Abstract
Mammalian Notch receptors require modification by fucose on epidermal growth factor-like (EGF) repeats of their extracellular domain to respond optimally to signal induction by canonical Notch ligands. Inactivation of the Golgi GDP-fucose transporter Slc35c1 in mouse or human does not cause marked defects in Notch signaling during development, and shows milder fucosylation defects than those observed in mice unable to synthesize GDP-fucose, indicating the existence of another mechanism for GDP-fucose transport into the secretory pathway. We show here that fibroblasts from mice or humans lacking Slc35c1 exhibit robust Notch signaling in co-culture signaling assays. A potential candidate for a second GDP-fucose transporter is the related gene Slc35c2. Overexpression of Slc35c2 reduces expression of the fucosylated epitopes Lewis X and sialylated Lewis X in CHO cells, indicating competition with Slc35c1. The fucosylation of a Notch1 EGF repeat fragment that occurs in the endoplasmic reticulum was increased in CHO transfectants overexpressing Slc35c2. In CHO cells with low levels of Slc35c2, both Delta1- and Jagged1-induced Notch signaling were reduced, and the fucosylation of a Notch1 fragment was also decreased. Immunofluorescence microscopy of rat intestinal epithelial cells and HeLa cells, and analysis of rat liver membrane fractions showed that Slc35c2 is primarily colocalized with markers of the cis-Golgi network and endoplasmic reticulum-Golgi intermediate compartment (ERGIC). The combined results suggest that Slc35c2 is either a GDP-fucose transporter that competes with Slc35c1 for GDP-fucose, or a factor that otherwise enhances the fucosylation of Notch and is required for optimal Notch signaling in mammalian cells.
Keywords: Endoplasmic Reticulum (ER), Glycosylation, Golgi, Notch Pathway, Notch Receptor, GDP-fucose Transport
Introduction
Notch in Drosophila and Notch1–4 in mammals are single transmembrane glycoproteins that regulate many cell fate decisions (1, 2). They contain up to 36 epidermal growth factor-like (EGF) repeats in the extracellular domain, a subset of which are modified with O-fucose attached to Ser or Thr by protein O-fucosyltransferase-1 (Pofut1)4 in mammals and OFUT1 in Drosophila (3). Targeted mutation of mouse Pofut1 (4, 5) or removal of Drosophila OFUT1 (6, 7) gives rise to global Notch signaling defects. Because loss of OFUT1 (8–11) or Pofut1 (5) may lead to reduced Notch expression at the cell surface, a requirement for fucose in Notch signaling has been investigated in mutants that cannot synthesize GDP-fucose. Lec13 Chinese hamster ovary (CHO) cells with reduced GDP-fucose (12–14), and mice with a null mutation in the FX gene (15, 16), provide evidence that fucose is required for optimal Notch signaling in mammals. This requirement may merely be to serve as the substrate of Fringe as proposed in Drosophila (9), but mice lacking all three Fringe genes are born and may survive for several months (17). Thus, it is important to determine the activities necessary for fucosylation of Notch. One activity that remains to be identified in mammals is the GDP-fucose transporter(s) necessary to provide GDP-fucose to Pofut1, a resident of the endoplasmic reticulum with a KDEL-like retrieval signal (18).
GDP-fucose, the donor substrate for all fucosyltransferases, is synthesized in the cytoplasm through a de novo pathway from mannose, or a salvage pathway that uses fucose from degraded glycoproteins or glycolipids or from exogenous l-fucose (19, 20). GDP-fucose transporters are required to transport GDP-fucose into the lumen of the secretory pathway for utilization as donor substrate by fucosyltransferases (21, 22). Mutations of the GDP-fucose transporter SLC35C1 in humans cause leukocyte adhesion deficiency type II (LADII) or congenital disorder of glycosylation type IIc characterized by severe immunodeficiency, mental retardation, and slow growth (23–26). Mice lacking Slc35c1 have a similar phenotype (27, 28). Importantly, neither mice nor humans lacking Slc35c1 have globally severe Notch signaling defects typical of the loss of Pofut1 that leads to embryonic lethality at mid-gestation (4, 5). Changes in T cell development that requires Notch1 signaling have not been reported in LADII patients nor mice lacking Slc35c1. In addition, cultured fibroblasts from LADII patients exhibit no reduction in the fucosylation of Notch1 ECD fragments, although their N-glycans lack fucose (29). In Drosophila, null mutants of the Drosophila Slc35c1 homologue have only mild Notch signaling deficiencies that are exacerbated at low temperatures (30). The combined observations suggest the existence of a GDP-fucose transport mechanism in addition to Slc35c1 that accomplishes the fucosylation of Notch receptors.
Sequence comparisons revealed a gene that clusters with SLC35C1 termed SLC35C2 in human (31), to be the most likely candidate for an alternative GDP-fucose transporter. Slc35c2 has about 22–23% identity and 37–38% similarity to Slc35c1 from Drosophila to human and is highly conserved between species. We previously reported that overexpression of Slc35c2 decreases expression of fucosylated epitopes termed Lewis X and sialyl-Lewis X in LEC11B CHO cells (32). A possible explanation is that Slc35c2 is a GDP-fucose transporter or transport facilitator that competes with Slc35c1 by directing GDP-fucose to an earlier secretory compartment where Notch is fucosylated. Pofut1 and OFUT1 have a KDEL-like sequence at their C terminus and are thought to cycle between the endoplasmic reticulum (ER) and the cis-Golgi network (8, 18). An alternative GDP-fucose transporter recently identified in Drosophila as important for Notch signaling is the homologue of human SLC35B4 (33). However, human SLC35B4 was shown to be a UDP-xylose/UDP-GlcNAc transporter (34). Most importantly, SLC35B4 was shown not to have GDP-fucose transport activity in vitro (34). Therefore, we investigated the effects of overexpression and reduced expression of mouse and CHO Slc35c2 on the fucosylation and activation of Notch signaling in mammalian cells. In this article we show that overexpressed Slc35c2 increases, whereas knockdown reduces, O-fucosylation of Notch1 EGF repeats, and that optimal Notch signaling requires Slc35c2 in mammalian cells.
EXPERIMENTAL PROCEDURES
Cells and Cell Culture
Parental CHO cells Pro−5 and Gat−2 (35) and LEC11B (Gat−LEC11.F2 (36)) were cultured at 37 °C in α-modified minimal essential medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS; Gemini, West Sacramento, CA) unless otherwise indicated. LADII patient and control human fibroblasts were kindly provided by Robert Haltiwanger (SUNY Stonybrook) (29), rat intestinal epithelial cells (IEC) cells were kindly provided by Len Augenlicht (Albert Einstein College of Medicine), and Slc35c1−/− and Slc35c1+/+ mouse embryonic fibroblasts (MEF) prepared from mouse embryos (27) were cultured at 37 °C in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% FBS (Gemini), penicillin, streptomycin, and glutamine (Millipore, Billerica, MA). L cells expressing Delta1 or Jagged1 were sorted for high ligand expression as described previously (14).
Plasmids
pCMV6-Kan/Neo-mSlc35c1 and pCMV6-Kan/Neo-mSlc35c2 were purchased from OriGene Technologies, Inc. (Rockville, MD). The open reading frames (ORF) of mSlc35c1 and mSlc35c2 were ligated into pCDNA3.1/Myc-His (Invitrogen) following excision by HindIII and XhoI. A CHO Slc35c2 cDNA ligated into pCR3.1 (Invitrogen) was described previously (32). Expression vectors containing CHO Fut6B or Fut9 cDNA and the Notch1 fragment EGF11–15MycHis6 were described previously (36–38). Tagged mouse Notch1 with 6 tandem Myc epitopes in place of the C-terminal PEST domain was kindly provided by Rafael Kopan (Washington University, St. Louis, MO) (39).
Slc35c1 PCR Genotyping
The genotype of Slc35c1 wild type and mutant MEFs was confirmed using primers: GFT-F1 (5′-GCG TTG CAA GTT CAG CCG AG-3′), GFT-R2 (5′-CCG TCG ACG GTA TCG ATA AGC-3′), and GFT-R1 (5′-GTG TGT TGG TCA AGA GTG TAA CCT ATG-3′) as described (27). GFT-F1 and GFT-R1 amplify a 2.3-kb product from the wild type allele and GFT-F1 and GFT-R2 amplify a 1.8-kb product from the mutant allele.
Generation of Anti-mouse Slc35c2 Polyclonal Antibody
An Slc35c2 C-terminal peptide (GEEEYFVTQGQQ) that is conserved in human, rat, and mouse was used to generate rabbit polyclonal antibodies at Proteintech Group Inc. (Chicago, IL) after keyhole limpet hemocyanin conjugation to the first glycine. Antisera were affinity purified using the same peptide on cyanogen bromide-activated Sepharose 4B (Sigma). The specificity of this antibody is shown in Fig. 7A and supplemental Fig. S2.
FIGURE 7.
Slc35c2 is primarily localized to Golgi in rat liver. A, a new polyclonal Ab against mouse and human Slc35c2 was tested on lysates of CHO cells expressing empty vector, mouse Slc35c1-Myc, mouse Slc35c2 with low transfection efficiency (C2(low)), or mouse Slc35c2-Myc and analyzed by Western blotting using anti-Slc35c2 C-terminal peptide antibody or anti-Myc. B, cell lysates from CHO cells transiently transfected with mouse Slc35c2 were treated with Endo H or PNGase F and analyzed by Western blotting using anti-Slc35c2 pAb. The membrane was stripped and re-probed with anti-Pofut1 C-terminal peptide pAb. Western blotting was performed using the Odyssey Infrared Imaging System. C, rat liver ER and Golgi fractions were assayed by immunoblotting using anti-Slc35c2 peptide or anti-PDI (ER marker) or anti-GM130 (Golgi marker) or anti-Pofut1 peptide antibodies. Western blotting was performed using the Odyssey Infrared Imaging System.
Fucose Metabolic Labeling
Duplicate cell cultures were plated at 1 × 106 cells/10-cm dish and transfected the next day using FuGENE 6 (Roche Applied Science) with 20 μg of Notch1 EGF11–15MycHis6. After 16 h of transfection, cells were metabolically labeled with 10 μCi/ml of l-[3H]fucose (American Radiolabeled Chemicals, Inc.) for 30 h. Notch1 EGF11–15MycHis6 was harvested by incubation with Ni2+ beads at 4 °C overnight with medium and cell lysate, washed, and eluted with 100 mm EDTA as described (38). Samples were analyzed by Western blot with anti-Myc monoclonal antibody 9E10 (1:200; Calbiochem, San Diego, CA) and autoradiography to detect [3H]fucose. Blots were quantitated by NIH Image J and the ratios of [3H]fucose incorporated relative to the anti-Myc signal were determined.
Targeted Knockdown of Slc35c2 in CHO Cells
Six different siRNAs targeting CHO Slc35c2 cDNA were generated using the pSilencer® siRNA Construction Kit (Ambion, Austin, TX). Parent Pro−5 CHO cells were transiently transfected with these siRNAs and Northern analyses were performed to determine the most active siRNA sequence. This was 5′-AAG TGG TGG TCT GAG TCA CCT-3′ that targets the 3′ UTR of CHO Slc35c2. This sequence was ligated into pSilencer2.1-U6 neo and pSilencer2.1-U6 hygro vectors and transfected either individually or in combination into Pro−5 CHO cells using Lipofectamine 2000 (Invitrogen). Stable transfectants were isolated by selection using G418 (neomycin) at 1.5 mg/ml of active concentration (Gemini, West Sacramento, CA), or hygromycin at 700 μg/ml (Calbiochem, Darmstadt, Germany), or both together as appropriate. Total RNA from stable transfectants was collected using TRIzol (Invitrogen) and cDNA was prepared using the SuperScriptTM III First-strand Synthesis System (Invitrogen). Semi-quantitative RT-PCR was performed to determine Slc35c2 transcripts using primers 5′-ACT TGG ATC CTC CTG TTC CTG CTG GCC CAC GGA T-3′ and 5′-ATC AGG ATC CTC CCA CCA AGA GTT GCC AGC AGT G-3′ and compared with GAPDH using primers PS800 5′-TGA ATT CAT TGA CCT CAA CTA CAT-3′ and PS801 5′-AGA ATT CTT ACT CCT TGG AGG CCA-3′ as control. Transfectants Slc35c2-KD1 and vector control have only pSilencer2.1-U6 hygro vector, whereas Slc35c2-KD2 contains both vectors.
Ligand-dependent Notch Signaling Assays
Notch signaling co-culture assays were performed as previously described (13, 14, 40). Briefly, duplicate cell cultures were plated at 2 × 105 cells per well of a six-well dish and, after ∼24 h, were cotransfected using FuGENE 6 (Roche Applied Science) with 0.2 μg of the Notch reporter plasmid TP-1 that carries eight copies of a RBP-Jκ DNA binding sequence driving a firefly luciferase reporter gene (41), 0.05 μg of pRL-TK, or 0.01 μg of pRL-CMV Renilla luciferase (Promega, Madison, WI), and 1.5 μg of pMIRB empty vector. After 16 h at 37 °C, 1.5–2 × 106 Jagged1/L, Delta1/L, or control L cells were overlaid. At 48 h after transfection, firefly and Renilla luciferase activities were quantitated in cell lysates using a dual luciferase assay (Promega, Madison, WI). Ligand-dependent Notch activation is expressed as the ratio of firefly:Renilla luciferase activity or as fold-activation in Ligand/L versus L cells.
Ligand-dependent Notch1 Activation Assay
For CHO cells, cultures were plated at 6 × 105 cells/60-mm dish and transfected the next day with 0.5 μg of pCS2-mNotch1 and 3 μg of pCS2 empty vector using FuGENE 6 (Roche Applied Science). After 16 h of transfection, 4.5 × 106 Jagged1/L, Delta1/L, or control L cells were overlaid. When the γ-secretase inhibitor (GSI) L-685,458 (42) was used, CHO cells were preincubated with 2 μm L-685,458 (Sigma) or DMSO for 1 h before ligand cells in medium containing 2 μm L-685,458 or DMSO were overlaid. After another 6 h, cells were lysed using 1× passive lysis buffer (Promega, Madison, WI). Lysates were resolved by SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membrane, and probed with anti-Notch1 antibody Val1744 (Cell Signaling Technology; 1:1000) for detection of cleaved, activated Notch1, followed by horseradish peroxidase (HRP)-conjugated secondary antibodies (Jackson ImmunoResearch). Bands were visualized using the enhanced chemiluminescence (SuperSignal kit, Pierce). Membranes were stripped and probed with anti-Myc monoclonal antibody 9E10 for detection of transfected full-length mNotch1-Myc. For human fibroblasts, experiments were performed essentially the same as for CHO cells except without transfection of mNotch1.
Western Blot Analysis
Cells in a six-well dish were washed 3 times with cold phosphate-buffered saline (PBS), pH 7.4, resuspended in 250 μl of 1× passive lysis buffer (Promega, Madison, WI) supplemented with 1× CompleteTM EDTA-free protease inhibitor (Roche Applied Science), vortexed, and incubated on ice for 10 min. Lysates were centrifuged at 6,000 × g for 5 min at 4 °C and the supernatant was collected. Protein concentration was determined using the Bio-Rad Dc protein assay kit (Bio-Rad) according to the manufacturer's instructions. Protein (∼60 μg) was separated by SDS-PAGE and transferred to a PVDF membrane. Blocking was performed in TBST (25 mm Tris-HCl, 125 mm NaCl, 0.1% Tween 20, pH 7.4) containing 5% nonfat dry milk. Primary and secondary antibodies were diluted in TBST containing 5% nonfat dry milk and incubated with the blot for 16 h at 4 °C and 2 h at room temperature, respectively. Proteins were visualized using enhanced chemiluminescence (SuperSignal kit; Pierce). Secondary antibodies were horseradish peroxidase (HRP)-conjugated IgG (Jackson ImmunoResearch). Rat liver membrane fractions were kindly provided by the late Dennis Shields (Albert Einstein College of Medicine). Mouse anti-GM130 mAb (1:500) was purchased from BD Transduction Laboratories. Mouse anti-PDI mAb (1:1,000) was obtained from Assay Designs (Ann Arbor, MI). Mouse anti-β-actin mAb (1:5,000) was purchased from AbCam (Cambridge, MA). Mouse anti-Myc mAb 9E10 was used at 1:200. Pofut1 C-terminal peptide antibody (Pofut1C) used at 1:1,000 was described previously (43). Alternatively, Western blotting was performed using the Odyssey Infrared Imaging System (LI-COR, Lincoln, NE). Briefly, membranes were blocked for 1 h at room temperature with Odyssey blocking buffer and then incubated overnight at 4 °C with primary antibodies in Odyssey blocking buffer containing 0.1% (v/v) Tween 20. After three washes with TBST, membranes were incubated for 1 h at room temperature with Alexa Fluor 680-conjugated anti-mouse IgG (Invitrogen; 1:10,000) and/or IRDye800-conjugated anti-rabbit IgG (Rockland, Gilbertsville, PA; 1:10,000) in Odyssey blocking buffer containing 0.1% (v/v) Tween 20 and 0.01% SDS. After washing twice with TBST and once with TBS, membranes were analyzed using an Odyssey Infrared Imager.
Glycosidase Digestions
Digestion of ∼60 μg of cell lysate was performed in denaturing buffer as recommended by the manufacturer containing Complete EDTA-free protease inhibitor (Roche Applied Science), 1 mm EDTA, and 15 μm pepstatin. Samples were heated at 55 °C for 20 min in digestion buffer before adding 1,000 units of PNGase F (New England Biolabs, Beverley, MA) or 0.01 unit of endoglycosidase H (Endo H; Roche Applied Science) and incubated at 37 °C for ∼18 h.
Flow Cytometry Analysis
Fluorescein-labeled Pisum sativum agglutinin (FITC-PSA; 5 μg/ml), fluorescein-labeled Phaseolus vulgaris leukoagglutinin (FITC-L-PHA; 10 μg/ml), biotinylated Aleuria aurantia lectin (AAL; 10 μg/ml), and phycoerythrin (PE) or fluorescein-streptavidin (PE-streptavidin or FITC-streptavidin; 10 μg/ml) were purchased from Vector Laboratories (Burlingame, CA). FITC-conjugated mouse anti-stage-specific embryonic antigen-1 (anti-SSEA-1 or anti-Lewis X; 1:100) mAb was obtained from Calbiochem. FITC- or PE-conjugated goat anti-mouse IgM were purchased from Invitrogen. Mouse IgM anti-SSEA-1 mAb (1:100) was prepared in the laboratory (36) from hybridoma cells kindly provided by Barbara Knowles (44). For flow cytometry, 5 × 105 cells were pelleted and washed once with cold PBS. Cells were incubated with primary antibodies for 1 h at room temperature in Hanks' balanced salt solution (Mediatech Inc., Herndon, VA) containing 1 mm CaCl2, 2% bovine serum albumin (BSA; fraction V Sigma), and 0.05% NaN3 (FACS buffer). After washing with FACS buffer three times, cells were incubated with PE- or FITC-conjugated secondary antibodies in FACS buffer for 30 min at room temperature. After three washings, cells were resuspended in FACS buffer without BSA and 2 μl of 7-amino-actinomycin D (BD Pharmingen) was added. Cells were subjected to flow cytometry using a FACScan (BD Biosciences) instrument. Data were analyzed using FlowJo software (Tree Star Inc., Ashland, OR) and cells that took up 7-amino-actinomycin D were excluded. Alternatively, cells were washed once with cold PBS and then fixed with 3% paraformaldehyde in PBS for 20 min at room temperature before adding primary antibody.
Immunofluorescence Microscopy
Cells were grown on 12-mm glass coverslips (Fisher Scientific) treated with polylysine and fixed with 3% paraformaldehyde in PBS. After incubation for 20 min at room temperature with blocking solution containing 0.5% BSA, 0.2% saponin, and 1% FBS in PBS, cells were incubated with primary antibodies in blocking solution for 1 h at room temperature in a moist chamber. Mouse anti-GM130 mAb (1:200) was purchased from BD Transduction Laboratories. Mouse anti-PDI mAb (1:400) and rabbit anti-PDI pAb (1:200) were obtained from Assay Designs (Ann Arbor, MI). Mouse anti-endoplasmic reticulum-Golgi intermediate compartment (ERGIC)-53 mAb (1:200) was purchased from ALEXIS Biochemicals. Mouse anti-Myc mAb 9E10 (1:100) was from Calbiochem. Rabbit anti-mannosidase II (ManII; 1:5000) was kindly provided by Kelly Moremen (University of Georgia) (45), and rabbit anti-β1,4-galactosyltransferase-I (β4GalT1; 1:100) pAb was kindly provided by Eric Berger (University of Zurich) (46). After washing with PBS six times, Alexa Fluor 488 goat anti-rabbit (1:400) and Alexa Fluor 568 (1:400) goat anti-mouse secondary antibodies (Invitrogen) were added and incubated for 1 h at room temperature. After washing with PBS, 200 ng/ml of 4′,6-diamidino-2-phenylindole (DAPI; Sigma) was used to stain cell nuclei. Samples were mounted on slides and imaged using Leica SP2 AOBS confocal microscope.
RESULTS
Ligand-induced Notch Signaling Is Fully Functional in the Absence of Slc35c1
MEFs were isolated from wild type and mutant embryos with targeted inactivation of the mouse Slc35c1 gene (27) and confirmed by genotyping (Fig. 1A). As expected, Slc35c1−/− MEFs did not bind the lectin AAL, which binds to terminal fucose residues (Fig. 1B). By contrast, both control and Slc35c1−/− MEFs bound L-PHA equivalently, showing that their respective complement of complex N-glycans was similar (Fig. 1C). A TP1-luciferase reporter assay was used to examine Notch signaling following co-culture with Delta1/L or Jagged1/L cells. Interestingly, both Delta1- and Jagged1-induced Notch signaling were increased about 2–3-fold in Slc35c1−/− compared with control MEFs (Fig. 1, D and E), although this might reflect a difference in endogenous Notch receptor levels that are difficult to detect in fibroblasts.
FIGURE 1.
Ligand-induced Notch signaling in Slc35c1−/− MEFs. A, PCR genotyping of Slc35c1−/− and Slc35c1+/+ MEFs was performed as described under “Experimental Procedures.” B, Slc35c1−/− (black line; C1−/−) and Slc35c1+/+ (gray line; C1+/+) MEFs were analyzed by flow cytometry for cell surface glycans containing a terminal fucose using biotinylated AAL. The shaded profile is the secondary antibody alone. C, flow cytometric analysis demonstrated equivalent binding of FITC-conjugated L-PHA to complex N-glycans in C1+/+ and C1−/− MEFs. The shaded profile is the unstained Slc35c1−/− MEFs. D, C1+/+ (white bars) and C1−/− (black bars) MEFs were transfected with TP-1 reporter and control plasmids and co-cultured with Delta1/L, Jagged1/L, or L cells. Notch signaling was determined as the ratio of firefly:Renilla luciferase activities. One representative experiment is shown. E, ligand-induced Notch signaling determined by fold-activation and normalized to 100% based on C1+/+. Error bars are S.E. (n = 3 independent experiments); *, p < 0.05 based on a one-tailed (Delta1/L); ***, p < 0.0001 based on a two-tailed (Jagged1/L) Student's t test.
Human fibroblasts with the T308R point mutation isolated from a LADII patient of Middle Eastern origin (29) were also examined for ligand-induced Notch signaling. First, severely reduced core and terminal fucosylation of N-glycans was confirmed in the LADII fibroblasts by flow cytometry using both the AAL and PSA fucose-binding lectins (Fig. 2, A and B). By contrast, both control and LADII fibroblasts exhibited similar binding of L-PHA showing that their complex N-glycan complement was similar (Fig. 2C). Because only low levels of TP-1 firefly luciferase vector could be transfected into human fibroblasts, endogenous Notch1 signaling was assayed by following the ligand-induced generation of cleaved, activated Notch1. LADII and control fibroblasts were co-cultured with Delta1- or Jagged1-expressing L cells or control L cells, and activation of Notch1 was detected by Western blot using an antibody against γ-secretase-cleaved Notch1 termed N1Val1744 (47). Enhanced activation of endogenous Notch1 in human fibroblasts was detected after stimulation by either Delta1/L or Jagged1/L cells compared with L cells (Fig. 2D). Although, LADII fibroblasts had more activated Notch1 than control fibroblasts when co-cultured with either of the Notch ligand-expressing cells, the Notch1 receptor was increased in amount by ligand stimulation of LADII cells compared with control cells (data not shown). Nevertheless, it is clear that LADII cells exhibited readily detectable Notch signaling that was inhibited by the γ-secretase inhibitor (GSI) L-685,458 (42) (Fig. 2E).
FIGURE 2.
Notch1 activation in LADII fibroblasts. A, LADII (black line) and control (gray line) fibroblasts were examined for binding of biotinylated-AAL by flow cytometry detected by FITC-streptavidin. The shaded profile is FITC-streptavidin alone. B, binding of FITC-conjugated PSA. The shaded profile is unstained LADII fibroblasts. C, binding of FITC-conjugated L-PHA. The shaded profile is unstained LADII fibroblasts. D, LADII and control fibroblasts were co-cultured with Delta1/L, Jagged1/L cells, or control ligand cells (L). After 6 h, lysates were analyzed by Western blot using anti-N1Val1744 antibody. Cleaved endogenous Notch1 (N1-ICD) was at 110 kDa. The asterisk indicates nonspecific bands on the same gel. The lower panel in D served as loading control. This is representative of 4 independent experiments. E, the experiment in D was performed for LADII fibroblasts incubated with 2 μm GSI L-685,458 or DMSO in the co-culture medium. Mouse β-actin was a loading control. This is a representative result from 2 independent experiments.
The combined data suggest that in the absence of Slc35c1, Notch receptors signal well following ligand stimulation. Because O-fucosylation of Notch in T308R LADII fibroblasts was previously shown to be unaffected by the mutation (29), and ligand-induced Notch signaling in both LADII fibroblasts and MEFs lacking Slc35c1 was robust, Slc35c2 was investigated as a potential GDP-fucose transport facilitator for Notch receptors.
Overexpression of Slc35c2 Reduces Some Fucosylation
We initially identified Slc35c2 by expression cloning as an inhibitor of Lewis X synthesis in LEC11B CHO cells that have a gain-of-function mutation that causes up-regulation of α1,3-fucosyltransferse 6B (Fut6B) (32). To confirm this phenotype in LEC11B cells expressing CHO Slc35c2 at more physiological levels, stable Slc35c2/LEC11B transfectants were used (32). It can be seen in Fig. 3A that LEC11B cells expressing Slc35c2 had lower amounts of the fucosylated Lewis X epitope detected by an anti-SSEA-1 mAb. However, there was no effect of overexpression of Slc35c2 on the α1,3-fucosyltransferase activity of LEC11B extracts (data not shown). Moreover, when CHO cells were transiently co-transfected with CHO fucosyltransferases Fut6B or Fut9 together with Slc35c2, a similar result was obtained. The degree of expression of Lewis X induced by either Fut6B or Fut9 was reduced when Slc35c2 was also overexpressed (Fig. 3, B and C). CHO Slc35c2 did not change the fucosylation of complex N-glycans as determined by PSA binding to Slc35c2/LEC11B cells (Fig. 3D).
FIGURE 3.
Slc35c2 inhibits some fucosylation. A, LEC11B cells stably expressing CHO Slc35c2 (C2; black line) or empty vector (Vec; gray line) were assayed for binding of mouse anti-SSEA-1 antibody by flow cytometry. The shaded profile is the secondary antibody alone. B and C, CHO cells transiently transfected with cDNAs encoding CHO Fut6B (B) or Fut9 (C) together with CHO Slc35c2 cDNA (C2; black line) or empty vector (Vec; gray line) were assayed for binding of mouse anti-SSEA-1 antibody by flow cytometry. The shaded profile is the secondary antibody alone. D, binding of FITC-conjugated PSA as in A. The shaded profile is unstained LEC11B cells.
Slc35c2 Enhances Fucosylation of Notch1 EGF11–15 in CHO Cells
To examine the effect of overexpressing Slc35c2 on the O-fucosylation of Notch1, a soluble fragment of the Notch1 extracellular domain containing EGF repeats 11–15 was used. This fragment has only one O-fucosylation site in EGF12 and no N-glycosylation site (38, 48). Notch1 EGF11–15 was transiently transfected into LEC11B cells stably expressing CHO Slc35c2 or empty vector. Cells were metabolically labeled for 30 h with l-[3H]fucose. Notch1 EGF11–15 in cell lysates and secretions were collected on Ni2+ beads. Western blot analysis with anti-Myc mAb and autoradiography to detect [3H]fucose were performed separately (Fig. 4A), and NIH Image J was used to quantitate the amount of [3H]fucose signal to the Notch fragment (Fig. 4B). Notch1 EGF11–15 from transfectants expressing Slc35c2 showed increased fucose labeling compared with cells transfected with empty vector. Although statistical significance was not reached, in all three experiments the ratio of [3H]Fuc:Myc for Slc35c2 transfectants was higher than control. Similar results were obtained in LEC11B cells stably expressing CHO Slc35c2 and mouse Notch1 EGF1–5.5 Thus, overexpression of Slc35c2 enhanced fucosylation of the Notch1 EGF11–15 fragment. However, overexpression of Slc35c2 did not correct the fucosylation defect in Slc35c1−/− MEFs, whereas overexpression of Slc35c1 alone or Slc35c1 and Slc35c2 together in Slc35c1−/− MEFs increased AAL binding (Fig. 4C). Neither Slc35c1 nor Slc35c2 overexpression changed the expression of complex N-glycans detected by FITC-conjugated L-PHA binding to Slc35c1−/− MEFs (Fig. 4D).
FIGURE 4.
Slc35c2 enhances O-fucosylation of a Notch1 fragment. A, Notch1 EGF11–15 was transfected into LEC11B CHO cells stably expressing CHO Slc35c2 or empty vector. Cells were metabolically labeled with l-[3H]fucose, and Notch1 EGF11–15 harvested by Ni2+ beads was analyzed by Western blot with anti-Myc mAb and autoradiography to detect [3H]fucose. B, blots were quantitated by NIH Image J and the relative ratios of [3H]fucose incorporated versus Myc signal are plotted. Error bar, S.E. (n = 3 independent experiments). Slc35c1−/− MEFs were transiently transfected with mouse Slc35c2 cDNA (C2; black line) or mouse Slc35c1 cDNA (C1; dashed line) or both Slc35c1 and Slc35c2 cDNA (C1+C2; dotted line) and analyzed for binding of biotinylated-AAL (C) or FITC-L-PHA (D). The shaded profile is untransfected Slc35c1−/− MEFs for biotinylated-AAL or unstained Slc35c1−/− MEFs for FITC-conjugated L-PHA, respectively.
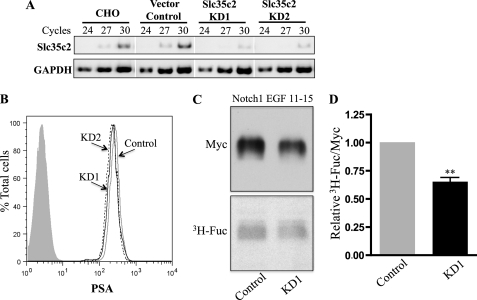
Knockdown of Slc35c2 Decreases the O-Fucosylation of Notch1
Because overexpression of Slc35c2 enhanced O-fucosylation of Notch1 EGF11–15, it was important to determine whether O-fucosylation of the Notch1 fragment would be relatively lower in cells with reduced Slc35c2. RNA interference was used to knockdown Slc35c2 in CHO cells. Vectors containing shRNA targeted against the 3′ UTR of CHO Slc35c2 were stably expressed and several transfectants were investigated. Semi-quantitative RT-PCR revealed a marked reduction of Slc35c2 transcripts in two isolates termed Slc35c2-KD1 and -KD2, and identified a vector control clone that had similar levels of Slc35c2 transcripts to CHO cells (Fig. 5A). The reduction of Slc35c2 transcripts did not affect the general fucosylation of N-glycans on the cell surface as shown by equivalent binding of PSA to Slc35c2 knockdown cell lines and vector control using flow cytometric analysis (Fig. 5B). Notch1 EGF11–15 was transiently expressed in Slc35c2 knockdown and control transfectants and cultured in [3H]fucose for 30 h. The Notch1 EGF11–15 fragment was collected from lysates and medium on Ni2+ beads and analyzed by Western blotting using anti-Myc mAb and fluorography (Fig. 5C). Signals were quantitated by NIH Image J, and the ratios of [3H]fucose:Myc were expressed relative to vector control (Fig. 5D). It can be seen that there was an ∼30% reduction of O-fucosylation on Notch1 EGF11–15 in the Slc35c2 knockdown line. The combined results suggest that Slc35c2 plays a role in the fucosylation of Notch receptors.
FIGURE 5.
Knockdown of Slc35c2 reduces Notch1 EGF11–15 fucosylation. A, RNA from CHO cells stably expressing shRNA targeting the CHO Slc35c2 3′ UTR was analyzed by semi-quantitative RT-PCR for Slc35c2 and GAPDH transcripts. B, flow cytometric analysis demonstrated equivalent binding of FITC-PSA to Slc35c2 knockdown cells (KD1, black line; KD2, dashed line) and vector control (Control; gray line). The shaded profile is unstained CHO cells. C, mouse Notch1 EGF11–15 expressed in Slc35c2-KD1 or vector control cells was labeled with l-[3H]fucose, collected on Ni2+ beads, and analyzed by Western blot using anti-Myc mAb and fluorography. D, NIH Image J was used to quantitate blots and the relative ratios of [3H]fucose per Notch1 fragment normalized to vector control were plotted. Error bar, S.E. (n = 3 independent experiments); **, p < 0.01, based on the two-tailed Student's t test.
Knockdown of Slc35c2 Decreases Notch Signaling
O-Fucosylation has been shown to be required for optimal Notch signaling in mammals (12, 14–16). To investigate whether a reduction in Slc35c2 in CHO cells affects Notch signaling, the TP1-luciferase reporter assay was performed in both Slc35c2 targeted lines and the vector control. Delta1- and Jagged1-induced Notch signaling were both reduced by up to 50% in both the Slc35c2 knockdown lines (Fig. 6, A and B). To investigate further, Notch signaling was assayed using the N1V1744 antibody against cleaved, activated Notch1 (47). In cells transiently expressing a mouse Notch1 cDNA with the C-terminal PEST domain replaced with the Myc6 tag (39), Delta1-induced endogenous and exogenous Notch1 activation was detected by the Val1744 antibody. The activation of transfected exogenous Notch1 generated a band of ∼70 kDa compared with the 110 kDa obtained for activated endogenous Notch1 (Fig. 6, C and D, and supplemental Fig. S1). Both endogenous and exogenous Notch1 activation were blocked by the GSI L-685,458 (Fig. 6D and supplemental Fig. S1). The activation of endogenous and exogenous Notch1 were reduced by ∼40–70% in cells with targeted knockdown of Slc35c2 (Fig. 6, C–E). The two different and independent assays show that optimal Notch signaling requires expression of Slc35c2 in mammalian cells.
FIGURE 6.
Knockdown of Slc35c2 reduces ligand-induced Notch signaling. A, Notch signaling in Slc35c2 knockdown lines (KD1 and KD2) and vector control (Control) using the TP1-luciferase and Renilla luciferase reporter assay. Activation of Notch signaling by Delta1/L or Jagged1/L was determined by the ratio of firefly:Renilla luciferase activities. The values are the average of duplicates from one representative experiment. B, relative activation of Notch signaling in Slc35c2-KD1 and -KD2 normalized to vector control. Error bars, S.E. (n = 4 independent experiments performed in duplicate); **, p < 0.01; ***, p < 0.001, based on the two-tailed Student's t test. C, mouse Notch1 with a C-terminal Myc tag (N1-Myc) was transiently transfected into Slc35c2 knockdown lines (KD1 and KD2) and vector control cells (Control). Delta1-induced Notch1 activation was analyzed by Western blot using N1Val1744 antibody and anti-Myc mAb. Activated Notch1 intracellular domains (N1 ICD) from both endogenous and exogenous N1 were detected. Results of a representative experiment are shown. D, the experiment in C was performed in the presence of 2 μm GSI L-685,458 or DMSO in co-culture medium with Delta1/L cells or control ligand cells (L). Both endogenous and introduced N1 ICDs were blocked by the GSI. E, relative expression of activated Notch1. The quantitation of signals from gels similar to C was performed using NIH Image J. The ratio of activated endogenous Notch1 to loading control (either β-actin or nonspecific band) was normalized to control; the ratio of activated exogenous Notch1 to transfected Notch1 was normalized to control. For endogenous N1 ICD, error bar represent S.E. (n = 4 independent experiments), ***, p < 0.001; for exogenous N1 ICD. Error bars represent S.E. (KD1, n = 5 independent experiments; KD2, n = 4 independent experiments); *, p < 0.05; **, p < 0.01, based on the two-tailed Student's t test.
Slc35c2 Colocalizes with the Golgi in Rat Liver
To investigate subcellular localization of Slc35c2, a polyclonal Ab was generated against the C terminus of mouse Slc35c2 (“Experimental Procedures”). To validate the specificity of this antibody, CHO cells were transiently transfected with mouse Slc35c2 with a C-terminal Myc tag, and cell lysates were analyzed by Western blot (Fig. 7A). Both the anti-Slc35c2 and the anti-Myc antibodies detected a band of the same size at ∼36 kDa under reducing conditions. Mouse Slc35c1 with a C-terminal Myc tag had a slightly lower molecular weight (Fig. 7A).
Slc35c2 has one potential N-glycosylation site, which might be used to predict the subcellular compartments it has traversed. Cell lysates from CHO cells expressing mouse Slc35c2-Myc were analyzed by Western blot using anti-Slc35c2 peptide antibody before and after treatment with Endo H or PNGase F. Endo H removes oligomannosyl and hybrid but not complex N-glycans, whereas PNGase F removes essentially all N-glycans. It can be seen in Fig. 7B that the molecular weight of mouse Slc35c2-Myc expressed in CHO cells did not change after either Endo H or PNGase F treatment. Evidence that both glycosidases were active was obtained by immunoblotting for Pofut1 using a C-terminal peptide antibody (43). The Pofut1 band shifted to the same low molecular weight after treatment with both glycosidases, consistent with a previous report that Pofut1 is sensitive to Endo H treatment (18). The specificity of the anti-Pofut1 peptide antibody was confirmed by showing that cell lysates from Pofut1−/− mouse ES cells (14) had no signal compared with control ES cells (data not shown).
Western analysis was used to determine the localization of Slc35c2 in rat liver ER and Golgi fractions (Fig. 7C). The purity of ER and Golgi fractions were reflected by the presence of most PDI in the ER fraction and of most GM130 in the Golgi fraction. Pofut1 colocalized with PDI in the ER fraction. Slc35c2 was found mostly in the Golgi fraction, although there appeared to be a weak band in the ER fraction. It appears that rat liver Slc35c2 is primarily localized in the Golgi compartment.
Slc35c2 Colocalizes with Golgi and ERGIC Markers by Immunofluorescence Microscopy
To examine subcellular localization of Slc35c2 by immunofluorescence microscopy, rat IEC (Fig. 8, A–F) or HeLa cells (Fig. 8, G–N) were transiently transfected with untagged mouse Slc35c2 cDNA or with mouse Slc35c2-Myc, fixed with 3% paraformaldehyde at 48 h after transfection and immunostained with anti-Slc35c2 peptide antibody or anti-Myc monoclonal antibody, along with antibodies against subcellular compartments, including PDI for ER, ERGIC-53 for ERGIC, GM130 for cis-Golgi, α-mannosidase II (ManII) for medial-Golgi, and β1,4-galactosyltransferase-I (β4GalT1) for trans-Golgi. Although Slc35c2 co-localized with cis-Golgi marker GM130 both in IEC and HeLa cells and medial-Golgi marker ManII in IEC (anti-ManII Ab did not give a signal in HeLa cells), it also co-localized with ERGIC-53 in HeLa cells. Slc35c2 did not co-localize with the ER marker PDI in most transfected cells. Quantitation of Slc35c2 subcellular localization was performed in IEC and HeLa cells transiently transfected with mouse Slc35c2 with or without a Myc tag. Among 100 transfected IEC or HeLa cells, Slc35c2 was found in a punctate staining pattern primarily in a Golgi-like compartment (Table 1). Although there were a small number of cells having an ER-like staining pattern, this was probably due to mislocalization caused by overexpression.
FIGURE 8.
Slc35c2 subcellular localization in IEC and HeLa cells. Expression of Slc35c2 in fixed rat IEC (A–F) or HeLa cells (G–N). Transfected Slc35c2 was detected in B, H, J, and L with anti-Slc35c2 pAb; Slc35c2-Myc was detected in D, F, and N with anti-Myc mAb. Other antibodies included anti-PDI (ER marker), anti-ERGIC-53 (ERGIC marker), anti-GM130 (cis-Golgi marker), anti-ManII (medial-Golgi marker), and anti-β4GalT1 (trans-Golgi marker). Images were taken using a Leica SP2 AOBS confocal microscope. Scale bars, 10 μm.
TABLE 1.
Quantitation of mouse Slc35c2 subcellular localization after transient transfection
IEC or HeLa cells were transiently transfected with mouse Slc35c2 cDNA with or without a C-terminal Myc tag. 100 transfected cells were counted to determine subcellular localization in the secretory pathway.
| ER | Golgi | ER and Golgi | |
|---|---|---|---|
| % | % | % | |
| IEC + mSlc35c2-Myc | 2 | 58 | 40 |
| IEC + mSlc35c2 | 0 | 71 | 29 |
| HeLa + mSlc35c2-Myc | 8 | 33 | 59 |
| HeLa + mSlc35c2 | 0 | 76 | 24 |
DISCUSSION
The O-fucose glycans of Notch are critical regulators of Notch signaling (49, 50). Thus it is important to understand all the factors required for their synthesis. In this article we identify Slc35c2 as a factor required for optimal Notch signaling in mammalian cells. We show that cells lacking the Golgi transporter Slc35c1 have robust Notch signaling, that overexpression of Slc35c2 increases the O-fucosylation of Notch1 EGF11–15, and that targeted knockdown of Slc35c2 reduces O-fucosylation of Notch and Notch signaling. Based on its sequence similarity to Slc35c1 (31), Slc35c2 would seem to be an obvious candidate for a GDP-fucose transporter that might provide GDP-fucose to Pofut1. Consistent with this are our findings that overexpression of Slc35c2 reduces fucosylation of N-glycans that occurs in the trans-Golgi, as shown previously (32) and in Fig. 3. The ER localization of Pofut1 reported previously (18) and shown by rat liver cell fractionation in Fig. 7 also suggested that there may be an ER GDP-fucose transporter. In fact, we tried several approaches to determine whether Slc35c2 has GDP-fucose transport activity in vitro. Both ER and Golgi membrane fractions isolated from LEC11B CHO cells stably expressing CHO Slc35c2 were assayed for GDP-fucose compared with UDP-Gal transport activity under varying conditions (21). However, although UDP-Gal transport was enriched ∼10-fold in Golgi versus ER vesicles as expected, no consistent evidence that Slc35c2 affected GDP-fucose transport activity was obtained in several independent experiments. Slc35c2 has also been tested for GDP-fucose transport activity in yeast microsomes, but it was toxic to yeast when highly expressed, and there was no GDP-fucose transport activity observed with low expression.6 Whereas a murine CMP-sialic acid transporter targeted to the Escherichia coli inner membrane using the N-terminal OmpA signaling sequence expressed CMP-sialic acid transport activity (51), our attempts to express mouse Slc35c2 with an N-terminal OmpA sequence in the E. coli inner membrane were not successful. However, a Mistic (membrane-integrating sequence for translation of integral membrane protein constructs (52, 53)) tag was able to target both Slc35c2 and Slc35c1 to E. coli inner membrane. Nevertheless, GDP-fucose transport activity was not detected in spheroplasts or membrane vesicles prepared from E. coli expressing either Slc35c2 or Slc35c1. Thus it remains to be shown whether Slc35c2 is a GDP-fucose transporter.
We report here that Slc35c2 facilitates the O-fucosylation of Notch and is required for optimal Notch signaling. One possibility is that Slc35c2 transiently mislocalizes Slc35c1 to the ER. However, Slc35c2 did not affect Slc35c1 Golgi localization in IEC when both were transiently transfected (supplemental Fig. S3). Furthermore, Slc35c2 did not inhibit rescue by Slc35c1 when both were co-expressed in Slc35c1−/− MEFs (Fig. 4C). Another possibility is that Slc35c2 may be a chaperone or other activity that functions, perhaps with an unrelated nucleotide sugar transporter, to promote the presentation of GDP-fucose to Pofut1, and potentially Pofut2, which is also an ER protein O-fucosyltransferase (54). Slc35c2 does not require Slc35c1, the only established mammalian GDP-fucose transporter, for its activity because cells lacking Slc35c1 activity exhibit Notch O-fucosylation (29) and robust Notch signaling (Figs. 1 and 2). Moreover, Slc35c2 is not mainly localized to the ER (Figs. 7 and 8). Based on colocalization with markers of ERGIC and Golgi, Slc35c2 appears to be located mainly in ERGIC and early Golgi compartments. However, Slc35c2 was also found in the ER and thus it may function throughout the secretory pathway. It appears unlikely, however, to function in concert with the ER GDP-fucose transporter recently identified in Drosophila (33). Drosophila Slc35b4 rescues Notch signaling-dependent expression of wg in the wing disc of an Slc35c1 mutant at low temperature. Homozygotes and hemizygotes of Slc35b4 mutations in Drosophila are lethal, although some of them survive until the third-instar larval stage (33). The expression of wg was abolished in these mutant wing discs. The mammalian homologue of this transporter, Slc35b4, has been shown not to transport GDP-fucose in a yeast microsome assay and to be localized to the Golgi (34). Drosophila Slc35b4 has a di-lysine retention/retrieval motif but this motif is not conserved in mammalian homologues. Alternatively, Slc35c2 might be induced to transport GDP-fucose by some other molecule. Although there is no example of a transporter chaperone to date, mammalian nucleotide sugar transporters may interact in the Golgi membrane and several of them, including Slc35b4 (34), transport more than one nucleotide sugar (55, 56). Although there is also no example of a transporter that transports both UDP- and GDP-sugars, chimeric transporters have been constructed that transport UDP-Gal and CMP-sialic acid (57–59). Although O-fucosylation of mouse Notch1 EGF29–36 appears to occur in the ER (18), we cannot rule out the possibility that Pofut1 might encounter GDP-fucose in the ERGIC or cis-Golgi compartments during recycling or that Pofut1 fucosylates Notch receptors in the ERGIC or cis-Golgi compartments rather than in the ER. A targeted mutation of Slc35c2 in the mouse will help to resolve these questions.
Supplementary Material
Acknowledgments
We thank Subha Sundaram and Huimin Shang for excellent technical assistance and Diana Popovici for preparing mouse embryo fibroblasts. We also thank those noted in the text for antibodies and cells and Nancy Carrasco for technical advice.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 95022 and RO1 36434 from the NCI (to P. S.) and NCI Grant PO1 13330 (to the Albert Einstein Cancer Center).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Y. Luo and R. S. Haltiwanger, personal communication.
H. Bakker, personal communication.
- Pofut1
- protein O-fucosyltransferase-1
- PE
- phycoerythrin
- SSEA-1
- stage-specific embryonic antigen-1
- ERGIC
- endoplasmic reticulum-Golgi intermediate compartment
- GSI
- γ-secretase inhibitor
- Fut6B
- α1,3-fucosyltransferse 6B
- Endo H
- endoglycosidase H
- IEC
- intestinal epithelial cell
- PDI
- protein disulfide isomerase
- ManII
- α-mannosidase II
- LADII
- leukocyte adhesion deficiency type II
- ER
- endoplasmic reticulum
- MEF
- mouse embryonic fibroblast
- PNGase
- peptide:N-glycosidase
- PSA
- Pisum sativum agglutinin.
REFERENCES
- 1.Artavanis-Tsakonas S., Rand M. D., Lake R. J. (1999) Science 284, 770–776 [DOI] [PubMed] [Google Scholar]
- 2.Kopan R., Ilagan M. X. (2009) Cell 137, 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y., Shao L., Shi S., Harris R. J., Spellman M. W., Stanley P., Haltiwanger R. S. (2001) J. Biol. Chem. 276, 40338–40345 [DOI] [PubMed] [Google Scholar]
- 4.Shi S., Stanley P. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5234–5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okamura Y., Saga Y. (2008) Mech. Dev. 125, 663–673 [DOI] [PubMed] [Google Scholar]
- 6.Okajima T., Irvine K. D. (2002) Cell 111, 893–904 [DOI] [PubMed] [Google Scholar]
- 7.Sasamura T., Sasaki N., Miyashita F., Nakao S., Ishikawa H. O., Ito M., Kitagawa M., Harigaya K., Spana E., Bilder D., Perrimon N., Matsuno K. (2003) Development 130, 4785–4795 [DOI] [PubMed] [Google Scholar]
- 8.Okajima T., Xu A., Lei L., Irvine K. D. (2005) Science 307, 1599–1603 [DOI] [PubMed] [Google Scholar]
- 9.Okajima T., Reddy B., Matsuda T., Irvine K. D. (2008) BMC Biol. 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sasaki N., Sasamura T., Ishikawa H. O., Kanai M., Ueda R., Saigo K., Matsuno K. (2007) Genes Cells 12, 89–103 [DOI] [PubMed] [Google Scholar]
- 11.Sasamura T., Ishikawa H. O., Sasaki N., Higashi S., Kanai M., Nakao S., Ayukawa T., Aigaki T., Noda K., Miyoshi E., Taniguchi N., Matsuno K. (2007) Development 134, 1347–1356 [DOI] [PubMed] [Google Scholar]
- 12.Moloney D. J., Panin V. M., Johnston S. H., Chen J., Shao L., Wilson R., Wang Y., Stanley P., Irvine K. D., Haltiwanger R. S., Vogt T. F. (2000) Nature 406, 369–375 [DOI] [PubMed] [Google Scholar]
- 13.Chen J., Moloney D. J., Stanley P. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13716–13721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stahl M., Uemura K., Ge C., Shi S., Tashima Y., Stanley P. (2008) J. Biol. Chem. 283, 13638–13651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou L., Li L. W., Yan Q., Petryniak B., Man Y., Su C., Shim J., Chervin S., Lowe J. B. (2008) Blood 112, 308–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waterhouse C., Johnson S., Phillipson M., Zbytnuik L., Petri B., Kelly M., Lowe J., Kubes P. (2010) Gastroenterology 138, 1079–1090 [DOI] [PubMed] [Google Scholar]
- 17.Moran J. L., Shifley E. T., Levorse J. M., Mani S., Ostmann K., Perez-Balaguer A., Walker D. M., Vogt T. F., Cole S. E. (2009) Dev. Dyn. 238, 1803–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo Y., Haltiwanger R. S. (2005) J. Biol. Chem. 280, 11289–11294 [DOI] [PubMed] [Google Scholar]
- 19.Ginsburg V. (1960) J. Biol. Chem. 235, 2196–2201 [PubMed] [Google Scholar]
- 20.Yurchenco P. D., Atkinson P. H. (1977) Biochemistry 16, 944–953 [DOI] [PubMed] [Google Scholar]
- 21.Puglielli L., Hirschberg C. B. (1999) J. Biol. Chem. 274, 35596–35600 [DOI] [PubMed] [Google Scholar]
- 22.Berninsone P. M., Hirschberg C. B. (2000) Curr. Opin. Struct. Biol. 10, 542–547 [DOI] [PubMed] [Google Scholar]
- 23.Etzioni A., Frydman M., Pollack S., Avidor I., Phillips M. L., Paulson J. C., Gershoni-Baruch R. (1992) N. Engl. J. Med. 327, 1789–1792 [DOI] [PubMed] [Google Scholar]
- 24.Hirschberg C. B. (2001) J. Clin. Invest. 108, 3–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lühn K., Wild M. K., Eckhardt M., Gerardy-Schahn R., Vestweber D. (2001) Nat. Genet. 28, 69–72 [DOI] [PubMed] [Google Scholar]
- 26.Lübke T., Marquardt T., Etzioni A., Hartmann E., von Figura K., Körner C. (2001) Nat. Genet. 28, 73–76 [DOI] [PubMed] [Google Scholar]
- 27.Hellbusch C. C., Sperandio M., Frommhold D., Yakubenia S., Wild M. K., Popovici D., Vestweber D., Gröne H. J., von Figura K., Lübke T., Körner C. (2007) J. Biol. Chem. 282, 10762–10772 [DOI] [PubMed] [Google Scholar]
- 28.Yakubenia S., Frommhold D., Schölch D., Hellbusch C. C., Körner C., Petri B., Jones C., Ipe U., Bixel M. G., Krempien R., Sperandio M., Wild M. K. (2008) Blood 112, 1472–1481 [DOI] [PubMed] [Google Scholar]
- 29.Sturla L., Rampal R., Haltiwanger R. S., Fruscione F., Etzioni A., Tonetti M. (2003) J. Biol. Chem. 278, 26727–26733 [DOI] [PubMed] [Google Scholar]
- 30.Ishikawa H. O., Higashi S., Ayukawa T., Sasamura T., Kitagawa M., Harigaya K., Aoki K., Ishida N., Sanai Y., Matsuno K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 18532–18537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishida N., Kawakita M. (2004) Pflugers Arch. 447, 768–775 [DOI] [PubMed] [Google Scholar]
- 32.Chen W., Tang J., Stanley P. (2005) Glycobiology 15, 259–269 [DOI] [PubMed] [Google Scholar]
- 33.Ishikawa H. O., Ayukawa T., Nakayama M., Higashi S., Kamiyama S., Nishihara S., Aoki K., Ishida N., Sanai Y., Matsuno K. (2010) J. Biol. Chem. 285, 4122–4129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashikov A., Routier F., Fuhlrott J., Helmus Y., Wild M., Gerardy-Schahn R., Bakker H. (2005) J. Biol. Chem. 280, 27230–27235 [DOI] [PubMed] [Google Scholar]
- 35.Stanley P., Caillibot V., Siminovitch L. (1975) Cell 6, 121–128 [DOI] [PubMed] [Google Scholar]
- 36.Zhang A., Potvin B., Zaiman A., Chen W., Kumar R., Phillips L., Stanley P. (1999) J. Biol. Chem. 274, 10439–10450 [DOI] [PubMed] [Google Scholar]
- 37.Patnaik S. K., Potvin B., Stanley P. (2004) J. Biol. Chem. 279, 49716–49726 [DOI] [PubMed] [Google Scholar]
- 38.Shi S., Ge C., Luo Y., Hou X., Haltiwanger R. S., Stanley P. (2007) J. Biol. Chem. 282, 20133–20141 [DOI] [PubMed] [Google Scholar]
- 39.Kopan R., Schroeter E. H., Weintraub H., Nye J. S. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 1683–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi S., Stahl M., Lu L., Stanley P. (2005) Mol. Cell. Biol. 25, 9503–9508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strobl L. J., Höfelmayr H., Stein C., Marschall G., Brielmeier M., Laux G., Bornkamm G. W., Zimber-Strobl U. (1997) Immunobiology 198, 299–306 [DOI] [PubMed] [Google Scholar]
- 42.Shearman M. S., Beher D., Clarke E. E., Lewis H. D., Harrison T., Hunt P., Nadin A., Smith A. L., Stevenson G., Castro J. L. (2000) Biochemistry 39, 8698–8704 [DOI] [PubMed] [Google Scholar]
- 43.Kim M. L., Chandrasekharan K., Glass M., Shi S., Stahl M. C., Kaspar B., Stanley P., Martin P. T. (2008) Mol. Cell. Neurosci. 39, 452–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Solter D., Knowles B. B. (1978) Proc. Natl. Acad. Sci. U.S.A. 75, 5565–5569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moremen K. W., Touster O., Robbins P. W. (1991) J. Biol. Chem. 266, 16876–16885 [PubMed] [Google Scholar]
- 46.Watzele G., Bachofner R., Berger E. G. (1991) Eur. J. Cell Biol. 56, 451–458 [PubMed] [Google Scholar]
- 47.Huppert S. S., Le A., Schroeter E. H., Mumm J. S., Saxena M. T., Milner L. A., Kopan R. (2000) Nature 405, 966–970 [DOI] [PubMed] [Google Scholar]
- 48.Shao L., Moloney D. J., Haltiwanger R. (2003) J. Biol. Chem. 278, 7775–7782 [DOI] [PubMed] [Google Scholar]
- 49.Haines N., Irvine K. D. (2003) Nat. Rev. Mol. Cell Biol. 4, 786–797 [DOI] [PubMed] [Google Scholar]
- 50.Stanley P. (2007) Curr. Opin. Struct. Biol. 17, 530–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maggioni A., von Itzstein M., Gerardy-Schahn R., Tiralongo J. (2007) Biochem. Biophys. Res. Commun. 362, 779–784 [DOI] [PubMed] [Google Scholar]
- 52.Kefala G., Kwiatkowski W., Esquivies L., Maslennikov I., Choe S. (2007) J. Struct. Funct. Genomics 8, 167–172 [DOI] [PubMed] [Google Scholar]
- 53.Roosild T. P., Greenwald J., Vega M., Castronovo S., Riek R., Choe S. (2005) Science 307, 1317–1321 [DOI] [PubMed] [Google Scholar]
- 54.Luo Y., Koles K., Vorndam W., Haltiwanger R. S., Panin V. M. (2006) J. Biol. Chem. 281, 9393–9399 [DOI] [PubMed] [Google Scholar]
- 55.Gerardy-Schahn R., Oelmann S., Bakker H. (2001) Biochimie 83, 775–782 [DOI] [PubMed] [Google Scholar]
- 56.Handford M., Rodriguez-Furlán C., Orellana A. (2006) Braz. J. Med. Biol. Res. 39, 1149–1158 [DOI] [PubMed] [Google Scholar]
- 57.Aoki K., Ishida N., Kawakita M. (2003) J. Biol. Chem. 278, 22887–22893 [DOI] [PubMed] [Google Scholar]
- 58.Aoki K., Ishida N., Kawakita M. (2001) J. Biol. Chem. 276, 21555–21561 [DOI] [PubMed] [Google Scholar]
- 59.Aoki K., Sun-Wada G. H., Segawa H., Yoshioka S., Ishida N., Kawakita M. (1999) J. Biochem. 126, 940–950 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.