Abstract
The disulfide relay system of the mitochondrial intermembrane space has been extensively characterized in Saccharomyces cerevisiae. It contains two essential components, Mia40 and Erv1. The genome of Arabidopsis thaliana contains a single gene for each of these components. Although insertional inactivation of Erv1 leads to a lethal phenotype, inactivation of Mia40 results in no detectable deleterious phenotype. A. thaliana Mia40 is targeted to and accumulates in mitochondria and peroxisomes. Inactivation of Mia40 results in an alteration of several proteins in mitochondria, an absence of copper/zinc superoxide dismutase (CSD1), the chaperone for superoxide dismutase (Ccs1) that inserts copper into CSD1, and a decrease in capacity and amount of complex I. In peroxisomes the absence of Mia40 leads to an absence of CSD3 and a decrease in abnormal inflorescence meristem 1 (Aim1), a β-oxidation pathway enzyme. Inactivation of Mia40 leads to an alteration of the transcriptome of A. thaliana, with genes encoding peroxisomal proteins, redox functions, and biotic stress significantly changing in abundance. Thus, the mechanistic operation of the mitochondrial disulfide relay system is different in A. thaliana compared with other systems, and Mia40 has taken on new roles in peroxisomes and mitochondria.
Keywords: Arabidopsis thaliana, Mitochondria, Peroxisomes, Protein Translocation, Subcellular Organelles, Biotic Stress, Disulfide Relay, Dual Targeting
Introduction
Characterization of the proteomes of mitochondria from Saccharomyces cerevisiae (yeast), mammals, and plants indicates that they contain from 1000 proteins in yeast to ∼2000 in higher organisms (1). As the coding capacity of mitochondria is limited to between 8 and approximately 50 proteins in yeast and plants, respectively (2), the majority of mitochondrial proteins are encoded by nuclear located genes, translated in the cytosol, and imported into mitochondria. The import of hundreds of different proteins is achieved by the combined action of a number of multisubunit, integral membrane protein complexes, known as translocases. These translocases work in conjunction with a variety of soluble components located in the cytosol, intermembrane space, and mitochondrial matrix, such as chaperones, peptidases, and assembly factors (3, 4).
The outer mitochondrial membrane contains the TOM complex (translocase of the outer membrane) and the sorting and assembly machinery; the latter is also known as the TOB complex (topogenesis of β-barrel proteins) (4). Almost all mitochondrial proteins are initially recognized by the outer membrane complex and are passed to other components depending on their final location. β-Barrel proteins of the outer membrane such as Tom40 and voltage-dependent anion channel protein (VDAC) are passed to the sorting and assembly machinery complex (5). Proteins imported via the general and carrier import pathways are passed to the translocases of the inner membrane 23 (TIM23) and 22 (TIM22), respectively (3, 4). Proteins are passed to the TIM23 complex directly from the outer membrane complex with the aid of Tim50 (6, 7) and also possibly Tim23, where the N-terminal region has been shown to transiently associate with the outer membrane (8). Proteins are transferred to the TIM22 complex via the aid of the small intermembrane space proteins Tim9 and -10 (3, 4). A number of variations of these main pathways can occur. These include the crossing over between pathways (9) and the utilization of different sorting processes. Proteins can be either stop-transfer-sorted or conservative-sorted, with the latter pathway using the Oxa1p translocase located on the inner mitochondrial membrane (10). A variety of other protein import pathways also exist, such as those utilized for the import of cytochrome c (11) and the small Tim proteins of the intermembrane space (12, 13).
The import pathway of the small intermembrane space proteins, such as Tim8, -9, -10, and -13, is the most recently described protein import pathway. Their import is achieved via the mitochondrial intermembrane space assembly machinery (MIA)2 that consists of Mia40 and Erv1 (3, 12, 14), both of which are essential proteins in yeast (15, 16). Characterization of the import pathway of Tim9 and -10 in yeast revealed that as little as nine amino acids are required to achieve transfer from the outer membrane complex on the outer membrane to Mia40, which acts as a receptor in the intermembrane space (17, 18). These proteins then undergo oxidative folding in the intermembrane space. Conserved cysteine residues in these proteins are oxidized by Mia40, which is subsequently oxidized by Erv1. Erv1 is oxidized by cytochrome c, which is in turn oxidized by cytochrome c oxidase, with molecular oxygen acting as the final electron acceptor (19).
As mitochondrial endosymbiosis occurred only once in evolutionary history (2), many features of mitochondrial biology are conserved across wide phylogenetic gaps. With respect to protein import into mitochondria, it is observed that whereas pore or channel-forming subunits of the membrane-bound translocases are well conserved, variability is often seen with other components of the import apparatus (20, 21). Thus, comparison of the plant import apparatus to that of yeast (and mammalian) systems reveals that although Tom40, Sam50, Tim17 and -23, Tim22, and Oxa1p are well conserved, other components differ to varying extents. Thus, none of the three functionally characterized protein import receptors of plant mitochondria is orthologous to yeast or mammalian protein import receptors (22). Furthermore, the mitochondrial processing peptidase is membrane-bound in plants compared with matrix located in yeast (23). In contrast, the presequence degrading peptidase is located in the matrix in plants, whereas its orthologous counterpart in yeast is located in the intermembrane space (24, 25).
To date no studies have been carried out on components of the MIA import pathway in plants. As an essential pathway in yeast, it might be expected to function in a similar manner in plants. We have analyzed the function of Mia40 and Erv1 in Arabidopsis thaliana with the aim of defining the essential components of the MIA pathway, their location, and the effects of inactivation.
EXPERIMENTAL PROCEDURES
cDNA Clones and Constructs
The chromosomal loci for AtMia40 (At5g23395) and AtErv1 (At1g49880) have been previously identified (26). The Oryza sativa (rice) Mia40 was identified as Os04g44550 by using AtMia40 to search the rice genome (27). cDNAs of AtMia40, AtErv1, and OsMia40 were cloned using gateway cloning (Invitrogen) into vectors expressing GFP as either N- or C-terminal fusions under the control of a constitutive promoter (28). The human Mia40 cDNA (AAH33775) was cloned as both N- and C-terminal fusions with GFP into pcDNA3 (Invitrogen) under the control of a constitutive promoter using standard cloning techniques (29, 30). The full cDNA of A. thaliana Tim9 (At3g46560), copper chaperone for CSD1 (Ccs1, At1g12520), copper/zinc superoxide dismutase 1 (CSD1, At1g08830), copper/zinc superoxide dismutase 3 (CSD3, At5g18100), and carbonic anhydrase 2 (CA2, At1g47260) were cloned into pDest14 (Invitrogen) using gateway-cloning techniques (Invitrogen) for in vitro transcription and translation. The cDNA clones encoding the following proteins have been described previously: AOX (X68702) (31), Pic (ABO16064) (32, 33), and NDC1 (At5g08740) (34).
GFP Subcellular Localizations
To determine the subcellular localization of AtMia40 and OsMia40 A. thaliana cell culture was transformed by biolistic transformation as previously described (28). GFP and RFP expression and targeting were visualized using a BX61 Olympus microscope (Olympus) using excitation wavelengths of 460/480 nm (GFP) and 535/555 nm (RFP) and emission wavelengths of 495–540 nm (GFP) and 570–625 nm (RFP). Subsequent images were captured using Cell imaging software as previously described (28). For the HsMia40, 143B osteosarcoma cells were plated onto 13-mm diameter glass coverslips and allowed to attach overnight. Cells were transfected using FuGENE HD with both GFP and RFP plasmids for 48 h and washed with Tris-buffered saline (5 mm Tris/HCl (pH 7.4), 20 mm NaCl). Cells were mounted in DABCO (1,4-diazabicyclo(2.2.2)octane)/polyvinyl alcohol medium. Images were acquired using an Olympus DP70 fluorescent inverted microscope as previously described (29).
T-DNA Insertion Lines
The following T-DNA insertion lines were obtained from the SALK collection (35) and genotyped by PCR to confirm homozygosity for the T-DNA insert: AtMia40 (At5g23395): SALK_044358 and AtErv1 (At1g498890): SALK_110883, SALK_131166, SALK_001649.
Organelle Purification
Mitochondria for in vitro import experiments were harvested from 20 g (fresh weight) of 14-day-old A. thaliana seedlings grown in liquid culture as previously described (22) with the BSA omitted from the last two wash steps. Typically 2–3 mg of mitochondrial protein was obtained. For immunodetection assays, highly purified mitochondria and peroxisomes were purified from 7-day-old A. thaliana cell suspension using free flow electrophoresis as described by (36).
In Vitro Import Studies
[35S]Met-labeled precursor proteins were synthesized using rabbit reticulocyte TNT in vitro transcription/translation lysate (Promega, Melbourne, Australia) as described previously (37). The use of equivalent quantities of mitochondria from different genotypes in import reactions was ensured by triplicate measurement of protein concentration with the Coomassie protein assay reagent (Thermo Scientific, Rockford, IL). Time course analysis of precursor protein import into intact mitochondria isolated from wild type (Col-0) or mutant plants was performed as described previously (22, 37). Proteinase K protected, mature, radiolabeled protein was quantified at each time point and normalized to the highest time point measurement for replicate experiments (22).
BN-PAGE Imports
For in vitro imports analyzed on BN-PAGE gels, import assays were performed as for SDS-PAGE except 250 μg of mitochondria were used per time point. After the import was carried out for the required time, mitochondria were pelleted at 20,000 × g for 5 min and then subjected to BN-PAGE according to the method described by Jänsch et al. (38). Mitochondrial proteins (250 μg) were solubilized with 5% (w/v) digitonin in a buffer containing 30 mm HEPES, 150 mm potassium acetate, and 10% (v/v) glycerol (pH 7.4) and incubated on ice for 15 min. Samples were centrifuged for 20 min at 15,000 × g, and Serva Blue G (0.2% (v/v) final) was added to the supernatant. Samples were loaded onto a 4.5–16% (v/v) gradient gel. After migration, gels were fixed in 40% (v/v) methanol, 10% (v/v) acetic acid, dried, and exposed as per SDS-PAGE gels.
Immunodetection of Proteins
Mitochondria and peroxisomes (25 μg) were resolved by SDS-PAGE, transferred to Hybond-C extra nitrocellulose membrane, and immunodetected as previously outlined (39). To generate antibodies to AtMia40, AtErv1, AtTim23-2 (At1g72750), AtSam50 (At3g11070), and the Rieske iron sulfur protein (RISP, At5g13430), recombinant proteins containing the full-length AtMia40 and AtErv1, amino acids 31–144 for AtTim23-2, the first 200 amino acids of AtSam50, and amino acids 21–151 of Rieske iron sulfur protein fused to an N-terminal His6 affinity purification tag were expressed in Escherichia coli strain BL21 (DE3)pLys (Stratagene, La Jolla, CA). The recombinant protein was purified by denaturing immobilized metal affinity chromatography (IMAC) using the Bio-Rad Profinia protein purification system. The resultant eluate was separated by SDS-PAGE, and the recombinant protein was extracted using a Bio-Rad Model 422 Electro-Eluter. Buffer exchange was performed using an Amicon Ultracel 5K centrifugal filter device (Millipore, Sydney, Australia) such that the antigen was re-suspended in PBS solution, recovering a total of 1 mg (AtErv1), 2 mg (AtMia40), 2 mg (AtTim23-2), 3 mg (AtSam50), and 3 mg of Rieske iron sulfur protein for inoculation. Four separate doses were administered to a rabbit at regular intervals over a 3-month period using standard protocols and Freund's complete adjuvant solution (40). Other antibodies used in this study have been described previously: 3-ketoacyl-CoA thiolase (Kat2) (41), Tim17-2 (42), Tom20-2, and Tom20-4 (22), Tom20-3, and Tom40 (28), coxII, ATP synthase, Ccs1, and CSD1 were obtained from Agrisera (Vännäs), voltage-dependent anion channel protein (VDAC; PM035) and E1α of pyruvate dehydrogenase (PM030) were obtained from Dr. Tom Elthon (University of Nebraska, Lincoln, NE), alternative oxidase (43), and NAD9 (44). The altered inflorescence meristem 1 (Aim1) antibody was obtained from Dr. Douglas Muench (University of Calgary).
Complex I Activity Assays
Complex I activity was measured on 140 μg of mitochondria in 1 ml of respiration buffer (0.3 m sucrose, 5 mm KH2PO4, 10 mm TES, 10 mm NaCl, 2 mm MgSO4, and 0.1% (w/v) BSA (pH 6.8)) obtained after freeze-thawing of the mitochondria using a Clark-type oxygen electrode (Hansatch Instrument). 1 mm deamino-NADH was added to the chamber, oxygen consumption was measured, and 2 mm CCCP was then added to ensure mitochondria were uncoupled. Rotenone (5 mm) was then added to inhibit complex I and to measure the rate of oxygen consumption of the alternative NADH dehydrogenases. Finally 0.1 mm of KCN was used to terminate the reaction. The activity of complex I was defined as the rotenone-sensitive rate of oxygen consumption, determined by subtracting the rate of oxygen consumption of the alternative NADH dehydrogenases from the total oxygen rate consumption.
Global Transcript Analysis
Analysis of the changes in transcript abundance between Col-0 and Δmia40 plants in 18-day-old A. thaliana seedlings was performed using Affymetrix GeneChipTM A. thaliana ATH1 Genome Arrays (Affymetrix, Santa Clara, CA). Green tissue from three seedlings was pooled for each biological replicate; Col-0 and Δmia40 tissue samples were collected in biological triplicate. For each replicate, total RNA was isolated from the leaves using the RNeasy Plant Mini Protocol (Qiagen, Clifton Hill, Australia) and quality-verified using a Bioanalyzer (Agilent Technologies, Palo Alto, CA), and spectrophotometric analysis was carried out to determine the A260:A280 and A260:A230 ratios. Preparation of labeled aRNA from 500 ng of total RNA (3′ IVT Express kit, Affymetrix) and target hybridization as well as washing, staining, and scanning of the arrays was carried out exactly as described in the Affymetrix GeneChipTM Expression Analysis Technical Manual using an Affymetrix GeneChip Hybridization Oven 640, an Affymetrix Fluidics Station 450, and an GeneChip Scanner 3000 7G at the appropriate steps.
Statistical Analysis
Data quality was assessed using GCOS 1.4 before CEL files were exported into AVADIS Prophetic (Version 4.3, Strand Genomics, San Francisco, CA) and Partek Genomics Suite software, Version 6.3 (Partek, St. Louis, MO) for further analysis. MAS5 normalization algorithms were carried out to generate present/absent calls across the arrays. Only those probe sets that were called present in at least two of three replicates in at least one genotype were included for further analysis. Ambiguous probe sets and bacterial controls were removed, resulting in a final data set of 12,551 probe identifiers. CEL files were also subjected to guanosine cytosine robust multi-array average normalization. Correlation plots were examined between all arrays using the scatter plot function; in all cases r ≥ 0.97 (data not shown). Guanosine cytosine robust multi-array average-normalized gene expression values were analyzed to identify differentially expressed genes by a regularized t test based on a Bayesian statistical framework using the software program Cyber-T (45). Cyber-T employs a mixture model-based methods described by Allison et al. (46) for the computation of the global false positive and false negative levels inherent in a DNA microarray experiment. The rates of false positives and false negatives as well as true positives and true negatives at any given p value threshold are estimated, i.e. a posterior probability of differential expression (PPDE) (p) value for each gene measurement and a PPDE (<p) value at any given p value threshold based on the experiment-wide global false positive level and the p value exhibited by that gene. There were 322 unique transcripts that were identified as significantly differentially expressed in the Δmia40 plants compared with Col-0 after false discovery rate correction at PPDE (<p) >0.95 (95% confidence interval) (supplemental Table 2). Functional categorization using GO-biological process annotations was performed on the total present set (12,551 transcripts) along with the 322 transcripts defined as differentially expressed (either positively or negatively). GO annotations (biological process) were obtained from the TAIR webpage. To determine any changes in distributions of different cellular locations, transcripts in the total present set and differentially expressed set were annotated based on their subcellular location: plastid, mitochondrial, peroxisomal, or other. Lists of genes encoding plastid and peroxisomal proteins were generated using the SUBA data base and included genes based on experimental determination (found by mass spectrometry or GFP profiling). The list of mitochondrial proteins used here has been described previously (47). The percentage distribution of each category was compared with that of the total present set using a χ2 test, and percentile distributions were considered to be significantly different at a 98% confidence interval. To gain a qualitative overview of changes in transcript abundance for Δmia40 plants compared with Col-0, the MapMan software was used (48). Only transcripts with significant changes after false discovery rate were displayed.
RESULTS
Erv1 Is an Essential Protein in A. thaliana, but Mia40 Is Dispensable
To determine the role of Mia40 and Erv1 in A. thaliana, T-DNA insertion lines that disrupt the genes and, thus, do not produce functional proteins were screened. In the case of Mia40, one line with a T-DNA insert that disrupted the gene was obtained from screening all lines annotated as having insertions in Mia40 (Δmia40) (Fig. 1A). The absence of the protein was confirmed by Western blot analysis against purified mitochondria from Col-0 and Δmia40 plants (see Fig. 3A). The lack of Mia40 had no discernable effect on growth (Fig. 1A), with normal seed set production observed compared with Col-0 growth under identical conditions. Even though a single insertional mutant could only be obtained for Mia40, the absence of the protein was confirmed by Western blotting, and it is the lack of expected phenotype that is reported. Furthermore, backcrossing and Southern blot analysis indicate a single insertion in the line characterized.
FIGURE 1.
T-DNA insertional inactivation of A. thaliana Mia40 and Erv1. A, shown is a picture of 25-day-old Col-0 and Δmia40 A. thaliana plants. B, shown is a representative picture of seeds from Col-0 and Δerv1 (heterozygous) self-fertilized plants. Arrows indicate aborted seed. C, shown is a schematic diagram of Erv1 proteins from different organisms. The gray region represents the most conserved region between different Erv1 sequences containing the redox center, the FAD binding site, and two conserved cysteine domains (CXXC and CX16C ). The location of the final cysteine pair differs between organisms. Yeast Erv1 and human Erv1 have it at the N terminus, and those for yeast Erv2, A. thaliana, and trypanosomes are at the C terminus. Yeast, S. cerevisiae; human, Homo sapiens; Arabidopsis, A. thaliana; trypanosomes, Trypanosoma brucei.
FIGURE 3.
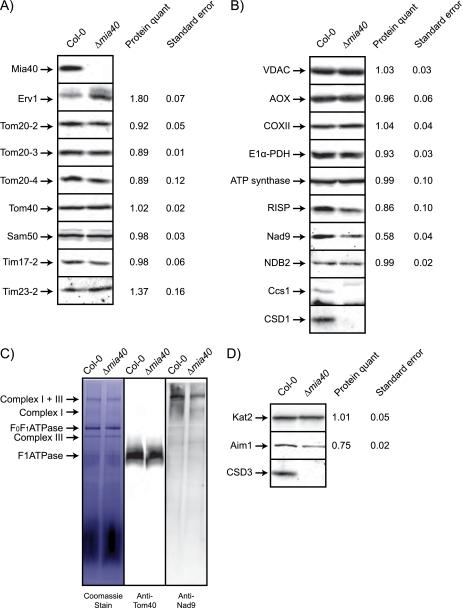
Western blot analysis to determine the affect of deleting A. thaliana Mia40 on mitochondrial and peroxisomal proteins and blue native gel analysis of Δmia40 mitochondria. A, Western blot analysis of mitochondria isolated from Col-0 and Δmia40 plants with a variety of antibodies raised against mitochondrial import components is shown. Numbers on the side represent the relative abundance of the protein in the Δmia40 mitochondria expressed relative to Col-0 (1); S.E. is also shown for three biological replicates. B, Western blot analysis of mitochondrial proteins is not involved in protein import. VDAC, voltage-dependent anion channel protein; AOX, alternative oxidase; E1α-PDH, E1α-subunit of pyruvate dehydrogenease; RISP, Rieske iron sulfur protein. C, blue native gel analysis of Col-0 and Δmia40 mitochondria is shown. Mitochondrial membrane complexes were separated by BN-PAGE and transferred onto a PVDF membrane. Membranes were stained in Coomassie and probed with antibodies against Tom40 and NAD9. D, shown is Western blot analysis of peroxisomes isolated from Col-0 and Δmia40 plants with a selection of antibodies raised against peroxisomal proteins.
In contrast, despite screening three T-DNA lines for Erv1, no homozygous T-DNA-inactivated lines for Erv1 could be obtained. In addition to back-crossing the T-DNA lines to remove any other T-DNA inserts or mutations that may be present, analysis of the seed from self- fertilized heterozygous plants (Erv1/erv1-T-DNA) consistently resulted in 25% of the seed being aborted for all lines analyzed (Fig. 1B). This is consistent with a lethal phenotype due to the absence of a functional gene encoding this protein.
The lethality of Δerv1 was not surprising as it is an essential protein in yeast. In yeast it is required for the import and assembly of small intermembrane space proteins (16, 49), specifically the small Tims that in turn are required for the import of carrier proteins to the inner membrane. They also play a role in β-barrel protein assembly in the outer membrane (3, 50). Therefore, a similar phenotype would be predicted for Δmia40 plants based on extensive studies in yeast and mammalian systems (12, 15, 51, 52). Examination of the sequences for both predicted proteins in A. thaliana revealed that they differed compared with their yeast orthologues. In the case of A. thaliana Mia40 it was a much smaller protein of 162 amino acids (supplemental Fig. 1A). The mammalian Mia40 protein is also shorter than the yeast orthologue, and the smaller “core” conserved region of Mia40 has been shown to be functional in the disulfide relay system (53). The only noticeable “unique”feature of the A. thaliana Mia40 was the presence of a putative peroxisomal PTS1 targeting signal, SKL, at the C terminus (54). Examination of the A. thaliana Erv1 protein sequence revealed that it differed in its arrangement of cysteine pairs to that of yeast and humans (supplemental Fig. 1, B and C). In all Erv1 sequences analyzed to date two pairs of cysteine motifs are conserved in all organisms, the CXXC and CX16C motifs. In regard to the third cysteine pair, yeast and human Erv1 proteins have an N-terminal CXXC motif, whereas A. thaliana has a C-terminal-located CXXXXC motif (Fig. 1C) (55). This cysteine motif shows distinct similarities to the yeast endoplasmic reticulum Erv2p (56) and the trypansome Erv1, both of which also have their third cysteine pair at the C terminus (Fig. 1C) (57). Phylogenetic analysis of all Erv1 and Mia40 protein sequences revealed that the plant proteins formed distinct groups (supplemental Fig. 2), noticeably the trypanosome Erv1 branches closest to the plant group, this group being the only group containing the third cysteine pair at the C-terminal end of the protein (Fig. 1C, supplemental Fig. 2).
Mia40 Is Targeted to Mitochondria and Peroxisomes in Plants
We fused A. thaliana Mia40 to GFP at its N and C termini to determine whether the predicted peroxisomal PTS1 targeting signal was functional. When GFP was fused to the N terminus, a pattern identical to peroxisomal-targeted RFP was seen (Fig. 2A, GFP-AtMIA40). As Mia40 has been characterized as an exclusively mitochondrial protein in yeast and mammalian systems (organisms that also contain peroxisomes), it was investigated if the peroxisomal targeting ability of A. thaliana Mia40 was a general feature of plant Mia40 proteins by testing the targeting of the rice Mia40. A similar result was observed in that rice Mia40 was targeted to peroxisomes when GFP was placed at its N terminus (Fig. 2A, GFP-OsMIA40).
FIGURE 2.
Determination of the targeting ability of Mia40 using GFP tagging, Western blots, and in vitro import assays. A, the localization of A. thaliana, rice, and human Mia40 was determined by fusing them to GFP at their N and C termini. A. thaliana and rice Mia40 GFP plasmids were transformed into A. thaliana suspension cells, and human Mia40 GFP plasmids were transfected into 143B osteosarcoma cells. For the A. thaliana suspension cell cultures, the RFP was targeted to mitochondria using the mitochondrial alternative oxidase targeting signal as a mitochondrial marker and RFP targeted to peroxisomes using a C-terminal SRL sequence as a peroxisomal marker. For the 143B osteosarcoma cells, RFP was targeted to mitochondria using the mitochondrial targeting signal from yeast cytochrome c oxidase 4 as a mitochondrial marker and RFP targeted to peroxisomes using a C-terminal SRL sequence as peroxisomal marker. B, Western blot analysis is shown of isolated mitochondria and peroxisomes with antibodies raised against A. thaliana Mia40, two peroxisomal markers, Kat2 and AIM1, and two mitochondrial markers, Tim17-2 and E1α of pyruvate dehydrogenase (E1α-PDH). C, in vitro import of radiolabeled Mia40 into isolated mitochondria is shown. Lane 1, precursor protein alone. Lane 2, precursor protein incubated with mitochondria under the conditions that support import into mitochondria. Lane 3, as lane 2 with proteinase K added after the incubation of precursor with mitochondria. Lanes 4 and 5, as lanes 2 and 3 with valinomycin added to the import assay before the addition of precursor protein. Lanes 6–9, as lanes 2–5 except that the mitochondrial outer membrane was ruptured after the incubation period with precursor protein but before the addition of proteinase K. Mit, mitochondria; Mit-OM, mitochondria with the outer membrane ruptured; PK, proteinase K; Val, valinomycin; p, precursor protein band; m*, inner membrane protected fragment of Tim23.
When GFP was fused to the C terminus of both A. thaliana and rice Mia40, a mitochondrial pattern of florescence was observed (Fig. 2A, top panel). Noticeably, the fluorescence produced ring like structures, which we have previously observed for mitochondrial proteins linked to GFP (22). They may represent GFP that has been targeted to mitochondria but not translocated across the outer membrane, possibly due to the lack of any pulling force by an intermembrane space protein such as Mia40. High background fluorescence in the cell cytosol was always evident with both A. thaliana and rice Mia40 with GFP fused to the C terminus despite testing in several cell types including onion, A. thaliana leaf, and root (data not shown). This high background fluorescence is not observed for a variety of other mitochondrial targeting signals and is unlikely to be a technical limitation of the system (28, 39). Additionally, the subcellular localization of Mia40 to both mitochondria and peroxisomes was confirmed using Western blotting on highly purified mitochondrial and peroxisomal fractions using an antibody raised against A. thaliana Mia40 (Fig. 2B). This antibody detected a specific protein in both highly purified mitochondria and peroxisomes purified by free flow electrophoresis (36). No Tim17-2 protein or E1α-subunit of pyruvate dehydrogenease (E1α-PDH) was detected in peroxisomes, indicating the purity of the peroxisomal fractions, thus verifying this novel peroxisomal location for Mia40 (Fig. 2B). The absence of significant peroxisome contamination in the mitochondrial fraction was confirmed using two peroxisomal markers, Kat2 and Aim1. Both markers show that the mitochondrial fraction was free of any significant peroxisome contamination. As A. thaliana Mia40 appeared to target GFP poorly to mitochondria, we investigated the import of A. thaliana Mia40 into isolated mitochondria (Fig. 2C). A. thaliana Mia40 was imported into a protease-protected location in a membrane potential-independent manner (Fig. 2C, lanes 1–5), consistent with a location in the intermembrane space. Rupture of the outer membrane before adding protease resulted in digestion of imported Mia40 (Fig. 2C, lanes 6–9). Import of A. thaliana Tim23 as a control verified that the membrane potential was collapsed as import of Tim23 was inhibited in the presence of valinomycin (Fig. 2C, lanes 1–5) and that the inner membrane was intact, as evidenced by the presence of a characteristic membrane-protected fragment of Tim23 upon the addition of protease when the outer membrane was ruptured (Fig. 2C, lanes 6–10).
Analysis of human Mia40 showed that it was only targeted to mitochondria when GFP was fused to the C-terminal end of human Mia40 (Fig. 2A, HsMIA40 GFP). Fusing GFP to the N-terminal end of human Mia40 resulted in no targeting of GFP as evidence by fluorescence throughout the cytoplasm (Fig. 2A, GFP HsMIA40). Notably, analysis of the targeting ability of A. thaliana Erv1 revealed that it is only targeted to mitochondria (data not shown).
Cellular Affects of Inactivation of Δmia40 in A. thaliana
As A. thaliana Mia40 is not an essential protein in A. thaliana, we investigated the abundance of various mitochondrial and peroxisomal proteins to gain insight into the function(s) of Mia40 (Fig. 3). Analysis of the abundance of a variety of protein import components in Δmia40 plants revealed that Erv1 and Tim23-2 had increased in abundance, but many other components were unchanged (Fig. 3A). Examination of a variety of other mitochondrial proteins revealed that although many were essentially unchanged in abundance, there were notable exceptions (Fig. 3B). First, there was a lack of CSD1, the intermembrane space located copper/zinc superoxide dismutase (58, 59), and the chaperone protein associated with CSD1, Ccs1, which plays a role in inserting copper into the active site of CSD1 (58, 59). The Nad9 subunit of complex I also decreased in abundance significantly (40%). The reduced amount of Nad9 on SDS-PAGE gels was confirmed by BN-PAGE (Fig. 3C), which indicated that the reduction of the amount of Nad9 was linked to a reduction in the amount of complex I in Δmia40 mitochondria. The capacity of complex I was measured using deamino NADH with mitochondria that had been subjected to freezing and thawing to allow deamino-NADH access to the matrix side of the inner membrane in the presence of CCCP to ensure unrestricted flow of electrons. The rotenone-sensitive deamino NADH-dependent oxygen consumption was 13.9 nmol of O2/min/mg of protein in mitochondria from wild type plants compared with 6.1 nmol of O2/min/mg of protein in mitochondria from Δmia40 plants. This indicated that complex 1 capacity was reduced by 44% in mitochondria from Δmia40 plants, consistent with the percentage reduction of the Nad9 protein from complex I.
The availability of antibodies to peroxisomal proteins in plants is limited. Nevertheless Western blot analysis using three antibodies revealed that two proteins were altered in abundance (Fig. 3D). Although levels of Kat2 were unchanged in abundance, CSD3 was completely absent in peroxisomes from Δmia40 plants, whereas Aim1 was reduced in abundance by 25%.
Analysis of Protein Import into Mitochondria of Δmia40 Plants
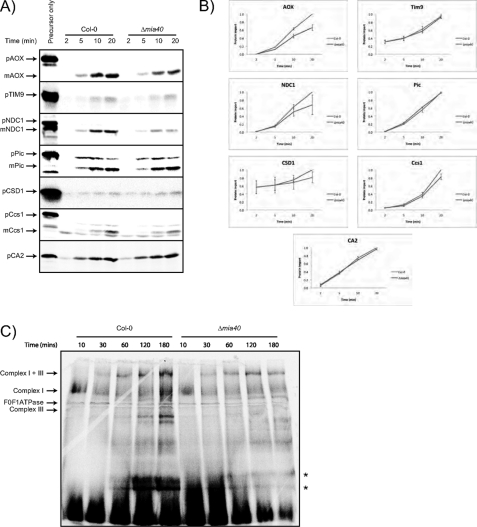
To investigate if the changes observed in the amount of mitochondrial proteins were due to changes in the rate of import or stability of imported proteins, in vitro import assays were carried out with a number of mitochondrial proteins into mitochondria isolated from wild type and Δmia40 plants (Fig. 4). A variety of precursor proteins were used; the alternative oxidase, alternative NAD(P)H dehydrogenase (NDC1), a subunit of complex 1 that has been labeled as CA2 (60, 61) along with CSD1 and Ccs1. Tim9 was used to test the uptake of small Tim proteins into the intermembrane space (17). Finally, we analyzed the import of the phosphate carrier (Pic), which is imported via the carrier import pathway utilizing the small Tim proteins of the intermembrane space (3, 4). Although the uptake of alternative oxidase and NDC1 was reduced to ∼50% compared with wild type, the import of CSD1 and CA2 was unaffected, with the import of the former slightly higher in mitochondria from Δmia40 plants. Import of Ccs1 was reduced by 20% in mitochondria from Δmia40 plants compared with wild type. Thus, although the abundance of some proteins was reduced, this did not result from alternations the rate of import. Note that as Nad9 is a mitochondrially encoded subunit in A. thaliana (62), its rate of import could not be tested.
FIGURE 4.
In vitro uptake assays into mitochondria isolated from Col-0 and Δmia40 plants. A, import of the alternative oxidase (mitochondrial (m) and (p) AOX), alternative NAD(P)H dehydrogenase (NDC1), Tim9, phosphate translocator (Pic), copper/zinc superoxide dismutase 1 (CSD1), Ccs1, and CA2 precursor proteins into isolated mitochondria is shown. p, precursor protein; m, mature protein, where indicated. B, shown is quantification of the rate of import of the various precursor proteins into mitochondria. The amounts of import into Col-0 mitochondria after 20 min was set to 1, and all other values are expressed in a relative manner. C, import of the complex I subunit CA2 into Col-0 and Δmia40 mitochondria and analyzed by BN-PAGE is shown.
The import of both Tim9 and Pic were unaffected (Fig. 4, A and B). The latter two proteins would be expected to be affected by Δmia40 A. thaliana mitochondria as Tim9 is a direct substrate of Mia40 in yeast and the import of carrier proteins (Pic) depends on the function of small Tim proteins. It has been previously demonstrated in plants that small intermembrane space proteins are involved in the import of carrier proteins (22). Thus, the lack of a phenotype for Δmia40 plants and the lack of an effect on protein import via the carrier import pathway suggests that Mia40 in A. thaliana does not play an essential role in the disulfide relay system for small Tim proteins as observed in yeast.
The reduction in the amounts of the proteins outlined above can be explained by either a failure to correctly assemble these proteins, altered expression of the genes that encode these proteins, or a combination of both. To investigate both of these possibilities, the assembly of proteins into mitochondria after import was investigated (Fig. 4C), and alterations in transcript abundance were analyzed (Fig. 5).
FIGURE 5.
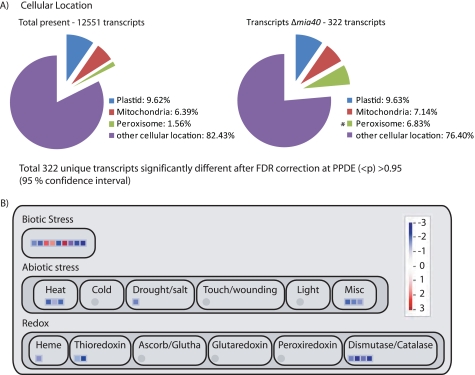
Analysis of the changes in transcriptome of A. thaliana Δmia40 plants. A, shown is the proportion of transcripts encoding proteins located in mitochondria, plastids, and peroxisomes in Col-0 and Δmia40 plants. B, shown is a MapMan visualization of changes in transcript abundance of genes encoding proteins involved in stress and redox metabolism. FDR, false discovery rate; PPDE, posterior probability of differential expression.
To investigate the assembly of protein after import, the assembly of a complex I protein was analyzed. Assembly of imported CA2 into complex I was investigated using BN-PAGE to assess the location of newly imported radiolabeled proteins, as previously carried out for a number of studies investigating assembly into multisubunit protein complexes in mitochondria (63, 64). It was evident that incorporation of CA2 into complex I, especially the supercomplex of complex I and III, was reduced in mitochondria from Δmia40 plants compared with wild type plants (Fig. 4C). In addition to observing reduced radiolabeling into this higher molecular mass complex, the intensity of radiolabeling at the lower regions of the gel was also altered (Fig. 4C). Although the intensity of radiolabeling at the lower regions was higher in mitochondria from Δmia40 plants compared with wild type at 10 and 30 min, it was substantially decreased at 120 and 180 min. This suggests that it is the assembly of subunits into complex I that is affected in mitochondria from Δmia40 plants compared with wild type plants. Furthermore, the unassembled, imported, radiolabeled CA2 appeared to be degraded in mitochondria from Δmia40 plants as evidenced by the decrease in the intensity of lower molecular mass products with time (Fig. 4C).
To gain further insight into the effects of deleting Mia40, the transcriptome of Δmia40 was compared with that of wild type plants. 322 transcripts were altered in abundance in the Δmia40 plants and were significantly overrepresented in the GO annotation-response to stress (biological processes functional categories), representing 8.4% of the changes observed compared with 5.7% in the genome. Furthermore, analysis of the predicted location of the proteins of genes whose transcripts changed in abundance revealed that proteins predicted to be located in peroxisomes were significantly overrepresented by 6.8% compared with the 1.5% observed in the genome (Fig. 5A). A MapMan overview of differentially expressed genes revealed that transcripts involved in responses to biotic stresses were overrepresented, including four superoxide dismutase genes (three copper zinc and one manganese) (Fig. 5B). Thus, overall peroxisomal function seemed to be affected to a greater extent than mitochondrial function at a transcript level. Specifically, transcripts from genes encoding CSD1, CSD2, and Ccs1 were decreased in abundance by more than 2-fold, with significant decreases also observed for CA1 (reduced −1.9-fold). This correlated with the decrease in protein observed by Western blotting.
DISCUSSION
Characterization of the functions of Mia40 in A. thaliana reveal conserved and novel functions compared with studies in yeast. The absence of CSD1 and Ccs1 in mitochondria from Δmia40 plants would be predicted from studies in yeast, and thus, this appears to be a conserved function of Mia40 across wide phylogenetic gaps. However, the decrease in amount and activity of complex I by ∼40% represents an additional role for Mia40 that has not been previously reported. Additionally, Mia40 does not play an essential role in the import and/or assembly of small Tim proteins into A. thaliana mitochondria, as evidenced by the fact that the carrier import pathway operated normally. Mia40 has also taken on additional novel roles in A. thaliana; it is also located in peroxisomes where it is required for the assembly of CSD3 and affects the abundance of a protein (Aim1) associated with β-oxidation of fatty acids. The changes in transcript abundance for many genes encoding peroxisomal proteins suggest that many more proteins are likely to be affected. Finally the absence of Mia40 results in an alteration of the basal transcriptome in A. thaliana. Global transcript analysis illustrated that for many of the proteins that decreased in abundance in Western blots, transcript abundances also decreased, suggesting that not only is Mia40 required for their assembly and stability, but that in its absence signals from the organelles result in these pathways being down-regulated.
The functions of Mia40 in A. thaliana differ considerably compared with those observed in yeast. Our results indicate that it is possible for the mitochondrial disulfide relay system to function without Mia40 in A. thaliana. This is consistent with the findings in trypanosomes, which also lack a Mia40 gene but still successfully import small Tim proteins into the intermembrane space (65). The different arrangement of cysteine pairs in Erv1 in A. thaliana (plants) and trypanosomes (57) may allow Erv1 in these organisms to function alone as a replacement of the Mia40/Erv1 system in yeast. Another possibility is that Erv1 functions in another, as yet unknown, pathway or with other uncharacterized components. Notably, the A. thaliana Erv1 protein could not complement a yeast erv1 mutant, indicating that it functions differently (supplemental Fig. 3), as observed previously (55). Furthermore, the amount of Erv1 significantly increased in A. thaliana Δmia40 plants, indicating that although A. thaliana Mia40 may not be an essential component for the Erv1 disulfide relay pathway in A. thaliana mitochondria, it may still participate in the process. Finally, the increase in Erv1 did not compensate for all of the functions of Mia40 in mitochondria from A. thaliana as both CSD1 and Ccs1 were absent in Δmia40 A. thaliana plants, in addition to the reduction in the amount and activity of complex I.
The reasons for this difference in the MIA pathway between yeast and plants is unclear, but in plants it may relate to the fact that the intermembrane space is also the location of ascorbate biosynthesis (26), and ascorbate can participate in oxidative protein folding (66). No change in total ascorbate was detected for Δmia40 plants (supplemental Fig. 4). Thus, either the plant Erv1 protein can function alone in the disulfide relay system, or if Mia40 is present, it is not essential and can be compensated for by an increase in the abundance of Erv1. It cannot be ruled out that other components that may be unique to the intermembrane space in plants replace the function of Mia40. Note that a calculation of the redox potentials of components of the mitochondrial disulfide relay system suggest that shuttling electrons from a protein substrate (redox potential −310 to −340 mV) via Mia40 (redox potential −200 mV) to Erv1 (redox potential −320 mV) presents a thermodynamically unfavorable reaction that may be overcome by a variety of mechanisms (57). Thus, the absence of Mia40 in this system does not present any mechanistic barrier to the operation of a disulfide relay.
Mia40 acts as a receptor for proteins imported in the intermembrane space in yeast (3). In A. thaliana (and by inference plants in general) the outermembrane receptor components differ from the yeast and mammalian systems. The plant outermembrane Tom20 and OM64 proteins are not orthologous to yeast Tom20 and Tom70 (22, 67–69). Although A. thaliana Mia40 may be orthologous to yeast Mia40, it differs in function (and location). Recently, in yeast it has been shown that Mia40 binds a specific nine-amino acid motif, intermembrane space-targeting signal (ITS) (70). A difference in this intermembrane space-targeting signal sequence may represent a different mode of import for plant intermembrane space proteins compared with yeast. However, when comparing the A. thaliana intermembrane space-targeting signal sequences to yeast, they show a high degree of similarity with at least one of the two crucial amino acids being identical across both species (70) (data not shown). This demonstrates that the binding region in the substrates of Mia40 is the same in yeast and A. thaliana, thus, indicating that this is unlikely to be the reason for the different functions between yeast and A. thaliana Mia40 proteins.
Although there was no deleterious phenotype associated with the deletion of Mia40, there were a number of changes in protein abundance in mitochondria and peroxisomes and a reduction in the import of some precursor proteins via the general import pathway. The abundances of Erv1 and Tim23-2 were increased, but no other significant changes in components of the mitochondrial protein import apparatus were observed. Some components showed a slight decrease, such as Tom20-2, Tom20-3, and Tom20-4 and the cytochrome bc1 complex, indicated by the reduction in the Rieske FeS protein. However, as inactivation of two of three Tom20 protein isoforms in A. thaliana does not greatly affect protein import via the general import pathway (22), these changes in Tom20 isoforms alone are unlikely to account for the observed changes in protein import.
A decrease in the Nad9 subunit of Complex I of the respiratory chain and a parallel decrease in complex I activity was observed in Δmia40 plants. The abundance of the other respiratory complexes did not differ from mitochondria from wild type plants. Additionally, no change was observed for the alternative oxidase at a protein or transcript level, indicating no general mitochondrial stress as a result of deletion of Mia40 (47). Finally, Δmia40 plants did not display any of the growth or developmental abnormalities associated with the absence of complex I that have been previously characterized in A. thaliana and tobacco (71, 72). As the import ability of a variety of proteins was largely unaffected, including several complex I subunits (CA2 and NDUFS8) (60), the reduction in the rate of protein import for some proteins is unlikely to be the cause for the changes in protein abundances observed. Previously we have characterized A. thaliana plants that have all functional Tom20 receptor components inactivated. Although import via the general import pathway was decreased by 80%, there was no detectable affects on plant growth or abundance of complex I or other respiratory chain complexes (22). Thus, the rate of protein import does not affect respiratory complex abundance in mitochondria in A. thaliana.
A study analyzing the phylogenetic distribution of proteins that are substrates for the intermembrane disulfide relay system revealed either two CX3C or two CX9C motifs with sizes between 9 and 18 kDa. From this study it was concluded that these proteins were an ancient family that are widespread in various eukaryotic lineages (74). The functions of many of these proteins (cmc1, cox17, cox19, pet 191, and som1) have been investigated in yeast and are associated with cytochrome c oxidase assembly and to a lesser extent in the cytochrome bc1 complex (12, 74). This is in contrast to what was observed here where cytochrome oxidase was unaffected, and the cytochrome bc1 complex was minimally affected. However, as yeast (S. cerevisiae) lack complex I, any role for Mia40 in complex I assembly (or stability) would not be uncovered using this system. It has been reported that mutations in the human orthologue of Erv1, GFER (growth factor, augmenter of liver regeneration) results in a reduction in activity of complex I, II, and IV activity (52). However in this study, although the absence of Mia40 results in an absence of Ccs1 in mitochondria, there does not appear to be any affect on assembly of cytochrome oxidase that might be associated with altered copper homeostasis and/or insertion into cytochrome oxidase.
The absence of Mia40 in A. thaliana also resulted in an alteration of the transcriptome. The majority of the changes observed were decreases in transcript abundance. Genes encoding peroxisomal proteins were disproportionally affected compared with those encoding mitochondrial or plastid proteins. This is consistent with the targeting and accumulation of Mia40 to peroxisomes. Several transcripts encoding peroxisomal proteins were down-regulated in Δmia40. Two short chain dehydrogenase/reductase (SDRa and SDRb) and Kat2 are involved in β-oxidation (75). The peroxisomal β-oxidation pathway in plants metabolizes saturated long chain fatty acids to supply energy during germination and seedling establishment (76). β-Oxidation not only metabolizes fatty acids but other substrates such as unsaturated fatty acids and hormone precursors may also be fed into this pathway (76, 77). It has been proposed that short chain dehydrogenase/reductase proteins in A. thaliana play an important role in this alternate form of the β-oxidation pathway in hormone metabolism, which also requires Kat2 to provide the thiolase activity (75). This suggests that Mia40 plays some role in fatty acid metabolism in peroxisomes, consistent with the decrease observed in Aim1 protein abundance.
Another interesting transcript, which decreased in abundance in Δmia40, was the homolog of the yeast Ccs1 (copper chaperone for superoxide dismutase 1 (Sod1)), AtCcs1. Mia40 in yeast has been shown to be required for the biogenesis of Ccs1 and Sod1 in the intermembrane space of mitochondria (73). In A. thaliana there are three Sod1-like proteins and one Ccs1-like protein. The three Sod1 proteins are located in plastids (CSD2), the cytosol (CSD1), and peroxisomes (CSD3), and all three have been presumed to be dependent on the one Ccs1 protein for proper function (58). Although no significant decreases were observed in the import of Ccs1 and CSD1 into mitochondria from Δmia40 plants, the lack of these proteins in mitochondria isolated from Δmia40 plants indicates that although Erv1 may be sufficient for import of these proteins, it cannot fulfill the role of Mia40 in assembly of Ccs1 and/or CSD1. Likewise, the absence of CSD3 in peroxisomes suggests that Mia40 is required for the assembly of Ccs1 in peroxisomes. Western blot analysis failed to detect Ccs1 in peroxisomes in A. thaliana, even from wild type plants, suggesting it is present in very low abundance.
Supplementary Material
Acknowledgments
We thank Prof. Harvey Millar and Dr. Holger Eubel for assistance with purification of peroxisomes.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Figs. 1–4.
- MIA
- mitochondrial intermembrane assembly
- CA2
- carbonic anhydrase 2
- Kat2
- 3-ketoacyl-CoA thiolase
- Aim1
- altered inflorescence meristem 1
- Sod1
- superoxide dismutase 1
- Ccs1
- copper chaperone for CSD1
- RFP
- red fluorescent protein
- BN
- blue native
- TES
- 2-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}ethanesulfonic acid.
REFERENCES
- 1.Elstner M., Andreoli C., Ahting U., Tetko I., Klopstock T., Meitinger T., Prokisch H. (2008) Mol. Biotechnol. 40, 306–315 [DOI] [PubMed] [Google Scholar]
- 2.Lang B. F., Gray M. W., Burger G. (1999) Annu. Rev. Genet. 33, 351–397 [DOI] [PubMed] [Google Scholar]
- 3.Chacinska A., Koehler C. M., Milenkovic D., Lithgow T., Pfanner N. (2009) Cell 138, 628–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neupert W., Herrmann J. M. (2007) Annu. Rev. Biochem. 76, 723–749 [DOI] [PubMed] [Google Scholar]
- 5.Walther D. M., Rapaport D. (2009) Biochim. Biophys. Acta 1793, 42–51 [DOI] [PubMed] [Google Scholar]
- 6.Geissler A., Chacinska A., Truscott K. N., Wiedemann N., Brandner K., Sickmann A., Meyer H. E., Meisinger C., Pfanner N., Rehling P. (2002) Cell 111, 507–518 [DOI] [PubMed] [Google Scholar]
- 7.Mokranjac D., Sichting M., Popov-Celeketić D., Mapa K., Gevorkyan-Airapetov L., Zohary K., Hell K., Azem A., Neupert W. (2009) Mol. Biol. Cell 20, 1400–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donzeau M., Káldi K., Adam A., Paschen S., Wanner G., Guiard B., Bauer M. F., Neupert W., Brunner M. (2000) Cell 101, 401–412 [DOI] [PubMed] [Google Scholar]
- 9.Pfanner N., Geissler A. (2001) Nat. Rev. Mol. Cell Biol. 2, 339–349 [DOI] [PubMed] [Google Scholar]
- 10.Preuss M., Ott M., Funes S., Luirink J., Herrmann J. M. (2005) J. Biol. Chem. 280, 13004–13011 [DOI] [PubMed] [Google Scholar]
- 11.Stuart R. A., Neupert W. (1990) Biochimie 72, 115–121 [DOI] [PubMed] [Google Scholar]
- 12.Hell K. (2008) Biochim. Biophys. Acta 1783, 601–609 [DOI] [PubMed] [Google Scholar]
- 13.Koehler C. M., Tienson H. L. (2009) Biochim. Biophys. Acta 1793, 139–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrmann J. M., Kauff F., Neuhaus H. E. (2009) Biochim. Biophys. Acta 1793, 71–77 [DOI] [PubMed] [Google Scholar]
- 15.Chacinska A., Pfannschmidt S., Wiedemann N., Kozjak V., Sanjuán Szklarz L. K., Schulze-Specking A., Truscott K. N., Guiard B., Meisinger C., Pfanner N. (2004) EMBO J. 23, 3735–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lisowsky T. (1992) Mol. Gen. Genet. 232, 58–64 [DOI] [PubMed] [Google Scholar]
- 17.Milenkovic D., Gabriel K., Guiard B., Schulze-Specking A., Pfanner N., Chacinska A. (2007) J. Biol. Chem. 282, 22472–22480 [DOI] [PubMed] [Google Scholar]
- 18.Milenkovic D., Ramming T., Müller J. M., Wenz L. S., Gebert N., Schulze-Specking A., Stojanovski D., Rospert S., Chacinska A. (2009) Mol. Biol. Cell 20, 2530–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riemer J., Bulleid N., Herrmann J. M. (2009) Science 324, 1284–1287 [DOI] [PubMed] [Google Scholar]
- 20.Kutik S., Stroud D. A., Wiedemann N., Pfanner N. (2009) Biochim. Biophys. Acta 1790, 409–415 [DOI] [PubMed] [Google Scholar]
- 21.Lister R., Hulett J. M., Lithgow T., Whelan J. (2005) Mol. Membr. Biol. 22, 87–100 [DOI] [PubMed] [Google Scholar]
- 22.Lister R., Carrie C., Duncan O., Ho L. H., Howell K. A., Murcha M. W., Whelan J. (2007) Plant Cell 19, 3739–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glaser E., Dessi P. (1999) J. Bioenerg. Biomembr. 31, 259–274 [DOI] [PubMed] [Google Scholar]
- 24.Glaser E., Nilsson S., Bhushan S. (2006) Biol. Chem. 387, 1441–1447 [DOI] [PubMed] [Google Scholar]
- 25.Kambacheld M., Augustin S., Tatsuta T., Müller S., Langer T. (2005) J. Biol. Chem. 280, 20132–20139 [DOI] [PubMed] [Google Scholar]
- 26.Lister R., Murcha M. W., Whelan J. (2003) Nucleic Acids Res. 31, 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 28.Carrie C., Kühn K., Murcha M. W., Duncan O., Small I. D., O'Toole N., Whelan J. (2009) Plant J. 57, 1128–1139 [DOI] [PubMed] [Google Scholar]
- 29.Davies S. M., Rackham O., Shearwood A. M., Hamilton K. L., Narsai R., Whelan J., Filipovska A. (2009) FEBS Lett. 583, 1853–1858 [DOI] [PubMed] [Google Scholar]
- 30.Hofmann S., Rothbauer U., Mühlenbein N., Baiker K., Hell K., Bauer M. F. (2005) J. Mol. Biol. 353, 517–528 [DOI] [PubMed] [Google Scholar]
- 31.Whelan J., Smith M. K., Meijer M., Yu J. W., Badger M. R., Price G. D., Day D. A. (1995) Plant Physiol. 107, 1469–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bathgate B., Baker A., Leaver C. J. (1989) Eur. J. Biochem. 183, 303–310 [DOI] [PubMed] [Google Scholar]
- 33.Murcha M. W., Elhafez D., Millar A. H., Whelan J. (2004) J. Mol. Biol. 344, 443–454 [DOI] [PubMed] [Google Scholar]
- 34.Elhafez D., Murcha M. W., Clifton R., Soole K. L., Day D. A., Whelan J. (2006) Plant Cell Physiol. 47, 43–54 [DOI] [PubMed] [Google Scholar]
- 35.Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., Shinn P., Stevenson D. K., Zimmerman J., Barajas P., Cheuk R., Gadrinab C., Heller C., Jeske A., Koesema E., Meyers C. C., Parker H., Prednis L., Ansari Y., Choy N., Deen H., Geralt M., Hazari N., Hom E., Karnes M., Mulholland C., Ndubaku R., Schmidt I., Guzman P., Aguilar-Henonin L., Schmid M., Weigel D., Carter D. E., Marchand T., Risseeuw E., Brogden D., Zeko A., Crosby W. L., Berry C. C., Ecker J. R. (2003) Science 301, 653–657 [DOI] [PubMed] [Google Scholar]
- 36.Eubel H., Meyer E. H., Taylor N. L., Bussell J. D., O'Toole N., Heazlewood J. L., Castleden I., Small I. D., Smith S. M., Millar A. H. (2008) Plant Physiol. 148, 1809–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whelan J., Hugosson M., Glaser E., Day D. A. (1995) Plant Mol. Biol. 27, 769–778 [DOI] [PubMed] [Google Scholar]
- 38.Jänsch L., Kruft V., Schmitz U. K., Braun H. P. (1996) Plant J. 9, 357–368 [DOI] [PubMed] [Google Scholar]
- 39.Carrie C., Murcha M. W., Kuehn K., Duncan O., Barthet M., Smith P. M., Eubel H., Meyer E., Day D. A., Millar A. H., Whelan J. (2008) FEBS Lett. 582, 3073–3079 [DOI] [PubMed] [Google Scholar]
- 40.Cooper H. M., Paterson Y. (2009) Current Protocols in Neuroscience, Chapter 5, Unit 5.5, pp. 5.5.1–5.5.10, John Wiley & Sons, Inc., Wiley Online Library [Google Scholar]
- 41.Footitt S., Cornah J. E., Pracharoenwattana I., Bryce J. H., Smith S. M. (2007) J. Exp. Bot. 58, 2959–2968 [DOI] [PubMed] [Google Scholar]
- 42.Murcha M. W., Elhafez D., Millar A. H., Whelan J. (2005) J. Biol. Chem. 280, 16476–16483 [DOI] [PubMed] [Google Scholar]
- 43.Elthon T. E., Nickels R. L., McIntosh L. (1989) Plant Physiol. 89, 1311–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamattina L., Gonzalez D., Gualberto J., Grienenberger J. M. (1993) Eur. J. Biochem. 217, 831–838 [DOI] [PubMed] [Google Scholar]
- 45.Baldi P., Long A. D. (2001) Bioinformatics 17, 509–519 [DOI] [PubMed] [Google Scholar]
- 46.Allison D. B., Cui X., Page G. P., Sabripour M. (2006) Nat. Rev. Genet. 7, 55–65 [DOI] [PubMed] [Google Scholar]
- 47.Van Aken O., Zhang B., Carrie C., Uggalla V., Paynter E., Giraud E., Whelan J. (2009) Mol. Plant 2, 1310–1324 [DOI] [PubMed] [Google Scholar]
- 48.Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L. A., Rhee S. Y., Stitt M. (2004) Plant J. 37, 914–939 [DOI] [PubMed] [Google Scholar]
- 49.Rissler M., Wiedemann N., Pfannschmidt S., Gabriel K., Guiard B., Pfanner N., Chacinska A. (2005) J. Mol. Biol. 353, 485–492 [DOI] [PubMed] [Google Scholar]
- 50.Hoppins S. C., Nargang F. E. (2004) J. Biol. Chem. 279, 12396–12405 [DOI] [PubMed] [Google Scholar]
- 51.Banci L., Bertini I., Cefaro C., Ciofi-Baffoni S., Gallo A., Martinelli M., Sideris D. P., Katrakili N., Tokatlidis K. (2009) Nat. Struct. Mol. Biol. 16, 198–206 [DOI] [PubMed] [Google Scholar]
- 52.Di Fonzo A., Ronchi D., Lodi T., Fassone E., Tigano M., Lamperti C., Corti S., Bordoni A., Fortunato F., Nizzardo M., Napoli L., Donadoni C., Salani S., Saladino F., Moggio M., Bresolin N., Ferrero I., Comi G. P. (2009) Am. J. Hum. Genet. 84, 594–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chacinska A., Guiard B., Müller J. M., Schulze-Specking A., Gabriel K., Kutik S., Pfanner N. (2008) J. Biol. Chem. 283, 29723–29729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma C., Reumann S. (2008) J. Exp. Bot. 59, 3767–3779 [DOI] [PubMed] [Google Scholar]
- 55.Levitan A., Danon A., Lisowsky T. (2004) J. Biol. Chem. 279, 20002–20008 [DOI] [PubMed] [Google Scholar]
- 56.Gerber J., Mühlenhoff U., Hofhaus G., Lill R., Lisowsky T. (2001) J. Biol. Chem. 276, 23486–23491 [DOI] [PubMed] [Google Scholar]
- 57.Endo T., Yamano K., Kawano S. (2010) Antioxid. Redox Signal. 13, 1359–1573 [DOI] [PubMed] [Google Scholar]
- 58.Chu C. C., Lee W. C., Guo W. Y., Pan S. M., Chen L. J., Li H. M., Jinn T. L. (2005) Plant Physiol. 139, 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kawamata H., Manfredi G. (2010) Antioxid. Redox. Signal 13, 1375–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klodmann J., Sunderhaus S., Nimtz M., Jänsch L., Braun H. P. (2010) Plant Cell 22, 797–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meyer E. H., Taylor N. L., Millar A. H. (2008) J. Proteome Res. 7, 786–794 [DOI] [PubMed] [Google Scholar]
- 62.Unseld M., Marienfeld J. R., Brandt P., Brennicke A. (1997) Nat. Genet. 15, 57–61 [DOI] [PubMed] [Google Scholar]
- 63.Lazarou M., Thorburn D. R., Ryan M. T., McKenzie M. (2009) Biochim. Biophys. Acta 1793, 78–88 [DOI] [PubMed] [Google Scholar]
- 64.Vögtle F. N., Schmidt O., Chacinska A., Pfanner N., Meisinger C. (2010) Methods Mol. Biol. 619, 425–436 [DOI] [PubMed] [Google Scholar]
- 65.Gentle I. E., Perry A. J., Alcock F. H., Likić V. A., Dolezal P., Ng E. T., Purcell A. W., McConnville M., Naderer T., Chanez A. L., Charrière F., Aschinger C., Schneider A., Tokatlidis K., Lithgow T. (2007) Mol. Biol. Evol. 24, 1149–1160 [DOI] [PubMed] [Google Scholar]
- 66.Bánhegyi G., Csala M., Szarka A., Varsányi M., Benedetti A., Mandl J. (2003) Biofactors 17, 37–46 [DOI] [PubMed] [Google Scholar]
- 67.Chew O., Lister R., Qbadou S., Heazlewood J. L., Soll J., Schleiff E., Millar A. H., Whelan J. (2004) FEBS Lett. 557, 109–114 [DOI] [PubMed] [Google Scholar]
- 68.Perry A. J., Hulett J. M., Likić V. A., Lithgow T., Gooley P. R. (2006) Curr. Biol. 16, 221–229 [DOI] [PubMed] [Google Scholar]
- 69.Perry A. J., Rimmer K. A., Mertens H. D., Waller R. F., Mulhern T. D., Lithgow T., Gooley P. R. (2008) Plant Physiol. Biochem. 46, 265–274 [DOI] [PubMed] [Google Scholar]
- 70.Sideris D. P., Petrakis N., Katrakili N., Mikropoulou D., Gallo A., Ciofi-Baffoni S., Banci L., Bertini I., Tokatlidis K. (2009) J. Cell Biol. 187, 1007–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meyer E. H., Tomaz T., Carroll A. J., Estavillo G., Delannoy E., Tanz S. K., Small I. D., Pogson B. J., Millar A. H. (2009) Plant Physiol. 151, 603–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vidal G., Ribas-Carbo M., Garmier M., Dubertret G., Rasmusson A. G., Mathieu C., Foyer C. H., De Paepe R. (2007) Plant Cell 19, 640–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reddehase S., Grumbt B., Neupert W., Hell K. (2009) J. Mol. Biol. 385, 331–338 [DOI] [PubMed] [Google Scholar]
- 74.Longen S., Bien M., Bihlmaier K., Kloeppel C., Kauff F., Hammermeister M., Westermann B., Herrmann J. M., Riemer J. (2009) J. Mol. Biol. 393, 356–368 [DOI] [PubMed] [Google Scholar]
- 75.Wiszniewski A. A., Zhou W., Smith S. M., Bussell J. D. (2009) Plant Mol. Biol. 69, 503–515 [DOI] [PubMed] [Google Scholar]
- 76.Baker A., Graham I. A., Holdsworth M., Smith S. M., Theodoulou F. L. (2006) Trends Plant Sci. 11, 124–132 [DOI] [PubMed] [Google Scholar]
- 77.Goepfert S., Poirier Y. (2007) Curr. Opin. Plant Biol. 10, 245–251 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.