Summary
CD4 T cells, also known as T helper (Th) cells, play an important role in orchestrating adaptive immune responses to various infectious agents. They are also involved in the induction of autoimmune and allergic diseases. Upon T cell receptor (TCR)-mediated cell activation, naïve CD4 T cells can differentiate into at least four major lineages, Th1, Th2, Th17 and iTreg cells, that participate in different types of immune responses. Networks of cytokines and transcription factors are critical for determining CD4 T cell fates and effector cytokine production. Here we review collaboration and cross-regulation between various essential cytokines in the activation / induction of key transcription factors during the process of Th cell differentiation towards these distinct lineages. We also discuss the interactions of key transcription factors at both genetic and protein levels and the function of the resulting network(s) in regulating the expression of effector cytokines.
Keywords: T helper cells, cytokines, transcription factors, T cell differentiation
Introduction
Upon activation by T cell receptor (TCR)- and cytokine-mediated signaling, naïve CD4 T cells may differentiate into at least four major types of T helper (Th) cells, Th1, Th2, Th17 and inducible T regulatory (iTreg) cells, that play a critical role in orchestrating adaptive immune responses to various microorganisms (1). They can be distinguished by their unique cytokine production profiles and their functions: Th1 cells predominantly produce interferon (IFN) γ and are important for protective immune responses to intracellular viral and bacterial infection; Th2 cells, by producing interleukin (IL)-4, IL-5, IL-9, IL-13 and IL-25, are critical for expelling extracellular parasites such as helminths; Th17 cells are responsible for controlling extracellular bacteria and fungi through their production of IL-17a, IL-17f and IL-22; inducible T regulatory (iTreg) cells, together with natural-occurring T regulatory (nTreg) cells, are important in maintaining immune tolerance, as well as in regulating lymphocyte homeostasis, activation and function.
The major determinant for Th cell differentiation is the cytokine milieu at the time of antigen encounter, although the nature of cognate antigen and its affinity to the TCR as well as the available co-stimulants, many of which regulate initial cytokine production, can influence Th cell fate. IL-12 and IFNγ are two important cytokines for Th1 differentiation. For Th2 differentiation, many cytokines including IL-4, IL-2, IL-7 and thymic stromal lymphopoietin (TSLP) may be involved. While transforming growth factor (TGF) β induces Th17 differentiation in the presence of IL-6, it also promotes iTreg cell differentiation when IL-2 but not IL-6 is available. In general, more than one cytokine is required for differentiation to any particular phenotype and cytokines that promote differentiation to one lineage may suppress adoption of other lineage fates. Furthermore, cells simultaneously or sequentially exposed to cytokine mixtures whose components that would normally induce different lineage fates may acquire a mixed Th phenotype. Thus, Th cell differentiation involves a complex cytokine network; this is particularly true in vivo.
Transcription factors are critical for Th cell differentiation and cytokine production. Cell fate determination in each lineage requires at least two types of transcription factors, the master regulators and the signal transducer and activator of transcription (STAT) proteins. The activity of the master regulators is controlled by their expression, whereas STAT proteins are activated by cytokines through post-transcriptional modifications such as phosphorylation. Some STAT proteins are responsible for inducing the expression of master regulators. In addition, the same STATs and the master regulators often collaborate in regulating cytokine production by directly acting on cytokine genes. The essential transcription factors of Th lineages are T-bet/Stat4 (Th1), GATA-3/STAT5 (Th2), RORγt/STAT3 (Th17), and Foxp3/STAT5 (iTreg), respectively.
Two factor-fate determination is an over-simplified model. Many other transcription factors are also involved in either regulating or fine-tuning the differentiation processes. In addition, transcription factors interact with each other at genetic and protein levels, forming a sophisticated transcriptional regulatory network.
The activation, differentiation and expansion of Th cells are tightly regulated by the activity and relative expression of specific transcription factors. Inappropriate regulation of the Th response to pathogens may lead to chronic infection, whereas an uncontrolled response causes excessive self-tissue damage. An undesirable activation of Th1, Th17 or Th2 cells can also result in organ-specific autoimmune diseases or allergic inflammatory diseases and asthma. Therefore, understanding the networks of cytokines and transcription factors during CD4 T cell differentiation is critical for diagnosis and treatment of many immune-related human diseases.
Networks of cytokines determines CD4 T cell fates
Cytokine-mediated positive feedback loops
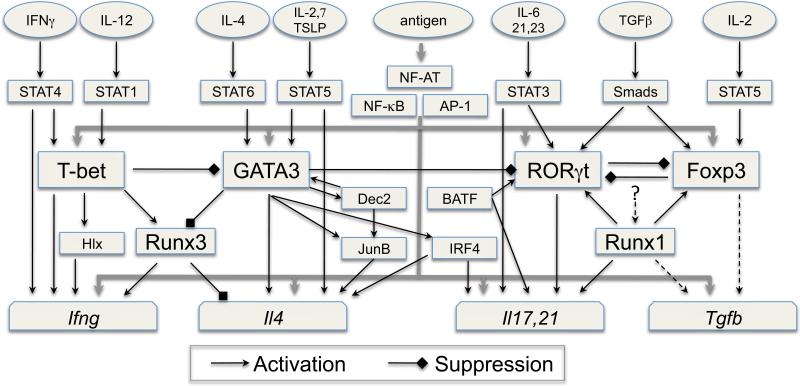
When stimulated by a cognate antigen presented by accessory cells in the presence of IL-4, naïve CD4 T cells differentiate into IL-4-producing Th2 cells (2-5). Similarly, IFNγ is important for the differentiation of IFNγ-producing Th1 cells (6) although IL-12 is also critical for this process (7). The combination of TGFβ and IL-6 was first shown to induce Th17 differentiation in vitro (8-10) but later it was reported that IL-21, a cytokine produced by Th17 cells, could replace IL-6 for the induction of Th17 differentation (11-13). The combination of TGFβ and IL-2 induces iTreg differentiation (14-18) and similar to nTregs, iTregs produce TGFβ. Therefore, an important principle for Th cell differentation is that one of the signature cytokines produced by each differentiated cell also plays a critical role in the induction of such cells potentially providing a powerful positive feedback loop. These “feedback” cytokines are IFNγ for Th1, IL-4 for Th2, IL-21 for Th17 and TGFβ for iTregs (Fig.1).
Fig. 1. The Network of transcription factors in CD4 T cells.
Together with transcription factors such as NF-AT, NF-κB and AP-1 activated and/or induced by TCR-mediated signaling, the activation of STAT proteins by different cytokines plays a critical role in inducing the expression of the lineage-specific master regulators T-bet (Th1), GATA3 (Th2), RORγt (Th17) and Foxp3 (Treg). The STAT proteins also collaborate with the master regulators, the transcription factors activated/induced by TCR and some secondary transcription factors, whose expression is controlled by the master regulators, for the induction of cytokine genes. Positive or negative regulation among these transcription factors occurs at the gene expression level and/or at the protein level through protein-protein interaction, forming a sophisticated transcriptional regulatory network during Th cell differentiation. The transcription factor complexes regulate chromatin remodeling and expression of the lineage-specific cytokines; one of the lineage-specific cytokines re-enforces further differention of such cells through a positive feedback loop (note: regulation of TGFβ expression may not be at transcriptional level).
To determine the importance of these potential auto-regulatory loops, knowing the pattern of cytokine production shortly after T cell activation of naïve CD4 T cells is very critical. This has been studied in considerable detail for in vitro Th2 differentiation. It has been shown that the strength of TCR signaling regulates initial cytokine production: low concentrations of cognate peptide induce IL-4-independent IL-4 production during the first 24 hours after T cell engagement whereas stimulation with high concentration of peptide suppresses “early” IL-4 and induces IFNγ (19-21). In addition to Th signature (feedback) cytokines, IL-2 and possibly the genetically adjacent IL-21 are also induced in naïve CD4 T cells upon T cell activation. Since IL-2 is important for initial IL-4 production (20), whereas IL-21 plays a role in inducing Th17 (11-13) and Tfh cells (22-24), regulation of the production of these cytokines in naïve T cells becomes an important subject to study.
Cytokine-mediated cross-regulation of Th cell differentiation
Cytokines that are important for Th cell differentiation towards one fate often suppress the development of other Th lineages. For example, IL-4, the important Th2 inducer, can suppress Th1, Th17 and Treg cell differentiation, partly due to STAT6-mediated upregulation of the transcriptional repressor, Gfi-1 (25, 26). TGFβ promotes the development of Th17 (8-10) or Treg cells (9, 14) but it suppresses Th1 and Th2 cell differentiation (27-29). While IL-2 is important for both Th2 and Treg cell differentiation, it represses Th17 cell differentiation (30). The cross-regulation among the cytokines may occur at transcriptional level when a transcriptional repressor or an inhibitor of signaling is induced. It is also likely that cytokines regulate each other's signaling by crosstalking through their shared signaling molecules. Furthermore, crosstalks between signaling triggered by TCR and cytokines exist. TCR activation transiently downregulate cytokine signaling (31); however, whether signals triggered by cytokines modulate TCR-mediated signal transduction is less clear.
Functions of cytokines at effector stages
At the effector stage of Th cell differentiation, each Th lineage preferentially expresses a distinct IL-1 family receptor, IL-18R1 for Th1, IL-33R (T1/ST2) for Th2 and IL-1R for Th17 (32). Stimulation of differentiated Th1 cells with IL-18 plus IL-12 induces TCR-independent IFNγ production (32-35). Similarly, IL-33 plus IL-2, IL-7 or TSLP induces IL-13 production in Th2 cells (32). IL-23 is important for full differentiation and maintenance of Th17 cells (36), although it is not involved in the initial differentiation stage due to lack of IL-23R expression on naive CD4 T cells (8, 9, 37). IL-1 plus IL-6, IL-21 or IL-23 induces IL-17 production in Th17 cells (32). TCR-independent effector cytokine production by Th cells induced by pro-inflammatory cytokines that are possibly produced by epithelial cells at infection sites suggests Th cells may function as innate-like effector cells when the appropriate cytokine milieu is created in their environment.
Cytokine networks: one or two cytokines cannot explain everything
As we have described, for each Th lineage differentiation, more than one cytokine is involved. For example, IL-4 alone is not suffient to induce Th2 differentiation in the absence of a STAT5 activator such as IL-2 (38, 39). TGFβ in combination with different cytokines can induce distinct Th cell fates: together with IL-2, TGFβ induces Treg differentation (15); with IL-6, it causes Th17 differentiation (8-10); with IL-4, it promotes the generation of IL-9-producing Th9 cells (40, 41). It is possible that TGFβ has other functions in the presence of other cytokines, such as IL-12 or IFNγ. IL-1 and TNFα are important inflammatory cytokines and IL-1 has been shown critical for the differentiation of Th17 cells, especially in humans (33, 42, 43). IL-1 is also a powerful adjuvant for a variety of Th responses by directly acting on CD4 T cells (44). Since IL-1 and tumor necrosis factor (TNF) α are produced at early stages of most immune responses, their effects on Th1, Th2 and Treg cell differentiation should also be carefully studied.
For in vitro cell differentiation, one can create a precise cytokine environment by adding exogeneous cytokines and antibodies to other cytokines; however, in vivo, naïve CD4 T cells may encounter different environments characterized by cytokines that are produced by accessory cells and by CD4 T cells themselves. Many of these cytokines have never been studied in detail in vitro for their effects on T cell differentiation and some of them may have opposite effects on a particular Th cell differentiation. Indeed, sequential exposure of both mouse and human CD4 T cells to IL-4 and IL-12 results in IL-4/IFNγ double-producing cells in vitro (45, 46). Therefore, in vivo Th cell differentiation is a complex process controlled by the network of cytokines.
A network of transcription factors determines CD4 T cell fates
Master regulators and their induction
As discussed above, cytokine signaling during activation of naïve CD4 T cells by cognate antigen plays a critical role in determining the fate of the responding CD4 T cells. STAT proteins are the major molecules that mediate cytokine signaling. Activated STAT molecules can induce the expression of the master regulators that determine Th cell fates. However, such induction does not occur without TCR signaling implying that the transcription factors activated by TCR, such as NFAT, NF-κB and AP-1, play important roles in regulating the expression of master regulators (Fig. 1).
T-bet
T-bet is regarded as the master regulator for Th1 cell differentiation and IFNγ production (47). T-bet deficient (Tbx21-/-) cells produce diminished but measurable amounts of IFNγ during in vitro culture and in in vivo responses to Leishmania major infection (48).
STAT1 activated by IFNγ has been shown to induce T-bet expression during Th1 differentiation in vitro (6, 49). Therefore, the IFNγ-STAT1-T-bet-IFNγ pathway serves as a powerful amplification mechanism for in vitro Th1 differentiation. In vivo, Stat1-/- CD4 T cells from T. gondii-infected mice express lower amounts of T-bet than that do infected wild-type mice; however, normal levels of IFNγ can be detected in the serum of Stat1-/- mice (50).
STAT4 activation by IL-12 is critical for Th1 responses both in vitro and in vivo (51-53) and the expression level of STAT4 is higher in Th1 than that in Th2 cells (54). The IL-12-STAT4 pathway is also responsible for up-regulating T-bet expression (54-56), although STAT4 may induce IFNγ production independently of T-bet. The relative importance of the IL-12/STAT4 and IFNγ/STAT1 pathways in inducing T-bet expression and IFNγ production in different in vivo Th1 models needs to be further studied.
It has been shown that TCR activation alone is sufficient to induce T-bet expression in human naïve CD4 T cells after 8-16 hr of stimulation; both IFNγ and IL-12 enhance such induction (57). Three DNase I hypersensitivity (HS) sites in the Tbx21 gene are induced during Th1 differentation, one of which is at Tbx21 promoter and is only weakly accessible in unstimulated naïve CD4 T cells. NFAT binds to the HS site that is located 7.5 Kb upstream of the transcription start site of Tbx21 whereas STAT4 binds to the HS site that is 12 Kb upstream of the start site. STAT1 has also been found to bind this distant enhancer to which STAT4 binds (55).
GATA3
GATA3 is the Th2 master regulator (45, 58-60) but it also plays important roles at multiple steps of CD4 T cell development (61). Th2 differentiation is completely abolished both in vitro and in vivo when GATA3 is conditionally deleted in peripheral CD4 T cells (45, 60). IL-4-mediated STAT6 activation is important for Th2 differentiation (62-64). A constitutively active form of STAT6 or tomoxifen-induced dimerization of a STAT6-estrogen receptor fusion protein induces GATA3 expression in the absence of IL-4 signaling (65, 66) suggesting that the IL-4-STAT6 pathway is necessary and sufficient for GATA3 upregulation in vitro when T cells are activated through TCR.
Although some in vivo Th2 responses such as that to Trichuris muris infection require the engagement of the IL-4-STAT6 pathway (67), STAT6-independent in vivo Th2 differentiation can also be obtained (68-72). Since the IL-4-independent Th2 response to Nippostrongylus brasiliensis still requires GATA3, this result suggests either that GATA3 can be upregulated by signaling pathways other than IL-4/STAT6 or that GATA3 upregulation is not essential for Th2 responses, with basal levels being sufficient under certain circumstances. Indeed, a constitutively activated STAT5 is able to induce IL-4-producing capacity without upregulating GATA3 expression (39), although this constitutively active STAT5 fails to induce IL-4-producing capacity in a Gata3-deficient setting.
Stimulation of naïve CD4 T cells with low-dose peptide induces TCR-driven IL-4-independent GATA3 expression (20). How such GATA3 induction is achieved is not certain. It has been shown that NF-κB (p50) is important for GATA3 expression (29). However, whether p50 directly regulates GATA3 especially at early stage of T cell activation has not been determined.
GATA3 may also induce its own expression when it reaches certain threshold (73). Our unpublished GATA3 ChIPseq data show that GATA3 can bind many sites within its gene body and up to 1 Mb downstream of Gata3 transcription start site suggesting that the regulatory elements for GATA3 expression may be far from each other. Furthermore, it has been recently reported that GATA3 and Dec2, another transcription factor, can form a positive regulatory loop during Th2 differentiation and that Dec2 binds to the Gata3 promoter (74). In the absence of Dec2, Th2 responses are diminished and there is a reduction of GATA3 and JunB expression. GATA3 and Dec2 may collaborate in JunB induction. It is not clear how Dec2 is initially induced but GATA3 seems to be dispensable for its induction in Th2 cells. Although STAT5 activation does not affect initial GATA3 induction, it is important for maintaining the expression of GATA3 in differentiated Th2 cells (32).
RORγt
RORγt is the master regulator for Th17 cells (75). RORγt-deficient mice produce diminished amounts of IL-17 and are partially resistant to EAE induction. TGFβ plus IL-6 induce RORγt in CD4 T cells that are being activated through their TCR. Three STAT3 activators, IL-6, IL-21 and IL-23, play critical roles in differentiation, amplification and maintenance of Th17 cells (8-13, 30, 76-79). STAT3 is required for the induction of RORγt and STAT3 directly binds to Rorc gene (80). Interestingly, BATF, a transcription factor belonging to the AP-1 family, is also necessary for RORγt induction (81). Runx1 has been reported to induce optimal RORγt expression (82). How TCR- and cytokine-mediated signaling regulate the expression and/or activation of BATF and Runx1 during Th17 differerentiation is not clear.
Foxp3
Foxp3 is the master regulator for both iTregs and nTregs (83-85). IPEX patients and Scurfy mice that carry mutations in Foxp3 have no or reduced functional Tregs (86-88). Naïve CD4 T cells stimulated through their TCR and TGFβR can develop into Foxp3+ Tregs (14). In humans, TCR activation is able to transiently induce Foxp3 expression consistent with the binding of NFAT and AP-1 at the promoter of FOXP3 gene (89, 90). In mice, collaboration between NFAT and Smad3, activated by TCR and TGFβ, respectively, is important for Foxp3 induction; NFAT and Smad3 interact with the conserved non-coding sequence (CNS) 1 located in the second intron of the Foxp3 gene (91). CNS1 deletion results in reduced Foxp3 induction in the periphery but not in the thymus (92). STAT5 activation is critical for Treg development and activated STAT5 directly binds to the Foxp3 promoter and to CNS2 in the 3’ region of the second intron (15-18).
The Foxp3/Runx1 complex is important for maintaining Foxp3 expression in nTregs through its binding to CNS2, suggesting that there is an auto-regulatory loop for Foxp3 expression (92). Indeed, the expression of a Foxp3 reporter in Foxp3-deficient mice was found to be unstable (93). Interestingly, CpG islands within Foxp3 CNS2 are methylated in iTregs and Foxp3 fails to bind to methylated CNS2 (92); this may explain why nTreg cells are more stable than iTregs in their expression of Foxp3. Our unpublished data suggest GATA3 also binds to Foxp3 CNS2; GATA3 deficiency in Tregs results in an unstable phenotype.
The NF-κB family member c-Rel is highly expressed in both nTregs and iTregs (94). c-Rel has been found to bind to CNS3 in intron 4 of the Foxp3 gene, the only CNS that is accessible in non-Tregs (92). In addition to its binding to the CNS3, c-Rel also binds to the Foxp3 promoter, and thus, c-Rel KO cells fail to give rise to Tregs (92, 95, 96).
Foxo proteins have been recently reported to play a critical role in the development of nTregs and in TGFβ-induced Foxp3 exprssion. Foxo binds to both the Foxp3 promoter and CNS2 and the Foxo-binding site near the transcription start site seems to be critical for the Foxp3 promoter activity (97, 98).
Foxp3 expression is not sufficient for expression of all the Treg functions; other transcription factors, such as Runx, are required. As discussed above, Foxp3 and Runx1 form a complex that is important for maintaining Foxp3 expression. In addition, this complex both positively regulates Treg-specific gene expression (99-102) and negatively regulates genes not expressed in Tregs, such as IL-2 (103). The Ikaros family protein Eos also regulates the repressive functions of Tregs possibly through protein-protein interactions with Foxp3 (104). Another Ikaros family protein, Helios, is expressed in nTregs but not in iTregs (105); however, the function of Helios in nTregs has not been determined. Some Treg functions, such as controlling Th2-related immune pathology, require the interaction of Foxp3 with IRF4 (106).
Cross-regulation among the key transcription factors
The adoption of one lineage fate during T cell differentiation is usually accompanied by suppression of the alternative fates the precursor could have assumed. Such suppression is mainly achieved by cross-regulation among transcription factors at both transcriptional and post-transcriptional levels.
For example, T-bet is not only important for Th1 differentiation, it is also critical for suppressing Th2 responses. Overexpressing T-bet suppresses GATA3 expression (56). By contract, in the absence of T-bet, GATA3 and IL-4 are upregulated even under a strong Th1-biased environment suggesting that T-bet normally suppresses Th2 differentiation (107). Consistent with this idea, Tbx21-/- mice have been reported to develop spontaneous airway hypersensitivity and T-bet-expressing CD4 T cells in asthmatic patients are reduced in frequency (108). T-bet also participates in suppressing Th17 responses since Tbx21-/- cells can produce IL-17 even when cultured under Th1 conditions. IL-17 production is further enhanced when both Eomes, another T-box family protein, and T-bet are absent. Eomes is responsible for T-bet-independent IFNγ production in CD8 T cells (109). Consequently, T-bet/Eomes double-deficient mice develop a wasting disease in response to LCMV infection owing to over-production of IL-17 (110). It has been recently reported that T-bet induction in IL-12-treated Th17 cells silences RORγt. T-bet is bound to a site near the Rorc second exon in IL-12-treated Th17 cells (111). However, our T-bet ChIPseq data showed that T-bet binds to a different site at Rorc locus in Th1 cells suggesting that repression of RORγt in Th1 cells may proceed by a different T-bet dependent pathway.
Not only does T-bet regulate other lineage genes at the transcriptional levels, it also controls the activity of other transcription factors through protein-protein interactions. Tyrosine-phosphorylated T-bet interacts with GATA3 and suppresses the function of GATA3 in inducing IL-5 production (112).
In addition, T-bet induces Runx3 in Th1 cells. Runx3 is an important transcriptional repressor that suppresses IL-4 production by directly binding to the Il4 locus (107, 113, 114). Runx3 has also been reported to repress GATA3 function possibly through its interaction with GATA3 protein (115).
GATA3 suppresses STAT4 expression in Th2 cells; GATA3-deficient CD4 T cells cultured under Th2-inducing conditions express higher levels of STAT4 than do wild-type Th2 cells (54, 107). We have recently reported that GATA3 interacts with Runx3 and inhibits Runx3-mediated IFNγ production (107). Both Runx3 and Eomes (109) are highly expressed in CD8 T cells. Indeed, Runx3 is responsible for Eomes upregulation and optimal IFNγ expression (116). Enforced expression of Runx3 in Th2 cells induces T-bet-independent Eomes and IFNγ expression (107). Deletion of the Gata3 gene from naïve CD4 or Th2 cells results in Eomes upregulation and IFNγ production independent of IL-12 and IFNγ signaling. The regulation of Eomes expression is complex since it is only expressed in a small subset of Th1 cells although all Th1 cells express high levels of Runx3 (107). T-bet and Eomes are inversely expressed in CD8 T cells over the course of a viral infection suggesting that T-bet may suppress Eomes expression (117). However, GATA3 and T-bet double-deficient CD4 T cells cultured under Th1-inducing condtions still fail to expess Eomes indicating that the expression of Eomes is either suppressed by IL-12 or activated by IL-4 (107). In addition, GATA3/Runx3/T-bet triple knockout “Th2” cells still express Eomes suggesting that Eomes can be induced independently of Runx3 and that this expression is normally inhibited by GATA3 in Th2 cells. Interestingly, IL-4 produced by NKT cells amplifies Eomes-expression in memory CD8 T cells (118). Therefore, IL-4 may enhance Eomes expression when GATA3 is present at low levels. GATA3 may also be involved in regulating Th17 responses since our unpublished data indicate that GATA3 directly binds to the Rorc locus and that deletion of Gata3 results in upregulation of RORγt expression.
Enforced expression of a constitutively active form of STAT5 in Th1 cells induces Th2 differentiation without upregulating GATA3. However, T-bet expression is suppressed in such cells suggesting that STAT5 signaling may affect Th1/Th2 differentiation by regulating the relative amount of GATA3 and T-bet, although it can directly act on the Il4 locus (39). STAT5 has also been shown to suppress Th17 cell differentiation through inhibiting RORγt upregualtion as well as IL-17 production (30).
Gfi-1, a transcription repressor that is induced by IL-4/STAT6 signaling, is important for Th2 cell growth (119). Interestingly, Gfi-1 suppresses Th1 (57), Th17 and iTreg differentiation (26). Ikaros is another transcriptional repressor that is involved in Th2 cell differentiation (120). A recent report further showed that Ikaros suppresses Th1 differentiation by inhibiting T-bet expression (121).
Whether or how RORγt suppress Th1 or Th2 differentiation has not been determined. However, RORγt overexpression in Th17 cells can prevent STAT4 activation and T-bet upregulation when such cells are treated with IL-12 (111). It has been recently shown that RORγt binds to the Foxp3 promoter and suppresses Foxp3 expression (122). IL-6, through STAT3 activation, has been shown to repress Foxp3 expression in Tregs (13, 78, 123-125). In addition, a dominant-negative STAT3 mutation in human result in the hyper-IgE syndrome (Job syndrome) suggesting that STAT3 activation, in addition to its role in Th17 induction, is also critical for suppressing Th2 responses (79, 126). TGFβ has been shown to inhibit both Th1 and Th2 differentiation, possibly through down-regulation of Th1 and Th2 master regulators, T-bet and GATA3 (27-29). TGFβ signaling also results in down-regulation of Gfi-1 expression (26). Since Gfi-1 suppresses both TGFβ-mediated Th17 and iTreg differentiation, downregulation of Gfi-1 by TGFβ is a critical step for TGFβ function. How TGFβ downregulates T-bet, GATA3 and Gfi-1 is not clear.
Foxp3 binds to both NFAT (127) and Runx1 (103), through which it suppresses IL-2 production by Treg cells. Foxp3 and RORγt co-expressing cells are found both in the gut and in TGFβ-treated cells in vitro. Foxp3 serves as an inhibitory factor for IL-17 production through its interaction with RORγt (123). T-bet is found to be co-expressed with Foxp3 (128) and our unpublished data suggest GATA3 is expressed by Foxp3+ cells. However, T-bet and GATA3 are not able to induce Th1 and Th2 cytokines in these Tregs, suggesting that Foxp3 suppresses their activity.
Transcription factor-mediated cytokine production
The main functions of Th cells are executed through production of effector cytokines. Master transcription factors collaborate with many other transcription factors including STAT proteins during the remodeling of cytokine loci and induction of cytokine transcription. Restimulation of differentiated Th cells through their TCR is one of the principal mechanims through thich these cells are induced to produce cytokines; transcription factors that are activated by TCR-mediated signaling, including NFAT, NF-κB and AP-1 participate in this process.
Chromatin remodeling and epigenetic modifications at cytokine loci presumably result from the upregualtion or activation of certain lineage specific transcription factors (129-131). Indeed, the master regulators GATA3 and T-bet have been shown to induce chromatin remodeling and epigenetic modifications at several cytokine loci (73, 132-134). Epigenetic modifications include DNA methylation at CpG islands, methylation and acetylation at various positions of the histones that pack DNA into nucleosomes.
Key transcription factors can affect epigenetic modifications through recruiting of or competing with chromatin remodeling complexes that bind to key regulatory elements. For example, T-bet recruits histone H3-K4 methyltransferase and H3-K27 demethylase complexes to the Ifng locus through its interaction with RbBp5 and JMJD3, respectively (135). Trimethylation of histone H3-K4 (H3K4me3) is usually associated with gene activation whereas trimethylation of histone H3-K27 (H3K27me3) is linked to gene repression. Genome-wide profiling of H3K4me3 and H3K27me3 by ChIPseq (chromatin immunoprecipitation followed by high-throughput sequencing) has confirmed that H3K4me3 modifications are present at each effector cytokine locus in cells that are capable of expressing that cytokine, whereas H3K27me3 modifications are found at loci of many cytokines that are not expressed in these cells (94). GATA3 has been reported to compete with a methyl CpG-binding domain protein-2 (MBD2) for binding to a particular methyl CpG in the Il4 locus and cells deficient in the DNA methyltrasferase Dnmt-1 or methyl-CpG binding domain protein 2 (MBD2) produce IL-4 without need for GATA3 upregulation (136, 137).
Epigenetic modifications on DNA or histones can affect the accessibility of the DNA and are associated with the acquisition of DNase I HS and with transcription factor binding. NFAT family proteins are commonly expressed and activated across all the lineages; however, their binding to the Il4 or the Ifng promoters are different in Th1 and Th2 cells because the accessibility of these two sites to NFAT is determined by epigenetic modifications at these cytokine loci (138). The induction of lineage-specific cytokine production is mediated by the binding of transcription factors to the cytokine loci at promoter and enhancer regions, many of which elements co-localize with DNase I hypersensitivity sites. Generally, these regions locate at CNSs containing multiple transcription factor binding sites.
Ifng
Many transcription factors, such as T-bet, STAT4, STAT5 and CTCF have been shown to bind to the promoter of Ifng (47, 114, 139-141). T-bet also collaborates with other transcription factors such as Hlx (142) and Runx3 (114), both of which are upregulated by T-bet, in inducing IFNγ production. Inhibition of Runx3 activity by a dominant-negative form of the factor or by gene deletion results in reduced IFNγ production by Th1 cells (107, 113, 116). Runx3 also binds to the Ifng promoter (107).
In addition to the promoter, the regulatory elements at the Ifng locus within ~140 Kb flanked by two insulators, that bind to CTCF, have been mapped and characterized through comprehensive epigenetic analyses and functional studies (129, 143). The T-bet responsive elements are identified at IfngCNS-22 and IfngCNS-34, CNSs located at 22 Kb and 34 Kb upstream of the Ifng transcription start site, respectively. Runx3 and STAT4 also bind to these CNSs (107, 111). IfngCNS-22 deletion results in redued expression of IFNγ in both T and NK cells (144). Our recent study showed that the Runx complex but not T-bet binds to IfngCNS-6, the site that has been shown to contain enhancer activity (145, 146). Interestingly, the Ifng locus has undergone substantial STAT4- and T-bet-independent chromatin remodeling at different CNSs even in Th17 cells, in which IFNγ is barely detectable. However, IfngCNS-6 remains inaccessible in Th17 cells suggesting this site plays a critical role in inducing IFNγ production (111). T-bet also synergizes with STAT4 to induce many other Th1-specific genes such as IL-18R1 and IL-12Rβ2, although the expression of the Th1-specific chemokine receptor, CXCR3, is T-bet-dependent but STAT4-independent (140).
Il4/Il13
Although many transcription factors, including c-Maf, JunB, IRF4 and NFAT, bind to the Il4 promoter (147-151), GATA3 does not bind to it consistent with the observation that deletion of Gata3 in established Th2 cells results in only modest reduction of IL-4 production (45). However, the expression of IL-5 and IL-13 completely depends on GATA3, consistent with the observations that GATA3 binds to the Il5 promoter (59) and to the CGRE region of the Il13 promoter (152) as reported earlier and confirmed by our recent unpublished GATA3 ChIPseq data.
Besides the DNase I hypersensitivity region in the Il4 promoter, which is designated HSI, other Il4 locus HSs have been identified in both Th1 and Th2 cells (153). HSII, located in the intron 2 of Il4 gene, has been shown to have strong enhancer activity (154). Our previous report showed that STAT5 binds to this enhancer in Th2 cells (39). Interestingly, histone H3 is K4 trimethylated in Th2 cells at HSII(94); H3K4me3 is a histone modification usually found in the promoter of an active gene. Our GATA3 ChIPseq data show that GATA3 also strongly binds to the HSII region. Collectively, these data indicate that the HSII region is critical for regulating IL-4 expression; however, this has not been tested genetically.
GATA3 expression and STAT5 activation are two critical events for IL-4 production and Th2 differentiation (155). STAT5 is activated by a variety of cytokines, among which are IL-2, IL-7 and TSLP, which act on CD4 T cells (156). Enhanced STAT5 signaling by enforced expression of a constitutively active form of STAT5a results in Th2 differentiation without upregulating GATA3 expression (39), but such Th2-inducing activity of STAT5 is lost in GATA3 KO cells indicating that GATA3 is required even if expressed at low levels (45). This suggests that high level of activated STAT5 may reduce the required level of GATA3 for the induction of IL-4 transcription. On the other hand, IL-4 cannot be induced without STAT5 activation even when GATA3 is highly expressed (38). Thus, both GATA3 and STAT5 are required for inducing Th2 cytokines. They may collaborate through directly binding to common cytokine loci such as the HSII site in the Il4 second intron. STAT5 ChIPseq (157) and GATA3 ChIPseq reveal overlapping binding patterns of these two molecules at Th2 cytokine loci. GATA3 and STAT5 also collaborate in inducing the Th2 specific cytokine receptor, T1/ST2, also known as IL-33R (32).
HSVa, located at 3’ end of the Il4 gene, is another enhancer whose activity is induced by TCR stimulation, consistent with NFAT binding to this site (158). GATA3 has also been reported to bind to HSVa; our GATA3 ChIPseq data confirm such binding. Interestingly, HSVa is a critical element in determing stochastic expression of IL-4 (159). Deletion of HSVa and HSV, which also contains a Notch/CSL binding site (160), results in reduced IL-4 production. HSIV, located within the Il4 gene, is accessible both in Th1 and Th2 cells. HSIV appears to serve as a silencer; deletion of this site results in de-repression of IL-4 production in Th1 cells (138). Suppression of IL-4 in Th1 cells is partly mediated by Runx3 binding to HSIV region (113, 114).
The expression of IL-4 and IL-13 are usually co-regulated since the genes encoding these two proteins are linked to each other. There are some regulatory elements, such as CNS1 (161), located in the intergenic region of Il4 and Il13, and the locus control region (LCR) (162, 163) in the 3’ of Rad50 gene body, that are important for both IL-4 and IL-13 expression.
Il17a/Il17f
Both RORγt and STAT3 bind to multiple sites in the Il17a/Il17f locus including the promoters of these cytokines (80, 82, 164, 165). Deletion of either Stat3 or Rorc abolishes IL-17 production whereas enforced expression of RORγt and a constitutively active STAT3 have a synergistic effect on IL-17 expression. Runx1 and NFAT also bind to the Il17a promoters (82, 164, 165). RORγt requires the coexpression of Runx1 to induce optimal IL-17 production in Th17 cells (82).
Besides the promoters, eight CNS sites have been identified within the Il17a/Il17f locus; all are associated with acetylated histone H3 in Th17 cells (166). CNS2 (also called CNS-5), ~5kb upstream of the Il17a transcription start site, contains ROR response elements. Both RORγt and Runx1 bind to CNS2 contributing to IL-17a induction (82). BATF, which is a critical AP-1 factor for RORγt induction and IL-17 production, directly binds to the CNS3 site of the Il17a/Il17f locus, ~10 Kb downstream of the Il17a transcription start site (81). Interestingly, the CNS3 site is also bound by the Gfi-1/LSD1 complex, resulting in the suppression of IL-17 expression (26). Furthermore, STAT4 binds to CNS3 when Th17 cells are treated with IL-12 (111). Our unpublished data also show that both GATA3 and T-bet bind to to CNS3 in Th2 and Th1 cells, respectively. These data suggest that CNS3 may serve as a switch for turning on or off IL-17 production. The importance of these CNS sites within the Il17a/Il17f locus has yet to be determined by genetically.
Transcription factors regulate heterogeneity and plasticity
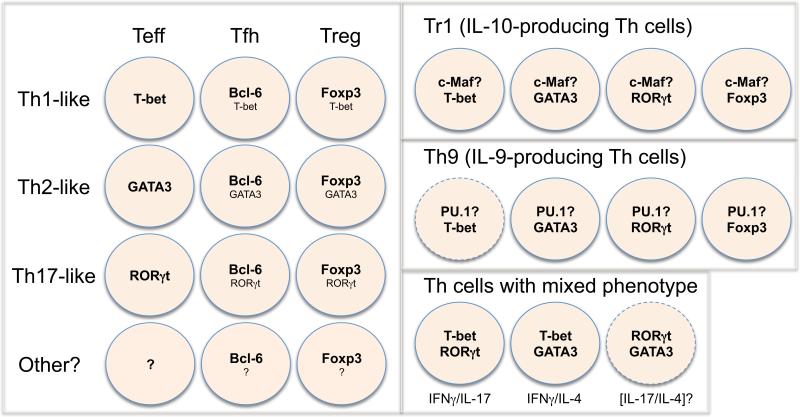
Th cells are more hetergenous and plastic than originally thought. Within each lineage, heterogeneity exists; some Th cells can be reprogrammed to express master regulators of other lineages. Such heterogeneity and plasticity may be explained by the finding that different transcription factors are co-expressed with the lineage's master regulator and sometimes even multiple master transcription factors are expressed in the same cell although at different levels (Fig. 2).
Fig. 2. Combinatorial expression of different transcription factors determines Th cell heterogeneity and plasticity.
Th cells are more heterogenous and plastic than originally thought. The heterogeneity and plasticity of these Th cells may be explained by the different combinations of expression of transcription factors in a single cell. Since many Tfh and Treg cells can exhibit Th1, Th2 or Th17-like phenotype, these cells may differentiate in parallel to conventional effector T (Teff) cells. For example, Th1-like Treg cells co-express Foxp3 and T-bet although the expression level of T-bet in such cells is lower than that in Th1 Teff cells. It has been reported that Tfh cells do not express T-bet, GATA3 or RORγt; however, “do not express” may actually be “express at lower level”. Alternatively, some Tfh cells may represent a different state of another unknown Teff cells whose master regulator is yet to be defined. Similarly, Tr1 (IL-10-producing cells) and Th9 (IL-9-producing cells) may represent certain subsets of Th1, Th2, Th17 or Treg cells, in which a transcription factor that is involved in IL-10 production (i.e. c-Maf) or in IL-9 production (i.e. PU.1) is co-expressed with other lineage-specific factors. Under certain circumstances, two cytokines of different lineages may be co-expressed in the same cells; this may be explained by the combinatorial expression of two master regulators (i.e. T-bet and GATA3 expression was found in IFNγ/IL-4 co-expressors).
Tfh Cells
Th1, Th2 and Th17 cells are three well-recognized effector CD4 T lineages. T follicular helper (Tfh) cells that reside in B cell follicle and are important for helping B cells make antibodies and for germinal center formation have been proposed to be a separate Th lineage (22-24). Alternatively, Tfh cells may be considered as a state of Th1, Th2 or Th17 cells since many recent reports have shown that Tfh cells can be generated during in vivo Th1, Th2 or Th17 responses and they express Th1-, Th2- or Th17-related cytokines (167-170). It is likely that some conventional Th1, Th2 or Th17 cells can develop into different Tfh cells when they receive appropriate signals from B cells; alternatively, cells may first adopt the Tfh phenotype and then further differentiate into distinct types of Tfh cells capable of producing Th1, Th2 or Th17 cytokines. The fact that, in the gut, Tfh cells can be derived from Tregs, suggests effector Th cells may also acquire Tfh function (171).
Stimulation of TCR-engaged CD4 T cells with IL-6 or IL-21 in the absence of TGFβ has been shown to induce Tfh cells that express IL-21 and CXCR5, the chemokine receptor responsible for cell homing to B cell follicle (23, 24, 172). In vivo, signals from the germinal center or B cells may be critical for Tfh cell differentiation (169, 170). The transcriptional repressor Bcl-6 is expressed in Tfh cells (173) and recent reports have shown that Bcl-6 is dispensable for Tfh cell differentiation (174-176). That endogenous Bcl-6 can be induced by enforced Bcl-6 expression indicates a powerful auto-regulatory positive feedback loop (174). Bcl-6 is responsible for the induction of Tfh-related molecules such as CXCR5 and PD-1 but it is not required for IL-21 production. Because there are Th1-, Th2- and Th17-like Tfh cells and the master regulators are absolutely required for effector cytokine prodution, i.e. GATA3 for IL-4, it is conceivable that Tfh cells may express T-bet, GATA3 or RORγt at a basal level in addition to Bcl-6 contributing to Th1, Th2 or Th17-like phenotype.
Tregs
More and more reports have shown that many transcription factors found in effector Th cells are also expressed in Tregs. Cells co-expressing RORγt and Foxp3 have been found in vivo both in mice and in humans (123). It has been proposed that such cells are precusors of Th17 and iTreg cells prior to their commitment to either lineage (123); however, it is now also clear that IL-6 can induce RORγt expression in Treg cells (124, 125). Interestingly, Foxp3+RORγt+ cells have regulatory functions (177, 178). Treg-specific deletion of Stat3 results in lose of the capacity of these Tregs to suppress Th17-related autoimmune diseases (179); however, whether STAT3 is responsible for upregulating RORγt and whether deletion of RORγt from Tregs will also induce Th17-mediated diseases is not certain. Some Treg cells express T-bet and these Treg cells may be critical for controlling Th1-related autoimmune diseases (128). Our unplished data suggest GATA3 is also expressed by Treg cells and that it plays an important role in maintaining Treg phenotype in an inflammatory environment. Therefore, Foxp3 can be co-expressed with T-bet, GATA3 and RORγt. It is likely that the ratio of Foxp3 expression to other transcription factors is critical for maintaining Treg function. Foxp3 may form transcription factor complexes with T-bet, GATA3 or RORγt so that the capacity of these factors to induce effector cytokines are inhibited while the expression of other genes usually found in the relevant effector cells, such as chemokine receptors, is preserved. The thresholds for master regulators to induce effector cytokines and to induce other molecules such as chemokine receptors may be quite different. Indeed, low levels of T-bet in Tregs are able to induce CXCR3 expression but not IFNγ production (128). Under cetain pathological or inflammatory situations, the expression of Th1, Th2 and Th17 master regulators may attain a certain threshold so that Foxp3 can no longer suppress the production of effector cytokines and such Tregs may convert into effector cells (180). Indeed, T-bet expression is upregulated in Tregs from mice infected with a lethal innoculum of Toxoplasma gondii; such Foxp3+ cells produce IFNγ (181). Interestingly, a Th1-like regulatory cell population expressing both T-bet and Foxp3 can produce IFNγ without losing its suppressive activity (182), but whether these two functions can be found in the same cell is not clear.
Many groups have reported other CD4 T cells such as Th3 (183), Th9 (40, 41), and Tr1 (184) cells that produce TGFβ, IL-9 and IL-10, respectively. Most of the Th3 cells are probably Foxp3-expressing Tregs. TGFβ-producing non-Tregs do exist, but the transcription factor(s) that regulates TGFβ expression has not be defined.
Th9 Cells
Addition of TGFβ and IL-4 during T cell activation induces Th9 cells (40, 41). A recent report showed that PU.1, a transcription factor selectively expressed in IL-4-non-producing Th2 cells (185), plays an important role in regulating IL-9 production although other factors also seem to be involved. Our unpublished data suggest that deletion of Gfi-1 from Th2 cells results in IL-9 production. Since PU.1 and Gfi-1 have been shown to antagonize each other in the determination of macrophage versus B cell fate (186), PU.1/Gfi-1 cross-regulation may also play an important role in regulating development of Th2 versus Th9 cells. Furthermore, our unpublished data indicate that GATA3 is required for the induction of IL-9-producing cells although it is expressed at lower levels in Th9 cells than that in Th2 cells. IL-9 production can also be induced in Th17 and Treg cells (187, 188) whose differentiation also involves TGFβ suggesting a transcription factor that is responsible for IL-9 production such as PU.1 may be coexpressed with other master regulators and this factor may be regulated by TGFβ signaling.
Tr1 cells
Most Th cells, including Th1, Th2, Th17 and Treg cells are able to produce IL-10 under certain circumstance; therefore, Tr1 cells may be a mixture of different cell types producing IL-10 and IL-10-inducing factors co-expressed with Th1, Th2, Th17 or Treg master regulators determine IL-10 production in different cell types. GATA3 contributes to IL-10 production in Th2 cells (our unpublished data) and Ikaros is also involved in IL-10 production (189). C-maf has been reported to regulate IL-10 expression in Th17 cells (190) and c-maf is also highly expressed in Th2 cells. These data suggest that Th2 cells may express a combination of IL-10-inducing factors and other transcription factor(s) that are responsible for IL-10 production in non-Th2 cells may also be found in Th2 cells.
Th Plasticity
Th cells are plastic. Both Th17 cells and Tregs cells can acquire the capacity to produce IFNγ if they are exposed to certain stimuli (94, 191). Recently, it has been reported that Th2 cells can also be taught to produce IFNγ without losing their capacity to produce IL-4 consistant with the co-expression of GATA-3 and T-bet in such cells, although their expression of GATA3 is lower than in “conventional” Th2 cells (192). The potential for “plastic” IFNγ-production may be imprinted in Th2, Th17 and Treg cells by bivalent histone H3 K4 and K27 trimethylation at the Tbx21 locus in these cells (94).
Thus, heterogeneity among Th cells of a given lienage is determined by expression of additional transcription factors besides the master regulators. Bivalent epigenetic modifications at the master regulator loci, such as Tbx21 and Gata3, may make it possible to induce these transcription factors in opposite lineages resulting in co-expression of multiple master regulators and change of their phenotypes. The relative amounts of these factors are important in determining the diversity and stability (or lack thereof) of the Th cell subsets.
Conclusions and perspectives
Naïve CD4 T cells have multiple fates that they may acquire when stimulated; the fate decision is regulated by the network of cytokines and transcription factors epressed in the activated cells. We are beginning to define the complexity of the cytokine and transcription factor networks as well as the diversity among Th cell subsets. There is still incertainty as to whether more Th cell subsets exist in vivo and, if so, how many. Reliable tools are needed to identify all the CD4 T cell subsets in vivo. Generation of multi-color indicator mice, in which the expression of all known lineage-specific master regulators are reported by different fluoresent proteins, would greatly benefit research on CD4 T cells.
The combination of cytokine signaling at different stages of T cell activation and the combination of transcription factors governing the differentiation processes are complex. A particular cytokine may have different function when it is combined with other cytokines. Since there are many cytokines being made at different levels during each immune response, fully understanding the differentiation process in vivo is difficult. Therefore, reconstitution of an in vivo differentiation in an in vitro culture remains a useful way to learn about cytokine networks. As our knowledge of the players grows, the conditions for in vitro differentiation cultures can be modified in such a way that it better represents an in vivo situation. Nonetheless, it must be recognized that many of the in vitro “networks” may be used rarely in vivo so that in vivo tests will be essential for a true understanding of the physiology and pathophysiology of CD4 T cell differentiation.
Due to the complexity of CD4 T cell population and their differentiation, the classical methods of studying individual factors one at a time, may be inefficient and sometimes misleading. This is because the function of any particular factor is always influenced by additional factors whose activation may vary under different conditions, and many biological responses are determined by quantitative changes in the expression of key factors. Therefore, a comparative analysis of different immune responses at a systemic level, in a quantitative way, is essential to yield a better understanding of the immune system.
Rapid advances in technology and genome informatics allow the performance of genome-wide studies. ChIPseq provide genome-wide maps of DNA binding sites of key transcription factors and transcription factor complexes. Profiling gene expression and mapping epigenetic modifications including DNA methylation, different histone modifications and appearance of DNase I hypersensitivity sites at a global level are essential to efficiently identify large numbers of cirtical cis-regulatory elements.
Quantitative measurements are also necessary to gain a complete picture of the transcriptional regulatory network and to understand how this network affects gene expression. The functions of many transcription factors depend on the amount of their expression relative to expression of other factors. Perturbation experiments, either knocking-down or enforced-expression in a titrated way, will yield valuable quantitative information on gene regulation.
Finally, mathematical models may be built to simulate the immune responses when the central pieces of the network including key transcription factors and crucial cis-regulatory elements of the target genes have been identified through genome-wide studies, and quantitative data sets are obtained through titrated pertubation of the key components.
Our ultimate goal of studying immune responses in mice is to help understand and treat human diseases. Indeed, some immune-related human diseases have been attributed to genetic mutations in key transcription factors, such as mutantions in FOXP3 that result in IPEX syndrome (87, 88) and STAT3 mutations that result in hyper-IgE syndrome associated with failure in Th17 responses to extracellular bacterial and fungal infections (79, 126). Many other immune-related diseases could also result from the mutations in particular cis-regulatory elements that are critical for the expression of key factors. Thus, understanding the molecular mechanisms, through which the network of transcription factors precisely control Th cell differentiation and Th cell heterogeneity, plasticity and stability, has great implications for understanding and treating a broad range of immune-related human diseases, including chronic viral, fungal, bacterial and parasitic infections, autoimmune diseases, allergic diseases and tumors.
Acknowledgments
This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health. We apoligize to these investigators whose work was not cited due to limitation of the space or our knowledge.
References
- 1.Zhu J, Paul WE. CD4 T cells: fates, functions, and faults. Blood. 2008;112:1557–69. doi: 10.1182/blood-2008-05-078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le Gros G, Ben-Sasson SZ, Seder R, Finkelman FD, Paul WE. Generation of interleukin 4 (IL-4)-producing cells in vivo and in vitro: IL-2 and IL-4 are required for in vitro generation of IL-4-producing cells. The Journal of experimental medicine. 1990;172:921–9. doi: 10.1084/jem.172.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swain SL, Weinberg AD, English M, Huston G. IL-4 directs the development of Th2-like helper effectors. J Immunol. 1990;145:3796–806. [PubMed] [Google Scholar]
- 4.Seder RA, Paul WE, Davis MM, Fazekas de St Groth B. The presence of interleukin 4 during in vitro priming determines the lymphokine-producing potential of CD4+ T cells from T cell receptor transgenic mice. The Journal of experimental medicine. 1992;176:1091–8. doi: 10.1084/jem.176.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsieh CS, Heimberger AB, Gold JS, O'Garra A, Murphy KM. Differential regulation of T helper phenotype development by interleukins 4 and 10 in an alpha beta T-cell-receptor transgenic system. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:6065–9. doi: 10.1073/pnas.89.13.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lighvani AA, et al. T-bet is rapidly induced by interferon-gamma in lymphoid and myeloid cells. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:15137–42. doi: 10.1073/pnas.261570598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsieh CS, Macatonia SE, Tripp CS, Wolf SF, O'Garra A, Murphy KM. Development of TH1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science. 1993;260:547–9. doi: 10.1126/science.8097338. [DOI] [PubMed] [Google Scholar]
- 8.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–89. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Bettelli E, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–8. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 10.Mangan PR, et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–4. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 11.Zhou L, et al. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nature immunology. 2007;8:967–74. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 12.Nurieva R, et al. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 2007;448:480–3. doi: 10.1038/nature05969. [DOI] [PubMed] [Google Scholar]
- 13.Korn T, et al. IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature. 2007;448:484–7. doi: 10.1038/nature05970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen W, et al. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. The Journal of experimental medicine. 2003;198:1875–86. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davidson TS, DiPaolo RJ, Andersson J, Shevach EM. Cutting Edge: IL-2 is essential for TGF-beta-mediated induction of Foxp3+ T regulatory cells. J Immunol. 2007;178:4022–6. doi: 10.4049/jimmunol.178.7.4022. [DOI] [PubMed] [Google Scholar]
- 16.Burchill MA, Yang J, Vogtenhuber C, Blazar BR, Farrar MA. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J Immunol. 2007;178:280–90. doi: 10.4049/jimmunol.178.1.280. [DOI] [PubMed] [Google Scholar]
- 17.Burchill MA, et al. Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity. 2008;28:112–21. doi: 10.1016/j.immuni.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao Z, et al. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007;109:4368–75. doi: 10.1182/blood-2006-11-055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Constant SL, Bottomly K. Induction of Th1 and Th2 CD4+ T cell responses: the alternative approaches. Annual review of immunology. 1997;15:297–322. doi: 10.1146/annurev.immunol.15.1.297. [DOI] [PubMed] [Google Scholar]
- 20.Yamane H, Zhu J, Paul WE. Independent roles for IL-2 and GATA-3 in stimulating naive CD4+ T cells to generate a Th2-inducing cytokine environment. The Journal of experimental medicine. 2005;202:793–804. doi: 10.1084/jem.20051304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tao X, Constant S, Jorritsma P, Bottomly K. Strength of TCR signal determines the costimulatory requirements for Th1 and Th2 CD4+ T cell differentiation. J Immunol. 1997;159:5956–63. [PubMed] [Google Scholar]
- 22.King C, Tangye SG, Mackay CR. T follicular helper (TFH) cells in normal and dysregulated immune responses. Annual review of immunology. 2008;26:741–66. doi: 10.1146/annurev.immunol.26.021607.090344. [DOI] [PubMed] [Google Scholar]
- 23.Vogelzang A, McGuire HM, Yu D, Sprent J, Mackay CR, King C. A fundamental role for interleukin-21 in the generation of T follicular helper cells. Immunity. 2008;29:127–37. doi: 10.1016/j.immuni.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Nurieva RI, et al. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29:138–49. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu J, Jankovic D, Grinberg A, Guo L, Paul WE. Gfi-1 plays an important role in IL-2-mediated Th2 cell expansion. Proc Natl Acad Sci U S A. 2006;103:18214–9. doi: 10.1073/pnas.0608981103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu J, et al. Down-regulation of Gfi-1 expression by TGF-beta is important for differentiation of Th17 and CD103+ inducible regulatory T cells. J Exp Med. 2009;206:329–41. doi: 10.1084/jem.20081666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorelik L, Fields PE, Flavell RA. Cutting edge: TGF-beta inhibits Th type 2 development through inhibition of GATA-3 expression. J Immunol. 2000;165:4773–7. doi: 10.4049/jimmunol.165.9.4773. [DOI] [PubMed] [Google Scholar]
- 28.Park IK, Letterio JJ, Gorham JD. TGF-beta 1 inhibition of IFN-gamma-induced signaling and Th1 gene expression in CD4+ T cells is Smad3 independent but MAP kinase dependent. Mol Immunol. 2007;44:3283–90. doi: 10.1016/j.molimm.2007.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorelik L, Constant S, Flavell RA. Mechanism of transforming growth factor beta-induced inhibition of T helper type 1 differentiation. The Journal of experimental medicine. 2002;195:1499–505. doi: 10.1084/jem.20012076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laurence A, et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26:371–81. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 31.Zhu J, et al. Transient inhibition of interleukin 4 signaling by T cell receptor ligation. J Exp Med. 2000;192:1125–34. doi: 10.1084/jem.192.8.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo L, et al. IL-1 family members and STAT activators induce cytokine production by Th2, Th17, and Th1 cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:13463–8. doi: 10.1073/pnas.0906988106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chung Y, et al. Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity. 2009;30:576–87. doi: 10.1016/j.immuni.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J, Zhu H, Murphy TL, Ouyang W, Murphy KM. IL-18-stimulated GADD45 beta required in cytokine-induced, but not TCR-induced, IFN-gamma production. Nature immunology. 2001;2:157–64. doi: 10.1038/84264. [DOI] [PubMed] [Google Scholar]
- 35.Robinson D, et al. IGIF does not drive Th1 development but synergizes with IL-12 for interferon-gamma production and activates IRAK and NFkappaB. Immunity. 1997;7:571–81. doi: 10.1016/s1074-7613(00)80378-7. [DOI] [PubMed] [Google Scholar]
- 36.McGeachy MJ, et al. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat Immunol. 2009;10:314–24. doi: 10.1038/ni.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annual review of immunology. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 38.Cote-Sierra J, et al. Interleukin 2 plays a central role in Th2 differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:3880–5. doi: 10.1073/pnas.0400339101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu J, Cote-Sierra J, Guo L, Paul WE. Stat5 activation plays a critical role in Th2 differentiation. Immunity. 2003;19:739–48. doi: 10.1016/s1074-7613(03)00292-9. [DOI] [PubMed] [Google Scholar]
- 40.Veldhoen M, et al. Transforming growth factor-beta ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nature immunology. 2008;9:1341–6. doi: 10.1038/ni.1659. [DOI] [PubMed] [Google Scholar]
- 41.Dardalhon V, et al. IL-4 inhibits TGF-beta-induced Foxp3+ T cells and, together with TGF-beta, generates IL-9+ IL-10+ Foxp3(-) effector T cells. Nature immunology. 2008;9:1347–55. doi: 10.1038/ni.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Acosta-Rodriguez EV, Napolitani G, Lanzavecchia A, Sallusto F. Interleukins 1beta and 6 but not transforming growth factor-beta are essential for the differentiation of interleukin 17-producing human T helper cells. Nature immunology. 2007;8:942–9. doi: 10.1038/ni1496. [DOI] [PubMed] [Google Scholar]
- 43.Wilson NJ, et al. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nature immunology. 2007;8:950–7. doi: 10.1038/ni1497. [DOI] [PubMed] [Google Scholar]
- 44.Ben-Sasson SZ, et al. IL-1 acts directly on CD4 T cells to enhance their antigen-driven expansion and differentiation. Proc Natl Acad Sci U S A. 2009;106:7119–24. doi: 10.1073/pnas.0902745106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu J, et al. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nature immunology. 2004;5:1157–65. doi: 10.1038/ni1128. [DOI] [PubMed] [Google Scholar]
- 46.Messi M, Giacchetto I, Nagata K, Lanzavecchia A, Natoli G, Sallusto F. Memory and flexibility of cytokine gene expression as separable properties of human T(H)1 and T(H)2 lymphocytes. Nat Immunol. 2003;4:78–86. doi: 10.1038/ni872. [DOI] [PubMed] [Google Scholar]
- 47.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–69. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 48.Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 2002;295:338–42. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- 49.Afkarian M, et al. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nature immunology. 2002;3:549–57. doi: 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- 50.Lieberman LA, Banica M, Reiner SL, Hunter CA. STAT1 plays a critical role in the regulation of antimicrobial effector mechanisms, but not in the development of Th1-type responses during toxoplasmosis. J Immunol. 2004;172:457–63. doi: 10.4049/jimmunol.172.1.457. [DOI] [PubMed] [Google Scholar]
- 51.Kaplan MH, Sun YL, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. 1996;382:174–7. doi: 10.1038/382174a0. [DOI] [PubMed] [Google Scholar]
- 52.Thierfelder WE, et al. Requirement for Stat4 in interleukin-12-mediated responses of natural killer and T cells. Nature. 1996;382:171–4. doi: 10.1038/382171a0. [DOI] [PubMed] [Google Scholar]
- 53.Cai G, Radzanowski T, Villegas EN, Kastelein R, Hunter CA. Identification of STAT4-dependent and independent mechanisms of resistance to Toxoplasma gondii. J Immunol. 2000;165:2619–27. doi: 10.4049/jimmunol.165.5.2619. [DOI] [PubMed] [Google Scholar]
- 54.Usui T, Nishikomori R, Kitani A, Strober W. GATA-3 suppresses Th1 development by downregulation of Stat4 and not through effects on IL-12Rbeta2 chain or T-bet. Immunity. 2003;18:415–28. doi: 10.1016/s1074-7613(03)00057-8. [DOI] [PubMed] [Google Scholar]
- 55.Yang Y, Ochando JC, Bromberg JS, Ding Y. Identification of a distant T-bet enhancer responsive to IL-12/Stat4 and IFNgamma/Stat1 signals. Blood. 2007;110:2494–500. doi: 10.1182/blood-2006-11-058271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Usui T, et al. T-bet regulates Th1 responses through essential effects on GATA-3 function rather than on IFNG gene acetylation and transcription. The Journal of experimental medicine. 2006;203:755–66. doi: 10.1084/jem.20052165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Placek K, et al. Integration of distinct intracellular signaling pathways at distal regulatory elements directs T-bet expression in human CD4+ T cells. J Immunol. 2009;183:7743–51. doi: 10.4049/jimmunol.0803812. [DOI] [PubMed] [Google Scholar]
- 58.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–96. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 59.Zhang DH, Cohn L, Ray P, Bottomly K, Ray A. Transcription factor GATA-3 is differentially expressed in murine Th1 and Th2 cells and controls Th2-specific expression of the interleukin-5 gene. J Biol Chem. 1997;272:21597–603. doi: 10.1074/jbc.272.34.21597. [DOI] [PubMed] [Google Scholar]
- 60.Pai SY, Truitt ML, Ho IC. GATA-3 deficiency abrogates the development and maintenance of T helper type 2 cells. Proc Natl Acad Sci U S A. 2004;101:1993–8. doi: 10.1073/pnas.0308697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ho IC, Tai TS, Pai SY. GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nature Reviews Immunology. 2009;9:125–35. doi: 10.1038/nri2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4:313–9. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 63.Shimoda K, et al. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature. 1996;380:630–3. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- 64.Takeda K, et al. Essential role of Stat6 in IL-4 signalling. Nature. 1996;380:627–30. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 65.Kurata H, Lee HJ, O'Garra A, Arai N. Ectopic expression of activated Stat6 induces the expression of Th2-specific cytokines and transcription factors in developing Th1 cells. Immunity. 1999;11:677–88. doi: 10.1016/s1074-7613(00)80142-9. [DOI] [PubMed] [Google Scholar]
- 66.Zhu J, Guo L, Watson CJ, Hu-Li J, Paul WE. Stat6 is necessary and sufficient for IL-4's role in Th2 differentiation and cell expansion. J Immunol. 2001;166:7276–81. doi: 10.4049/jimmunol.166.12.7276. [DOI] [PubMed] [Google Scholar]
- 67.Else KJ, Finkelman FD, Maliszewski CR, Grencis RK. Cytokine-mediated regulation of chronic intestinal helminth infection. The Journal of experimental medicine. 1994;179:347–51. doi: 10.1084/jem.179.1.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jankovic D, Kullberg MC, Noben-Trauth N, Caspar P, Paul WE, Sher A. Single cell analysis reveals that IL-4 receptor/Stat6 signaling is not required for the in vivo or in vitro development of CD4+ lymphocytes with a Th2 cytokine profile. J Immunol. 2000;164:3047–55. doi: 10.4049/jimmunol.164.6.3047. [DOI] [PubMed] [Google Scholar]
- 69.Finkelman FD, et al. Stat6 regulation of in vivo IL-4 responses. J Immunol. 2000;164:2303–10. doi: 10.4049/jimmunol.164.5.2303. [DOI] [PubMed] [Google Scholar]
- 70.Min B, et al. Basophils produce IL-4 and accumulate in tissues after infection with a Th2-inducing parasite. The Journal of experimental medicine. 2004;200:507–17. doi: 10.1084/jem.20040590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Voehringer D, Shinkai K, Locksley RM. Type 2 immunity reflects orchestrated recruitment of cells committed to IL-4 production. Immunity. 2004;20:267–77. doi: 10.1016/s1074-7613(04)00026-3. [DOI] [PubMed] [Google Scholar]
- 72.van Panhuys N, et al. In vivo studies fail to reveal a role for IL-4 or STAT6 signaling in Th2 lymphocyte differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12423–8. doi: 10.1073/pnas.0806372105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ouyang W, et al. Stat6-independent GATA-3 autoactivation directs IL-4-independent Th2 development and commitment. Immunity. 2000;12:27–37. doi: 10.1016/s1074-7613(00)80156-9. [DOI] [PubMed] [Google Scholar]
- 74.Yang XO, et al. Requirement for the basic helix-loop-helix transcription factor Dec2 in initial TH2 lineage commitment. Nat Immunol. 2009;10:1260–6. doi: 10.1038/ni.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ivanov II, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–33. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 76.Harris TJ, et al. Cutting edge: An in vivo requirement for STAT3 signaling in TH17 development and TH17-dependent autoimmunity. J Immunol. 2007;179:4313–7. doi: 10.4049/jimmunol.179.7.4313. [DOI] [PubMed] [Google Scholar]
- 77.Mathur AN, et al. Stat3 and Stat4 direct development of IL-17-secreting Th cells. J Immunol. 2007;178:4901–7. doi: 10.4049/jimmunol.178.8.4901. [DOI] [PubMed] [Google Scholar]
- 78.Yang XO, et al. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. The Journal of biological chemistry. 2007;282:9358–63. doi: 10.1074/jbc.C600321200. [DOI] [PubMed] [Google Scholar]
- 79.Milner JD, et al. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 2008;452:773–6. doi: 10.1038/nature06764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Durant L, et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 2010;32:605–15. doi: 10.1016/j.immuni.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schraml BU, et al. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature. 2009;460:405–9. doi: 10.1038/nature08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang F, Meng G, Strober W. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nature immunology. 2008;9:1297–306. doi: 10.1038/ni.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nature immunology. 2003;4:330–6. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 84.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–61. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 85.Williams LM, Rudensky AY. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nature immunology. 2007;8:277–84. doi: 10.1038/ni1437. [DOI] [PubMed] [Google Scholar]
- 86.Brunkow ME, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nature genetics. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 87.Patel DD. Escape from tolerance in the human X-linked autoimmunity-allergic disregulation syndrome and the Scurfy mouse. The Journal of clinical investigation. 2001;107:155–7. doi: 10.1172/JCI11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wildin RS, et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nature genetics. 2001;27:18–20. doi: 10.1038/83707. [DOI] [PubMed] [Google Scholar]
- 89.Mantel PY, et al. Molecular mechanisms underlying FOXP3 induction in human T cells. J Immunol. 2006;176:3593–602. doi: 10.4049/jimmunol.176.6.3593. [DOI] [PubMed] [Google Scholar]
- 90.Tran DQ, Ramsey H, Shevach EM. Induction of FOXP3 expression in naive human CD4+FOXP3 T cells by T-cell receptor stimulation is transforming growth factor-beta dependent but does not confer a regulatory phenotype. Blood. 2007;110:2983–90. doi: 10.1182/blood-2007-06-094656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, Tone M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat Immunol. 2008;9:194–202. doi: 10.1038/ni1549. [DOI] [PubMed] [Google Scholar]
- 92.Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–12. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gavin MA, et al. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445:771–5. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- 94.Wei G, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–67. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Isomura I, et al. c-Rel is required for the development of thymic Foxp3+ CD4 regulatory T cells. The Journal of experimental medicine. 2009;206:3001–14. doi: 10.1084/jem.20091411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ruan Q, et al. Development of Foxp3(+) regulatory t cells is driven by the c-Rel enhanceosome. Immunity. 2009;31:932–40. doi: 10.1016/j.immuni.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ouyang W, Beckett O, Ma Q, Paik JH, Depinho RA, Li MO. Foxo proteins cooperatively control the differentiation of Foxp3(+) regulatory T cells. Nature immunology. 2010 doi: 10.1038/ni.1884. [DOI] [PubMed] [Google Scholar]
- 98.Harada Y, Elly C, Ying G, Paik JH, Depinho RA, Liu YC. Transcription factors Foxo3a and Foxo1 couple the E3 ligase Cbl-b to the induction of Foxp3 expression in induced regulatory T cells. The Journal of experimental medicine. 2010 doi: 10.1084/jem.20100004. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bruno L, et al. Runx proteins regulate Foxp3 expression. The Journal of experimental medicine. 2009;206:2329–37. doi: 10.1084/jem.20090226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rudra D, Egawa T, Chong MM, Treuting P, Littman DR, Rudensky AY. Runx-CBFbeta complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nature immunology. 2009;10:1170–7. doi: 10.1038/ni.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kitoh A, et al. Indispensable role of the Runx1-Cbfbeta transcription complex for in vivo-suppressive function of FoxP3+ regulatory T cells. Immunity. 2009;31:609–20. doi: 10.1016/j.immuni.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 102.Klunker S, et al. Transcription factors RUNX1 and RUNX3 in the induction and suppressive function of Foxp3+ inducible regulatory T cells. The Journal of experimental medicine. 2009;206:2701–15. doi: 10.1084/jem.20090596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ono M, et al. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature. 2007;446:685–9. doi: 10.1038/nature05673. [DOI] [PubMed] [Google Scholar]
- 104.Pan F, et al. Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science. 2009;325:1142–6. doi: 10.1126/science.1176077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Thornton AM, et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol. 2010;184:3433–41. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zheng Y, et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–6. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yagi R, et al. The transcription factor GATA3 actively represses RUNX3 protein-regulated production of interferon-gamma. Immunity. 2010;32:507–17. doi: 10.1016/j.immuni.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Finotto S, et al. Development of spontaneous airway changes consistent with human asthma in mice lacking T-bet. Science. 2002;295:336–8. doi: 10.1126/science.1065544. [DOI] [PubMed] [Google Scholar]
- 109.Pearce EL, et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–3. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 110.Intlekofer AM, et al. Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science. 2008;321:408–11. doi: 10.1126/science.1159806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mukasa R, et al. Epigenetic Instability of Cytokine and Transcription Factor Gene Loci Underlies Plasticity of the T Helper 17 Cell Lineage. Immunity. 2010 doi: 10.1016/j.immuni.2010.04.016. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hwang ES, Szabo SJ, Schwartzberg PL, Glimcher LH. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science. 2005;307:430–3. doi: 10.1126/science.1103336. [DOI] [PubMed] [Google Scholar]
- 113.Naoe Y, et al. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbf beta binding to the Il4 silencer. J Exp Med. 2007;204:1749–55. doi: 10.1084/jem.20062456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Djuretic IM, Levanon D, Negreanu V, Groner Y, Rao A, Ansel KM. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nature immunology. 2007;8:145–53. doi: 10.1038/ni1424. [DOI] [PubMed] [Google Scholar]
- 115.Kohu K, et al. The Runx3 transcription factor augments Th1 and down-modulates Th2 phenotypes by interacting with and attenuating GATA3. J Immunol. 2009;183:7817–24. doi: 10.4049/jimmunol.0802527. [DOI] [PubMed] [Google Scholar]
- 116.Cruz-Guilloty F, et al. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. The Journal of experimental medicine. 2009;206:51–9. doi: 10.1084/jem.20081242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Intlekofer AM, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nature immunology. 2005;6:1236–44. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 118.Weinreich MA, Takada K, Skon C, Reiner SL, Jameson SC, Hogquist KA. KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity. 2009;31:122–30. doi: 10.1016/j.immuni.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhu J, et al. Growth factor independent-1 induced by IL-4 regulates Th2 cell proliferation. Immunity. 2002;16:733–44. doi: 10.1016/s1074-7613(02)00317-5. [DOI] [PubMed] [Google Scholar]
- 120.Quirion MR, Gregory GD, Umetsu SE, Winandy S, Brown MA. Cutting edge: Ikaros is a regulator of Th2 cell differentiation. J Immunol. 2009;182:741–5. doi: 10.4049/jimmunol.182.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Thomas RM, et al. Ikaros silences T-bet expression and interferon-gamma production during T helper 2 differentiation. The Journal of biological chemistry. 2010;285:2545–53. doi: 10.1074/jbc.M109.038794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Burgler S, Mantel PY, Bassin C, Ouaked N, Akdis CA, Schmidt-Weber CB. RORC2 Is Involved in T Cell Polarization through Interaction with the FOXP3 Promoter. J Immunol. 2010;184:6161–9. doi: 10.4049/jimmunol.0903243. [DOI] [PubMed] [Google Scholar]
- 123.Zhou L, et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–40. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xu L, Kitani A, Fuss I, Strober W. Cutting edge: regulatory T cells induce CD4+CD25-Foxp3- T cells or are self-induced to become Th17 cells in the absence of exogenous TGF-beta. J Immunol. 2007;178:6725–9. doi: 10.4049/jimmunol.178.11.6725. [DOI] [PubMed] [Google Scholar]
- 125.Yang XO, et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008;29:44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Minegishi Y, et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448:1058–62. doi: 10.1038/nature06096. [DOI] [PubMed] [Google Scholar]
- 127.Wu Y, et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126:375–87. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 128.Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nature immunology. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nature Reviews Immunology. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 130.Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity. 2006;24:369–79. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 131.Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annual review of immunology. 2006;24:607–56. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- 132.Mullen AC, et al. Role of T-bet in commitment of TH1 cells before IL-12-dependent selection. Science. 2001;292:1907–10. doi: 10.1126/science.1059835. [DOI] [PubMed] [Google Scholar]
- 133.Seki N, et al. IL-4-induced GATA-3 expression is a time-restricted instruction switch for Th2 cell differentiation. J Immunol. 2004;172:6158–66. doi: 10.4049/jimmunol.172.10.6158. [DOI] [PubMed] [Google Scholar]
- 134.Lee HJ, et al. GATA-3 induces T helper cell type 2 (Th2) cytokine expression and chromatin remodeling in committed Th1 cells. The Journal of experimental medicine. 2000;192:105–15. doi: 10.1084/jem.192.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Miller SA, Huang AC, Miazgowicz MM, Brassil MM, Weinmann AS. Coordinated but physically separable interaction with H3K27-demethylase and H3K4-methyltransferase activities are required for T-box protein-mediated activation of developmental gene expression. Genes Dev. 2008;22:2980–93. doi: 10.1101/gad.1689708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Makar KW, Perez-Melgosa M, Shnyreva M, Weaver WM, Fitzpatrick DR, Wilson CB. Active recruitment of DNA methyltransferases regulates interleukin 4 in thymocytes and T cells. Nature immunology. 2003;4:1183–90. doi: 10.1038/ni1004. [DOI] [PubMed] [Google Scholar]
- 137.Hutchins AS, et al. Gene silencing quantitatively controls the function of a developmental trans-activator. Molecular cell. 2002;10:81–91. doi: 10.1016/s1097-2765(02)00564-6. [DOI] [PubMed] [Google Scholar]
- 138.Ansel KM, et al. Deletion of a conserved Il4 silencer impairs T helper type 1-mediated immunity. Nature immunology. 2004;5:1251–9. doi: 10.1038/ni1135. [DOI] [PubMed] [Google Scholar]
- 139.Shi M, Lin TH, Appell KC, Berg LJ. Janus-kinase-3-dependent signals induce chromatin remodeling at the Ifng locus during T helper 1 cell differentiation. Immunity. 2008;28:763–73. doi: 10.1016/j.immuni.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Thieu VT, et al. Signal transducer and activator of transcription 4 is required for the transcription factor T-bet to promote T helper 1 cell-fate determination. Immunity. 2008;29:679–90. doi: 10.1016/j.immuni.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sekimata M, et al. CCCTC-binding factor and the transcription factor T-bet orchestrate T helper 1 cell-specific structure and function at the interferon-gamma locus. Immunity. 2009;31:551–64. doi: 10.1016/j.immuni.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mullen AC, et al. Hlx is induced by and genetically interacts with T-bet to promote heritable T(H)1 gene induction. Nature immunology. 2002;3:652–8. doi: 10.1038/ni807. [DOI] [PubMed] [Google Scholar]
- 143.Schoenborn JR, et al. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nature immunology. 2007;8:732–42. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hatton RD, et al. A distal conserved sequence element controls Ifng gene expression by T cells and NK cells. Immunity. 2006;25:717–29. doi: 10.1016/j.immuni.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 145.Lee DU, Avni O, Chen L, Rao A. A distal enhancer in the interferon-gamma (IFN-gamma) locus revealed by genome sequence comparison. The Journal of biological chemistry. 2004;279:4802–10. doi: 10.1074/jbc.M307904200. [DOI] [PubMed] [Google Scholar]
- 146.Shnyreva M, et al. Evolutionarily conserved sequence elements that positively regulate IFN-gamma expression in T cells. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12622–7. doi: 10.1073/pnas.0400849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Agarwal S, Avni O, Rao A. Cell-type-restricted binding of the transcription factor NFAT to a distal IL-4 enhancer in vivo. Immunity. 2000;12:643–52. doi: 10.1016/s1074-7613(00)80215-0. [DOI] [PubMed] [Google Scholar]
- 148.Kim JI, Ho IC, Grusby MJ, Glimcher LH. The transcription factor c-Maf controls the production of interleukin-4 but not other Th2 cytokines. Immunity. 1999;10:745–51. doi: 10.1016/s1074-7613(00)80073-4. [DOI] [PubMed] [Google Scholar]
- 149.Li B, Tournier C, Davis RJ, Flavell RA. Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. The EMBO journal. 1999;18:420–32. doi: 10.1093/emboj/18.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lohoff M, et al. Dysregulated T helper cell differentiation in the absence of interferon regulatory factor 4. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:11808–12. doi: 10.1073/pnas.182425099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rengarajan J, Mowen KA, McBride KD, Smith ED, Singh H, Glimcher LH. Interferon regulatory factor 4 (IRF4) interacts with NFATc2 to modulate interleukin 4 gene expression. The Journal of experimental medicine. 2002;195:1003–12. doi: 10.1084/jem.20011128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yamashita M, et al. Identification of a conserved GATA3 response element upstream proximal from the interleukin-13 gene locus. The Journal of biological chemistry. 2002;277:42399–408. doi: 10.1074/jbc.M205876200. [DOI] [PubMed] [Google Scholar]
- 153.Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–75. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- 154.Hural JA, Kwan M, Henkel G, Hock MB, Brown MA. An intron transcriptional enhancer element regulates IL-4 gene locus accessibility in mast cells. J Immunol. 2000;165:3239–49. doi: 10.4049/jimmunol.165.6.3239. [DOI] [PubMed] [Google Scholar]
- 155.Zhu J, Yamane H, Cote-Sierra J, Guo L, Paul WE. GATA-3 promotes Th2 responses through three different mechanisms: induction of Th2 cytokine production, selective growth of Th2 cells and inhibition of Th1 cell-specific factors. Cell research. 2006;16:3–10. doi: 10.1038/sj.cr.7310002. [DOI] [PubMed] [Google Scholar]
- 156.Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by gamma(c) family cytokines. Nature Reviews Immunology. 2009;9:480–90. doi: 10.1038/nri2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Liao W, et al. Priming for T helper type 2 differentiation by interleukin 2-mediated induction of interleukin 4 receptor alpha-chain expression. Nature immunology. 2008;9:1288–96. doi: 10.1038/ni.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Solymar DC, Agarwal S, Bassing CH, Alt FW, Rao A. A 3' enhancer in the IL-4 gene regulates cytokine production by Th2 cells and mast cells. Immunity. 2002;17:41–50. doi: 10.1016/s1074-7613(02)00334-5. [DOI] [PubMed] [Google Scholar]
- 159.Guo L, Hu-Li J, Paul WE. Probabilistic regulation of IL-4 production in Th2 cells: accessibility at the Il4 locus. Immunity. 2004;20:193–203. doi: 10.1016/s1074-7613(04)00025-1. [DOI] [PubMed] [Google Scholar]
- 160.Amsen D, Blander JM, Lee GR, Tanigaki K, Honjo T, Flavell RA. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell. 2004;117:515–26. doi: 10.1016/s0092-8674(04)00451-9. [DOI] [PubMed] [Google Scholar]
- 161.Mohrs M, et al. Deletion of a coordinate regulator of type 2 cytokine expression in mice. Nature immunology. 2001;2:842–7. doi: 10.1038/ni0901-842. [DOI] [PubMed] [Google Scholar]
- 162.Lee GR, Fields PE, Griffin TJ, Flavell RA. Regulation of the Th2 cytokine locus by a locus control region. Immunity. 2003;19:145–53. doi: 10.1016/s1074-7613(03)00179-1. [DOI] [PubMed] [Google Scholar]
- 163.Lee GR, Spilianakis CG, Flavell RA. Hypersensitive site 7 of the TH2 locus control region is essential for expressing TH2 cytokine genes and for long-range intrachromosomal interactions. Nature immunology. 2005;6:42–8. doi: 10.1038/ni1148. [DOI] [PubMed] [Google Scholar]
- 164.Yang XO, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chen Z, et al. Selective regulatory function of Socs3 in the formation of IL-17-secreting T cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8137–42. doi: 10.1073/pnas.0600666103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Akimzhanov AM, Yang XO, Dong C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. The Journal of biological chemistry. 2007;282:5969–72. doi: 10.1074/jbc.C600322200. [DOI] [PubMed] [Google Scholar]
- 167.Bauquet AT, et al. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nature immunology. 2009;10:167–75. doi: 10.1038/ni.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Reinhardt RL, Liang HE, Locksley RM. Cytokine-secreting follicular T cells shape the antibody repertoire. Nature immunology. 2009;10:385–93. doi: 10.1038/ni.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.King IL, Mohrs M. IL-4-producing CD4+ T cells in reactive lymph nodes during helminth infection are T follicular helper cells. The Journal of experimental medicine. 2009;206:1001–7. doi: 10.1084/jem.20090313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zaretsky AG, Taylor JJ, King IL, Marshall FA, Mohrs M, Pearce EJ. T follicular helper cells differentiate from Th2 cells in response to helminth antigens. The Journal of experimental medicine. 2009;206:991–9. doi: 10.1084/jem.20090303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tsuji M, et al. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer's patches. Science. 2009;323:1488–92. doi: 10.1126/science.1169152. [DOI] [PubMed] [Google Scholar]
- 172.Breitfeld D, et al. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. The Journal of experimental medicine. 2000;192:1545–52. doi: 10.1084/jem.192.11.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Chtanova T, et al. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. J Immunol. 2004;173:68–78. doi: 10.4049/jimmunol.173.1.68. [DOI] [PubMed] [Google Scholar]
- 174.Nurieva RI, et al. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–5. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Johnston RJ, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–10. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yu D, et al. The Transcriptional Repressor Bcl-6 Directs T Follicular Helper Cell Lineage Commitment. Immunity. 2009;31:457–68. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 177.Tartar DM, et al. FoxP3+RORgammat+ T helper intermediates display suppressive function against autoimmune diabetes. J Immunol. 2010;184:3377–85. doi: 10.4049/jimmunol.0903324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lochner M, et al. In vivo equilibrium of proinflammatory IL-17+ and regulatory IL-10+ Foxp3+ RORgamma t+ T cells. The Journal of experimental medicine. 2008;205:1381–93. doi: 10.1084/jem.20080034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Chaudhry A, et al. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–91. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zhou X, et al. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nature immunology. 2009;10:1000–7. doi: 10.1038/ni.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Oldenhove G, et al. Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity. 2009;31:772–86. doi: 10.1016/j.immuni.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Stock P, Akbari O, Berry G, Freeman GJ, Dekruyff RH, Umetsu DT. Induction of T helper type 1-like regulatory cells that express Foxp3 and protect against airway hyper-reactivity. Nature immunology. 2004;5:1149–56. doi: 10.1038/ni1122. [DOI] [PubMed] [Google Scholar]
- 183.Chen Y, Kuchroo VK, Inobe J, Hafler DA, Weiner HL. Regulatory T cell clones induced by oral tolerance: suppression of autoimmune encephalomyelitis. Science. 1994;265:1237–40. doi: 10.1126/science.7520605. [DOI] [PubMed] [Google Scholar]
- 184.Groux H, et al. A CD4+ T-cell subset inhibits antigen-specific T-cell responses and prevents colitis. Nature. 1997;389:737–42. doi: 10.1038/39614. [DOI] [PubMed] [Google Scholar]
- 185.Chang HC, et al. PU.1 expression delineates heterogeneity in primary Th2 cells. Immunity. 2005;22:693–703. doi: 10.1016/j.immuni.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 186.Spooner CJ, Cheng JX, Pujadas E, Laslo P, Singh H. A recurrent network involving the transcription factors PU.1 and Gfi1 orchestrates innate and adaptive immune cell fates. Immunity. 2009;31:576–86. doi: 10.1016/j.immuni.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Nowak EC, et al. IL-9 as a mediator of Th17-driven inflammatory disease. The Journal of experimental medicine. 2009;206:1653–60. doi: 10.1084/jem.20090246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lu LF, et al. Mast cells are essential intermediaries in regulatory T-cell tolerance. Nature. 2006;442:997–1002. doi: 10.1038/nature05010. [DOI] [PubMed] [Google Scholar]
- 189.Umetsu SE, Winandy S. Ikaros is a regulator of Il10 expression in CD4+ T cells. J Immunol. 2009;183:5518–25. doi: 10.4049/jimmunol.0901284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Xu J, et al. c-Maf regulates IL-10 expression during Th17 polarization. J Immunol. 2009;182:6226–36. doi: 10.4049/jimmunol.0900123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lee YK, et al. Late developmental plasticity in the T helper 17 lineage. Immunity. 2009;30:92–107. doi: 10.1016/j.immuni.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Hegazy AN, et al. Interferons direct Th2 cell reprogramming to generate a stable GATA-3(+)T-bet(+) cell subset with combined Th2 and Th1 cell functions. Immunity. 2010;32:116–28. doi: 10.1016/j.immuni.2009.12.004. [DOI] [PubMed] [Google Scholar]