Abstract
Lectin-like transcript 1 (LLT1) encoded by CLEC2D gene is a C-type lectin-like molecule interacting with human CD161 (NKR-P1A) receptor expressed by natural killer cells and subsets of T cells. Using RT-PCR and sequencing, we identified several CLEC2D alternatively spliced transcript variants generated by exon skipping. In addition to the reported transcript variants 1 (LLT1) and 2, we identified a novel splice variant 4 and transcripts coding for putative soluble proteins. CLEC2D transcripts were detected primarily in hematopoietic cell lines and were found to be co-induced by the same activation signals. Although very low amounts of putative soluble CLEC2D protein isoforms could be produced by transfectants, CLEC2D isoforms 2 and 4 were efficiently expressed. By contrast to LLT1, which was detected on the cell surface, isoform 2 and 4 remained in the endoplasmic reticulum where they formed homodimers or heterodimers with LLT1. They failed to interact with CD161, leaving LLT1 as the sole ligand for this receptor. CLEC2D therefore uses gene splicing to generate protein isoforms that are structurally distinct and that have different biological activities.
Keywords: Cell Surface Receptor, Gene Expression, Immunology, Lymphocyte, MHC
Introduction
Multiple receptors are expressed on the surface of natural killer (NK)2 cells and are required for target recognition and effector functions. Among them, a group of receptors displays C-type lectin-like domains and is encoded within the NK gene complex on chromosome 6 in mice and chromosome 12 in humans. They are called C-type lectin-like because they have lost the Ca2+-dependent carbohydrate recognition domain present in most conventional C-type lectins while maintaining partial structural similarities (1). They interact with ligands through protein-protein interaction rather than binding carbohydrates as do conventional C-type lectins. Interestingly, it was observed that genes coding for some of these receptors and their respective ligands co-segregated in the NK gene complex. This is the case for KLRB1 genes coding for the NKR-P1 family of receptors located in close proximity to CLEC2D genes coding for osteoclast inhibitory lectin molecules also named C-type lectin related (Clr) in mice and rats or lectin-like transcript 1 (LLT1) in humans. In mice, the inhibitory NKR-P1B/D was described to bind to Clrb, and the activating NKR-P1F was described to bind to Clrg and/or Clrx (2–5). The functions of these interactions remain largely unknown. Clrb was found ubiquitously expressed and may count among the MHC-independent NK missing self-recognition mechanisms. Clrg and Clrx expression patterns are more restricted. Their interaction with NKR-P1F induced an activating signal, but further studies are required to understand their role in vivo. Similarly, NKR-P1/Clr interactions have been identified in rats (6). In addition, we and others identified LLT1 as the ligand of human CD161 receptor (NKR-P1A) (7, 8). Expression of LLT1 on transfectants inhibited NK cell cytotoxicity and IFN-γ secretion. Its role on T cells remains controversial. It has been shown that CD161 triggered a co-stimulatory signal on T cells and NKT cells (7, 9, 10) or inhibited TNF-α secretion by CD8+ T cells (8). In an attempt to further characterize the physiological role played by LLT1/CD161 interaction, we sought to characterize LLT1 expression profile. By doing so, we identified several alternatively spliced transcript variants encoded by the CLEC2D gene. We demonstrated that although several CLEC2D protein isoforms can be expressed, only LLT1 bound to CD161.
EXPERIMENTAL PROCEDURES
Cell Lines and Primary Cells
The following human cell lines were maintained in complete RPMI 1640 or Iscove's modified Dulbecco's medium (Lonza, Levallois-Perret Cedex/Paris, France) supplemented with 10% fetal calf serum (Pierce), penicillin (100 IU/ml), and streptomycin (100 μg/ml) (Lonza): 293T embryonic kidney cell line; C1R, 721.45, 721.221, JY, Col., and Lep. B cell lines; THP-1 acute monocytic leukemia; Jurkat acute T cell leukemia; Raji Burkitt lymphoma; and U373 glioblastoma-astrocytoma. 293T or C1R cells stably expressing LLT1 or CD161 were described previously (7). 293T-AICL stable transfectants were obtained by cell sorting of EGFP+ electroporated 293T cells transduced with full-length AICL cloned into pIRES2-EGFP vector (Clontech). Human peripheral blood mononuclear cells (PBMCs) were separated by Ficoll-Paque Plus density gradient centrifugation (GE Healthcare) from blood purchased from the Etablissement Français du Sang. Monocytes, B cells, and NK cells were isolated by positive magnetic selection with anti-CD14, anti-CD19, or anti-CD56 microbeads (Miltenyi Biotec, Paris, France), respectively, and further sorted by flow cytometry on a FACSVantage (Becton Dickinson, Le Pont de Claix Cedex, France) using anti-CD3, -CD56, -CD19, and -CD14 mAbs (BD Biosciences, Le Pont de Claix, France). T cells were directly sorted by flow cytometry as CD3+, CD3+CD4+, or CD3+CD8+ cells using anti-CD3, -CD4, and -CD8 mAbs (BD Biosciences). Purity was typically >92–99%. Monocyte-derived dendritic cells (DCs) were generated from positively selected CD14+ monocytes cultured for 5 days in the presence of 200 units/ml IL-4 (BD Biosciences) and 50 ng/ml GM-CSF (PeproTech, Neuilly-sur-Seine, France). The cells were matured by the addition of 1 μg/ml LPS (Sigma) or a mixture of cytokines (BD Biosciences) including IL-1β (10 ng/ml), IL-6 (10 ng/ml), TNF-α (10 ng/ml), and prostaglandin E2 (1000 units/ml) over 2 days. DC maturation was monitored by the acquisition of CD83 expression. CD69-expressing activated T cells were obtained after stimulation of polyclonal human T cells with 5 ng/ml phorbol 12-myristate 13-acetate and 500 ng/ml ionomycin (Sigma) for 4 h.
Cloning of CLEC2D Alternatively Spliced Transcript Variants: RT-PCR and Real Time RT-PCR
Total RNA were extracted with TRI® reagent (Euromedex, Souffelweyersheim, France), contaminating genomic DNA were removed using RQ1 DNase (Promega, Charbonnieres, France), and RNA was precipitated using lithium chloride (Ambion, Applied Biosystems, Courtaboeuf Cedex, France). cDNAs were synthesized by reverse transcription using SuperScript II reverse transcriptase (Invitrogen) and random hexamer primers (Roche Applied Science) in the presence of RNasin Plus (Promega). cDNAs were used to clone CLEC2D transcript variants and to perform standard and real time RT-PCR.
For the cloning, the following primers were used: LLT1-F, 5′-ATGCATGACAGTAACAATGTGG-3′ and LLT1-R, 5′-TAGTTGGGGCTTTGCTGTAA-3′ (Eurogentec, Angers, France). PCR conditions were as follows: 30 cycles of denaturation at 95 °C for 45 s, annealing at 60 °C for 45 s, and extension at 72 °C for 1 min. PCR products were purified, subcloned into pGEMTeasy according to the manufacturer's instructions (Promega), and sequenced.
For RT-PCR, 30 cycles of denaturation at 95 °C for 45 s, annealing at 60 °C for 45 s, and extension at 72 °C for 1 min were performed using the following primers: for CLEC2D variant 1, F1–2, 5′-GCTTCCAGGAACTGAATTTC-3′ and R1, 5′-CCCAGGATAGGAAACTGTC-3′; for CLEC2D variant 2, F1–2, 5′-GCTTCCAGGAACTGAATTTC-3′ and R2, 5′-CCCAGGATAGGAAACCATGA-3′; for CLEC2D variant 4, F4, 5′-CCTGCAAAGCCAGGTTG-3′ and R4, 5′-CCAGGATAGGAAACCAGTT-3′; and for β-actin: Actin-F, 5′-GGCATGGGTCAGAAGGATT-3′ and Actin-R, 5′-TTCTCCATGTCGTCCCAGTT-3′.
For real time RT-PCR, amplification (45 cycles of denaturation at 95 °C for 10 s, annealing at 60 °C for 10 s, and extension at 72°c for 10 s) was monitored using SYBR green on a Light-Cycler (Roche Applied Science). The following primers were used: for CLEC2D variant 1: 5′-TTCCTATCCTGGGAGCAGGA-3′ and 5′-GACATGTATATCTGATTTGGAACAA-3′; for CLEC2D variant 2: 5′-GAGCAGAGAACAAGGCCAAC-3′ and 5′-CCCAGGATAGGAAACCATGA-3′; and for β-actin: Actin-F and Actin-R as described above. Transcript levels were expressed relative to β-actin.
Constructs and Transduction
Full-length CLEC2D variants 2 and 4 were cloned into RSV5.neo containing C-terminal c-Myc and hexahistidine tag sequences. This vector and control RSV5-CLEC2A were generously provided by A. Steinle and described previously (11). Transient expression was obtained by electroporation of 293T and 293T-LLT1 cells (250 V, 500 microfarads). CLEC2D variant 5 and the sequence coding for the extracellular domain of LLT1 (aa 60–191) were cloned into pSecTag2hygroA containing c-Myc and hexahistidine tag sequences (Invitrogen). COS cells were transiently transfected using a DEAE-dextran method, and soluble LLT1 was detected in the supernatant after 3 days of culture.
Multimers were generated as described previously (7). The extracellular domains of CLEC2D isoforms 2 (aa 60–194) and 4 (aa 60–132) or CLEC2A (aa 52–174) were cloned into pcDNA3 (Invitrogen) in-frame with the signal sequence of human CD5 and the Fc portion of human IgG1. Dimers were produced in 293T, and multimeric complexes were generated as described previously (7).
Anti-LLT1 Antibodies
Anti-LLT1 mAbs were generated by immunization of RBF mice (Charles River, Chatillon sur Chalaronne, France) three times with Hela-LLT1 cells followed by a boost with recombinant LLT1-Fc fusion proteins (7). Three days later, the splenocytes were fused with Fox-NY myeloma cells (ATCC, Manassas, VA). Hybridoma's supernatants were screened for production of LLT1 specific mAbs by flow cytometry and ELISA. Two clones were characterized: 2F1 and 4F68. Anti-LLT1 MAB3480 and 4C7 were purchased from R & D Systems (Lille, France) and Abnova (Abnova, Interchim, Montluçon, France), respectively.
Biochemistry and Western Blot
The cells were lysed in 1% Igepal CA-630 (Sigma), 40 mm Tris-HCl, pH 8, 150 mm NaCl, 5 mm EDTA, 5 mm iodoacetamide, 2 mm PMSF, and protease inhibitor mixture (Complete Mini tablet; Roche Applied Science) at 4 °C for 30 min. Whole cell lysates were treated with endoglycosidase H (Endo H) (New England Biolabs, OZYME, Saint Quentin Yvelines Cedex, France) or peptide N-glycosidase F (PNGase F) (Roche Applied Science) according to the manufacturer's instructions. For immunoprecipitations, the cells were lysed in 0.5% Igepal, 50 mm n-octylglucoside (Sigma), 40 mm Tris-HCl, pH 8, 150 mm NaCl, 5 mm EDTA, 5 mm iodoacetamide, 2 mm PMSF, and protease inhibitor mixture (Complete Mini tablet; Roche Applied Science) at 4 °C for 30 min. CLEC2D isoforms 2 and 4 were immunoprecipitated using anti-Myc 9E10 mAb (generated in house) and protein A-Sepharose (Sigma). The proteins were loaded on 12% SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad). After saturation, the membranes were incubated with either anti-LLT1 2F1 or anti-Myc 9E10 mAb followed by HRP-conjugated F(ab′)2 fragment goat anti-mouse IgG (Jackson ImmunoResearch Laboratories) or anti-β-actin mAb (Sigma) followed by HRP-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories). Binding of the mAbs was revealed using SuperSignal West Femto (Pierce).
Flow Cytometry
Untransfected and transfected 293T or 293T-LLT1 cells were stained with isotype mIgG1 MOPC-21 (Sigma), anti-Myc 9E10, or anti-LLT1 4F68 mAb followed by allophycocyanin-conjugated goat anti-mouse Ig (BD Biosciences). The anti-AICL 7GA mAb was kindly provided by A. Steinle (12). Anti-CD3 and anti-CD69 mAbs were purchased from BD Biosciences. 293T-LLT1 and 293T-CD161 were incubated with CLEC2D multimers as described previously (7). Fluorescence was analyzed on a FACSCalibur cytometer with the CellQuest Pro Software (BD Biosciences).
ELISA
Fc fusion proteins (10 μg/ml in PBS) were coated onto a Maxisorp ELISA plate (Nunc, VWR International S.A.S., Fontenay-sous-Bois Cedex, France) and blocked with PBS, 0.1% Tween, 5% BSA. Primary antibodies were incubated at 5 μg/ml in PBS, 0.1% BSA for 2 h at room temperature followed by HRP-conjugated F(ab′)2 fragment goat anti-mouse IgG (Jackson ImmunoResearch Laboratories) for 1 h at room temperature. Peroxidase activity was detected using SIGMAFASTTM OPD according to the manufacturer's instructions (Sigma).
Homology Modeling of CLEC2D Isoforms and Structural Representation
To obtain qualitative structural insight, homology models of the three CLEC2D isoforms have been built. For CLEC2D isoform 1 (191 aa), the CD69 structure that has the Protein Data Bank code 3HUP (114-aa overlap, 62.3% conserved) (13) was chosen as template for the homology model, based on homology with positions 73–186 of CLEC2D isoform 1. For CLEC2D isoform 2 (194 aa), identification of a template was more challenging. The CD69 structure, 3HUP (79-aa overlap, 69.6% conserved), only has homology to positions 73–151 of isoform 2. However, NKG2A structure, chain F, code 3CDG (102-aa overlap, 49.0% conserved) (14), shows homology to aa 75–175 of CLEC2D isoform 2 and was chosen as template. For CLEC2D isoform 4 (132 aa), KLRG1 structure chain C, code 3FF8 (57-aa overlap, 49.1% conserved) (15) shows homology to aa 75–131 and was chosen as template. The homology models were generated using MOE (version 2009.10, Chemical Computing Group, Köln, Germany) by aligning sequences with templates and subsequently building the models using built in procedures. All of the structure representations were drawn with PyMol (The PyMOL Molecular Graphics System, version 1.2r3pre, Schrödinger, LLC., Camberley, UK).
RESULTS
Identification of Alternatively Spliced Transcript Variants of CLEC2D
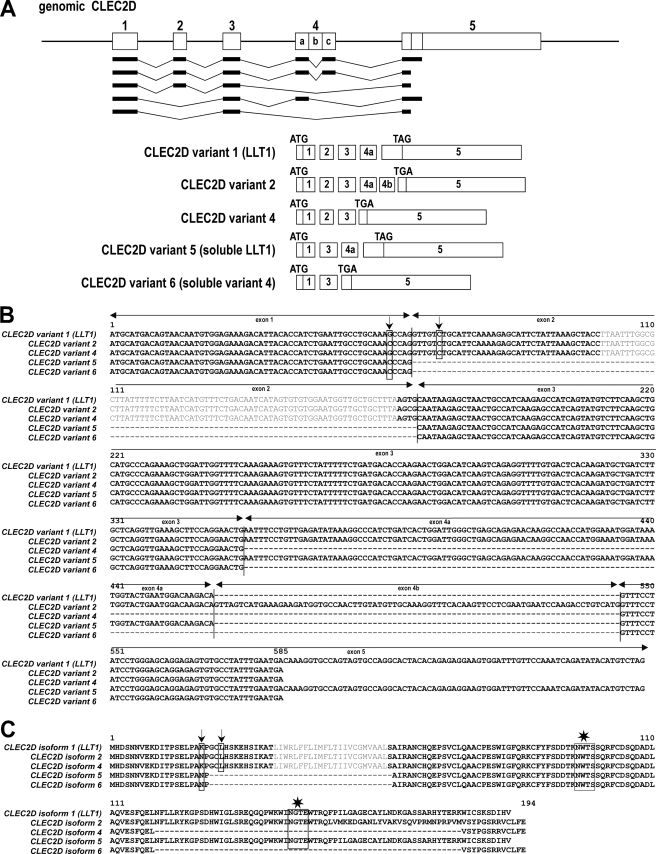
RT-PCR using primers encompassing the predicted coding region of full-length LLT1 was carried out on total RNA from PBMCs. Surprisingly, instead of the expected 599-bp band, we detected several bands between 400 and 800 bp. CLEC2D gene is composed of five exons separated by four introns (Fig. 1A). Three alternative transcripts have been described in the expressed sequence tag databases: one coding for LLT1 (isoform 1), one for isoform 2, and a third one being a nonsense RNA decay candidate (originally named isoform 3). As shown in Fig. 1A, CLEC2D variants 1 and 2 differ in their use of splice donor and acceptor sites within exon 4 leading to an additional sequence called 4b in variant 2. This modifies the reading frame in exon 5 and thus the stop codon usage. To assess whether the detected bands were transcripts generated from alternative splicing, we purified and sequenced them. Using this approach, we identified at least five alternatively spliced CLEC2D variants generated by exon skipping (Fig. 1, A and B). In addition to the previously reported variants 1 and 2, we also identified a novel variant 4 predicted and annotated by Ensembl gene build as CLEC2D-202 that has been attributed the GenBankTM accession number FN813349. This variant 4 is composed of exons 1, 2, 3, and 5 (Fig. 1A). Two other variants were sequenced and resulted from the excision of exon 2 that led to an in-frame deletion of the entire transmembrane region. One coded for a putative soluble form of LLT1 (variant 5, GenBankTM accession number FN813350), and the other one coded for a putative soluble form of newly identified variant 4 (variant 6, GenBankTM accession number FN13351). In agreement with the single-nucleotide polymorphism databases, we also found two nonsynonymous single-nucleotide polymorphisms resulting in N19K and L23V substitutions (Fig. 1, B and C).
FIGURE 1.
Identification of CLEC2D alternatively spliced transcript variants. A, schematic representation of CLEC2D transcript variants. The exons are numbered, and initiation codons (ATG) and stop codons (TAG or TGA) are indicated. B, alignment of nucleotide sequences of cloned CLEC2D transcripts. The positions of single-nucleotide polymorphisms are shown by the arrows. C, alignment of predicted CLEC2D protein isoforms. Polymorphisms are indicated by the arrows, and predicted N-glycosylation sites are indicated by the stars. Transmembrane segments in plain text were predicted using the TMHMM2 program.
The putative CLEC2D protein isoforms were aligned (Fig. 1C). They all code for type II transmembrane or soluble proteins and carry one or two N-linked glycosylation motifs. Interestingly, whereas the N-terminal domains are identical, the C-terminal portions of the sequences coding for the extracellular domains differ significantly. LLT1 and CLEC2D isoform 2 possess C-type lectin-like domains between amino acids 75 and 185 and between amino acids 75–192, respectively, whereas CLEC2D isoform 4 has lost this domain.
Expression of CLEC2D Alternatively Spliced Transcript Variants in Human Cells
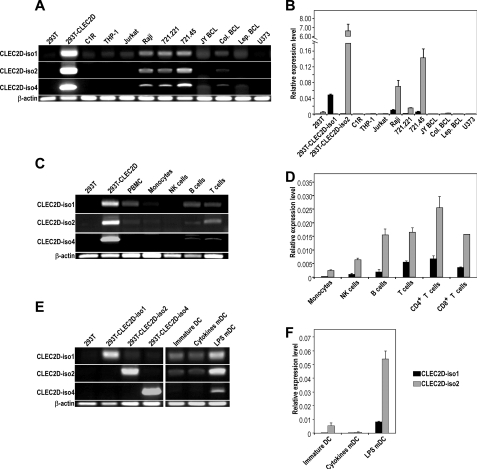
Given the multiple existing alternatively spliced transcript variants, we next assessed in which cell types they could be detected and whether they were differentially expressed. To do so, we set up standard and real time RT-PCR experiments using primers primarily overlapping exon-exon junctions to specifically amplify each transcript. A specific amplification yielding a single band was obtained for CLEC2D variant 1 (LLT1) and variant 2 but not for CLEC2D variant 4. The latter was then only studied by standard RT-PCR where we could discriminate it through its size. We first analyzed total RNA derived from a panel of cell lines. Interestingly, we observed that the three CLEC2D splice variants were consistently detected in the same cell lines. However, their expression was restricted primarily to cells of the B lineage, including the Burkitt lymphoma Raji and the mutant and parental B cell lines 721.221 and 721.45 (Fig. 2, A and B). We could not detect CLEC2D transcript variants in a panel of tumor cell lines, osteoblast cell lines, fibroblast, and myoblasts (data not shown). We then used total RNA derived from PBMCs of healthy donors. In peripheral blood, the three variants were detected primarily in T and B lymphocytes, were found at lower levels in NK cells, and were almost absent from monocytes (Fig. 2, C and D). Quantification of mRNA expression by real time RT-PCR showed that the relative amount of CLEC2D variant 2 was higher than CLEC2D variant 1 (2–14-fold increase). LLT1 was previously reported to be expressed upon activation, following specific Toll-like receptor stimulation (16). Similarly, we found CLEC2D variant 1 (LLT1) transcripts but also CLEC2D variants 2 and 4 in LPS-matured monocyte-derived DCs and not in immature DCs or DCs matured with a mixture of cytokines (Fig. 2, E and F). Collectively, these data indicated that CLEC2D gene splicing generates several transcripts that are expressed by the same cells and are regulated by the same stimuli.
FIGURE 2.
Relative amount of CLEC2D spliced transcript variants in cell lines and PBMCs. A, C, and E, CLEC2D transcript variants 1, 2, and 4 were amplified by RT-PCR. B, D, and F, CLEC2D transcript variants 1 and 2 were quantified by real time RT-PCR. A and B, in the indicated cell lines. C and D, in the indicated cell populations isolated from peripheral blood of a representative healthy donor. E and F, in immature and mature monocyte-derived DCs. The results are representative of two to four independent experiments.
Identification of CLEC2D Protein Isoforms and Specificity of Anti-LLT1 mAbs
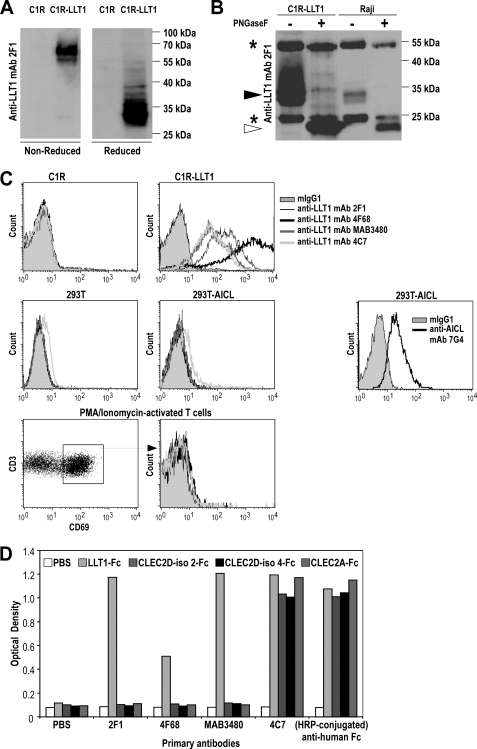
We next assessed whether CLEC2D alternatively spliced transcript variants could be translated in human cells. To do so, two novel anti-LLT1 mAbs were generated from immunization of mice with LLT1-expressing cells and LLT1-Fc fusion proteins (7). Clone 2F1 identified LLT1 on Western blot as a 66–70-kDa protein under nonreducing conditions and 33–35-kDa protein under reducing conditions, consistent with LLT1 expressed as a disulfide-bonded homodimer (Fig. 3A). Clone 4F68 immunoprecipitated LLT1 from C1R-LLT1 transfectants and the Burkitt lymphoma Raji (Fig. 3B). Removal of N-linked glycosylations by PNGase F treatment yielded a protein of ∼22 kDa corresponding to the predicted molecular mass of LLT1 (Fig. 3B). Clone 4F68 was found to be the most efficient mAb detecting LLT1 by flow cytometry on C1R-LLT1 transfectants when compared with 2F1 and commercially available MAB3480 and 4C7 mAbs (Fig. 3C). Interestingly, competition assays showed that none of the anti-LLT1 mAbs competed with 4F68, and only MAB3480 partially decreased 2F1 staining (data not shown). Both 2F1 and 4F68 did not bind to AICL and CD69, the closest homologues of LLT1, when tested on AICL-expressing transfectants or CD69+ activated T cells (Fig. 3C). mAbs 2F1 and 4F68 also specifically recognized plate-bound recombinant LLT1-Fc and not the other CLEC2D isoform-Fc proteins or CLEC2A-Fc. A similar binding pattern was obtained with MAB3480 mAb. By contrast, 4C7 mAb detected the three recombinant CLEC2D protein isoforms and CLEC2A, demonstrating that this antibody is not specific of LLT1 (Fig. 3D). Its cross-reactivity with CLEC2A was confirmed by staining U937 cells reported to express CLEC2A but not LLT1 (data not shown).
FIGURE 3.
Characterization of anti-LLT1 mAbs. A, homodimers and monomers of LLT1 detected in C1R-LLT1 cells by Western blot with anti-LLT1 mAb 2F1 under nonreducing and reducing conditions, respectively. B, LLT1 detection by Western blot using 2F1 mAb following immunoprecipitation with anti-LLT1 4F68, digestion with PNGase F when indicated, and loading on an SDS-PAGE under reducing conditions. The black arrowhead indicates monomers of LLT1, and the white arrowhead indicates deglycosylated monomers of LLT1. The asterisks indicate heavy and light chains of 4F68 mAb used to immunoprecipitate LLT1. C, the specificity of anti-LLT1 mAbs was analyzed by flow cytometry. They stained C1R-LLT1 cells but not C1R cells, AICL-transfected 293T cells, and CD69+ activated T cells. AICL expression was shown using anti-AICL mAb 7G4. D, binding of anti-LLT1 mAbs to plate-bound CLEC2D isoforms and CLEC2A-Fc fusion proteins. The results are representative of three independent experiments.
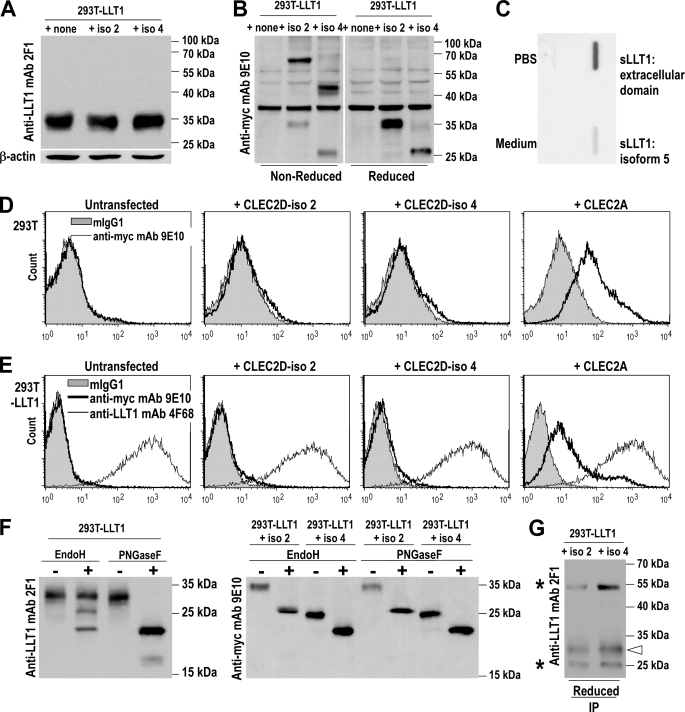
To check expression of the CLEC2D isoforms, the various CLEC2D transcripts were cloned into expression vectors containing a c-Myc and hexahistidine tag at the C terminus, except for full-length CLEC2D variant 1 (LLT1) cloned into pIRES2-EGFP (7). Expression of CLEC2D protein isoforms 2 and 4 was confirmed by Western blot analysis of whole cell lysates of 293T and 293T-LLT1 transfected cells. In co-transfections with LLT1, 2F1 exclusively detected LLT1 (Fig. 4A), whereas CLEC2D isoform 2 and 4 proteins were respectively detected at 70 and 52 kDa under nonreducing conditions and 35 and 26 kDa under reducing conditions using anti-Myc mAb 9E10 (Fig. 4B). Homodimers of isoforms 2 and 4 were also detected in 293T cells (data not shown). These findings demonstrated that in addition to LLT1, CLEC2D isoforms 2 and 4 were expressed as proteins and could form homodimers.
FIGURE 4.
Expression of CLEC2D protein isoforms in transfectants. 293T cells were transfected with CLEC2D isoform 1 (LLT1) alone or together with CLEC2D isoforms 2 and 4 or with CLEC2A. A, LLT1 alone was detected by Western blot analysis of whole cell lysates using 2F1 mAb under reducing conditions. β-Actin was used as internal control. B, CLEC2D isoforms 2 and 4 were detected on whole cell lysates using anti-Myc 9E10 mAb. A nonspecific band was detected in all samples at ∼38 kDa. C, supernatants of transfected COS cells were analyzed for the presence of soluble LLT1 by slot-blot using anti-Myc 9E10 mAb. D and E, cell surface expression of CLEC2D protein isoforms was analyzed by flow cytometry on 293T cells (D) or 293T-LLT1 cells (E) transfected with CLEC2D isoforms or CLEC2A. LLT1 expression was monitored using anti-LLT1 4F68 mAb and the other CLEC2D isoforms and CLEC2A using anti-Myc 9E10 mAb. F, CLEC2D protein isoform expression on whole cell lysates following Endo H or PNGase F treatment. G, LLT1 expression after immunoprecipitation of CLEC2D isoforms 2 and 4 with anti-Myc, revealed using 2F1 mAb. The white arrow indicates monomers of LLT1 under reducing conditions, and asterisks indicate the heavy and light chains of the anti-Myc mAb used to immunoprecipitate. The results are representative of two to four independent experiments.
The sequencing of transcripts had shown that excision of exon 2 generated alternatively spliced transcripts coding for putative soluble CLEC2D isoforms. We therefore measured the presence of soluble LLT1 in the supernatant of transiently transfected COS cells by slot-blot using anti-Myc 9E10 (Fig. 4C) or anti-LLT1 2F1 (data not shown). We consistently found very low amounts of soluble LLT1 in the supernatant of cells transfected with the expression vector containing the splice variant 5, whereas a significant amount of soluble LLT1 was produced from a vector coding only for the extracellular domain of LLT1. These data suggested that excision of exon 2 is not sufficient to allow the production of soluble CLEC2D protein isoforms.
CLEC2D Isoforms 2 and 4 Remain in the Endoplasmic Reticulum (ER)
Because CLEC2D protein isoforms 2 and 4 were detected in whole cell lysates, we investigated whether they were expressed at the cell surface. Flow cytometry analysis of transiently transfected 293T cells using anti-Myc mAb did not reveal any cell surface expression of CLEC2D isoforms 2 and 4, whereas CLEC2A cloned in the same expression vector was expressed (Fig. 4D). We next tested whether their cell surface expression required the presence of LLT1, similarly to CD94/NKG2A efficiently expressed when both molecules are present in a single cell (17). As shown in Fig. 4E, CLEC2D isoforms 2 and 4 were still not expressed on the surface of a transfected 293T-LLT1 clone, whereas the presence of LLT1 did not prevent expression of CLEC2A. The level of expression of LLT1 was also not affected by the presence of any of the CLEC2D isoforms or CLEC2A. To assess whether CLEC2D isoforms 2 and 4 remained in the ER, we treated whole cell lysates of co-transfectants with Endo H. Two forms of LLT1, one resistant and one sensitive, were identified, consistent with LLT1 trafficking to the cell surface. The Endo H-sensitive form yielded two bands corresponding to partial and complete or nearly complete removal of sugars as compared with PNGase F-treated samples (Fig. 4F). By contrast, CLEC2D isoforms 2 and 4 were entirely Endo H-sensitive, yielding a band at the level of PNGase F-treated samples (Fig. 4F). These data indicated that CLEC2D isoforms 2 and 4 remained in the ER and that a majority formed homodimers (Fig. 4B). However, because of the homology between CLEC2D isoforms in domains generally involved in homodimerization, we also monitored the presence of heterodimers. CLEC2D isoforms 2 and 4 were immunoprecipitated from co-transfectants using anti-Myc mAb, and LLT1 was detected by Western blot (Fig. 4G). This suggested that heterodimers could also be formed. Interestingly, the LLT1 form co-immunoprecipitated with CLEC2D isoforms 2 and 4 had an apparent molecular mass of 33 kDa, whereas LLT1 is generally detected as a smear between 33 and 35 kDa. This observation is consistent with differential glycosylations that could be correlated with acquisition of sugars while migrating to the Golgi and to the cell surface. The heterodimers would therefore contain the ER-resident form of LLT1.
CLEC2D Protein Isoform 1 (LLT1) Is the Only Ligand of CD161 Receptor
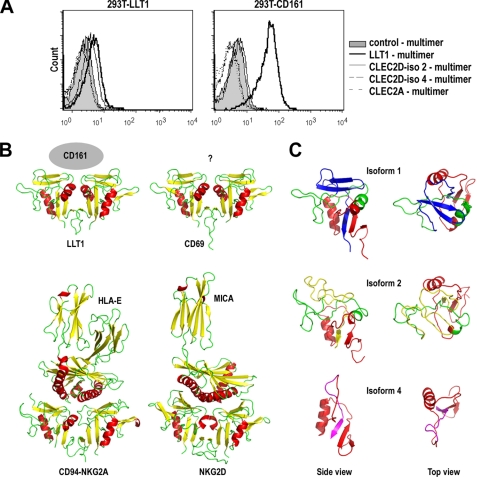
We and others previously demonstrated that LLT1 is the ligand of the human CD161 (NKR-P1A) receptor (7, 8). To assess whether the other CLEC2D isoforms could also bind to this receptor, we generated multimers of recombinant soluble CLEC2D isoform-Fc fusion proteins complexed to protein A and tested their binding to 293T-CD161 cells (7). As shown in Fig. 5A, LLT1 multimer bound to 293T-CD161 but not to control 293T-LLT1 cells. By contrast, multimers of CLEC2D isoform 2 and 4 failed to bind to both 293T-CD161 and 293T-LLT1 cells. In addition, we could not confirm the binding of CLEC2A (also named PILAR) to CD161 using a CLEC2A-Fc multimer (Fig. 5A) in contrast to Huarte et al. (18) but in agreement with Spreu et al. (19). These results demonstrated that LLT1 is the sole CLEC2D isoform that binds to CD161. This is consistent with a lack of homology between CLEC2D isoforms in the extracellular domain at the C terminus of the proteins (Fig. 1C). To better understand the structural basis for the specificity of the binding to CD161, we generated structural representations of the three CLEC2D isoforms. The model for LLT1 was generated based on the crystal structure of CD69, the closest homologue of LLT1 (13, 20). As shown in Fig. 5B, the structure of LLT1 resembles those of CD69 but also of CD94/NKG2A heterodimers and NKG2D homodimers that have been crystallized with their respective ligands HLA-E and MICA (14, 21, 22). Using this model, we can speculate that the interface between LLT1 and CD161 is located at the level of the β-strands and the loop between them formed by residues CAYLN-DKGA-SSAR in LLT1. The models for CLEC2D isoforms 2 and 4 were generated based on the crystal structure of NKG2A and KLRG1, respectively (15). As shown in Fig. 5C, their overall structures differ significantly from LLT1, providing insights for their failure to bind to CD161.
FIGURE 5.
CLEC2D protein isoforms 2 and 4 and CLEC2A do not interact with CD161. A, CLEC2D isoforms or CLEC2A multimers binding to 293T-CD161 or control 293T-LLT1 analyzed by flow cytometry. The results are representative of four independent experiments. B, structure comparison of CLEC2D isoform 1 (LLT1) dimer structure model with x-ray crystal structures of the extracellular domains of CD69 dimer (3HUP), CD94/NKG2A dimer in complex with HLA-E (3CDG), and NKG2D dimer in complex with MICA (1HYR). The side views of ribbon representations are shown with β-strands colored in yellow and α-helices in red. C, single domain side and top view of CLEC2D isoform 1, 2, and 4 structure models. The models have been colored with the following scheme: red (isoform 1, 2, and 4 shared sequence), green (isoform 1 and 2 shared sequence), blue (isoform 1-specific sequence), yellow (isoform 2-specific sequence), and magenta (isoform 3-specific sequence).
DISCUSSION
It is reported that 92–94% of genes in the human genome produce transcripts that are alternatively spliced, thus generating a variety of exon combinations from a single primary transcript (23, 24). Alternative splicing is a key factor increasing cellular and functional complexity. Indeed, alternative transcripts are very often differentially expressed between cells or tissues and display different functions. In this study, we have shown that CLEC2D encodes multiple transcripts through alternative splicing. This finding is consistent with Northern blot analysis, which previously identified several transcripts of 5, 4.5, 2, and 0.9 kilobases in total RNA from a human NK cell line YT (25). In addition to the reported CLEC2D variant 1 (LLT1) and variant 2, we have cloned and sequenced three additional variants, one coding for a transmembrane protein isoform 4 and two coding for putative soluble isoforms. We found that only the transmembrane protein isoforms were efficiently produced. The putative soluble proteins display the first 20 aa at the N terminus of the cytoplasmic domain followed by the extracellular domain. Cloning of variant 5 into an expression vector in front or not of a signal sequence did not lead to efficient expression of soluble LLT1, whereas cloning of the extracellular domain did. This result suggested that the 20 N-terminal aa prevented efficient production of soluble CLEC2D protein isoforms. Interestingly, a similar observation has been made for CLEC2A (11). It is therefore unlikely that a soluble LLT1 generated from alternative splicing could compete with membrane-bound LLT1. It remains to be assessed whether soluble LLT1 could be generated from proteolysis similarly to MICA (26).
The characterization of CLEC2D variants is of importance when assessing LLT1 transcription profile and was not taken into account in previous studies (16, 27). Our data showed that all the CLEC2D transcript variants seemed to be expressed in the same cells but with variable amounts of variants 1 and 2, underscoring the need to use specific primers. We also showed that CLEC2D isoforms 2 and 4 could be efficiently expressed as proteins, and we therefore monitored the specificity of the anti-LLT1 mAbs. The anti-LLT1 mAbs 2F1 and 4F68 that we have developed, together with MAB3480, were found to be specific of LLT1 and did not bind to isoforms 2 and 4 or to other C-type lectin-like molecules with close homology to LLT1 such as AICL, CD69, or CLEC2A. By contrast, we report that the 4C7 mAb is not specific and recognizes an epitope shared between all of the CLEC2D isoforms and CLEC2A.
CLEC2D encodes LLT1 (variant 1), which has been reported to bind to CD161 receptor, thus regulating NK and T cell functions. Alternative gene splicing has been shown to be used in some cases to generate inhibitory variants, i.e. IL-4 or IL-2 variants competing with IL-4 and IL-2 (28, 29). We therefore assessed whether the other CLEC2D isoforms could bind to CD161 and regulate LLT1/CD161 interaction. Using multimeric complexes, we demonstrated that CLEC2D isoform 2 and 4 do not bind to CD161. This is consistent with the lack of homology observed between these three CLEC2D isoforms in their extracellular domain and was highlighted by the structural models we generated. Residues that are specific to LLT1 were found to form an area similar to those involved in ligand interaction in related C-type lectins such as CD94/NKG2A and NKKG2D (14, 21, 22), thus reinforcing the conclusion that among CLEC2D isoforms, LLT1 is the sole ligand of CD161. In addition, it is important to note that we could not confirm the binding of CLEC2A to CD161.
Finally, CLEC2D isoforms 2 and 4 were not expressed at the cell surface but localized in the ER forming homodimers or heterodimers with LLT1. Their presence did not seem to modulate LLT1 cell surface expression. Further studies need to be undertaken to identify their functional role.
Taken together, we showed that CLEC2D encodes multiple alternatively spliced transcripts co-expressed in the same cell types and induced upon the same signals of activation but that only CLEC2D isoform 1 (LLT1) is expressed on the cell surface. In addition we demonstrated that only CLEC2D isoform 1 (LLT1) is able to bind CD161 receptor, suggesting that other CLEC2D isoforms display different biological activities remaining to be determined. Finally, we could not confirm that CLEC2A (PILAR) is a ligand of CD161 but highlighted that 4C7 mAb is not specific of LLT1 and cross-reacts with CLEC2A.
Acknowledgments
We thank Elisabeth Galsgaard and Nicolai Wagtmann for helpful discussion and Helle Hald, Anders Moselund, and Anne Lazzari for technical assistance.
This work was supported by grants from Novo Nordisk S/A, Agence Nationale de Recherches sur le SIDA, and Ensemble contre le SIDA.
- NK
- natural killer
- LLT1
- lectin-like transcript 1
- Clr
- C-type lectin related
- AICL
- activation-induced C-type lectin
- PBMC
- peripheral blood mononuclear cell
- DC
- dendritic cell
- ER
- endoplasmic reticulum
- aa
- amino acid(s)
- Endo H
- endoglycosidase H
- PNGase F
- peptide N-glycosidase F.
REFERENCES
- 1.Weis W. I., Taylor M. E., Drickamer K. (1998) Immunol. Rev. 163, 19–34 [DOI] [PubMed] [Google Scholar]
- 2.Carlyle J. R., Jamieson A. M., Gasser S., Clingan C. S., Arase H., Raulet D. H. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 3527–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iizuka K., Naidenko O. V., Plougastel B. F., Fremont D. H., Yokoyama W. M. (2003) Nat. Immunol. 4, 801–807 [DOI] [PubMed] [Google Scholar]
- 4.Aust J. G., Gays F., Mickiewicz K. M., Buchanan E., Brooks C. G. (2009) J. Immunol. 183, 106–116 [DOI] [PubMed] [Google Scholar]
- 5.Tian W., Nunez R., Cheng S., Ding Y., Tumang J., Lyddane C., Roman C., Liou H. C. (2005) Cell. Immunol. 234, 39–53 [DOI] [PubMed] [Google Scholar]
- 6.Kveberg L., Dai K. Z., Westgaard I. H., Daws M. R., Fossum S., Naper C., Vaage J. T. (2009) Eur. J. Immunol. 39, 541–551 [DOI] [PubMed] [Google Scholar]
- 7.Aldemir H., Prod'homme V., Dumaurier M. J., Retiere C., Poupon G., Cazareth J., Bihl F., Braud V. M. (2005) J. Immunol. 175, 7791–7795 [DOI] [PubMed] [Google Scholar]
- 8.Rosen D. B., Bettadapura J., Alsharifi M., Mathew P. A., Warren H. S., Lanier L. L. (2005) J. Immunol. 175, 7796–7799 [DOI] [PubMed] [Google Scholar]
- 9.Exley M., Garcia J., Balk S. P., Porcelli S. (1997) J. Exp. Med. 186, 109–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pozo D., Valés-Gómez M., Mavaddat N., Williamson S. C., Chisholm S. E., Reyburn H. (2006) J. Immunol. 176, 2397–2406 [DOI] [PubMed] [Google Scholar]
- 11.Spreu J., Kienle E. C., Schrage B., Steinle A. (2007) Immunogenetics 59, 903–912 [DOI] [PubMed] [Google Scholar]
- 12.Welte S., Kuttruff S., Waldhauer I., Steinle A. (2006) Nat. Immunol. 7, 1334–1342 [DOI] [PubMed] [Google Scholar]
- 13.Kolenko P., Skálová T., Vanek O., Stepánková A., Dusková J., Hasek J., Bezouska K., Dohnálek J. (2009) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65, 1258–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petrie E. J., Clements C. S., Lin J., Sullivan L. C., Johnson D., Huyton T., Heroux A., Hoare H. L., Beddoe T., Reid H. H., Wilce M. C., Brooks A. G., Rossjohn J. (2008) J. Exp. Med. 205, 725–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y., Hofmann M., Wang Q., Teng L., Chlewicki L. K., Pircher H., Mariuzza R. A. (2009) Immunity 31, 35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosen D. B., Cao W., Avery D. T., Tangye S. G., Liu Y. J., Houchins J. P., Lanier L. L. (2008) J. Immunol. 180, 6508–6517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brooks A. G., Posch P. E., Scorzelli C. J., Borrego F., Coligan J. E. (1997) J. Exp. Med. 185, 795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huarte E., Cubillos-Ruiz J. R., Nesbeth Y. C., Scarlett U. K., Martinez D. G., Engle X. A., Rigby W. F., Pioli P. A., Guyre P. M., Conejo-Garcia J. R. (2008) Blood 112, 1259–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spreu J., Kuttruff S., Stejfova V., Dennehy K. M., Schittek B., Steinle A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 5100–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Natarajan K., Sawicki M. W., Margulies D. H., Mariuzza R. A. (2000) Biochemistry 39, 14779–14786 [DOI] [PubMed] [Google Scholar]
- 21.Kaiser B. K., Pizarro J. C., Kerns J., Strong R. K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6696–6701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li P., Morris D. L., Willcox B. E., Steinle A., Spies T., Strong R. K. (2001) Nat. Immunol. 2, 443–451 [DOI] [PubMed] [Google Scholar]
- 23.Pan Q., Shai O., Lee L. J., Frey B. J., Blencowe B. J. (2008) Nat. Genet. 40, 1413–1415 [DOI] [PubMed] [Google Scholar]
- 24.Wang E. T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S. F., Schroth G. P., Burge C. B. (2008) Nature 456, 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boles K. S., Barten R., Kumaresan P. R., Trowsdale J., Mathew P. A. (1999) Immunogenetics 50, 1–7 [DOI] [PubMed] [Google Scholar]
- 26.Salih H. R., Rammensee H. G., Steinle A. (2002) J. Immunol. 169, 4098–4102 [DOI] [PubMed] [Google Scholar]
- 27.Roth P., Mittelbronn M., Wick W., Meyermann R., Tatagiba M., Weller M. (2007) Cancer Res. 67, 3540–3544 [DOI] [PubMed] [Google Scholar]
- 28.Atamas S. P., Choi J., Yurovsky V. V., White B. (1996) J. Immunol. 156, 435–441 [PubMed] [Google Scholar]
- 29.Tsytsikov V. N., Yurovsky V. V., Atamas S. P., Alms W. J., White B. (1996) J. Biol. Chem. 271, 23055–23060 [DOI] [PubMed] [Google Scholar]