Introduction
Blood feeding mosquitoes, including the dengue and yellow fever vector Aedes aegypti, transmit many of the world's deadliest diseases. Such diseases have resurged in developing countries and are also emerging as clear threats for epidemic outbreaks in developed countries. Recent mosquito genome projects have stimulated increased interest in the potential for arthropod-borne disease control by genetic manipulation of vector insects. Targets of particular interest include genes that regulate development. However, although the Ae. aegypti genome project uncovered homologues of many known developmental regulatory genes, extremely little is known about the genetic regulation of development in Ae. aegypti or other vector mosquitoes. Analysis of the expression and function of individual developmental genes must now be completed. This chapter describes methodology for culturing, collection and fixation of developing tissues, analysis of gene and protein expression, and knockdown of genes during Ae. aegypti development. The techniques described will permit detailed analyses of the functions of developmental regulatory genes and the selective inhibition of such genes during Ae. aegypti development. This methodology, much of which is applicable to other mosquito species, is useful to both the comparative development and vector research communities.
Background information
Ae. aegypti (Diptera: Culicidae) is a vector mosquito of medical importance. This species has a cosmotropical distribution between 20° S and 30° N latitudes and is found throughout most tropical to subtropical world regions, where it exhibits a preference for human habitats bearing standing water. Ae. aegypti adults (Fig. 1), which resemble the Asian tiger mosquito Ae. albopictus, are a medium-sized mosquito approximately 4-7 mm in length. Adults have white scales on the dorsal surface of their thorax and a dark brown to black abdomen that may possess white scales. Tarsal segments of the hind legs have white basal bands that form what appear to be stripes (Carpenter and LaCasse, 1955).
Figure 1.
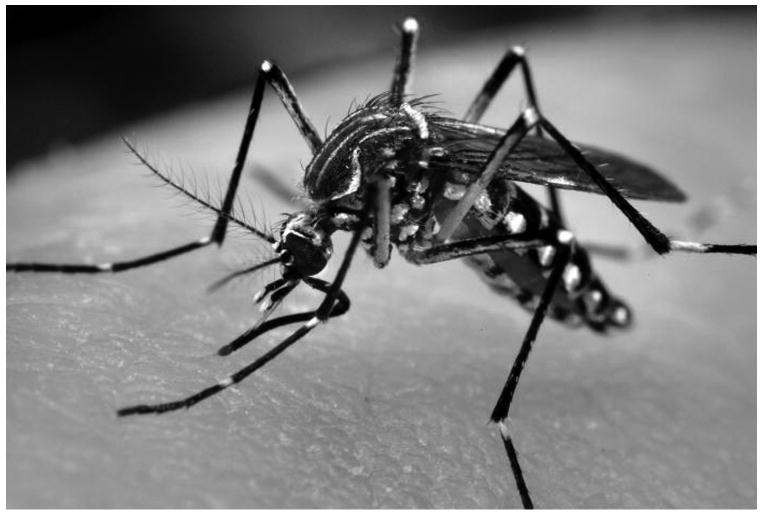
Adult Ae. aegypti female. Ae. aegypti adults have white scales on the dorsal surface of their thorax as well as a dark brown/black abdomen that may also possess white scales. The tarsal segments of the hind legs have white basal bands. Photo Credit: James Gathany, CDC.
Males and females feed on plant nectar, but females possess piercing-sucking mouthparts adapted for acquiring vertebrate blood meals prior to oviposition. Females blood feed mainly at dusk and dawn and primarily on human hosts. Adult females can produce up to five batches of eggs during their lifetime and lay 100-200 eggs per batch. Although eggs are typically laid on damp surfaces, eggs can survive desiccation for several months (Clements, 1999). Mosquitoes in the tribe Aedini living in temperate areas typically overwinter in a diapaused egg stage that is resistant to freezing and which lasts until spring, when they hatch as larvae. The four larval stages, as well as the pupal stage, are aquatic. Much of the larval stages, which last at least four days, is spent at the water surface, as larvae need to breathe, but larvae swim below the surface when they feed on organic particulate matter such as algae and other microscopic organisms in their aquatic habitats. Following the fourth instar, Ae. aegypti enter a mobile, non-feeding pupal stage which lasts approximately two days. The adult life span of this insect varies depending on environmental conditions, but ranges from two weeks to a month (Christophers, 1960; Clements, 1999; Foster and Walker, 2002).
Ae. aegypti is commonly referred to as the yellow fever or dengue fever mosquito, as it is responsible for transmission of both diseases. Prior to 1900, Ae. aegypti had largely escaped recognition as a world species. However, following the turn of the 20th century discovery that Ae. aegypti and other mosquitoes transmit human disease, numerous collections of Ae. aegypti from many tropical regions were established (Christophers, 1960). Ae. aegypti is the primary vector for the yellow fever virus, which infects 200,000 people and results in 30,000 deaths annually. Yellow fever occurs primarily in tropical regions of Africa, as well as in parts of South America. The severity of this illness ranges from a self-limited febrile illness to severe hepatitis with hemorrhagic fever. Ae aegypti is also the primary mosquito vector for the transmission of dengue viral infections, which can result in dengue fever, a nonspecific febrile illness which is the most widespread and significant arboviral disease in the world. Dengue virus is also the etiological agent of dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS), which are severe and sometimes fatal forms of the disease. Dengue is a resurgent disease worldwide in the tropics, and DHF has been reported from 18 countries in the Americas. Dengue virus is presently a threat to over 2.5 billion people, with an annual incidence of approximately 50 million cases and 500,000 cases of DHF/DSS resulting in ∼24,000 deaths annually (http://www.cdc.gov).
A number of factors contribute to the present inability to prevent or control mosquito-borne diseases. Although an effective vaccine for yellow fever exists, vaccination distribution is problematic, and outbreaks often occur in regions where vaccinations are not available. A vaccine for dengue does not exist, and dengue epidemics are becoming larger and more frequent. In addition to poor progress in vaccine development and distribution, the emergence of insecticide resistance in mosquito populations, a general lack of support for mosquito control programs, and increased global travel contribute to the prevalence of diseases transmitted by mosquito vectors (http://www.cdc.gov). It is therefore increasingly critical to study the biology of vector mosquitoes such as Ae. aegypti. In particular, the developmental biology of vector mosquitoes, the focus of this chapter, has been grossly understudied.
The Ae. aegypti genome project (Nene et al., 2007) revealed homologues of many known developmental regulatory genes. Characterization of the function of these genes could reveal novel strategies for vector control. However, studying the development of this species has been a challenge. Christophers (1960), indicated that “the eggs of Ae. aegypti are not the most suitable form on which to study mosquito embryology.” In fact, in his book, which remains the most comprehensive description of Ae. aegypti biology to date, he offered a description of Culex molestus, rather than Ae. aegypti development. The chorion and serosal cuticle of Ae. aegypti, which serve as barriers to fixatives, probes, and antibodies, has made working with this species a challenge in the past. However, we offer here a series of protocols that make analysis of Ae. aegypti development manageable. Given the many known advantages of this system (Severson et al., 2004a), these protocols, in combination with recent advances in the study of genetics and genomics in this species (Chen et al., 2008), are helping to establish Ae. aegypti as an emerging model organism for vector mosquito development.
Sources and Husbandry
Ae. aegypti is relatively simple to grow in culture (Protocol 1, Fig. 2) and has therefore been the subject of a wide range of laboratory investigations. Other mosquito species are often difficult to adapt to laboratory culture and require constant maintenance following successful introduction to the lab. This is because the life cycles of most mosquito species are continuous, with eggs typically being laid on the water surface and hatching soon thereafter. In contrast, Ae. aegypti preferentially oviposit away from the water surface and are tolerant to desiccation, an adaptation which allows for collection of eggs on artificial substrates such as paper towels and subsequent storage for several months. Stored eggs can later be induced to hatch in deoxygenated water. Furthermore, mosquitoes collected in the field can readily adapt to laboratory rearing conditions and can even successfully reproduce following single pair matings (Severson et al., 2004a). Given the advantages of this system, many strains have been reared in laboratory settings. Although the University of Notre Dame initially served as the World Health Organization's repository for Ae. aegypti strains, the ever growing number of strains are no longer maintained in a central location, but can be acquired from the individual laboratories in which they are studied.
Figure 2.
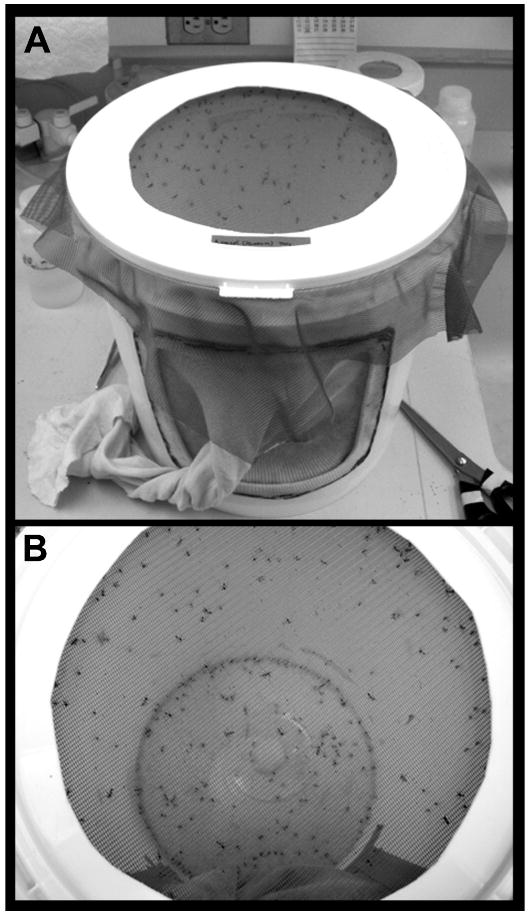
Mosquito rearing cage. Methodology for culturing mosquitoes is described in Protocol 1. The cotton sleeve on the cage (seen in A) permits human access and manipulations within the cage. The netted top of the cage (top down view shown in B) allows for blood feeding prior to egg collections.
Related Species
Sequencing of the Ae. aegypti (Nene et al., 2007) and Anopheles gambiae (Holt et al., 2002) genomes was an important advancement, as these species represent significant members of two medically important mosquito subfamilies, Culicinae and Anophelinae. Arbovirus and lymphatic filariasis transmission is largely associated with Culicinae, while Anopheline mosquitoes are the primary vectors for malaria transmission. The Culex quinquefasciatus (dominant lymphatic filariasis vector) genome is also now available at Vectorbase (http://cquinquefasciatus.vectorbase.org). Completion of these three mosquito genomes is allowing for global characterization of sequence conservation and structure through comparative and functional analyses (reviewed by Chen et al., 2008). While many commonalities exist, significant differences in the biological characteristics and genomic structures of these mosquitoes have been observed (Knudson et al., 2002; Rai and Black, 1999; Severson et al., 2004b).
In terms of studying the developmental biology of mosquitoes, Ae. vexans is probably the most carefully described Aedine mosquito (Moretti and Larsen, 1973). Although these descriptions will be of use to those studying Ae. aegypti development, the genome of Ae. vexans has not yet been sequenced. As we embark upon genetic characterization of Ae. aegypti development, studies in Drosophila melanogaster, a well-established model for insect development, and Tribolium castaneum, an emerging model (Brown et al., 2009), will be useful, as the genomes of these species have been sequenced. Mosquito homologues of genes that regulate fruit fly development are being characterized (see below). The process of generating transgenic mosquitoes has been adapted from approaches developed in Dr. melanogaster (see below). Techniques for fixation and preparation of developing tissues (Protocol 2, Fig. 3) developmental analyses of gene (Protocol 3, Fig. 4) and protein expression (Protocol 4, Fig. 5) in Ae. aegypti described here were adapted from Drosophila protocols (Patel, 1994, 1996). The technique for RNA inhibition (RNAi) in Ae. aegypti included here (Protocol 5) draws from successful RNAi studies performed in Tr. castaneum (Brown et al., 2009).
Figure 3.
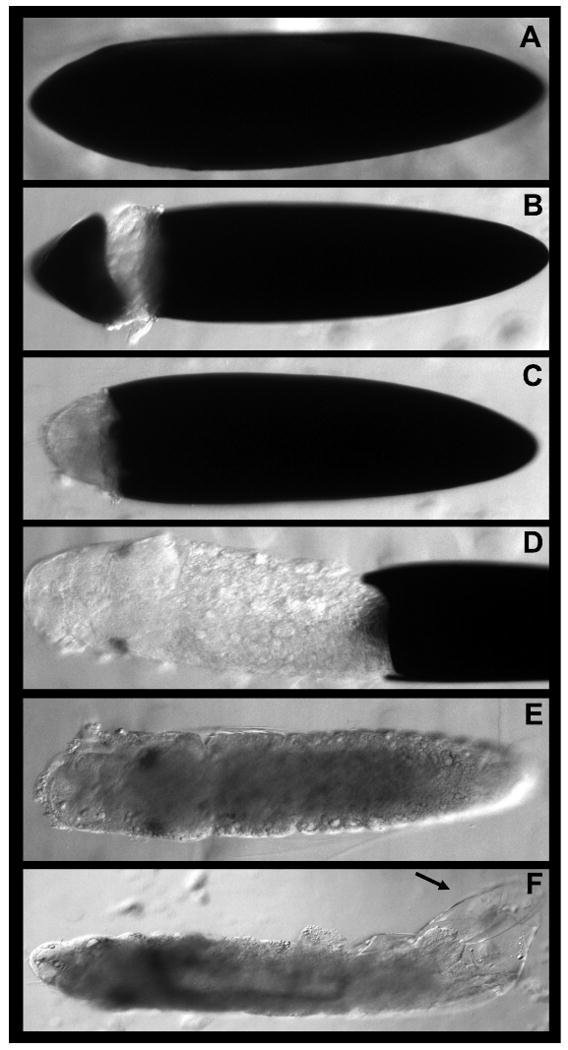
Dissection methodology for Ae. aegypti embryos. Methods for fixing and dissecting mosquito embryos are included in Protocol 2. The process of dissecting a fixed embryo is shown in A-F (see text for details). It is critical to remove the black endochorion (A-D) and transparent serosal cuticle (arrowhead in F) in order to successfully perform gene and protein expression assays.
Figure 4.

Expression of axon guidance genes in Ae. aegypti embryos. (A) fra and casein kinase (B, C) are expressed ventrally in the developing nerve cord at 55 hrs. of development. Lateral views of whole-mount embryos stained with the accompanying in situ methodology (Protocol 3) are shown in A and B (anterior is oriented left). A filleted nerve cord is shown in C (anterior is oriented up).
Figure 5.
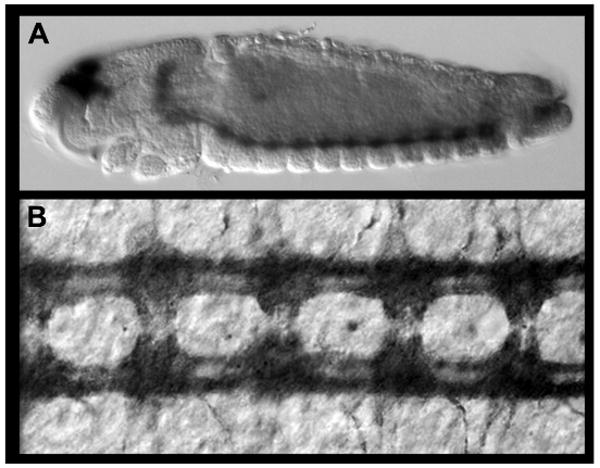
Protein expression analysis in Ae. aegypti during embryonic development. (A,B) Acetylated tubulin expression labels the axons of the developing nerve cord. A lateral view of a 55 hr whole-mount embryo stained with the accompanying immunohistochemistry methodology (Protocol 4) is shown in A (anterior is oriented left). A filleted nerve cord from a 55 hr embryo is shown in B (anterior is oriented up).
Uses of the Ae. aegypti Model System
A model for mosquito-pathogen interactions
Given the relative ease of rearing Ae. aegypti in culture (Protocol 1), this species has been the subject of a wide range of laboratory investigations ranging from analysis of its morphology, physiology, genetics, vector competence, and molecular evolution (Clements, 1992; Severson et al., 2004a, Chen et al., 2008). In particular, analysis of Ae. aegypti has provided broad insight into the study of mosquito/pathogen interactions. Following a blood meal from an infected host, the pathogen must survive internal defense mechanisms which aim to recognize and destroy foreign entities. Variability in vector competence is attributable to genetic differences, and it is therefore critical to study vector competence in genetically well characterized mosquito species such as Ae. aegypti (see below). Use of Ae. aegypti has permitted genetic analysis of the basis of dengue vector competence. Furthermore, although Ae. aegyti is not a vector for human malarial parasites, it is a vector for the avian malarial parasite Plasmodium gallinaceum, and is thus serving as a model for the genetic basis of vector competence for malaria parasites. Likewise, although Ae. aegypti is not a natural vector for lymphatic filariasis, the Liverpool strain of Ae. aegypti is susceptible to Brugia malayi and Wucheria bancrofti, two filarial nematode strains which infect humans. Ae. aegypti can therefore also be used as a model for the genetic basis of vector competence to filarial worms (reviewed by Severson et al., 2004a and Chen et al., 2008).
Overview of Ae. aegypti Development
Recent mosquito genome projects (Holt et al., 2002; Nene et al., 2007) have stimulated increased interest in the potential for arthropod-borne disease control by genetic manipulation of vector insects. Targets of particular interest include genes that regulate development. Although the Ae. aegypti genome project uncovered homologues of many known developmental regulatory genes, extremely little is known about the genetic regulation of development in Ae. aegypti or other vector mosquitoes. Raminani and Cupp published descriptions of Ae. aegypti early embryology (1975) and organogenesis (1978). Their studies, which provide a firm basis for future studies which will examine developmental gene function in Ae. aegypti, are summarized below and in Table 1.
TABLE 1.
Developmental Events in Aedes aegypti Embryogenesis (after Raminani and Cupp, 1978). The timing of major embryonic developmental events is indicated in hrs after egg laying (AEL) at 25° C.
| Age (hrs AEL) |
Event |
|---|---|
| 0.5 | Polar body formation |
| 1 | Fertilization |
| 1-2 | 1st and 2nd mitotic divisions |
| 3 | Formation of pole cells and primary yolk cells |
| 4 | Peripheral migration of cleavage nuclei |
| 5 | Pole cells outside the blastoderm |
| 6 | Syncitial blastoderm |
| 8 | Cellularization of blastoderm |
| 9 | Germ band formation; secondary yolk cells |
| 11 | Pole cell entry into the blastoderm |
| 12 | Initiation of gastrulation; Migration of presumptive mesodermal cells, elongation of the germ band |
| 14 | Amnion and serosa formation |
| 15 | Segmentation of the embryo begins; Ventral nerve cord formation begins. |
| 18 | Labral lobe formation |
| 19 | Anterior midgut-stomodaeal and posterior midgut-proctodaeal invaginations form. |
| 25 | Antennae, mandibles and maxillae begin to form; anterior migration of the mouthparts begins. |
| 30 | Vitelline membrane present; germ band retraction begins; labial lobes appear. |
| 35 | Blastokinesis and dorsal closure begin; foregut differentiation and formation of the midgut begins; formation of the brain and gonads initiates. |
| 40 | Differentiation of the suboesophageal ganglion and tracheal system starts. |
| 45 | Hindgut differentiation begins; formation of the retrocerebral complex and stomodaeal nervous system initiates. |
| 50 | Breakdown of the amnion and serosa; oenocytes and fat cells are formed; muscles of the head and mouthparts begins to form. |
| 60 | Differentiation of trunk musculature |
| 65 | Heart formation |
| 75 | Formation of the hatching spine begins. |
| 96 | Embryogenesis completed |
Embryonic development of Ae. aegypti (analyzed at 25° C) proceeds as follows: Fusion of the female and male pronuclei occurs approximately one hour following oviposition. The first and second mitotic divisions occur before the end of the second hour of development. Asynchronous nuclear divisions follow and result in the formation of a syncytial blastoderm by 6 hrs. of development. The germ band forms by 9 hrs., and cellularization of the blastoderm is complete by 10 hrs. of development. Columnar cells of the ventral blastoderm form the embryo proper. The amnion arises from ventrolateral cells, and dorsal blastodermal cells form the serosa. Pole cells, which begin to form at 3 hrs., remain outside the blastoderm until 11 hrs., at which time they migrate with the germ band as dorsal elongation occurs. Gastrulation initiates at 12 hrs. of development. Segments are visible at 15 hrs. and are well defined by 20 hrs. (Raminani and Cupp, 1975). Germ band shortening initiates at 30 hrs. and is complete by 40 hrs. A vitelline membrane forms by 30 hrs. of development, and lysis of the amnion and serosa is complete by 55 hrs. Dorsal closure is complete by 65 hrs. (Raminani and Cupp, 1975).
Raminani and Cupp (1978) also documented major events in organogenesis. Ventral nerve cord formation initiates at 15 hrs. of development. Labral development initiates at 18 hrs., and the labrum is visible as a distinct lobe by 35 hrs. Posterior midgut-proctodeal and anterior midgut-stomodael invaginations form at 19 hrs. Antennal rudiments, the mandible, and maxillae are visible by 25 hrs, and the labium can be observed at 30 hrs. Foregut differentiation, midgut, brain, and gonad formation occurs at 35 hrs. At 40 hrs., tracheal system differentiation initiates. Differentiation of the hindgut commences at 45 hrs. Head and mouthpart muscles begin to form at 50 hrs., while trunk musculature differentiates at 60 hrs. The heart is formed by 65 hrs. Formation of the hatching spine begins at 75 hrs., and embryogenesis is complete by 96 hrs.
Ae. aegypti and Dr. melanogaster development are compared in Table 2. The relative timing of major developmental events in both species is noted. While the relative order of developmental events is well conserved, embryogenesis proceeds more rapidly in Dr. melanogaster. This comparison will be useful for those who are interested in studying a particular aspect of mosquito development, or who wish to analyze the function of Ae. aegypti homologues of fly developmental genes.
TABLE 2.
Temporal Comparison of Major Developmental Events in Dr. melanogaster and Ae. aegypti Embryogenesis. The relative timing of major developmental events, measured in hrs after egg laying (AEL) at 25° C, is noted for both species. The relevant Drosophila stage is also indicated. While major developmental events and their relative timing is generally well conserved, embryogenesis proceeds more rapidly in Dr. melanogaster.
| Event |
Dr. melanogaster Age (hours AEL; stage) |
Ae. aegypti Age (hours AEL) |
|---|---|---|
| Cleavage | 0-1.2; 1-4 | 4 |
| Pole cells form | 1.2; 3 | 3 |
| Syncitial blastoderm | 1.5-2.2; 4 | 6 |
| Cellularization of blastoderm | 2.5; 5 | 8 |
| Onset of gastrulation | 3; 6 | 12 |
| Midgut formation begins | 3.1; 7 | 35 |
| Germ band extension | 3.3; 8 | 12 |
| Nerve cord formation begins | 3.75; 9 | 15 |
| Foregut differentiation | 4.25; 10 | 35 |
| Tracheal system differentiation | 5.25; late 10 | 40 |
| Brain formation initiates | 5.3; 11 | 35 |
| Germ band retraction | 7.25; 12 | 30 |
| Segment formation evident | 7.25; 12 | 15 |
| Dorsal closure begins | 9.25; 13 | 35 |
| Gonad formation begins | 11.4; 15 | 35 |
| Completion of dorsal closure | 13; 15 | 65 |
A model for vector mosquito developmental genetics
Although we have made great advances in understanding developmental genetics in Drosophila, comparatively little is known about the genetic basis for development in mosquitoes and other arthropods. Studying developmental genetics in mosquitoes will surely benefit the evolutionary development research community, as data collection from non-model arthropods has been an important aspect of the recent “evo-devo” revolution. Additionally, detailed characterization of mosquito development and genes that regulate this process could precipitate new strategies for vector control.
To date, only a handful of Ae. aegypti developmental genes have been studied. Several groups have used a candidate gene approach to identify mosquito homologues of Drosophila gene products that localize to the future germ cells of the fly embryo. Expression of the nanos (Calvo et al., 2005; Adelman et al., 2007) and oskar (Goltsev et al., 2004, Juhn and James, 2006) genes has been analyzed in several mosquito species, including Ae. aegypti. The expression profiles of these genes are generally well conserved, however some differences in their temporal and spatial distributions have been noted (reviewed by Chen et al., 2008). These studies were completed, at least in part, through efforts to develop a drive system that will quickly spread and fix antipathogen effector genes in natural populations (Juhn and James, 2006). To this end, Adelman et al. (2007) showed that nanos control sequences demonstrated promise as part of a transposable element-based gene drive system. Thus, an evo-devo approach was applied in an effort to develop translational strategies for vector control.
In recent years, we have begun analysis of nervous system development in Ae. aegypti (Simanton et al., 2009; Clemons et al., in preparation). Analysis of mosquito nervous system development will lead to a better understanding of the developmental basis of motor function, sensory processing, and behavior, key aspects of mosquito host location. Our analyses suggest that the process of embryonic nerve cord formation, as well as the function of the axon guidance molecules Netrin (Simanton et al., 2009) and its receptor Frazzled/DCC (Clemons et al., in preparation) are conserved between Drosophila and Ae. aegypti. We are beginning to extend our analyses of axon guidance gene function to the developing olfactory system. The olfactory system is of particular interest to the vector community, as location of human hosts is an olfactory-driven behavior.
While detailed and thorough analysis of all aspects of mosquito development is critical, analysis of tissues that are vital to host location and the spread of infection is of particular importance. Mosquitoes rely on their olfactory systems for location of human hosts. During blood feeding, disease causing viruses and parasites are ingested and encounter the mosquito midgut epithelium, where they replicate. The infection spreads to secondary sites in the mosquito body prior to infecting the salivary glands. Following salivary gland infection, female mosquitoes are competent for disease transmission for the duration of their lives (reviewed by Catteruccia, 2007). Although analysis of mosquito olfactory, gut, and salivary gland development could have important applications for disease control, little is known about the development of these tissues in mosquitoes. We are in the process of identifying molecular markers for these tissues and are beginning to study the genetic regulation of their development.
Genetics, Genomics, and Associated Resources
Ae. aegypti is genetically one of the best characterized insect species. The development of genetic markers in Ae. aegypti has facilitated the tracking of groups of genes or genome segments in relation to a phenotype of interest. Linkage maps were initially generated from isozyme and mutant marker loci (Munsterman and Craig, 1979). In recent years, genetic studies in Ae. aegypti have benefitted from the construction of DNA-based marker technology which has resulted in the construction of genetic maps. Restriction fragment length polymorphisms (RFLPs) were used in the development of a genetic map consisting of 53 loci covering 134 cM (Severson et al., 1993). This map was expanded through the inclusion of single nucleotide polymorphisms (SNPs), single strand conformational polymorphisms (SSCP), and microsatellite (SSR) markers, to cover 154 loci over 205 cM (Severson et al., 2002, Chambers et al., 2007). A genetic map consisting of 96 loci and spanning 168 cM was also prepared through use of SSCP analysis of random amplified polymorphic DNA (RAPD) markers (Antolin et al., 1996). SSCP technology was applied toward the development of additional genetic markers from cDNA sequences (Bosio et al., 2000; Fulton et al., 2001). More recently, Lovin et al. (2009) showed that the Ae. aegypti genome is populated with single copy polymorphic microsatellite loci which are suitable for genetic and population studies (Hemme et al., 2010).
David Severson coordinated a whole genome shotgun sequencing effort to determine the complete annotated genome sequence of Ae. aegypti. Preliminary annotations were developed by two independent groups (The Institute for Genomic Research and VectorBase), and a final annotated sequence was reported (Nene et al., 2007). All information for the Ae. aegypti genome project is accessible at Vectorbase (http://aaegypti.vectorbase.org/index.php). AaegL1.2, an update to the initial gene set (AaegL1.1) was released by Vectorbase in 2009. The genome sequence assembly consists of 4,758 scaffolds spanning 1,310 million basepairs (Mbp). 15,988 genes are estimated; rules and conventions for naming genetic features in members of the family Culicidae are described (VectorBase). In recent years, VectorBase has initiated a community annotation system, a microarray and gene expression repository, and has continued to develop software infrastructure and tools for interrogating stored sequence data of Ae. aegypti and other arthropod vectors (Lawson et al., 2009).
Technical Approaches
Generation of transgenics
The recent completion of the Ae. aegypti genome project (Nene et al., 2007) is facilitating studies aiming to expand the current understanding of mosquito biology. Such investigations may ultimately identify new targets of vector control, lead to development of strategies for introduction of refractory traits in natural populations, or help to control the size of mosquito populations (Catteruccia, 2007). The potential for arthropod-borne disease control by genetic manipulation of vector insects has been recognized and advocated for four decades (Knipling et al., 1968). The identification of genes involved in host-seeking, blood meal acquisition and digestion, reproduction, susceptibility/refractoriness to pathogens, and insecticide resistance is expected to yield novel approaches for the control of mosquito-borne illnesses (Chen et al., 2008). Several genetic control strategies for mosquitoes have been proposed, including: sterile male release programs, cytoplasmic incompatibility, hybrid sterility, and population replacement. However, while several of these strategies are promising, they have not yet been implemented successfully. Recently, considerable efforts are being directed toward applying transgenesis technology to arthropod-borne disease control, with an emphasis on population replacement (Adelman et al., 2002; Moreira et al., 2002; Robinson et al., 2004, Chen et al., 2008). A major focus of this research is to use these techniques to generate genetically-modified mosquitoes carrying transgenes that disrupt their ability to interact with hosts and transmit pathogens. These transgenic mosquitoes would be introduced into natural mosquito populations. Successful implementation of such strategies will require intensive prerelease and post-release investigations of mosquito ecology and population biology (Scott et al., 2002).
Successful integration of transgenes into the germline of Ae. aegypti (reviewed by Adelman et al., 2002; Lobo et al., 2006, Chen et al., 2008) has been accomplished with class II transposable elements, including Hermes, mariner, and piggybac. A high-efficiency germ-line transformation protocol has been described (Lobo et al., 2006), and a helpful video protocol documenting the microinjection process is available (Jasinskiene et al., 2007). Transgenes are introduced through microinjecting mosquito embryos at syncitial blastoderm stages. Fluorescent protein markers such as GFP are typically used to monitor transgene insertion. Such methodology has generated transgenic mosquitoes that may prove useful for the control of mosquito-borne illnesses, including a recently described female-specific flightless phenotype strategy for mosquito control (Fu et al., 2010).
Functional analysis of genes
RNA interference (RNAi) is proving to be a useful method for functional analysis of genes in Ae. aegypti (Brown and Catteruccia, 2006). For analysis in adults, dsRNA is typically injected into female abdomens; a video protocol illustrating this methodology is available (Luna et al., 2007). Blitzer et al. (2005) used RNAi knockdown strategies for functional analysis in larvae. A protocol for siRNA-mediated knockdown of genes in embryos is included here (Protocol 5). These methodologies are yielding information about genes that are important for combating mosquito-borne illnesses. For example, silencing of the Ae. aegypti cactus genes was found to promote developmental arrest and death of the avian malaria parasite, Plasmodium gallinaceum (Zou et al., 2008). Xi et al. (2008) used RNAi strategies to functionally assess the role of the Ae. aegypti Toll pathway in regulating resistance to dengue virus.
Analysis of Developmental Genetics in Ae. aegypti
In recent years, evolutionary developmental biologists have applied knowledge of developmental genetics in Drosophila melanogaster, a genetically tractable model organism, to better understand development of other arthropods. These efforts typically involve the tedious process of cloning and assigning homology to genes from non-model organisms in which functional analyses are not always possible. Sequencing of the Tribolium castenium genome and the development of strategies for examining gene function have greatly enhanced analysis of developmental genetics in the red flour beetle (Brown et al., 2009). It is anticipated that the Ae. aegypti genome sequence, in combination with the development of techniques for analyzing gene expression and function during development, will allow Ae. aegypti to emerge as an excellent model for vector mosquito development. This chapter describes methodology for culturing and egg collection (Protocol 1), fixation and dissection (Protocol 2, Fig. 3), analysis of gene (Protocol 3, Fig. 4) and protein (Protocol 4, Fig. 5) expression, and knockdown of genes (Protocol 5) during Ae. aegypti development. The techniques described will permit detailed analysis of the function of developmental regulatory genes and the selective inhibition of such genes in Ae. aegypti. These studies and methodology, much of which are applicable to other mosquito species, are useful to both the comparative development and vector research communities.
Acknowledgments
We thank Frank Collins for advice, encouragement, and use of equipment. We are extremely grateful to Sun Longhua (Malcolm Frasier lab) who taught us to microinject mosquito embryos and has served as an expert microinjection consultant. Finally, we thank Nipam Patel for advice and for his Drosophila immunohistochemistry and in situ hybridization protocols which were adapted here for use in Ae. aegypti. Kristopher Kast and Caitlin Jacowski were supported by the University of Notre Dame College of Science Summer Undergraduate Research Fellowship program. NIH/NIAID Award R01 AI 081795-01, NIH/NINDS Award R15 NS 048904-0, and an IUSM Research Support Funds Grant (all to MDS) and NIH/NIAID Award RO1 AI 059342 (to DWS) have funded Ae. aegypti projects in our laboratories.
Footnotes
Conflicts of interest: none declared
References and Resources
- Adelman ZN, Jasinskiene N, James AA. Development and applications of transgenesis in the yellow fever mosquito, Aedes aegypti. Mol Biochem Parasitol. 2002;121:1–10. doi: 10.1016/s0166-6851(02)00028-2. [DOI] [PubMed] [Google Scholar]
- Adelman ZN, Jasinskiene N, Onal S, Juhn J, Ashikyan A, Salampessy M, MacCauley T, James AA. Nanos gene control DNA mediates developmentally regulated transposition in the yellow fever mosquito Aedes aegypti. Proc Natl Acad Sci U S A. 2007;104:9970–9975. doi: 10.1073/pnas.0701515104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antolin MF, Bosio CF, Cotton J, Sweeney W, Strand MR, Black WC., 4th Intensive linkage mapping in a wasp (Bracon hebetor) and a mosquito (Aedes aegypti) with single-strand conformation polymorphism analysis of random amplified polymorphic DNA markers. Genetics. 1996;143:1727–1738. doi: 10.1093/genetics/143.4.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosio CF, Fulton RE, Salasek ML, Beaty BJ, Black WC., 4th Quantitative trait loci that control vector competence for dengue-2 virus in the mosquito Aedes aegypti. Genetics. 2000;156:687–698. doi: 10.1093/genetics/156.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AE, Catteruccia F. Toward silencing the burden of malaria: progress and prospects for RNAi-based approaches. Biotechniques. 2006 Suppl:38–44. doi: 10.2144/000112117. [DOI] [PubMed] [Google Scholar]
- Brown SJ, Shippy TD, Miller S, Bolognesi R, Beeman RW, Lorenzen MD, Bucher G, Wimmer EA, Klingler M. The red flour beetle, Tribolium castaneum (Coleoptera): a model for studies of development and pest biology. CSH Protoc. 2009 doi: 10.1101/pdb.emo126. pdb emo126. [DOI] [PubMed] [Google Scholar]
- Calvo E, Walter M, Adelman ZN, Jimenez A, Onal S, Marinotti O, James AA. Nanos (nos) genes of the vector mosquitoes, Anopheles gambiae, Anopheles stephensi and Aedes aegypti. Insect Biochem Mol Biol. 2005;35:789–798. doi: 10.1016/j.ibmb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Carpenter SJ, Lacasse WJ. Mosquitoes of North America (North of Mexico) University of California Press; Berkeley, CA: 1955. [Google Scholar]
- Catteruccia F. Malaria vector control in the third millennium: progress and perspectives of molecular approaches. Pest Manag Sci. 2007;63:634–640. doi: 10.1002/ps.1324. [DOI] [PubMed] [Google Scholar]
- Chambers EW, Meece JK, McGowan JA, Lovin DD, Hemme RR, Chadee DD, McAbee K, Brown SE, Knudson DL, Severson DW. Microsatellite isolation and linkage group identification in the yellow fever mosquito Aedes aegypti. J Hered. 2007;98:202–210. doi: 10.1093/jhered/esm015. [DOI] [PubMed] [Google Scholar]
- Chen XG, Mathur G, James AA. Gene expression studies in mosquitoes. Adv Genet. 2008;64:19–50. doi: 10.1016/S0065-2660(08)00802-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophers SR. Aedes aegypti, The yellow fever mosquito: its life history, bionomics, and structure. Cambridge University Press; Cambridge, UK: 1960. [Google Scholar]
- Clements A. The Biology of Mosquitoes: Volume 1: Development, Nutrition and Reproduction. Chapman and Hall; New York, NY: 1999. [Google Scholar]
- Foster WA, Walker ED. Mosquitoes (Culicidae) In: Mullen GA, Durden LA, editors. Medical and Veterinary Entomology. Academic Press; San Diego, CA: 2002. pp. 203–262. [Google Scholar]
- Fu G, Lees RS, Nimmo D, Aw D, Jin L, Gray P, Berendonk TU, White-Cooper H, Scaife S, Kim Phuc H, et al. Female-specific flightless phenotype for mosquito control. Proc Natl Acad Sci U S A. 2010;107:4550–4554. doi: 10.1073/pnas.1000251107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton RE, Salasek ML, DuTeau NM, Black WC., 4th SSCP analysis of cDNA markers provides a dense linkage map of the Aedes aegypti genome. Genetics. 2001;158:715–726. doi: 10.1093/genetics/158.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goltsev Y, Fuse N, Frasch M, Zinzen RP, Lanzaro G, Levine M. Evolution of the dorsalventral patterning network in the mosquito, Anopheles gambiae. Development. 2007;134:2415–2424. doi: 10.1242/dev.02863. [DOI] [PubMed] [Google Scholar]
- Hemme RR, Thomas CL, Chadee DD, Severson DW. Influence of urban landscapes on population dynamics in a short-distance migrant mosquito: evidence for the dengue vector Aedes aegypti. PLoS Negl Trop Dis. 2010;4:e634. doi: 10.1371/journal.pntd.0000634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JM, Wides R, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Jasinskiene N, Juhn J, James A. Microinjection of A. aegypti Embryos to obtain Transgenic Mosquitoes. Journal of Visualized Experiments. 2007;5:219. doi: 10.3791/219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhn J, James AA. Oskar gene expression in the vector mosquitoes, Anopheles gambiae and Aedes aegypti. Insect Mol Biol. 2006;15:363–372. doi: 10.1111/j.1365-2583.2006.00655.x. [DOI] [PubMed] [Google Scholar]
- Juhn J, Marinotti O, Calvo E, James AA. Gene structure and expression of nanos (nos) and oskar (osk) orthologues of the vector mosquito, Culex quinquefasciatus. Insect Mol Biol. 2008;17:545–552. doi: 10.1111/j.1365-2583.2008.00823.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipling EF, Laven H, Craig GB, Pal R, Kitzmiller JB, Smith CN, Brown AW. Genetic control of insects of public health importance. Bull World Health Organ. 1968;38:421–438. [PMC free article] [PubMed] [Google Scholar]
- Knudson DL, Brown SE, Severson DW. Culicine genomics. Insect Biochem Mol Biol. 2002;32:1193–1197. doi: 10.1016/s0965-1748(02)00082-6. [DOI] [PubMed] [Google Scholar]
- Lawson D, Arensburger P, Atkinson P, Besansky NJ, Bruggner RV, Butler R, Campbell KS, Christophides GK, Christley S, Dialynas E, et al. VectorBase: a data resource for invertebrate vector genomics. Nucleic Acids Res. 2009;37:D583–D587. doi: 10.1093/nar/gkn857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo NF, Clayton JR, Fraser MJ, Kafatos FC, Collins FH. High efficiency germ-line transformation of mosquitoes. Nat Protoc. 2006;1:1312–1317. doi: 10.1038/nprot.2006.221. [DOI] [PubMed] [Google Scholar]
- Lovin DD, Washington KO, deBruyn B, Hemme RR, Mori A, Epstein SR, Harker BW, Streit TG, Severson DW. Genome-based polymorphic microsatellite development and validation in the mosquito Aedes aegypti and application to population genetics in Haiti. BMC Genomics. 2009;10:590. doi: 10.1186/1471-2164-10-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna BM, Juhn J, James AA. Injection of dsRNA into female A. aegypti mosquitos. J Vis Exp. 2007;5:215. doi: 10.3791/215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Arias A. Development and patterning of the larval epidermis of Drosophila. In: Bate M, Martinex Arias A, editors. The development of Drosophila melanogaster. I. Cold Spring Harbor Laboratory Press; Plainview, NY: 1993. pp. 517–608. [Google Scholar]
- Moreira LA, Ghosh AK, Abraham EG, Jacobs-Lorena M. Genetic transformation of mosquitoes: a quest for malaria control. Int J Parasitol. 2002;32:1599–1605. doi: 10.1016/s0020-7519(02)00188-1. [DOI] [PubMed] [Google Scholar]
- Moretti LJ, Larsen JR. Embryology of Aedes vexans. In: Horsfall WR, Fowler HW, Moretti LJ, Larsen JR, editors. Bionomics and embryology of the inland flood water mosquito aedes vexans. University of Illinois Press; Champaign, IL: 1973. pp. 137–206. [Google Scholar]
- Munstermann L, Craig GB., Jr Genetics of Aedes aegypti: updating the linkage map. J Hered. 1979;70:291–296. [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi Z, Megy K, Grabherr M, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel N. In situ hybridization to whole mount Drosophila embryos. In: Krieg PA, editor. A laboratory guide to RNA: isolation, analysis, and synthesis. Wiley-Liss; New York, NY: 1996. pp. 357–370. [Google Scholar]
- Patel NH. Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. Methods Cell Biol. 1994;44:445–487. doi: 10.1016/s0091-679x(08)60927-9. [DOI] [PubMed] [Google Scholar]
- Rai KS, Black WC., 4th Mosquito genomes: structure, organization, and evolution. Adv Genet. 1999;41:1–33. doi: 10.1016/s0065-2660(08)60149-2. [DOI] [PubMed] [Google Scholar]
- Raminani LN, Cupp EW. Embryology of Aedes aegypti (L.) (Diptera: Culicidae): organogenesis. Int J Insect Morphol & Embryol. 1978;7:273–296. [Google Scholar]
- Raminani LN, Cupp EW. Early embryology of Aedes aegypti (L.) (Diptera: Culicdae) Int J Insect Morphol Embryol. 1975;4:517–528. [Google Scholar]
- Robinson AS, Franz G, Atkinson PW. Insect transgenesis and its potential role in agriculture and human health. Insect Biochem Mol Biol. 2004;34:113–120. doi: 10.1016/j.ibmb.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Scott TW, Takken W, Knols BG, Boete C. The ecology of genetically modified mosquitoes. Science. 2002;298:117–119. doi: 10.1126/science.298.5591.117. [DOI] [PubMed] [Google Scholar]
- Severson DW, Knudson DL, Soares MB, Loftus BJ. Aedes aegypti genomics. Insect Biochem Mol Biol. 2004a;34:715–721. doi: 10.1016/j.ibmb.2004.03.024. [DOI] [PubMed] [Google Scholar]
- Severson DW, DeBruyn B, Lovin DD, Brown SE, Knudson DL, Morlais I. Comparative genome analysis of the yellow fever mosquito Aedes aegypti with Drosophila melanogaster and the malaria vector mosquito Anopheles gambiae. J Hered. 2004b;95:103–113. doi: 10.1093/jhered/esh023. [DOI] [PubMed] [Google Scholar]
- Severson DW, Meece JK, Lovin DD, Saha G, Morlais I. Linkage map organization of expressed sequence tags and sequence tagged sites in the mosquito, Aedes aegypti. Insect Mol Biol. 2002;11:371–378. doi: 10.1046/j.1365-2583.2002.00347.x. [DOI] [PubMed] [Google Scholar]
- Simanton W, Clark S, Clemons A, Jacowski C, Farrell-VanZomeren A, Beach P, Browne WE, Duman-Scheel M. Conservation of arthropod midline netrin accumulation revealed with a cross-reactive antibody provides evidence for midline cell homology. Evol Dev. 2009;11:260–268. doi: 10.1111/j.1525-142X.2009.00328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z, Ramirez JL, Dimopoulos G. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog. 2008;4:e1000098. doi: 10.1371/journal.ppat.1000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Shin SW, Alvarez KS, Bian G, Kokoza V, Raikhel AS. Mosquito RUNX4 in the immune regulation of PPO gene expression and its effect on avian malaria parasite infection. Proc Natl Acad Sci U S A. 2008;105:18454–18459. doi: 10.1073/pnas.0804658105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Useful web resources
- http://aaegypti.vectorbase.org/index.php Vectorbase website, which includes sequence data, expression data, images, and other additional resources pertaining to Ae. aegypti.
- http://www.cdc.gov/Dengue/ CDC website on dengue, which includes travel/outbreak notices, prevention, symptoms, epidemiology and statistics, entomology and ecology, clinical/laboratory guidance, education and training.
- http://www.cdc.gov/ncidod/dvbid/yellowfever/ CDC website for yellow fever, which includes information about prevention, vaccination, transmission, symptoms, diagnostic testing, health care provider links and references, and travel.


