Abstract
There are considerable racial disparities in prostate cancer risk, with a 60% higher incidence rate among African American (AA) men compared with European American (EA) men, and a 2.4 fold higher mortality rate in AA men than in EA men. Recently, studies have implicated several African-ancestry associated prostate cancer susceptibility loci on chromosome 8q24. In the current study, we performed admixture mapping in AA men from two independent case-control studies of prostate cancer to confirm the 8q24 ancestry association and also identify other genomic regions that may harbor prostate cancer susceptibility genes. A total of 482 cases and 261 controls were genotyped for 1,509 ancestry informative markers across the genome. The mean estimated individual admixture proportions were 20% European and 80% African. The most significant observed increase in European ancestry occurred at rs2141360 on chromosome 7q31 in both the case-only (p=0.0000035) and case-control analyses. The most significant observed increase in African ancestry across the genome occurred at a locus on chromosome 5q35 identified by SNPs rs7729084 (case-only analysis: p=0.002), and rs12474977 (case-control analysis: p=0.004), which are separated by 646 kb and were adjacent to one another on the panel. On chromosome 8, rs4367565 was associated with the greatest excess African ancestry in both the case-only and case-control analyses (case-only and case-control p=0.02), confirming previously reported African-ancestry associations with chromosome 8q24. In conclusion, we confirmed ancestry associations on 8q24, and identified additional ancestry-associated regions potentially harboring prostate cancer susceptibility loci.
Keywords: Prostate Cancer, Admixture Mapping, Ancestry, PODXL, DOCK4
Introduction
Prostate cancer (MIM #176807) is the most common cancer among American men, affecting 1 in 6 of the male population. The American Cancer Society estimates that about 186,300 men in the United States will be diagnosed with prostate cancer in 2008 and about 28,660 men will die of this disease. Prostate cancer is the second leading cause of cancer death in men. There are considerable racial disparities in prostate cancer risk, with a 60% higher incidence rate among African American men compared with White men, and a 2.4 fold higher mortality rate in African American men than in White men (Jemal et al. 2008). In addition to African American race, established risk factors for prostate cancer include age and family history. It is estimated that highly penetrant inherited prostate cancer susceptibility genes contribute to approximately five percent of all prostate cancer cases, but they play a more significant role in early-onset disease. Family history studies support the role of genes in cancer risk, with an estimated 4.5 fold increase in risk among men with an affected brother, and an estimated 2.3 fold increase in risk in men with an affected father (Cerhan et al. 1999).
Multiple genome-wide linkage scans for prostate cancer genes have yielded few consistent results (Easton et al. 2003; Schaid 2004). A recently published report from a genome wide admixture mapping study of prostate cancer in 1,597 African American men detected a susceptibility region on chromosome 8q24 (Freedman et al. 2006). Multiple follow-up studies using both admixture and traditional mapping methods have confirmed linkage to this region, (Ghoussaini et al. 2008; Gudmundsson et al. 2007; Haiman et al. 2007; Robbins et al. 2007; Schumacher et al. 2007; Suuriniemi et al. 2007; Wang et al. 2007; Yeager et al. 2007) and three out of four loci in this region have been shown to be independently associated with prostate cancer risk in African American populations (Haiman et al. 2007). In the current study, our objective was to confirm the 8q24 ancestry association and identify other genomic regions associated with prostate cancer susceptibility using admixture mapping methods in African-American prostate cancer cases and controls.
Materials and Methods
Study subjects
Subjects included in this project are from two independent sources. The first was a case-control study of prostate cancer, Gene-Environment Interaction in Prostate Cancer (GECAP) (Rybicki et al. 2006). All cases were diagnosed with adenocarcinoma of the prostate within two years of the date of enrollment, either of Caucasian or African-American race, less than 75 years of age, living in the metropolitan Detroit tri-county area, and used Henry Ford Health System (HFHS) for their primary care. Controls were randomly selected from the same HFHS population base from which cases were drawn and frequency matched to cases on race and five-year age stratum for a 3:1 case:control ratio; there were more cases than controls because the GECAP study was designed primarily for case-only analyses. From the original study population, African-American cases and controls were included in the admixture analyses. The second source of subjects was men who participated in one of three case-control studies at Howard University (HU). The HU population consists of unrelated African American men affected and unaffected with prostate cancer, previously described by Robbins et al. (2007). All affected men were histologically diagnosed with adenocarcinoma of the prostate. Subjects were recruited over 4 years in the Washington DC area from a Howard University Hospital urology practice and/or from ongoing free prostate cancer screening programs at the Howard University Cancer Center. Clinical evaluations, including PSA levels were determined for all prostate cancer cases and all controls. Included in these studies were African American men 40 to 90 years of age. All healthy unaffected volunteers had PSA levels <2.5ng/ml and normal DREs.
For all study participants, standardized demographic and medical history were collected, and all DNA used for genotyping was extracted from blood samples drawn at the time of interview by a trained phlebotomist. All study subjects provided informed, signed consent, and study protocols were approved by institutional Internal Review Boards.
Genotyping
Primers for a panel of 1,509 SNPs informative for West African versus European ancestry for use on the Illumina Bead Station platform were provided by one of the authors (David Reich). After removing SNPs with poor genotype call rates (n=188), 1321 SNPs were available for analysis. Earlier versions of this panel were used in the Freedman et al (2006) prostate cancer admixture mapping study, and the panel genotyped is now available as a standard product from Illumina (http://www.illumina.com/pages.ilmn?ID=235). For quality control in our study, DNA samples from 30 Centre d’Etude du Polymorphisme Humain (CEPH) individuals were included so that their genotype results could be compared with those publicly available through HapMap. Concordance was excellent for the markers we used for analysis.
Admixture Mapping
Data were analyzed using the ADMIXMAP statistical program, http://homepages.ed.ac.uk/pmckeigu/admixmap/ (Hoggart et al. 2003; Hoggart et al. 2004). ADMIXMAP uses a hybrid of Bayesian and traditional modeling to compare observed vs. expected ancestry across the genome. Both case-only and case-control approaches were implemented. All models were adjusted for age at diagnosis for cases and age at study entry for controls. We report Z scores and associated p-values for the case-only analyses. The Z score is a test statistic for association with ancestry at each locus, based on comparing the observed and expected proportions of gene copies at each locus. In our models, a positive Z-score indicates excess African Ancestry, and a negative Z-score indicates excess European ancestry. Prior allele frequencies for the African and European ancestral populations were provided by David Reich based on genotypes from the West African and European populations previously reported in Smith et al. (2004) and in the International HapMap project (http://www.hapmap.org/). Analyses were performed in the entire sample, stratified by age, and stratified by prostate cancer aggressiveness within the HFHS samples. We defined aggressive prostate cancer as Gleason sum ≥ 4+3 or stage ≥ T3 (regional or distant).
Results
Of the 776 samples genotyped, a total of 482 cases and 261 controls were eligible for inclusion in the analyses. 33 samples (4.4%) were excluded because they failed one of the standard data quality checks described in previous reports (Freedman et al. 2006; Reich et al. 2005). Briefly, samples were removed if they had a much lower genotyping completeness than other samples used in the analysis based on an empirical inspection of the distribution, if they were duplicates or first degree relatives of other samples in the analysis, if they had greater than about 85% European origin from our analysis, or if they had one parent of entirely European origin. The mean estimated individual admixture proportions after removal of the outlier samples were 20% European and 80% African. Patient characteristics overall and by study site are described in Table 1. Equal numbers of cases were derived from each source, and a greater proportion of the controls came from HU. Cases from HU were significantly older than cases from HFHS, and controls from HU were significantly younger than controls from HFHS. Median PSA in the HFHS cases was significantly lower than the median PSA in the HU cases, and there was no difference between the two sites with respect to median PSA in the controls.
Table 1.
Participant characteristics, overall and by study site
| All Sites | Henry Ford Health System | Howard University | P value | |
|---|---|---|---|---|
| Total | ||||
| Cases | 482 | 241 | 241 | |
| Controls | 261 | 92 | 169 | |
| Mean Age | ||||
| Cases | 63.8 | 61.5 | 66.1 | <0.0001a |
| Controls | 58.2 | 60.9 | 56.7 | 0.0004a |
| Median PSA | ||||
| Cases | 6.0 | 5.6 | 7.8 | 0.0007b |
| Controls | 0.9 | 0.8 | 0.9 | 0.6787b |
t-test comparing HFHS and HU
Mann-Whitney U-test comparing HFHS and HU
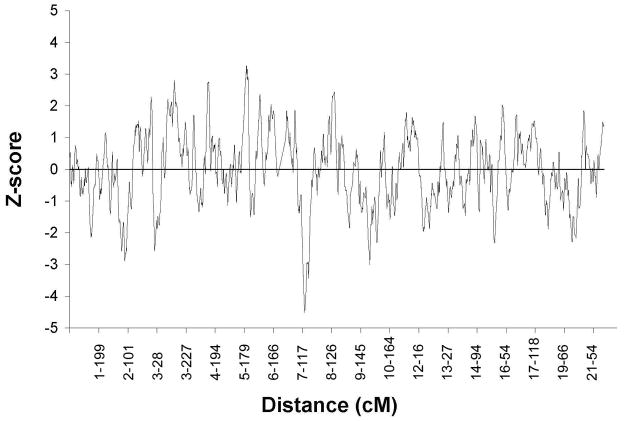
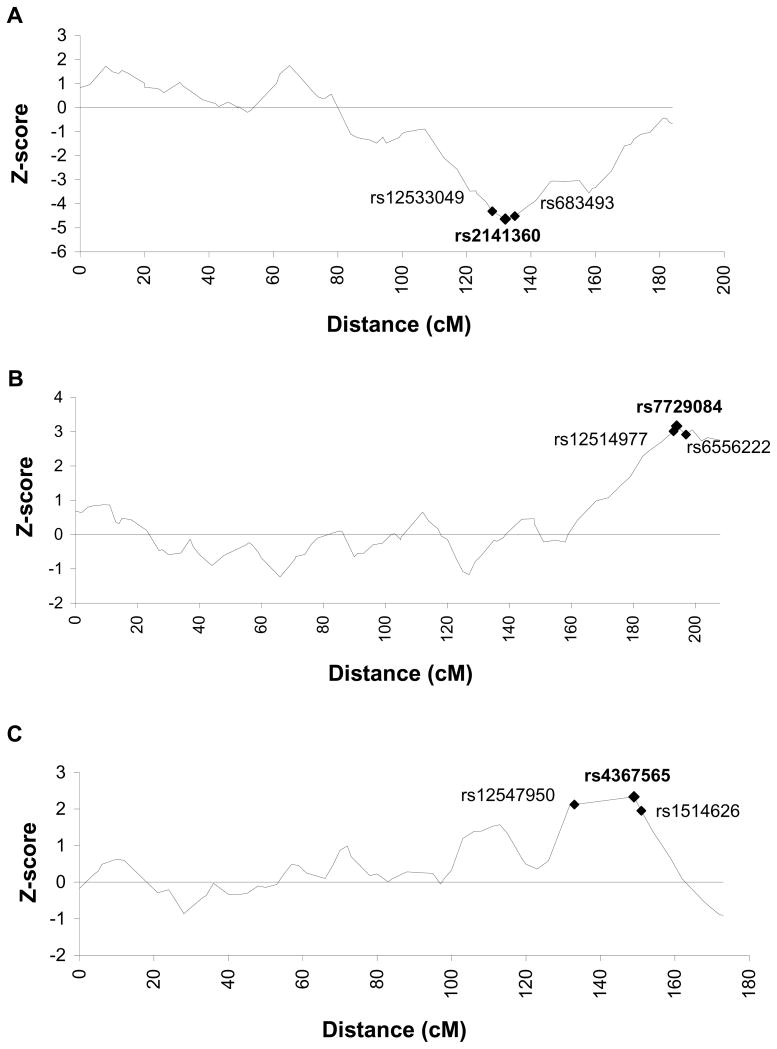
The age-adjusted genomewide z-scores for the case-only analyses are presented in Figure 1, and information about markers at local peaks with |Z| >3.0 is summarized in Table 2. In general, results from the case-control analyses did not differ greatly from those obtained using the more powerful case-only approach, therefore only case-only results are presented unless discrepancies in inferences from the two methods occurred. The greatest observed increase in ancestry occurred at marker rs2141360 on chromosome 7 (Figure 2a), where excess European ancestry was observed (case-only p=0.0000036). The greatest observed increase in African ancestry across the genome occurred at rs7729084 on Chromosome 5q35 in the case-only analysis (p=0.0016, Figure 2b), and at marker rs12474977 in the case-control analysis (p=0.0039). Note that marker rs12474977 is 646 kb centromeric to marker rs7729084, and the two markers were adjacent to each other on the panel. The marker on Chromosome 8 that was associated with the greatest excess African ancestry in both the case-only (Figure 2c) and case-control analyses was rs4367565 (case-only and case-control p<0.02).
Figure 1.
Genomewide admixture mapping results in 482 men with prostate cancer, adjusted for age
Table 2.
Summary of SNPs at regional ancestry association peaks with |Z| >3.0
| Entire Sample | Age<60 | Age≥60 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Locus Name | Locationa | Positionb | Excess Ancestral Population | Z Score | p value | Z Score | p value | Z Score | p value |
| rs6724395 | 2p14 | 69799822 | European | −2.666 | 0.0077 | −4.195 | 0.000027 | −0.381 | 0.7029 |
| rs7729084 | 5q35.2 | 174314186 | African | 3.164 | 0.0016 | 2.704 | 0.0068 | 2.221 | 0.0263 |
| rs692843 | 5q35.2 | 176188252 | African | 3.058 | 0.0022 | 3.116 | 0.0018 | 2.036 | 0.0418 |
| rs2141360 | 7q31.31 | 120847409 | European | −4.636 | 0.0000035 | −3.172 | 0.0015 | −3.311 | 0.0009 |
| rs683493 | 7q31.33 | 125592537 | European | −4.516 | 0.0000063 | −3.338 | 0.0008 | −2.994 | 0.0028 |
| rs4623785 | 10p13 | 12784342 | European | −3.116 | 0.0018 | −1.523 | 0.1279 | −2.551 | 0.0107 |
dbSNP build 129
HG18 genome build
Figure 2.
Admixture mapping results from select chromosomes in 482 African American men with prostate cancer, adjusted for age. (A) Chromosome 7 (B) Chromosome 5 (C) Chromosome 8
When subjects were stratified by age (<60 or ≥60), both age categories supported an association with rs2141360 on chromosome 7q31 (Table 2). Conversely, support for rs4367565 on chromosome 8q24 was greater in men in the older age group than in the younger age group (Z=2.063, p=0.039 vs. Z=1.513, p=0.13). Overall, the greatest ancestry association observed in the men < age 60 was at rs6724395 on chromosome 2p14, in a region of excess European ancestry (Z=−4.195, p=0.000027). The greatest excess African ancestry in men < age 60 occurred at rs692842, approximately 1.9 Mb from the peak on chromosome 5 in the unstratified sample. Among men ≥ age 60, the greatest excess European ancestry was observed at rs2141360 on chromosome 7q31 (Z=−3.311, p=0.00090); note that this is also where a regional peak in the entire sample occurred. In this same subsample, the greatest excess African ancestry was observed at rs9288952 on chromosome 3 (Z=3.208, p=0.0013).
Within the HFHS subjects, we were able to stratify on prostate cancer aggressiveness in 238 cases with available clinical information. In the 134 men with aggressive disease, the strongest ancestry association was at the same marker on chromosome 7q31.31 (Z=−4.782, p=0.0000017), however no association with this marker was observed in the 104 men with non-aggressive disease (Z=−1.561, p=0.12, data not shown).
Discussion
Results from the current study confirm the African ancestry association with the region on chromosome 8q detected by Freedman et al. (2006), with greatest excess African ancestry on chromosome 8 at rs4367565 (Z=2.37, p-0.018). This finding is also consistent with several other confirmatory studies (Ghoussaini et al. 2008; Gudmundsson et al. 2007; Haiman et al. 2007; Robbins et al. 2007; Schumacher et al. 2007; Suuriniemi et al. 2007; Wang et al. 2007; Yeager et al. 2007). Robbins et al. (2007) report the greatest association at rs7008482 (p=0.0005), which is flanked in our study by rs12547950 (Z=2.13, p-0.033) ~2.7 Mb centromeric and the local peak at rs437565, 5.3 Mb telomeric.
Additionally, ancestry associations with regions on chromosomes 7 and 5 were detected in our study population. Of note, our best evidence for genome-wide ancestry association overall and within the aggressive cancer group resides in a region on chromosome 7q31.31 which contains the PODXL (Podocalyxin-Like, MIM *607679) gene. Association between this region and high Gleason grade was first identified by Witte et al. (2000) at 7q32.3 with high Gleason grade, with a peak ~2 cM centromeric to D7S1804, and evidence for this association was strengthened by the 2003 expanded study (Witte et al. 2003). Subsequently, association with high Gleason score was confirmed in a German population (Paiss et al. 2003) and by an allelic imbalance study (Neville et al. 2002). The PODXL gene, located within this region, was implicated in aggressive prostate cancer risk by Casey et al. (2006), and the variant most associated with prostate cancer was very rare in the Yoruban population (<1%). A second candidate gene in this region is DOCK4 (Dedicator of Cytokinesis 4, MIM *602632) which is involved in regulating intercellular junctions and cell migration, and also has been shown to harbor a missense mutation in prostate cancer cells resulting in defective Rap1 activation (Yajnik et al. 2003).
The greatest association with African ancestry occurred on 5q35 at marker rs7729084. This region also demonstrated the greatest association with African Ancestry on chromosome 5, albeit not statistically significant, in the Freedman study (2006). Witte et al. (2000) first observed linkage to a ~26 cM region on 5q31.3–33.3 associated with high Gleason score, with peak linkage occurring halfway between D5S1480 and D5S820 (p=0.0004); this region is slightly centromeric to the 5q35.2 region observed to have excess African ancestry in our sample. Association between this region on chromosome 5 and Gleason grade has been confirmed by several other genomewide linkage scans (Goddard et al. 2001; Schaid et al. 2007; Slager et al. 2006), including a follow-up study by Witte et al. (2003).
While we are encouraged by the consistency between our results and multiple confirmatory findings on chromosome 8,(Ghoussaini et al. 2008; Gudmundsson et al. 2007; Haiman et al. 2007; Robbins et al. 2007; Schumacher et al. 2007; Suuriniemi et al. 2007; Wang et al. 2007; Yeager et al. 2007) including one that included the HU samples also used in our study (Robbins et al. 2007), the study is limited by a relatively small sample size to detect loci with smaller effects, particularly after stratifying by age and disease aggressiveness. Of the 22 SNPs included in the Robbins paper, 16 (including all that achieved statistical significance) fell between 2 flanking markers (rs12547950 and rs4367565) in our paper. Z scores of the HFHS samples and HU samples were very similar at these flanking markers: 2.20 and 1.30 at rs12547950 and 1.58 and 2.09 at rs4367565 respectively. When the HU samples were excluded from the analyses, the peak on chromosome 8 in the HFHS samples is at rs6994682 (Z=2.52), approximately 30 centimorgans centromeric to the combined peak rs4367565. We were unable to examine the specific prostate-cancer associated regions identified within this genomic locus (Haiman et al. 2007; Robbins et al. 2007) because of inadequate SNP density. A final limitation was that the ADMIXMAP program does not yet have the capacity to examine the X chromosome reliably. Previously published ADMIXMAP power calculations established that a Z score of 4.27 with a corresponding p-value of 10−5 is the threshold for statistical significance (Hoggart et al. 2004). We were not able to achieve this level of power for the loci on chromosomes 8 and 5. However the locus on chromosome 7 far exceeded this criterion level of genome-wide significance.
In conclusion, we confirmed the previous African ancestry associations with a genomic locus on 8q24, and identified a possible second locus on Chromosome 7q31 associated with excess European ancestry in this region. Fine mapping with a higher density of ancestry informative marker density in this region is needed to narrow the defined locus and better target future candidate gene studies.
Acknowledgments
This project was supported by the U.S. Department of Defense W81XWH 06-1-0181, NIH/NIEHS 5 R01 ES11126-03, NCI 1U54CA091431-010001, Wayne State University Research Enhancement Program, and the Barbara Ann Karmanos Cancer Institute.
References
- Casey G, Neville PJ, Liu X, Plummer SJ, Cicek MS, Krumroy LM, Curran AP, McGreevy MR, Catalona WJ, Klein EA, Witte JS. Podocalyxin variants and risk of prostate cancer and tumor aggressiveness. Hum Mol Genet. 2006;15:735–41. doi: 10.1093/hmg/ddi487. [DOI] [PubMed] [Google Scholar]
- Cerhan JR, Parker AS, Putnam SD, Chiu BC, Lynch CF, Cohen MB, Torner JC, Cantor KP. Family history and prostate cancer risk in a population-based cohort of Iowa men. Cancer Epidemiology, Biomarkers & Prevention. 1999;8:53–60. [PubMed] [Google Scholar]
- Easton DF, Schaid DJ, Whittemore AS, Isaacs WJ. Where are the prostate cancer genes? - A summary of eight genome wide searches. Prostate. 2003;57:261–269. doi: 10.1002/pros.10300. [DOI] [PubMed] [Google Scholar]
- Freedman ML, Haiman CA, Patterson N, McDonald GJ, Tandon A, Waliszewska A, Penney K, Steen RG, Ardlie K, John EM, Oakley-Girvan I, Whittemore AS, Cooney KA, Ingles SA, Altshuler D, Henderson BE, Reich D. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci U S A. 2006;103:14068–73. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoussaini M, Song H, Koessler T, Al Olama AA, Kote-Jarai Z, Driver KE, Pooley KA, Ramus SJ, Kjaer SK, Hogdall E, DiCioccio RA, Whittemore AS, Gayther SA, Giles GG, Guy M, Edwards SM, Morrison J, Donovan JL, Hamdy FC, Dearnaley DP, Ardern-Jones AT, Hall AL, O’Brien LT, Gehr-Swain BN, Wilkinson RA, Brown PM, Hopper JL, Neal DE, Pharoah PD, Ponder BA, Eeles RA, Easton DF, Dunning AM. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst. 2008;100:962–6. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard KA, Witte JS, Suarez BK, Catalona WJ, Olson JM. Model-free linkage analysis with covariates confirms linkage of prostate cancer to chromosomes 1 and 4. Am J Hum Genet. 2001;68:1197–206. doi: 10.1086/320103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, Rafnar T, Bergthorsson JT, Agnarsson BA, Baker A, Sigurdsson A, Benediktsdottir KR, Jakobsdottir M, Xu J, Blondal T, Kostic J, Sun J, Ghosh S, Stacey SN, Mouy M, Saemundsdottir J, Backman VM, Kristjansson K, Tres A, Partin AW, Albers-Akkers MT, Godino-Ivan Marcos J, Walsh PC, Swinkels DW, Navarrete S, Isaacs SD, Aben KK, Graif T, Cashy J, Ruiz-Echarri M, Wiley KE, Suarez BK, Witjes JA, Frigge M, Ober C, Jonsson E, Einarsson GV, Mayordomo JI, Kiemeney LA, Isaacs WB, Catalona WJ, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–7. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, Waliszewska A, Neubauer J, Tandon A, Schirmer C, McDonald GJ, Greenway SC, Stram DO, Le Marchand L, Kolonel LN, Frasco M, Wong D, Pooler LC, Ardlie K, Oakley-Girvan I, Whittemore AS, Cooney KA, John EM, Ingles SA, Altshuler D, Henderson BE, Reich D. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–44. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggart CJ, Parra EJ, Shriver MD, Bonilla C, Kittles RA, Clayton DG, McKeigue PM. Control of confounding of genetic associations in stratified populations. Am J Hum Genet. 2003;72:1492–1504. doi: 10.1086/375613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggart CJ, Shriver MD, Kittles RA, Clayton DG, McKeigue PM. Design and analysis of admixture mapping studies. Am J Hum Genet. 2004;74:965–78. doi: 10.1086/420855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- Neville PJ, Conti DV, Paris PL, Levin H, Catalona WJ, Suarez BK, Witte JS, Casey G. Prostate cancer aggressiveness locus on chromosome 7q32-q33 identified by linkage and allelic imbalance studies. Neoplasia (New York) 2002;4:424–31. doi: 10.1038/sj.neo.7900254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiss T, Worner S, Kurtz F, Haeussler J, Hautmann RE, Gschwend JE, Herkommer K, Vogel W. Linkage of aggressive prostate cancer to chromosome 7q31–33 in German prostate cancer families. Eur J Hum Genet. 2003;11:17–22. doi: 10.1038/sj.ejhg.5200898. [DOI] [PubMed] [Google Scholar]
- Reich D, Patterson N, De Jager PL, McDonald GJ, Waliszewska A, Tandon A, Lincoln RR, DeLoa C, Fruhan SA, Cabre P, Bera O, Semana G, Kelly MA, Francis DA, Ardlie K, Khan O, Cree BA, Hauser SL, Oksenberg JR, Hafler DA. A whole-genome admixture scan finds a candidate locus for multiple sclerosis susceptibility. Nat Genet. 2005;37:1113–8. doi: 10.1038/ng1646. [DOI] [PubMed] [Google Scholar]
- Robbins C, Torres JB, Hooker S, Bonilla C, Hernandez W, Candreva A, Ahaghotu C, Kittles R, Carpten J. Confirmation study of prostate cancer risk variants at 8q24 in African Americans identifies a novel risk locus. Genome Res. 2007;17:1717–22. doi: 10.1101/gr.6782707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybicki BA, Neslund-Dudas C, Nock NL, Schultz LR, Eklund L, Rosbolt J, Bock CH, Monaghan KG. Prostate cancer risk from occupational exposure to polycyclic aromatic hydrocarbons interacting with the GSTP1 Ile105Val polymorphism. Cancer Detect Prev. 2006;30:412–22. doi: 10.1016/j.cdp.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid DJ. The complex genetic epidemiology of prostate cancer. Hum Mol Genet. 2004;13(Spec No 1):R103–21. doi: 10.1093/hmg/ddh072. [DOI] [PubMed] [Google Scholar]
- Schaid DJ, Stanford JL, McDonnell SK, Suuriniemi M, McIntosh L, Karyadi DM, Carlson EE, Deutsch K, Janer M, Hood L, Ostrander EA. Genome-wide linkage scan of prostate cancer Gleason score and confirmation of chromosome 19q. Hum Genet. 2007;121:729–35. doi: 10.1007/s00439-007-0368-5. [DOI] [PubMed] [Google Scholar]
- Schumacher FR, Feigelson HS, Cox DG, Haiman CA, Albanes D, Buring J, Calle EE, Chanock SJ, Colditz GA, Diver WR, Dunning AM, Freedman ML, Gaziano JM, Giovannucci E, Hankinson SE, Hayes RB, Henderson BE, Hoover RN, Kaaks R, Key T, Kolonel LN, Kraft P, Le Marchand L, Ma J, Pike MC, Riboli E, Stampfer MJ, Stram DO, Thomas G, Thun MJ, Travis R, Virtamo J, Andriole G, Gelmann E, Willett WC, Hunter DJ. A common 8q24 variant in prostate and breast cancer from a large nested case-control study. Cancer Res. 2007;67:2951–6. doi: 10.1158/0008-5472.CAN-06-3591. [DOI] [PubMed] [Google Scholar]
- Slager SL, Zarfas KE, Brown WM, Lange EM, McDonnell SK, Wojno KJ, Cooney KA. Genome-wide linkage scan for prostate cancer aggressiveness loci using families from the University of Michigan Prostate Cancer Genetics Project. Prostate. 2006;66:173–9. doi: 10.1002/pros.20332. [DOI] [PubMed] [Google Scholar]
- Smith MW, Patterson N, Lautenberger JA, Truelove AL, McDonald GJ, Waliszewska A, Kessing BD, Malasky MJ, Scafe C, Le E, De Jager PL, Mignault AA, Yi Z, De The G, Essex M, Sankale JL, Moore JH, Poku K, Phair JP, Goedert JJ, Vlahov D, Williams SM, Tishkoff SA, Winkler CA, De La Vega FM, Woodage T, Sninsky JJ, Hafler DA, Altshuler D, Gilbert DA, O’Brien SJ, Reich D. A high-density admixture map for disease gene discovery in african americans. Am J Hum Genet. 2004;74:1001–13. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suuriniemi M, Agalliu I, Schaid DJ, Johanneson B, McDonnell SK, Iwasaki L, Stanford JL, Ostrander EA. Confirmation of a positive association between prostate cancer risk and a locus at chromosome 8q24. Cancer Epidemiol Biomarkers Prev. 2007;16:809–14. doi: 10.1158/1055-9965.EPI-06-1049. [DOI] [PubMed] [Google Scholar]
- Wang L, McDonnell SK, Slusser JP, Hebbring SJ, Cunningham JM, Jacobsen SJ, Cerhan JR, Blute ML, Schaid DJ, Thibodeau SN. Two common chromosome 8q24 variants are associated with increased risk for prostate cancer. Cancer Res. 2007;67:2944–50. doi: 10.1158/0008-5472.CAN-06-3186. [DOI] [PubMed] [Google Scholar]
- Witte JS, Goddard KAB, Conti DV, Elston RC, Lin JL, Suarez BK, Broman KW, Burmester JK, Weber JL, Catalona WJ. Genomewide scan for prostate cancer-aggressiveness loci. American Journal of Human Genetics. 2000;67:92–99. doi: 10.1086/302960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte JS, Suarez BK, Thiel B, Lin J, Yu A, Banerjee TK, Burmester JK, Casey G, Catalona WJ. Genome-wide scan of brothers: replication and fine mapping of prostate cancer susceptibility and aggressiveness loci. Prostate. 2003;57:298–308. doi: 10.1002/pros.10304. [DOI] [PubMed] [Google Scholar]
- Yajnik V, Paulding C, Sordella R, McClatchey AI, Saito M, Wahrer DC, Reynolds P, Bell DW, Lake R, van den Heuvel S, Settleman J, Haber DA. DOCK4, a GTPase activator, is disrupted during tumorigenesis. Cell. 2003;112:673–84. doi: 10.1016/s0092-8674(03)00155-7. [DOI] [PubMed] [Google Scholar]
- Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee N, Wang Z, Welch R, Staats BJ, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Gelmann EP, Tucker M, Gerhard DS, Fraumeni JF, Jr, Hoover R, Hunter DJ, Chanock SJ, Thomas G. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–9. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]