Abstract
C-reactive protein (CRP) is a phylogenetically conserved protein; in humans, it is present in the plasma and at sites of inflammation. At physiological pH, native pentameric CRP exhibits calcium-dependent binding specificity for phosphocholine. In this study, we determined the binding specificities of CRP at acidic pH, a characteristic of inflammatory sites. We investigated the binding of fluid-phase CRP to six immobilized proteins: complement factor H, oxidized low-density lipoprotein, complement C3b, IgG, amyloid β, and BSA immobilized on microtiter plates. At pH 7.0, CRP did not bind to any of these proteins, but, at pH ranging from 5.2 to 4.6, CRP bound to all six proteins. Acidic pH did not monomerize CRP but modified the pentameric structure, as determined by gel filtration, 1-anilinonaphthalene-8-sulfonic acid-binding fluorescence, and phosphocholine-binding assays. Some modifications in CRP were reversible at pH 7.0, for example, the phosphocholine-binding activity of CRP, which was reduced at acidic pH, was restored after pH neutralization. For efficient binding of acidic pH-treated CRP to immobilized proteins, it was necessary that the immobilized proteins, except factor H, were also exposed to acidic pH. Because immobilization of proteins on microtiter plates and exposure of immobilized proteins to acidic pH alter the conformation of immobilized proteins, our findings suggest that conformationally altered proteins form a CRP-ligand in acidic environment, regardless of the identity of the protein. This ligand binding specificity of CRP in its acidic pH-induced pentameric state has implications for toxic conditions involving protein misfolding in acidic environments and favors the conservation of CRP throughout evolution.
Keywords: Inflammation, Innate Immunity, Ligand Binding Protein, Pattern Recognition Receptor, Protein-Protein Interactions, C-reactive Protein
Introduction
C-reactive protein (CRP)4 is an evolutionarily conserved protein. CRP is found in all animals, from invertebrates to vertebrates, where it has been sought. Human CRP is a pentameric protein comprised of five identical, non-covalently associated 23,028-Da subunits (reviewed in Refs. 1–3). At physiological pH, CRP is best known for its Ca2+-dependent binding to phosphocholine (PCh)-containing substances such as pneumococcal C-polysaccharide (PnC) (4). Each CRP subunit binds two Ca2+ ions with a KD value of 30 to 60 μm (5, 6). Binding of CRP to some nuclear proteins, enzymatically modified low-density lipoprotein and phosphoethanolamine-conjugated materials has also been reported (7–11). The Ca2+- and PCh-binding sites are located on each subunit of CRP, and the available data suggest that these sites participate in the binding of CRP to most of its known ligands. Under certain conditions, CRP dissociates to generate the monomeric form of CRP (mCRP), which can be distinguished from pentameric CRP using specific monoclonal antibodies (12–17).
In normal healthy individuals, the median concentration of CRP in the serum is 0.8 μg/ml; in acute phase, the concentration increases to 500 μg/ml or more (18). CRP is also found at the sites of inflammation, both in humans and experimental animals (19–23). The deposition of CRP at the sites of inflammation is independent of the circulating concentration of CRP. For example, CRP is deposited at atherosclerotic lesions although the circulating concentration of CRP increases only minimally in atherosclerosis (8, 22–24). Due to the deposition of CRP at the sites of inflammation, the concentration of CRP at the sites of inflammation is not known. The mechanism proposed for the deposition of CRP at the sites of inflammation is largely based on the PCh-binding and other ligand-binding properties of CRP, which occur at physiological pH. Because the pH at the sites of inflammation may be acidic (25–35), in this study, we investigated the binding specificities of CRP at acidic pH to define the functions of CRP at the sites of inflammation.
Factor H is a complement regulatory protein that protects host cells from complement attack (36). Factor H can also protect pathogens from complement attack if the pathogens can recruit factor H (37, 38). We explored CRP-factor H interactions because of their possible involvement in the anti-pneumococcal functions of CRP observed in murine models of pneumococcal infection. Human CRP protects mice from lethality following infection with Streptococcus pneumoniae type 3 (39–42). Because type 3 pneumococci bind factor H (38) and because CRP also binds factor H under certain conditions (43–45), it has been proposed that CRP may bind to pneumococci-bound factor H and contribute to anti-pneumococcal function of CRP (46, 47). We explored CRP-factor H interactions also because of their possible involvement in age-related macular degeneration. If CRP could bind to factor H, then it could be important for age-related macular degeneration due to the implications of factor H in this disease (48).
Oxidized low density lipoprotein (oxLDL) is an atherogenic form of LDL. It enters macrophages to form foam cells that contribute to the development of atherosclerosis, which is an inflammatory disease (49). CRP is localized with LDL in atherosclerotic lesions present in humans and experimental animal models (8, 22, 23). Because the pH of the atherosclerotic lesions may be acidic (28–32), in this study, we also investigated the effects of acidic pH on CRP-oxLDL interactions.
In addition to factor H and oxLDL, we randomly selected four other proteins: complement component C3 fragment C3b, IgG, amyloid β fragment 1–38 (Aβ), and BSA, to explore the binding reactivities of CRP at acidic pH. We expected that CRP, at acidic pH, might bind to factor H, oxLDL, C3b, IgG, and Aβ, but would not bind to BSA. Surprisingly, we found that CRP, at acidic pH, bound to all six proteins including BSA, as if CRP recognized a general pattern created by these proteins that were immobilized to microtiter plates and also exposed to acidic pH at 37 °C.
EXPERIMENTAL PROCEDURES
Purification of CRP, Factor H, LDL, and IgG
Native CRP was purified from pleural fluid in three steps: Ca2+-dependent affinity chromatography on a PCh-conjugated-Sepharose column (Pierce), followed by anion-exchange chromatography on a MonoQ column (GE Healthcare), and gel filtration on a Superose 12 column (GE Healthcare), as described previously (50). On the day of the experiments, CRP was re-purified by gel filtration on a Superose 12 column (GE Healthcare) to remove any form of modified CRP that might have been generated due to storage of CRP. Re-purified CRP was stored in 10 mm Tris-HCl, pH 7.2, 150 mm NaCl (TBS) containing 0.1 mm CaCl2 at 4 °C and was used within 2 weeks.
Factor H was purified from normal human plasma as described previously (51). Purified factor H was greater than 97% homogenous by SDS-PAGE with an apparent molecular mass of 155 kDa in its reduced form. Three different extinction coefficient values have been reported for factor H (1%) at 280 nm using a 1-cm light path: 12.4, 16.2, and 19.5 (51–53). The concentration of factor H was determined using all three extinction coefficient values.
LDL (1.019 < d < 1.063) was isolated from human plasma by sequential ultracentrifugation, as described previously (54). IgG was purified from human serum by caprylic acid precipitation, as described previously (55), followed by affinity chromatography on a protein A-Sepharose column (Pierce) and anion-exchange chromatography on a MonoQ column (GE Healthcare).
Preparation of OxLDL
OxLDL was prepared by treating LDL with 20 μm CuCl2 for 24 h at 37 °C, as described previously (56, 57). Oxidation was arrested with 0.5 mm EDTA. OxLDL was dialyzed against PBS/Chelex 100 resin (Bio-Rad), filter sterilized, and stored in the dark at 4 °C. Oxidation of LDL was evaluated by agarose gel electrophoresis: oxLDL had 35-fold higher RF values compared with native LDL. The degree of oxidation was also determined by TBARS assay: the oxLDL resulted in 16–22 nmol of MDA/mg of protein. The protein concentration in the oxLDL preparations was measured using a protein assay kit (Bio-Rad).
Construction, Expression, and Purification of P115A Mutant CRP
Site-directed mutagenesis of CRP to substitute Pro115 with Ala was carried out using the mutagenic oligonucleotide 5′-TCACCCTGGCCTTCCCAT, as described previously (58). The P115A mutant CRP cDNA was expressed in COS cells and purified from the culture supernatant as described above for native CRP.
Protein Ligand-binding Assay
In the protein ligand-binding assays, we determined the binding of CRP to factor H, oxLDL, human C3b (Calbiochem), IgG, Aβ fragment 1–38 (catalog number A0189, Sigma), and BSA (catalog number A0281, Sigma). Microtiter wells were coated with factor H (2 μg/ml based on the extinction coefficient value of 12.4; 1.5 μg/ml based on the extinction coefficient value of 16.2; 1.3 μg/ml based on the extinction coefficient value of 19.5), oxLDL (10 μg/ml), C3b (5 μg/ml), IgG (10 μg/ml), Aβ (2 μg/ml), or BSA (10 μg/ml), in TBS, overnight at 4 °C. The unreacted sites in the wells were blocked with TBS containing 0.5% gelatin. CRP (native or P115A mutant), diluted in TBS containing 0.1% gelatin, 0.02% Tween 20, and 0.1 mm CaCl2 (TBS-Ca), was added in duplicate wells, and incubated for 2 h at 37 °C. As mentioned in the figure legends, this step in some assays were performed in TBS-Ca with pH ranging from 7.0 to 4.6, in some assays this step was performed at room temperature (25 °C), in some assays performed in 2 mm CaCl2, and in some assays performed in 1 mm EDTA. After the CRP incubation step, the wells were washed with TBS-Ca. Immunoaffinity purified polyclonal rabbit anti-CRP antibody (1 μg/ml), diluted in TBS-Ca, was used (1 h at 37 °C) to detect bound CRP. Immunoaffinity purified polyclonal rabbit anti-CRP antibody was purified from the rabbit anti-human CRP antiserum (Sigma) by affinity chromatography on a CRP-conjugated agarose column prepared by using the AminoLink Immobilization kit (Pierce), as described previously (11). In some assays, monoclonal anti-CRP antibodies (mAb) were used to detect bound CRP. HRP-conjugated donkey anti-rabbit IgG (GE Healthcare) and HRP-conjugated goat anti-mouse IgG (Thermo Scientific), diluted in TBS-Ca, were used (1 h at 37 °C) as secondary antibodies. Color was developed and absorbance was read at 405 nm in a microtiter plate reader (Molecular Devices).
To determine the effects of PCh (Sigma) on the binding of CRP to immobilized proteins, the protein ligand-binding assays were performed as mentioned above, except that CRP was added to the wells in the presence of PCh. The pH of the buffers was readjusted to the desired value after addition of all other components to TBS.
PnC-binding Assay
Binding activity of CRP for PCh was evaluated by using PnC (Statens Serum Institut) as the ligand. Microtiter wells were coated with 10 μg/ml of PnC in TBS overnight at 4 °C. The unreacted sites in the wells were blocked with TBS containing 0.5% gelatin. CRP diluted in TBS-Ca (pH 7.2 and 4.6) was added in duplicate wells. As mentioned in the figure legends, in some assays, CRP was first diluted in TBS-Ca, pH 4.6, incubated for 2 h at 37 °C, and then the pH was readjusted to 7.2 before adding CRP to the wells. After incubating the plates for 2 h at 37 °C, the wells were washed with TBS-Ca. Three different anti-CRP antibodies were used (1 h at 37 °C) to detect bound CRP: a polyclonal anti-CRP antibody and two anti-CRP mAb, HD2.4 and 3H12. HRP-conjugated donkey anti-rabbit IgG and HRP-conjugated goat anti-mouse IgG, diluted in TBS-Ca, were used (1 h at 37 °C) as the secondary antibodies. Color was developed and the absorbance was read at 405 nm in a microtiter plate reader.
Gel Filtration
Gel filtration analysis of CRP at pH 4.6 was carried out on a Superose 12 column. The column was equilibrated with TBS, pH 4.6, containing 0.1 mm CaCl2 before applying CRP to the column, and eluted with the same buffer at a flow rate of 0.25 ml/min. CRP at pH 7.2 was used to determine the elution volume of native pentameric CRP. Sixty fractions (0.25 ml) were collected and protein was measured to locate the elution volume of CRP from the column.
1-Anilinonaphthalene-8-sulfonic Acid-binding Fluorescence Assay
The hydrophobic fluorescent probe 1-anilinonaphthalene-8-sulfonic acid (ANS) was purchased from AnaSpec, Inc. The ANS-binding fluorescence assays were performed as described previously (59, 60) to investigate the structural changes in CRP at pH 4.6. CRP (50 μg/ml) in TBS containing 0.1 mm CaCl2, at various pH, was mixed with ANS at a final concentration of 100 μm. The fluorescence intensity of the binding of ANS to CRP was measured using the excitation and emission wavelengths of 390 and 460 nm, respectively, in a spectrofluorometer (Fluostar Galaxy, BMG Lab Technologies).
RESULTS
All experiments were performed three times and comparable results were obtained each time. Results of a representative experiment are shown in the figures where the raw data (A405) were used to plot the curves.
At Acidic pH, CRP Binds to a Variety of Other Proteins Immobilized to Microtiter Plates
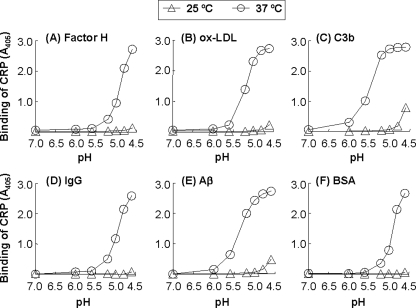
Fig. 1 shows the results of protein ligand-binding assays in which we determined the binding of CRP to immobilized factor H, oxLDL, C3b, IgG, Aβ, and BSA, as a function of pH (7.0–4.6), at 25 and 37 °C. As shown in Fig. 1A, at 25 °C, CRP did not bind to factor H at any pH. However, at 37 °C, CRP bound to factor H but only when the pH was decreased from 7.0 to 5.2; and the binding increased when the pH was further lowered to 4.6. Similar results were obtained with oxLDL (Fig. 1B). At 25 °C, CRP did not bind to oxLDL at any pH. At 37 °C, CRP bound to oxLDL when the pH was decreased from 7.0 to 5.2; and binding increased when the pH was further lowered to 4.8. As shown in Fig. 1C, at 25 °C, CRP bound to C3b only at pH 4.6. At 37 °C, CRP bound to C3b when the pH was decreased from 7.0 to 6.0; and the binding was increased when the pH was further lowered to 5.2. As shown in Fig. 1D, at 25 °C, CRP did not bind to IgG at any pH. At 37 °C, CRP bound to IgG when the pH was decreased from 7.0 to 5.2; and binding increased when the pH was further lowered to 4.6. As shown in Fig. 1E, at 25 °C, CRP bound to Aβ only at pH 4.6. At 37 °C, CRP bound to Aβ when the pH was decreased from 7.0 to 5.6; and binding increased when the pH was further lowered to 5.0. We have previously reported that CRP binds to Aβ at acidic pH (61); we included it here with the detailed investigation of the CRP-Aβ interaction for completeness. As shown in Fig. 1F, at 25 °C, CRP did not bind to BSA at any pH. At 37 °C, CRP bound to BSA when the pH was decreased from 7.0 to 5.2; and binding increased when the pH was further lowered to 4.6.
FIGURE 1.
Binding of CRP to six different immobilized proteins as a function of pH and temperature. Results of a protein ligand-binding assay are shown. Microtiter wells were coated with factor H, oxLDL, C3b, IgG, Aβ, and BSA. The unreacted sites in the wells were blocked with gelatin. CRP (10 μg/ml), diluted in TBS-Ca, pH 7.0–4.6, was then added to the wells. One set of wells was incubated at 25 °C and another at 37 °C, for 2 h. Bound CRP was detected by using a rabbit polyclonal anti-CRP antibody and HRP-conjugated donkey anti-rabbit IgG. The absorbance of the developed color was read at 405 nm and plotted on the y axis.
In control experiments, in which only gelatin-coated wells were used, CRP did not bind to gelatin at acidic pH. When only the immobilized proteins, and not CRP, were treated with acidic pH and the assay was performed at pH 7.0, CRP did not bind to immobilized proteins (data not shown). These results indicated that CRP acquired the capability to bind to immobilized proteins when both CRP and immobilized proteins were exposed to acidic pH. We have not yet investigated the mechanism of action of temperature in the binding of CRP to immobilized ligands at acidic pH.
The Binding of CRP to Immobilized Proteins at Acidic pH Is Not a Nonspecific Protein-Protein Interaction
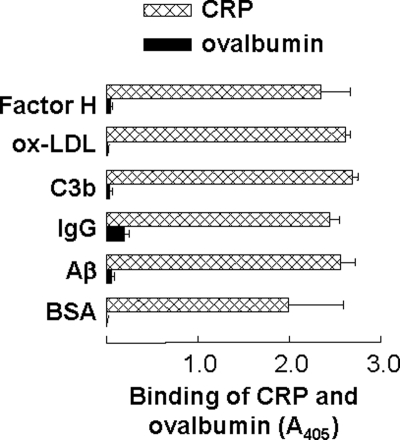
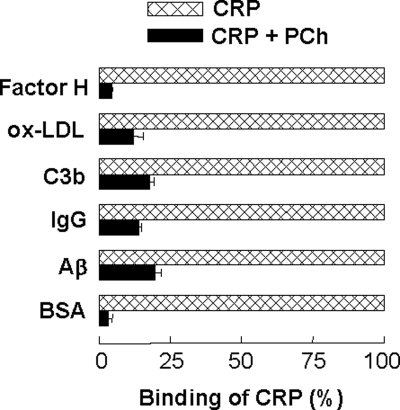
To show that the binding of CRP to immobilized proteins at acidic pH was not a nonspecific protein-protein interaction, our first approach was to use two other proteins, chicken ovalbumin (Sigma) and rabbit IgG, in addition to CRP, and determine their binding to all six immobilized proteins at acidic pH. Ovalbumin did not bind to any of the six immobilized proteins at pH 4.6 (Fig. 2). As shown in supplemental Fig. S1, there was no effect of acidic pH on the binding of rabbit IgG to five of the six immobilized proteins. Whatever background absorbance (A405) was noted at pH 7.0 remained unchanged at pH 4.6. Our second approach was to use PCh with CRP in the protein ligand-binding assays. We reasoned that if binding of CRP to immobilized proteins at acidic pH was nonspecific and involved completely denatured and fragmented CRP, then the binding should not be affected by the presence of PC with CRP. As shown in Fig. 3, PCh inhibited the binding of CRP to all six immobilized proteins at pH 4.6. Combined data suggested that binding of CRP to immobilized proteins at acidic pH was a function of CRP and not a nonspecific protein-protein interaction occurring at acidic pH.
FIGURE 2.
Comparison of the binding of chicken ovalbumin and CRP to immobilized proteins at acidic pH. Microtiter wells were coated with factor H, oxLDL, C3b, IgG, Aβ, and BSA. The unreacted sites in the wells were blocked with gelatin. CRP (10 μg/ml) and ovalbumin (10 μg/ml), diluted in TBS-Ca, pH 4.6, were added to the wells and incubated at 37 °C for 2 h. Bound CRP was detected as described in the legend to Fig. 1. Bound ovalbumin was detected by using a goat anti-chicken egg albumin IgG (MP Biomedicals) and HRP-conjugated bovine anti-goat IgG (Santa Cruz Biotechnology). The absorbance of the developed color was read at 405 nm and plotted on the x axis. Results are shown as mean ± S.D. of three independent experiments.
FIGURE 3.
Inhibition of the binding of CRP to immobilized proteins by PCh at acidic pH. Results of a protein ligand-binding assay are shown. Microtiter wells were coated with factor H, oxLDL, C3b, IgG, Aβ, and BSA. The unreacted sites in the wells were blocked with gelatin. CRP (10 μg/ml), diluted in TBS-Ca, pH 4.6, with and without 10 mm PCh, was then added to the wells, and incubated at 37 °C for 2 h. Bound CRP was detected as described in the legend to Fig. 1. The binding of CRP to each immobilized protein in the absence of PCh was taken as 100%. Results are shown as mean ± S.D. of three independent experiments.
Acidic pH Does Not Monomerize CRP but Modifies the Pentameric Structure
We evaluated the effects of acidic pH on the structure of CRP by using gel filtration and ANS-binding fluorescence assay. Using gel filtration, we determined whether CRP remained pentameric at pH 4.6 when incubated for 2 h at 37 °C. As shown in supplemental Fig. S2, the elution profiles of CRP at pH 7.2 and 4.6 were similar. The peak in the elution profile at pH 4.6 was lower than the peak in the elution profile at pH 7.2 because the amount of CRP injected into the column at pH 4.6 was less than the amount of CRP injected at pH 7.2. The elution profile of CRP, which was preincubated at pH 4.6 for 2 h at 37 °C and then the pH was neutralized was also similar to the elution profile of CRP at pH 7.2. These data indicated that CRP remained pentameric at pH 4.6.
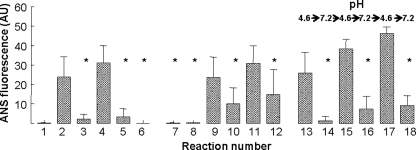
Using ANS-binding fluorescence assays, we determined whether the pentameric structure of CRP was modified at pH 4.6 (Fig. 4). When CRP, at pH 7.2, was incubated with ANS, negligible fluorescence was observed (reaction 1). The binding of ANS to CRP at pH 4.6 was increased compared with that at pH 7.2 (compare reactions 2 and 4 with 1) as reflected by the increase in fluorescence. When the pH of CRP solutions, which were preincubated at pH 4.6 for 2 h at either 25 or 37 °C, was neutralized, the binding of ANS to CRP was not increased compared with that at pH 7.2 (compare reactions 3 and 5 with 1). No fluorescence was observed when we assessed the binding of ANS to CRP at pH 7.2 in the absence of Ca2+ and in the presence of 1 mm EDTA (reaction 6), suggesting that the increased fluorescence seen in reactions 2 and 4 was dependent on the acidic pH and was not only due to the possible detachment of Ca2+ from CRP at acidic pH.
FIGURE 4.
Effects of acidic pH on the pentameric structure of CRP. ANS-binding fluorescence of CRP at pH 7.2 and 4.6 are shown. Reaction 1, CRP at pH 7.2; reaction 2, CRP at pH 4.6 preincubated at 25 °C for 2 h; reaction 3, after incubating CRP at pH 4.6 at 25 °C for 2 h, the pH was neutralized; reaction 4, CRP at pH 4.6 preincubated at 37 °C for 2 h; reaction 5, after incubating CRP at pH 4.6 at 37 °C for 2 h, the pH was neutralized. Reaction 6, CRP at pH 7.2 in the presence of 1 mm EDTA; reaction 7, CRP at pH 7.2; reaction 8, CRP with 10 mm PC at pH 7.2; reaction 9, CRP at pH 4.6 preincubated at 25 °C for 2 h; reaction 10, CRP with 10 mm PC at pH 4.6 preincubated at 25 °C for 2 h; reaction 11, CRP at pH 4.6 preincubated at 37 °C for 2 h; reaction 12, CRP with 10 mm PC at pH 4.6 preincubated at 37 °C for 2 h. Reactions 13–18, CRP at pH 4.6 was first incubated at 37 °C for 2 h. After removing an aliquot, the pH was neutralized. After removing another aliquot, the pH was readjusted to 4.6 and CRP was again incubated at 37 °C for 2 h. After removing an aliquot, the pH was neutralized again. This process was continued until three neutralizations of pH. Results are shown as mean ± S.D. of three independent experiments. * indicates p < 0.05 when compared with reaction 4.
Because, as shown in Fig. 3, PCh was found to inhibit binding of CRP to immobilized proteins, we next determined if PCh inhibited the acidic pH-induced structural change in CRP (reactions 7–12). The increase in the fluorescence of CRP at pH 4.6 (reactions 9 and 11) was inhibited by the presence of 10 mm PCh with CRP (compare reactions 10 and 12 with 9 and 11). These results indicated that PCh-bound CRP was resistant to acidic pH and that the acidic pH-induced modification of the pentameric structure of CRP was required for the binding of CRP to immobilized proteins. To determine the reversibility of the acidic pH-induced changes in the structure of CRP, we sequentially alternated the pH of a CRP solution between 4.6 and 7.2 (reactions 13–18). Again, the ANS fluorescence of CRP did not increase when CRP was at pH 7.2 and increased when the same CRP solution was at pH 4.6. Combined data from gel filtration and ANS-binding assays suggested that, at acidic pH, CRP was not monomerized, CRP was not completely denatured, the pentameric structure of CRP was modified, and at least some modifications were reversible.
The Acidic pH Reversibly Modifies the PCh-binding Site of CRP
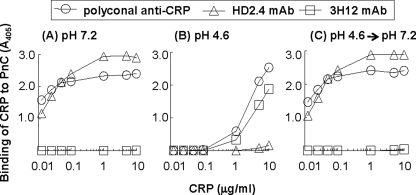
We next determined the effects of acidic pH on the PCh-binding site of CRP using a PnC-binding assay (Fig. 5). A change in the PnC-binding activity would reflect a change in the PCh-binding site of CRP. We first used the polyclonal anti-CRP antibody to detect PnC-bound CRP. At pH 7.2 (Fig. 5A), CRP bound efficiently, even at 12.5 ng/ml, to PnC. However, at pH 4.6 (Fig. 5B), the PCh-binding activity of CRP was drastically reduced. At least 1.0 μg/ml of CRP was required to detect some binding of CRP to PnC. Surprisingly, when the acidic pH of CRP was neutralized to 7.2 (Fig. 5C), the PCh-binding activity of CRP was restored. These results indicated that acidic pH modified the PCh-binding site of CRP, but the modification was reversible.
FIGURE 5.
Reversible decrease in the PC-binding activity of CRP at acidic pH. Results of a PnC-binding assay are shown. Microtiter wells were coated with PnC. The unreacted sites in the wells were blocked with gelatin. CRP was then added to the wells and incubated at 37 °C for 2 h. Bound CRP was detected using a polyclonal anti-CRP antibody, anti-CRP mAbs HD2.4 and 3H12, and corresponding HRP-conjugated secondary antibodies. The absorbance of the developed color was read at 405 nm and plotted on the y axis. A, CRP was in TBS-Ca, pH 7.2. B, CRP was in TBS-Ca, pH 4.6, and incubated at 37 °C for 2 h before adding to the wells. C, as in B, except that the pH was neutralized before adding CRP to the wells.
We next determined whether CRP remained pentameric once it was bound to PnC at pH 4.6, by using anti-CRP mAb HD2.4 and anti-mCRP mAb 3H12 (Fig. 5). The HD2.4 mAb recognizes native pentameric CRP (62). The 3H12 mAb does not recognize native pentameric CRP because the epitope of 3H12 is hidden in the intersubunit contact zones of the pentamer; it recognizes modified and monomeric forms of CRP (15, 63). At pH 7.2 (Fig. 5A), PnC-bound CRP could be detected by using HD2.4 mAb, but not 3H12 mAb, indicating that the native pentameric structure of CRP was maintained after binding to PnC. However, at pH 4.6 (Fig. 5B), PnC-bound CRP could be detected by using 3H12 mAb, but not HD2.4 mAb. At pH 4.6, the HD2.4 reactivity was lost and 3H12 reactivity was gained, indicating monomerization of CRP after binding to PnC at pH 4.6. Surprisingly, when the acidic pH of CRP was neutralized (Fig. 5C), the HD2.4 reactivity of PnC-bound CRP was recovered, and the 3H12 reactivity was lost. These results indicated that CRP, after binding to PnC at pH 4.6, was either monomerized or remained pentameric but the structure was further modified. These results also confirmed that acidic pH-mediated change in the PCh-binding site of CRP was reversible.
Except for Factor H, the Exposure of Immobilized Proteins to Acidic pH Is Also Required for CRP to Bind
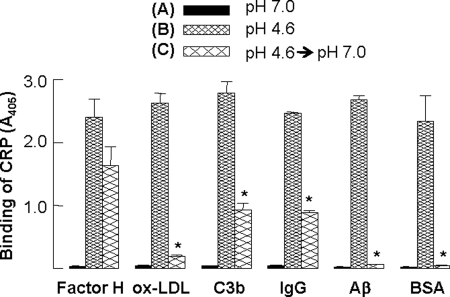
Next, we evaluated the effects of pH neutralization on the protein ligand-binding specificity of CRP (Fig. 6). CRP, at pH 7.0, did not bind to any of the six immobilized proteins. At pH 4.6, CRP bound to all six immobilized proteins. When the pH of the CRP solution, which was 4.6 and incubated for 2 h at 37 °C, was neutralized, and the binding assay was performed at pH 7.0, CRP still bound to factor H, indicating that the factor H-binding site formed on CRP by acidic pH treatment was not abolished after pH neutralization. This finding also suggested that the acidic pH was required for CRP to be capable of binding to factor H, but, the acidic pH was not required for factor H to be a CRP-binding protein. When the pH of the CRP solution, which was 4.6 and incubated for 2 h at 37 °C, was neutralized, and the binding assay was performed at pH 7.0, the binding ability of CRP to oxLDL, C3b, IgG, Aβ, and BSA was either lost or significantly reduced, suggesting that exposure of both CRP and these immobilized proteins to acidic pH was required for the binding of CRP.
FIGURE 6.
Reversible protein ligand-binding activity of CRP at acidic pH. Microtiter wells were coated with factor H, oxLDL, C3b, IgG, Aβ, and BSA. The unreacted sites in the wells were blocked with gelatin. CRP (10 μg/ml) was then added to the wells and incubated at 37 °C for 2 h. Bound CRP was detected as described in the legend to Fig. 1. A, CRP was in TBS-Ca, pH 7.2. B, CRP was in TBS-Ca, pH 4.6, and incubated at 37 °C for 2 h before adding to the wells. C, as in B, except that the pH was neutralized before adding CRP to the wells. Results are shown as mean ± S.D. of three independent protein ligand-binding assays. * indicates p < 0.006 between B and C.
The Structure of CRP Is Changed Further after Binding to Immobilized Proteins at pH 4.6
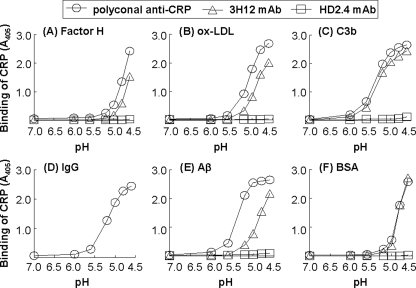
Because, as shown in Fig. 5, we found that CRP expressed a mCRP epitope after binding to PnC at pH 4.6, we next determined whether CRP also expressed the mCRP epitope after binding to immobilized protein ligands. Fig. 7 shows the results of a protein ligand-binding assay in which we detected the binding of CRP to factor H, oxLDL, C3b, IgG, Aβ, and BSA at 37 °C, as a function of pH (7.0–4.6), by using anti-CRP mAb HD2.4 and 3H12. Polyclonal anti-CRP antibody was used as a control; the results with the polyclonal antibody were similar to that shown in Fig. 1. The mAb 3H12 efficiently recognized CRP bound to all immobilized proteins. The mAb HD2.4 did not recognize CRP bound to immobilized proteins. With the IgG-coated wells (Fig. 7D), assays using mAb could not be performed due to cross-reactivities between different antibodies and due to high background color development in the wells. These results indicated that CRP, after binding to immobilized proteins at acidic pH, was either monomerized or remained pentameric but the structure was partially modified in a way to allow CRP residues involved in the intersubunit contact zone to be antigenically expressed.
FIGURE 7.
Structural change in CRP after binding to immobilized proteins at acidic pH. Microtiter wells were coated with factor H, oxLDL, C3b, IgG, Aβ, and BSA. CRP (10 μg/ml), diluted in TBS-Ca, pH 7.0–4.6, was then added to the wells and incubated at 37 °C for 2 h. Bound CRP was detected using three different anti-CRP antibodies: a polyclonal anti-CRP antibody, anti-CRP mAb HD2.4, or anti-CRP mAb 3H12. After addition of appropriate HRP-conjugated secondary antibodies, the absorbance of the developed color was read at 405 nm and plotted on the y axis.
The Binding of CRP to Immobilized Proteins Is Independent of the Ca2+-binding Site of CRP
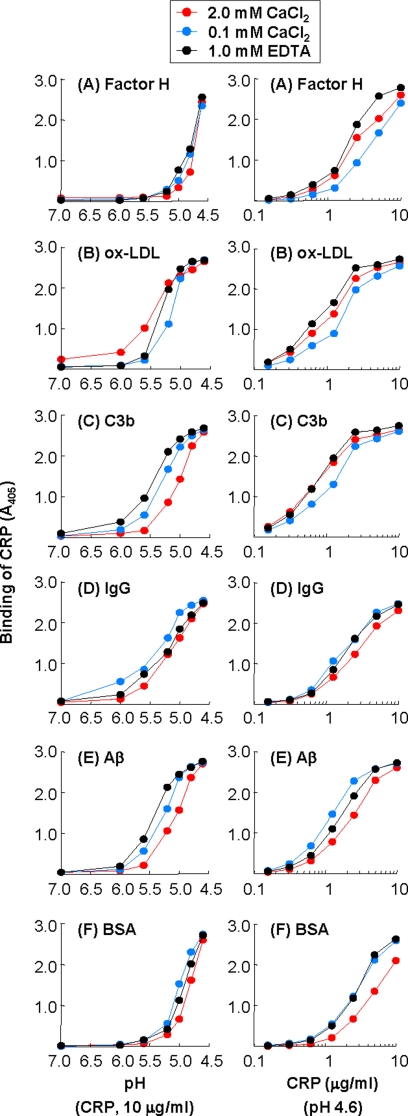
To define the binding site on CRP for immobilized proteins, we first investigated the requirement of Ca2+ for the binding of CRP to immobilized proteins. We compared the binding of CRP that was diluted in EDTA buffer with that of CRP that was diluted in TBS-Ca to immobilized proteins as a function of pH (Fig. 8A–F, left panel). As shown, the binding curves of CRP in the presence of Ca2+ (either 2 or 0.1 mm) or EDTA were similar for all six proteins. Next, we performed a CRP dose-response experiment at pH 4.6, in the presence of Ca2+ or EDTA. As shown (Fig. 8, A–F, right panels), the binding curves of CRP in the presence of Ca2+ (either 2 or 0.1 mm) or EDTA were similar for all six proteins. These results indicate that the binding of CRP to immobilized proteins was independent of the Ca2+-binding site of CRP.
FIGURE 8.
Effects of Ca2+ on the binding of CRP to immobilized proteins at acidic pH. Microtiter wells were coated with factor H, oxLDL, C3b, IgG, Aβ, and BSA. Three preparations of CRP were used: two with Ca2+ (2 mm and 0.1 mm) and one with EDTA. Left panels, CRP (10 μg/ml), diluted in TBS-Ca, pH 7.0–4.6, with Ca2+ (2 mm and 0.1 mm) or EDTA, was added to the wells and incubated at 37 °C for 2 h. Right panels, increasing concentrations of CRP, diluted in TBS-Ca, pH 4.6, with Ca2+ (2 mm and 0.1 mm) or EDTA, was added to the wells and incubated at 37 °C for 2 h. In all panels, bound CRP was detected as described in the legend to Fig. 1.
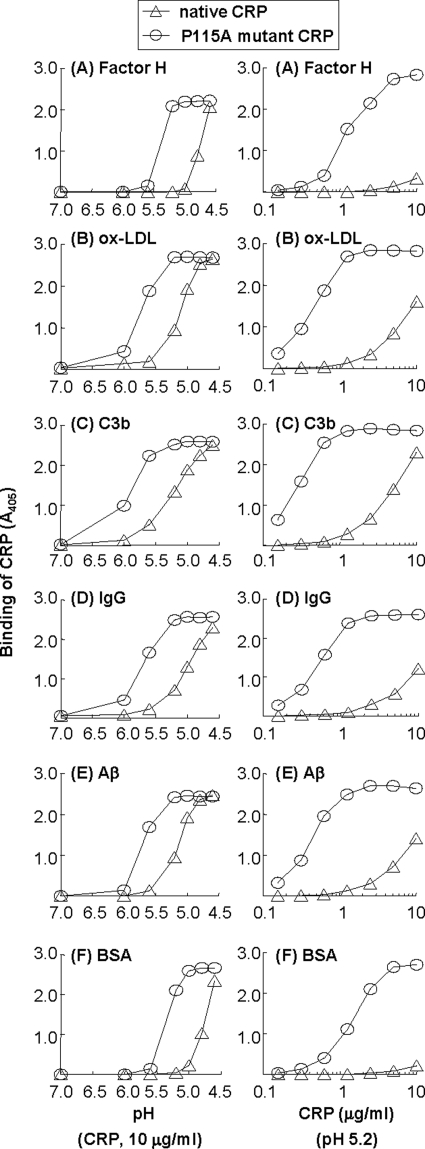
Substitution of Pro115 with Ala in CRP Reduces the Requirement of Acidic Buffer by 1 pH Unit and Enhances the Binding of CRP to Immobilized Proteins
Based on the data suggesting that the binding of CRP to immobilized proteins was independent of the Ca2+-binding site of CRP but dependent on the modification of pentameric structure of CRP, we hypothesized that the site in CRP for binding to immobilized proteins was hidden at pH 7.0 and exposed by acidic pH-mediated loosening of pentameric CRP. Accordingly, we focused on amino acids participating in the intersubunit interactions in the CRP pentamer. In this study, we selected one such amino acid, Pro115, and mutated it to Ala, with the aim to generate a CRP mutant that does not bind to immobilized proteins at acidic pH. The P115A mutant CRP was expressed well in COS cells and could be purified by the same method used for native CRP. The elution profile of the P115A mutant CRP from the gel filtration column was identical to that of native CRP (data not shown). Similarly, the PCh-binding activity of the P115A mutant CRP was identical to that of native CRP (data not shown). Thus, the mutation of Pro115 to Ala did not affect the overall pentameric structure of CRP.
We first compared the binding of purified P115A mutant CRP to immobilized ligands with that of native CRP as a function of pH (Fig. 9, A–F, left panel). As shown, the P115A mutant CRP bound to all six immobilized proteins. However, for comparable binding of native and P115A mutant CRP to any of the six proteins, the requirement of acidic buffer by P115A mutant CRP was decreased by at least 1 pH unit compared with that required by native CRP. We also performed a CRP dose-response experiment at pH 5.2 and compared binding of the P115A mutant CRP to immobilized ligands with that of native CRP (Fig. 9, A–F, right panel). The P115A mutant CRP was ∼10-fold more efficient than native CRP in binding to all six immobilized proteins.
FIGURE 9.
Binding of native and P115A mutant CRP to immobilized proteins at acidic pH. Microtiter wells were coated with factor H, oxLDL, C3b, IgG, Aβ, and BSA. Left panels, native and P115A mutant CRP (10 μg/ml), diluted in TBS-Ca, pH 7.0–4.6, was added to the wells and incubated at 37 °C for 2 h. Right panels, increasing concentrations of native and P115A mutant CRP, diluted in TBS-Ca, pH 5.2, was added to the wells and incubated at 37 °C for 2 h. In all panels, bound CRP was detected as described in the legend to Fig. 1.
Because P115A mutant CRP required a buffer less acidic than that required by native CRP for comparable binding to immobilized ligands, we hypothesized that the substitution of Pro115 to Ala changed the structure of CRP. We evaluated the change in the structure of CRP caused by substitution of Pro115 to Ala using ANS-binding fluorescence assays. As shown in supplemental Fig. S3, at acidic pH, binding of ANS to the P115A mutant CRP was more efficient than binding of ANS to the native CRP. The structural change in the P115A mutant CRP at pH 5.2 was similar to the structural change in native CRP at pH 4.6, which supports the finding that P115A mutant CRP required a buffer less acidic than required by native CRP for comparable binding to immobilized ligands.
DISCUSSION
In this study, we investigated the binding of fluid-phase CRP to six proteins: factor H, oxLDL, C3b, IgG, Aβ, and BSA, which were immobilized on microtiter wells. Our major findings were: 1) at pH 7.0, CRP did not bind to any of these proteins; however, at acidic pH, CRP bound to all six proteins. 2) The acidic pH did not monomerize CRP but modified the pentameric structure. Some modifications were reversible at pH 7.0. 3) For CRP to bind to immobilized proteins, exposure of both CRP and immobilized proteins to acidic pH was required, except in the case of factor H. 4) The binding was unaffected by the presence or absence of Ca2+. 5) Substitution of Pro115, an amino acid that participates in intersubunit interactions in the CRP pentamer, with Ala reduced the requirement of acidic pH by 1 unit and enhanced the binding of CRP to all six immobilized proteins.
We used CRP at 10 μg/ml (∼100 nm) or lower concentrations in the binding assays. We used Ca2+ at a concentration of 0.1 mm, which was ∼1000-fold molar excess of CRP concentration. However, we also used 2 mm Ca2+ in a key experiment (Fig. 8). There was no difference in the results obtained with 0.1 or 2 mm Ca2+ when 10 μg/ml of CRP was used. Also, for two reasons we did not investigate binding of fluid-phase CRP to fluid-phase protein ligands. First, our aim was to investigate the binding of CRP to deposited proteins. Second, we thought it unlikely that CRP interacted with other proteins while in circulation.
The following observations suggested that the binding of CRP to immobilized proteins was a specific binding activity of CRP. 1) At least two other proteins, rabbit IgG and chicken ovalbumin, did not bind to any of the six immobilized proteins at acidic pH. 2) CRP bound to immobilized proteins in a CRP concentration-dependent and pH-dependent manner. 3) There was variation in the strengths of acidic pH required for the binding of CRP to each individual immobilized protein. 4) The binding of CRP to all six immobilized proteins was inhibited by PCh. 5) The binding of CRP was drastically affected by mutating only a single amino acid in CRP.
Although in the absence of a ligand, CRP remained pentameric at acidic pH, perhaps this was not the case once CRP was bound to the ligand. The data obtained with the use of mAb specific for modified forms of CRP indicated that CRP, after binding to PnC or protein ligands at acidic pH, was either monomerized or remained pentameric but the structure was partially modified in a way to express intersubunit contact residues that are hidden in the intact pentameric structure. These results are consistent with the previously published reports that CRP monomerizes after binding to membranes and activated platelets (15, 17). These results also explain the successful detection of CRP deposited in vivo at the sites of inflammation even using mCRP-specific mAb in immunohistochemistry (17). It is possible that mCRP remains bound to the ligand once it is formed after binding to an immobilized ligand, and, therefore, mCRP may be considered a ligand-bound by-product of CRP.
The acidic pH could be found at localized inflammatory sites or in extracellular spaces where damaged cells are present (35). Although there have been many unsuccessful attempts to determine the pH of tissues in vivo, an in vitro study determined that activated macrophages can acidify the microenvironment between the cells to pH 3.6, by proton generation (27, 28). One example of a localized inflammatory site where the pH could be acidic is atherosclerotic lesions. Atherosclerosis is an inflammatory disease and in areas in which inflammation takes place, the extracellular pH decreases due to hypoxia, lactate generation, and proton generation (27–31). Atherosclerotic lesions may therefore have a transient decrease in pH to acidic levels. Other examples of a localized inflammatory site where the pH could be acidic are the synovial fluid in various joint diseases and tumors (25, 26, 33, 34).
Binding of CRP to all six immobilized proteins including BSA at acidic pH was surprising. To interpret these results, we first considered that the immobilized protein ligands were also exposed to acidic pH when CRP, diluted in acidic pH buffer, was added to the microtiter wells. We hypothesize that CRP binds to a common structure formed by all proteins after immobilization and exposure to acidic pH. Because the exposure of acidic pH to immobilized proteins causes conformational alteration of proteins, in addition to deposition and aggregation of proteins on the walls of the microtiter plates, we conclude that, at acidic pH, CRP binds to deposited, aggregated, and conformationally altered proteins, as we have proposed previously (61). Our data suggest that conformationally altered proteins, regardless of the identity of the protein, form a CRP ligand. It is possible that the temperature of 37 °C also contributed to the conversion of immobilized proteins into a CRP ligand. This ligand binding specificity of CRP may play a beneficial role not only in atherosclerosis where LDL gets conformationally altered, but also at other inflammatory sites, such as synovial fluid in various joint diseases, where local inflamed tissue becomes acidic and IgG antibodies present in the immune complexes may be conformationally altered (25, 26). The binding of CRP to oxLDL and IgG at these sites would not reflect the specificity of CRP for these two ligands but instead highlight the ability of CRP to bind to conformationally altered proteins. Additionally, the binding specificity of CRP for conformationally altered proteins raises the possibility that CRP may also bind to aggregates of misfolded proteins and amyloids at the sites where the pH is acidic. Indeed, CRP immunoreactivity has been demonstrated in the brains of Alzheimer patients (64–66).
It has been reported previously that, at acidic pH, CRP also binds to extracellular matrix protein fibronectin (67–70). Fibronectin is a multifunctional glycoprotein and is highly expressed in several tumors that are sites of acidic pH (26, 71). The interaction of CRP with fibronectin does not require Ca2+ and the maximal interaction occurs at pH 5.0. Our current data suggest that the binding of CRP to fibronectin at acidic pH is due to the binding specificity of acidic pH-treated CRP for conformationally altered proteins.
Because all the previously known ligand-binding sites on CRP were active at physiological pH and because our data suggested that the binding of CRP to immobilized proteins at acidic pH was independent of the Ca2+-binding site of CRP, we hypothesized that the binding site on CRP for immobilized proteins required the participation of amino acids that were hidden in the intersubunit regions in native pentameric CRP. The majority of the intersubunit contacts in CRP involve the loop containing amino acids 115–123 on one subunit interacting with a variety of residues in the next subunit. For example, the side chain of Pro115 in one subunit interacts with Trp205 of the adjacent subunit and the main chain of Pro115 interacts with Tyr40 (72, 73). The data obtained with the P115A CRP mutant did not reveal the binding site on CRP for immobilized proteins, but, these data suggested that acidic pH changes the intersubunit region perhaps from a closed to an open conformation and that the change in CRP induced by acidic pH could be mimicked in part by mutating Pro115.
The exact mechanism of action of acidic pH on the structure and function of CRP is not clear. Here, by using gel filtration and ANS fluorescence, we show that acidic pH changes the pentameric structure of CRP but does not monomerize CRP. The acidic pH-dependent conformational change in CRP has also been shown previously by using circular dichroism and fluorescence spectra analyses (74). Another previous report indicated that acidic pH does not monomerize CRP but changes the conformation of the pentameric structure and that some acidic pH-induced conformationally altered CRP pentamers form decamers (75). Thus, it is possible that the binding specificities that we found in this study were due to acidic pH-induced formation of CRP decamers. Alternatively, because we found that the binding of CRP to immobilized ligands at acidic pH was unaffected by the presence of Ca2+, it seems reasonable to assume that acidic pH protonates CRP, perhaps at the Ca2+-binding site, and induces the detachment of one or both Ca2+ from CRP. The effects of loss of Ca2+ on the structure of CRP combined with the direct effects of acidic pH on the structure of CRP might have contributed to acidic pH-dependent binding specificity of CRP.
Initially, the aim of our study was to explore the binding of CRP to factor H and oxLDL. The results showed that even if we had not used these two proteins, we would have arrived at the same conclusions. However, our finding that CRP does not bind to immobilized factor H at physiological pH is consistent with other reports showing that native pentameric CRP does not bind to immobilized factor H (76–78). One recent study reported that CRP bound to factor H at physiological pH. In this study (45), CRP, when added at concentrations of 1.4 mg/ml to factor H but not at lower concentrations, bound to factor H at physiological pH. We do not have an explanation for what structural changes occurred in CRP molecules or how the function of CRP molecules was changed when CRP was present at high concentrations. In our studies, we used CRP at 10 μg/ml or less and did not observe binding of CRP to factor H at physiological pH. We did not use CRP at higher concentrations because it has been shown that CRP, in the purified form, at high concentrations such as 1.4 mg/ml, has a tendency to self-aggregate (79–81). Whether such self-aggregation of CRP at high concentrations occurs in body fluids in vivo is not known. Also, we used ELISA-based assays, which can detect CRP even if 100 ng of CRP was bound to the ligand-coated microtiter wells. If we had used 1.4 mg/ml of CRP in our assays, then assuming that 0.0001% (that is, 140 ng/ml) of this CRP was modified and assuming that modified CRP binds to factor H at physiological pH, we would have detected the binding of modified CRP, and not of native pentameric CRP, to the ligand coated on the wells. Considering the sensitivity and limitations of ELISA-based assays, we felt that using CRP at low concentrations was more appropriate to investigate the ligand-binding properties of CRP molecules in our assays. Combined data suggest that if factor H is deposited at an inflammatory site or on a pathogen and if the pH of the environment is acidic, then pentameric CRP can also bind to factor H.
The binding of native pentameric CRP to oxLDL has been investigated previously in many laboratories. Some reports indicated that CRP bound to oxLDL at physiological pH (82–84). Another study showed that CRP did not bind to oxLDL (14). Our data suggest that CRP should be able to bind to oxLDL present in atherosclerotic lesions after a partial change in structure induced by a localized decrease in extracellular pH. Without this structural change that allows binding to oxLDL, CRP was not atheroprotective in murine models of atherosclerosis (85, 86). It follows that if indeed CRP requires an acidic environment to function in vivo, then it would be necessary to achieve acidic pH at the sites of inflammation in animal models of human diseases. This could be a rate-limiting step in the bioactivity of CRP; without sufficient acidity, a possible effect of CRP may be missed.
We conclude that pH regulates some functions of CRP. Acidic pH transforms native pentameric CRP into another pentameric form, which displays two binding specificities: one for factor H and another for a ligand made up of conformationally altered proteins irrespective of the identity of the protein. The second binding specificity of CRP may be responsible for the deposition of CRP at the sites of inflammation with acidic pH. These bioactivities are consistent with CRP being a component of the innate immune system, which confers pattern recognition function. These bioactivities of CRP also support the hypothesis that CRP has been conserved throughout evolution for protection against disease and toxicity caused by protein misfolding and conformationally altered proteins in acidic milieu.
Supplementary Material
Acknowledgment
We thank Dr. William Stone, Department of Pediatrics, ETSU, for use of the spectrofluorometer.
This work was supported, in whole or in part, by National Institutes of Health Grants R01HL071233 (to A. A.) and R01DK35081 (to M. K. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- CRP
- pentameric C-reactive protein
- mCRP
- monomeric C-reactive protein
- LDL
- low density lipoprotein
- oxLDL
- oxidized LDL
- Aβ
- amyloid β protein fragment 1–38
- PCh
- phosphocholine
- PnC
- pneumococcal C-polysaccharide
- ANS
- 1-anilinonaphthalene-8-sulfonic acid.
REFERENCES
- 1.Singh S. K., Suresh M. V., Voleti B., Agrawal A. (2008) Ann. Med. 40, 110–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black S., Kushner I., Samols D. (2004) J. Biol. Chem. 279, 48487–48490 [DOI] [PubMed] [Google Scholar]
- 3.Volanakis J. E. (2001) Mol. Immunol. 38, 189–197 [DOI] [PubMed] [Google Scholar]
- 4.Volanakis J. E., Kaplan M. H. (1971) Proc. Soc. Exp. Biol. Med. 136, 612–614 [DOI] [PubMed] [Google Scholar]
- 5.Kinoshita C. M., Ying S. C., Hugli T. E., Siegel J. N., Potempa L. A., Jiang H., Houghten R. A., Gewurz H. (1989) Biochemistry 28, 9840–9848 [DOI] [PubMed] [Google Scholar]
- 6.Christopeit T., Gossas T., Danielson U. H. (2009) Anal. Biochem. 391, 39–44 [DOI] [PubMed] [Google Scholar]
- 7.Marnell L., Mold C., Du Clos T. W. (2005) Clin. Immunol. 117, 104–111 [DOI] [PubMed] [Google Scholar]
- 8.Bhakdi S., Torzewski M., Klouche M., Hemmes M. (1999) Arterioscler. Thromb. Vasc. Biol. 19, 2348–2354 [DOI] [PubMed] [Google Scholar]
- 9.Singh S. K., Suresh M. V., Prayther D. C., Moorman J. P., Rusiñol A. E., Agrawal A. (2008) J. Immunol. 180, 4316–4322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwalbe R. A., Dahlbäck B., Coe J. E., Nelsestuen G. L. (1992) Biochemistry 31, 4907–4915 [DOI] [PubMed] [Google Scholar]
- 11.Agrawal A., Simpson M. J., Black S., Carey M. P., Samols D. (2002) J. Immunol. 169, 3217–3222 [DOI] [PubMed] [Google Scholar]
- 12.Potempa L. A., Siegel J. N., Fiedel B. A., Potempa R. T., Gewurz H. (1987) Mol. Immunol. 24, 531–541 [DOI] [PubMed] [Google Scholar]
- 13.Verma S., Szmitko P. E., Yeh E. T. H. (2004) Circulation 109, 1914–1917 [DOI] [PubMed] [Google Scholar]
- 14.Ji S. R., Wu Y., Potempa L. A., Qiu Q., Zhao J. (2006) Int. J. Biochem. Cell Biol. 38, 648–661 [DOI] [PubMed] [Google Scholar]
- 15.Ji S. R., Wu Y., Zhu L., Potempa L. A., Sheng F. L., Lu W., Zhao J. (2007) FASEB J. 21, 284–294 [DOI] [PubMed] [Google Scholar]
- 16.Boncler M., Watała C. (2009) Acta Biochim. Pol. 56, 17–31 [PubMed] [Google Scholar]
- 17.Eisenhardt S. U., Habersberger J., Murphy A., Chen Y. C., Woollard K. J., Bassler N., Qian H., von Zur Muhlen C., Hagemeyer C. E., Ahrens I., Chin-Dusting J., Bobik A., Peter K. (2009) Circ. Res. 105, 128–137 [DOI] [PubMed] [Google Scholar]
- 18.Pepys M. B., Hirschfield G. M. (2003) J. Clin. Invest. 111, 1805–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kushner I., Kaplan M. H. (1961) J. Exp. Med. 114, 961–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krijnen P. A., Ciurana C., Cramer T., Hazes T., Meijer C. J., Visser C. A., Niessen H. W., Hack C. E. (2005) J. Clin. Pathol. 58, 382–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson P. T., Betts K. E., Radeke M. J., Hageman G. S., Anderson D. H., Johnson L. V. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17456–17461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reynolds G. D., Vance R. P. (1987) Arch. Pathol. Lab. Med. 111, 265–269 [PubMed] [Google Scholar]
- 23.Torzewski M., Rist C., Mortensen R. F., Zwaka T. P., Bienek M., Waltenberger J., Koenig W., Schmitz G., Hombach V., Torzewski J. (2000) Arterioscler. Thromb. Vasc. Biol. 20, 2094–2099 [DOI] [PubMed] [Google Scholar]
- 24.Kushner I., Samols D., Magrey M. (2010) Arthritis Care Res. 62, 442–446 [DOI] [PubMed] [Google Scholar]
- 25.Treuhaft P. S., McCarty D. J. (1971) Arthritis Rheum. 14, 475–484 [DOI] [PubMed] [Google Scholar]
- 26.Wike-Hooley J. L., Haveman J., Reinhold H. S. (1984) Radiother. Oncol. 2, 343–366 [DOI] [PubMed] [Google Scholar]
- 27.Silver I. A., Murrills R. J., Etherington D. J. (1988) Exp. Cell Res. 175, 266–276 [DOI] [PubMed] [Google Scholar]
- 28.Leake D. S. (1997) Atherosclerosis 129, 149–157 [DOI] [PubMed] [Google Scholar]
- 29.Björnheden T., Levin M., Evaldsson M., Wiklund O. (1999) Arterioscler. Thromb. Vasc. Biol. 19, 870–876 [DOI] [PubMed] [Google Scholar]
- 30.Naghavi M., John R., Naguib S., Siadaty M. S., Grasu R., Kurian K. C., van Winkle W. B., Soller B., Litovsky S., Madjid M., Willerson J. T., Casscells W. (2002) Atherosclerosis 164, 27–35 [DOI] [PubMed] [Google Scholar]
- 31.Haka A. S., Grosheva I., Chiang E., Buxbaum A. R., Baird B. A., Pierini L. M., Maxfield F. R. (2009) Mol. Biol. Cell 20, 4932–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sneck M., Kovanen P. T., Oörni K. (2005) J. Biol. Chem. 280, 37449–37454 [DOI] [PubMed] [Google Scholar]
- 33.Andersson S. E., Lexmüller K., Johansson A., Ekström G. M. (1999) J. Rheumatol. 26, 2018–2024 [PubMed] [Google Scholar]
- 34.Andreev O. A., Dupuy A. D., Segala M., Sandugu S., Serra D. A., Chichester C. O., Engelman D. M., Reshetnyak Y. K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7893–7898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lardner A. (2001) J. Leukocyte Biol. 69, 522–530 [PubMed] [Google Scholar]
- 36.Pangburn M. K. (2000) Immunopharmacology 49, 149–157 [DOI] [PubMed] [Google Scholar]
- 37.Mold C., Kingzette M., Gewurz H. (1984) J. Immunol. 133, 882–885 [PubMed] [Google Scholar]
- 38.Jarva H., Janulczyk R., Hellwage J., Zipfel P. F., Björck L., Meri S. (2002) J. Immunol. 168, 1886–1894 [DOI] [PubMed] [Google Scholar]
- 39.Mold C., Nakayama S., Holzer T. J., Gewurz H., Du Clos T. W. (1981) J. Exp. Med. 154, 1703–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yother J., Volanakis J. E., Briles D. E. (1982) J. Immunol. 128, 2374–2376 [PubMed] [Google Scholar]
- 41.Szalai A. J., Briles D. E., Volanakis J. E. (1995) J. Immunol. 155, 2557–2563 [PubMed] [Google Scholar]
- 42.Suresh M. V., Singh S. K., Ferguson D. A., Jr., Agrawal A. (2006) J. Immunol. 176, 4369–4374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sjöberg A. P., Trouw L. A., Clark S. J., Sjölander J., Heinegård D., Sim R. B., Day A. J., Blom A. M. (2007) J. Biol. Chem. 282, 10894–10900 [DOI] [PubMed] [Google Scholar]
- 44.Laine M., Jarva H., Seitsonen S., Haapasalo K., Lehtinen M. J., Lindeman N., Anderson D. H., Johnson P. T., Järvelä I., Jokiranta T. S., Hageman G. S., Immonen I., Meri S. (2007) J. Immunol. 178, 3831–3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okemefuna A. I., Nan R., Miller A., Gor J., Perkins S. J. (2010) J. Biol. Chem. 285, 1053–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suresh M. V., Singh S. K., Ferguson D. A., Jr., Agrawal A. (2007) J. Immunol. 178, 1158–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agrawal A., Suresh M. V., Singh S. K., Ferguson D. A., Jr. (2008) Endocr. Metab. Immune Disord. Drug Targets 8, 231–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skerka C., Zipfel P. F. (2008) Vaccine 26, I9–14 [DOI] [PubMed] [Google Scholar]
- 49.Galkina E., Ley K. (2009) Annu. Rev. Immunol. 27, 165–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh S. K., Suresh M. V., Hammond D. J., Jr., Rusiñol A. E., Potempa L. A., Agrawal A. (2009) Clin. Chim. Acta 406, 151–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pangburn M. K., Müller-Eberhard H. J. (1983) Biochemistry 22, 178–185 [DOI] [PubMed] [Google Scholar]
- 52.Okemefuna A. I., Nan R., Gor J., Perkins S. J. (2009) J. Mol. Biol. 391, 98–118 [DOI] [PubMed] [Google Scholar]
- 53.Hakobyan S., Harris C. L., Tortajada A., Goicochea de Jorge E., García-Layana A., Fernández-Robredo P., Rodríguez de Córdoba S., Morgan B. P. (2008) Invest. Ophthalmol. Vis. Sci. 49, 1983–1990 [DOI] [PubMed] [Google Scholar]
- 54.Havel R. J., Eder H. A., Bragdon J. H. (1955) J. Clin. Invest. 34, 1345–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steinbuch M., Audran R. (1969) Arch. Biochem. Biophys. 134, 279–284 [DOI] [PubMed] [Google Scholar]
- 56.Brown A. J., Leong S. L., Dean R. T., Jessup W. (1997) J. Lipid Res. 38, 1730–1745 [PubMed] [Google Scholar]
- 57.Rusiñol A. E., Yang L., Thewke D., Panini S. R., Kramer M. F., Sinensky M. S. (2000) J. Biol. Chem. 275, 7296–7303 [DOI] [PubMed] [Google Scholar]
- 58.Agrawal A., Volanakis J. E. (1994) J. Immunol. 152, 5404–5410 [PubMed] [Google Scholar]
- 59.Chapeaurouge A., Johansson J. S., Ferreira S. T. (2002) J. Biol. Chem. 277, 16478–16483 [DOI] [PubMed] [Google Scholar]
- 60.De Filippis V., de Laureto P. P., Toniutti N., Fontana A. (1996) Biochemistry 35, 11503–11511 [DOI] [PubMed] [Google Scholar]
- 61.Singh S. K., Hammond D. J., Jr., Beeler B. W., Agrawal A. (2009) Clin. Chim. Acta 409, 143–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kilpatrick J. M., Kearney J. F., Volanakis J. E. (1982) Mol. Immunol. 19, 1159–1165 [DOI] [PubMed] [Google Scholar]
- 63.Ying S. C., Shephard E., de Beer F. C., Siegel J. N., Harris D., Gewurz B. E., Fridkin M., Gewurz H. (1992) Mol. Immunol. 29, 677–687 [DOI] [PubMed] [Google Scholar]
- 64.Iwamoto N., Nishiyama E., Ohwada J., Arai H. (1994) Neurosci. Lett. 177, 23–26 [DOI] [PubMed] [Google Scholar]
- 65.Duong T., Nikolaeva M., Acton P. J. (1997) Brain Res. 749, 152–156 [DOI] [PubMed] [Google Scholar]
- 66.McGeer E. G., Yasojima K., Schwab C., McGeer P. L. (2001) Neurobiol. Aging 22, 843–848 [DOI] [PubMed] [Google Scholar]
- 67.Salonen E. M., Vartio T., Hedman K., Vaheri A. (1984) J. Biol. Chem. 259, 1496–1501 [PubMed] [Google Scholar]
- 68.Tseng J., Mortensen R. F. (1988) Mol. Immunol. 25, 679–686 [DOI] [PubMed] [Google Scholar]
- 69.Tseng J., Mortensen R. F. (1989) Exp. Cell Res. 180, 303–313 [DOI] [PubMed] [Google Scholar]
- 70.Suresh M. V., Singh S. K., Agrawal A. (2004) J. Biol. Chem. 279, 52552–52557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ruoslahti E. (1999) Adv. Cancer Res. 76, 1–20 [DOI] [PubMed] [Google Scholar]
- 72.Srinivasan N., White H. E., Emsley J., Wood S. P., Pepys M. B., Blundell T. L. (1994) Structure 2, 1017–1027 [DOI] [PubMed] [Google Scholar]
- 73.Shrive A. K., Cheetham G. M., Holden D., Myles D. A., Turnell W. G., Volanakis J. E., Pepys M. B., Bloomer A. C., Greenhough T. J. (1996) Nat. Struct. Biol. 3, 346–354 [DOI] [PubMed] [Google Scholar]
- 74.Miyazawa K., Inoue K. (1990) J. Immunol. 145, 650–654 [PubMed] [Google Scholar]
- 75.Blizniukov O. P., Kozmin L. D., Falikova V. V., Martynov A. I., Tishchenko V. M. (2003) Biofizika 48, 844–852 [PubMed] [Google Scholar]
- 76.Mold C., Gewurz H., Du Clos T. W. (1999) Immunopharmacology 42, 23–30 [DOI] [PubMed] [Google Scholar]
- 77.Hakobyan S., Harris C. L., van den Berg C. W., Fernandez-Alonso M. C., de Jorge E. G., de Cordoba S. R., Rivas G., Mangione P., Pepys M. B., Morgan B. P. (2008) J. Biol. Chem. 283, 30451–30460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mihlan M., Stippa S., Józsi M., Zipfel P. F. (2009) Cell Death Differ. 16, 1630–1640 [DOI] [PubMed] [Google Scholar]
- 79.Wang H. W., Wu Y., Chen Y., Sui S. F. (2002) Int. J. Mol. Med. 9, 665–671 [PubMed] [Google Scholar]
- 80.Blizniukov O. P., Kozmin L. D., Falikova V. V., Martynov A. I., Tischenko V. M. (2003) Mol. Biol. 37, 912–918 [PubMed] [Google Scholar]
- 81.Okemefuna A. I., Stach L., Rana S., Buetas A. J., Gor J., Perkins S. J. (2010) J. Biol. Chem. 285, 1041–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chang M. K., Binder C. J., Torzewski M., Witztum J. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13043–13048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Van Tits L., de Graaf J., Toenhake H., van Heerde W., Stalenhoef A. (2005) Arterioscler. Thromb. Vasc. Biol. 25, 717–722 [DOI] [PubMed] [Google Scholar]
- 84.Singh U., Dasu M. R., Yancey P. G., Afify A., Devaraj S., Jialal I. (2008) J. Lipid Res. 49, 1015–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tennent G. A., Hutchinson W. L., Kahan M. C., Hirschfield G. M., Gallimore J. R., Lewin J., Sabin C. A., Dhillon A. P., Pepys M. B. (2008) Atherosclerosis 196, 248–255 [DOI] [PubMed] [Google Scholar]
- 86.Torzewski M., Reifenberg K., Cheng F., Wiese E., Küpper I., Crain J., Lackner K. J., Bhakdi S. (2008) Thromb. Haemost. 99, 196–201 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.