9 acetylation sites were mutated, the SIE reporter activity was comparable to that seen with STAT3/CBP alone, suggesting that deacetylated survivin loses its ability to inhibit STAT3 transcriptional activity and that survivin acetylation at multiple sites may coordinately modulate STAT3 oncogenic activity.
To identify STAT3 gene targets repressed by survivin129K, we cotransfected cDNAs encoding survivin129K, STAT3, and CBP into HEK293T cells and performed quantitative RT-PCR on the mRNA generated from the transfectants, using primers to BCLXL, MYC, CyclinD1, MUC1, MMP9, VEGF, p53, and MCL1 genes commonly targeted by STAT3. Survivin129K repressed expression of BCLXL and MCL1 but did not affect the remaining targets (Fig. 6C), suggesting its involvement as a tumor suppressor acting specifically in mitochondria-mediated survival pathways regulated by STAT3. In support of this data, IL-6-treated Coriell lymphoblasts encoding 129K also repressed expression of BCLXL and CyclinD1, relative to that observed in Coriell lymphoblasts encoding 129E (not shown).
Neither STAT3 Acetylation nor Its Phosphorylation Is Required for DNA Binding of the STAT3/Survivin Complex
Cytokine-activated STAT3 is phosphorylated at Tyr-705 and acetylated at Lys-685, within the C-terminal SH2-transcriptional activation domain (27), the same domain that binds survivin. Both modifications are required for STAT3 dimerization (16, 28). To determine if either of these modifications is required for formation of the STAT3/survivin complex, we transfected HEK293T cells with phosphorylation (Y705F) and acetylation (K685R) defective mutants (16), and compared the binding of these mutant complexes to the SIE with that of the wild-type STAT3/survivin129K complex. In this analysis, both the phosphorylation and acetylation mutants were recovered from STAT3/survivin/SIE complexes (Fig. 6D), suggesting that neither modification is necessary for the formation of this complex. Interestingly, the STAT3 K685R/survivin complex bound with even greater affinity to the SIE than did the wild-type STAT3 homodimer complex, suggesting that STAT3 acetylation at Lys-685 modulates STAT3/survivin stability.
Survivin Does Not Inhibit STAT3 Dimerization or Its Acetylation
To determine the structural requirements for dimerization of STAT3 within the STAT3/survivin complex, we cotransfected HEK293T cells with cDNAs encoding a MYC-tagged STAT3, a Flag-tagged STAT3, and a MYC-tagged survivin 129K construct. Flag-STAT3 was present together with MYC-STAT3 in the STAT3/survivin complex (Fig. 7A), indicating that STAT3 binds to DNA as a dimer and that survivin does not inhibit the dimerization process. To determine if survivin affects the STAT3/CBP interaction, we co-transfected HEK293T cells with cDNAs encoding Flag-STAT3, CBP, and MYC-survivin. No differences in STAT3 acetylation were observed either in the presence or absence of survivin (Fig. 7B), suggesting that survivin does not inhibit CBP-mediated STAT3 acetylation. Taken together, our results support a model in which survivin, acetylated at Lys-129 by CBP, binds to STAT3 within the nucleus at target gene promoters containing the SIE element, where it represses STAT3 transcriptional activity.
FIGURE 7.
Survivin does not inhibit STAT3 homodimerization or acetylation. A, lysates from HEK293T cells transfected with cDNAs encoding Flag- and MYC-STAT3, with or without MYC-survivin were immunoprecipitated with anti-Flag then immunoblotted with anti-MYC or anti-Flag. B, lysates from HEK293T cells transfected with cDNAs encoding Flag-STAT3, HA-CBP, and MYC-survivin were immunoprecipitated with anti-Flag then immunoblotted with a pan-acetylation (Ac) antibody.
DISCUSSION
The diverse functions of the survivin protein are determined by its interactions with key partner proteins, located within distinct intracellular compartments (5, 6). Survivin bears a nuclear export signal within its homodimerization domain at amino acids 89–98 (11), allowing it to bind to the export carrier protein CRM1 and shuttle between the nuclear and cytoplasmic compartments (11, 12). Previous studies showed that survivin associates with CRM1 as a monomer (12), with mutation of the survivin homodimerization interface resulting in enhanced CRM1 binding and nuclear export (12). Our findings reveal that post-translational modification by CBP-mediated acetylation at survivin Lys-129 facilitates survivin homodimerization, thus inhibiting its interaction with CRM1. By contrast, deacetylation at this locus enhances survivin/CRM1 heterodimerization, apparently due to the loss of survivin homodimerization.
The polymorphism at the 129K locus shifts the nuclear-to-cytoplasmic survivin protein ratio toward an increase in the cytoplasmic component, thus increasing the amount of survivin available to block caspase activation and inhibit apoptosis. If the 129E protein is expressed as the major form of survivin in some tumor cells, as suggested in a neuroblastoma cell line here, its expression would likely confer resistance to cell death and correlate clinically with a poor response to therapy. This shift in nuclear-to-cytoplasmic ratio due to a change in acetylation state offers one possible explanation for the immunohistochemical findings observed in previous clinical studies that suggest that nuclear expression of survivin confers a good prognosis, while cytoplasmic expression correlates with a poor prognosis (10).
To date, survivin function within the nucleus has been exclusively defined by its expression during mitosis, where it binds a chromosomal passenger complex of proteins and aids in chromosomal movement and cytokinesis (9). However, immunofluorescence and immunohistochemical studies in tumor cells provide evidence that the protein is also expressed during interphase (5, 6) and suggest that it may have other functions within this compartment. Our results identify a new function for nuclear survivin, as a cofactor for the STAT3 transcription factor, regulating its activity along selective target gene promoters. Indeed, nuclear survivin represses STAT3-mediated transcription of at least two STAT3 signature genes in tumor cells (29) that play an important role in resistance to apoptosis in many tumor types. As the survivin gene itself is a target of STAT3 activation (30), the negative effect of survivin on STAT3 transcriptional activity suggests a negative feedback control loop for survivin expression, which is likely an important control mechanism that exists to regulate its activity during normal homeostasis.
Constitutive activation of STAT3 is observed in 40–60% of all human tumors, including breast, ovarian, lung, prostate, lymphomas, among others (31). The current known mechanisms of STAT3 inhibition include dephosphorylation, inhibition of JAK activation by SOCS proteins (32), and abrogation of DNA binding by the protein inhibitor of STAT activation (PIAS) (33). Unlike PIAS, which dissociates STAT3 from DNA (33), survivin can terminate STAT3 transcriptional activity by forming a complex with STAT3-bound DNA in the presence or absence of CBP, presumably by altering the transcriptional active conformation of STAT3 along its promoter. This mechanism may present a novel means to therapeutic intervention in STAT3-driven tumors.
In summary, our findings show that the dynamic equilibrium between survivin acetylation and deacetylation determines whether the protein forms homodimers or whether it heterodimerizes with CRM1. Subsequently, its subcellular localization is determined and as a consequence, whether it inhibits transactivation of the STAT3 oncoprotein. Modification by acetylation may allow survivin to act as double-edged sword where it either enhances tumor cell growth and survival by inactivating caspases in the cytoplasm or inhibits this growth by terminating STAT3 activity in oncogenic transcription in the nucleus. These results may have implications for the development of novel therapeutic strategies of STAT3-activated tumors and may also implicate this acetylation site as a novel susceptibility locus in some tumors.
Acknowledgments
We thank the Taplin Biological Mass Spectrometry at Harvard University, Cell Biology Dept. for performing the mass spectrometry. We thank Novus Biological Laboratories for manufacturing the acetylated (129) survivin antibody. We thank John Gilbert for editorial assistance.
This work was supported, in whole or in part, by National Institutes of Health Cancer Research Center Grant P20RR017695-6A2 (to R. A. A.) and by a Center for Excellence in Women's Health/Brown University and Women and Infants Hospital Award (to R. A. A.).
- IAP
- inhibitor of apoptosis protein
- HAT
- histone acetyltransferase
- CBP
- CREB-binding protein
- NES
- nuclear export signal
- TSA
- trichostatin A.
REFERENCES
- 1.Ambrosini G., Adida C., Altieri D. C. (1997) Nat. Med. 3, 917–921 [DOI] [PubMed] [Google Scholar]
- 2.Uren A. G., Wong L., Pakusch M., Fowler K. J., Burrows F. J., Vaux D. L., Choo K. H. (2000) Curr. Biol. 10, 1319–1328 [DOI] [PubMed] [Google Scholar]
- 3.Jiang Y., de Bruin A., Caldas H., Fangusaro J., Hayes J., Conway E. M., Robinson M. L., Altura R. A. (2005) J. Neurosci. 25, 6962–6970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang Y., Nishimura W., Devor-Henneman D., Kusewitt D., Wang H., Holloway M. P., Dohi T., Sabo E., Robinson M. L., Altieri D. C., Sharma A., Altura R. A. (2008) Diabetes 57, 2718–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altieri D. C. (2008) Nat. Rev. Cancer. 8, 61–70 [DOI] [PubMed] [Google Scholar]
- 6.Altieri D. C. (2008) Oncogene 27, 6276–6284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dohi T., Xia F., Altieri D. C. (2007) Mol. Cell. 27, 17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fortugno P., Wall N. R., Giodini A., O'Connor D. S., Plescia J., Padgett K. M., Tognin S., Marchisio P. C., Altieri D. C. (2002) J. Cell. Sci. 115, 575–585 [DOI] [PubMed] [Google Scholar]
- 9.Lens S. M., Vader G., Medema R. H. (2006) Curr. Opin. Cell. Biol. 18, 616–622 [DOI] [PubMed] [Google Scholar]
- 10.Li F., Yang J., Ramnath N., Javle M. M., Tan D. (2005) Int. J. Cancer. 114, 509–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stauber R. H., Rabenhorst U., Rekik A., Engels K., Bier C., Knauer S. K. (2006) Traffic 7, 1461–1472 [DOI] [PubMed] [Google Scholar]
- 12.Engelsma D., Rodriguez J. A., Fish A., Giaccone G., Fornerod M. (2007) Traffic 8, 1495–1502 [DOI] [PubMed] [Google Scholar]
- 13.Verdecia M. A., Huang H., Dutil E., Kaiser D. A., Hunter T., Noel J. P. (2000) Nat. Struct Biol. 7, 602–608 [DOI] [PubMed] [Google Scholar]
- 14.Muchmore S. W., Chen J., Jakob C., Zakula D., Matayoshi E. D., Wu W., Zhang H., Li F., Ng S. C., Altieri D. C. (2000) Mol. Cell. 6, 173–182 [PubMed] [Google Scholar]
- 15.Becker S., Groner B., Müller C. W. (1998) Nature 394, 145–151 [DOI] [PubMed] [Google Scholar]
- 16.Yuan Z. L., Guan Y. J., Chatterjee D., Chin Y. E. (2005) Science 307, 269–273 [DOI] [PubMed] [Google Scholar]
- 17.Arora V., Cheung H. H., Plenchette S., Micali O. C., Liston P., Korneluk R. G. (2007) J. Biol. Chem. 282, 26202–26209 [DOI] [PubMed] [Google Scholar]
- 18.Caldas H., Jiang Y., Holloway M. P., Fangusaro J., Mahotka C., Conway E. M., Altura R. A. (2005) Oncogene 24, 1994–2007 [DOI] [PubMed] [Google Scholar]
- 19.Haynes P. A., Fripp N., Aebersold R. (1998) Electrophoresis 19, 939–945 [DOI] [PubMed] [Google Scholar]
- 20.Keller A., Eng J., Zhang N., Li X. J., Aebersold R. (2005) Mol. Syst. Biol. 1, 2005.0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye S., Dhillon S., Ke X., Collins A. R., Day I. N. (2001) Nucleic Acids Res. 29, E88–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X. J., Seto E. (2008) Mol. Cell. 31, 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sadoul K., Boyault C., Pabion M., Khochbin S. (2008) Biochimie 90, 306–312 [DOI] [PubMed] [Google Scholar]
- 24.Wang Y. H., Tsay Y. G., Tan B. C., Lo W. Y., Lee S. C. (2003) J. Biol. Chem. 278, 25568–25576 [DOI] [PubMed] [Google Scholar]
- 25.Scroggins B. T., Robzyk K., Wang D., Marcu M. G., Tsutsumi S., Beebe K., Cotter R. J., Felts S., Toft D., Karnitz L., Rosen N., Neckers L. (2007) Mol. Cell. 25, 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wegenka U. M., Buschmann J., Lütticken C., Heinrich P. C., Horn F. (1993) Mol. Cell. Biol. 13, 276–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Darnell J. E., Jr. (1997) Science 277, 1630–1635 [DOI] [PubMed] [Google Scholar]
- 28.Sasse J., Hemmann U., Schwartz C., Schniertshauer U., Heesel B., Landgraf C., Schneider-Mergener J., Heinrich P. C., Horn F. (1997) Mol. Cell. Biol. 17, 4677–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alvarez J. V., Febbo P. G., Ramaswamy S., Loda M., Richardson A., Frank D. A. (2005) Cancer Res. 65, 5054–5062 [DOI] [PubMed] [Google Scholar]
- 30.Mahboubi K., Li F., Plescia J., Kirkiles-Smith N. C., Mesri M., Du Y., Carroll J. M., Elias J. A., Altieri D. C., Pober J. S. (2001) Lab. Invest. 81, 327–334 [DOI] [PubMed] [Google Scholar]
- 31.Frank D. A. (2007) Cancer Lett. 251, 199–210 [DOI] [PubMed] [Google Scholar]
- 32.Naka T., Narazaki M., Hirata M., Matsumoto T., Minamoto S., Aono A., Nishimoto N., Kajita T., Taga T., Yoshizaki K., Akira S., Kishimoto T. (1997) Nature 387, 924–929 [DOI] [PubMed] [Google Scholar]
- 33.Chung C. D., Liao J., Liu B., Rao X., Jay P., Berta P., Shuai K. (1997) Science 278, 1803–1805 [DOI] [PubMed] [Google Scholar]