Abstract
The architecture of both phenotypic variation and reproductive isolation are important problems in evolutionary genetics. The nematode genus Caenorhabditis includes both gonochoristic (male/female) and androdioecious (male/hermaprodite) species. However, the natural genetic variants distinguishing reproductive mode remain unknown, and nothing is known about the genetic basis of postzygotic isolation in the genus. Here we describe the hybrid genetics of the first Caenorhabditis species pair capable of producing fertile hybrid progeny, the gonochoristic Caenorhabditis sp. 9 and the androdioecious C. briggsae. Though many interspecies F1 arrest during embryogenesis, a viable subset develops into fertile females and sterile males. Reciprocal parental crosses reveal asymmetry in male-specific viability, female fertility, and backcross viability. Selfing and spermatogenesis are extremely rare in XX F1, and almost all hybrid self-progeny are inviable. Consistent with this, F1 females do not express male-specific molecular germline markers. We also investigated three approaches to producing hybrid hermaphrodites. A dominant mutagenesis screen for self-fertile F1 hybrids was unsuccessful. Polyploid F1 hybrids with increased C. briggsae genomic material did show elevated rates of selfing, but selfed progeny were mostly inviable. Finally, the use of backcrosses to render the hybrid genome partial homozygous for C. briggsae alleles did not increase the incidence of selfing or spermatogenesis relative to the F1 generation. These hybrid animals were genotyped at 23 loci, and significant segregation distortion (biased against C. briggsae) was detected at 13 loci. This, combined with an absence of productive hybrid selfing, prevents formulation of simple hypotheses about the genetic architecture of hermaphroditism. In the near future, this hybrid system will likely be fruitful for understanding the genetics of reproductive isolation in Caenorhabditis.
THE genetic basis of phenotypic diversity is an important, albeit poorly understood phenomenon. Caenorhabditis nematodes provide a system that can address such an issue. Caenorhabditis elegans can act as an excellent point of reference for comparative development studies (Félix 2007; Lin et al. 2009; Schulze and Schierenberg 2009), and the variation in reproductive mode within Caenorhabditis is an alluring subject for such investigations (Haag 2005). Some Caenorhabditis species are gonochoristic (male/female), whereas others are androdioecious (male/hermaphrodite; Figure 1). Hermaphrodites and females are somatically similar, but while females only make oocytes, hermaphrodites briefly undergo spermatogenesis before switching to oogenesis (Ellis and Schedl 2006). This striking interspecies difference is not only discrete and easily scored, but is also of great consequence for reproductive strategies and population genetics.
Figure 1.—
C. briggsae and C. sp. 9 differ in reproductive mode. (A) A young C. sp. 9 JU1325 adult female. The most proximal germ cell is an oocyte (oo), and no sperm cells are present. (B) A young C. briggsae VT847 adult hermaphrodite. Mature sperm (sp) are the most proximal population of germ cells, followed by developing spermatocytes (spc) and oocytes (oo). Bars, 100 μm. spth, spermatheca; v, vulva.
Many studies have addressed the evolution of germline sex determination in Caenorhabditis. Phylogenetic analyses suggest that the trait has evolved convergently in this lineage multiple times (Cho et al. 2004; Kiontke et al. 2004). Consistent with this, differences in the presence and functions of germline sex determination genes have been uncovered between the convergently evolved C. elegans and C. briggsae (Nayak et al. 2005; Hill et al. 2006; Guo et al. 2009). Similarities in germline sex determination between gonochoristic and androdioecious Caenorhabditis species have also been found (Haag and Kimble 2000; Chen et al. 2001; Haag et al. 2002). Remarkably, reverse genetic manipulations can cause a C. remanei female to produce activated sperm and lay self-progeny (Baldi et al. 2009). However, despite these successes, there has been little progress in identifying the historical causative genetic differences distinguishing hermaphrodites from their female ancestors. Indeed, because the exact cause of the sperm-to-oocyte switch in C. elegans remains elusive (Ellis and Schedl 2006) candidate-gene approaches to understanding the evolution of this trait in other Caenorhabditis species are problematic. The female–hermaphrodite species pairs studied thus far have been quite diverged from each other (Haag and Kimble 2000; Cutter 2008). Here, we explore the possibility that a more closely related mixed-mode species pair might open the door to traditional genetic trait mapping via hybrids.
In addition to the evolution of novel forms, another long-standing problem in biology is the genetic basis of postzygotic reproductive isolation. Indeed, the literature on interspecies hybrids is vast in Drosophila (Orr 2005) and other taxa (Presgraves 2010). Recent advances in Drosophila (Presgraves et al. 2003; Brideau et al. 2006; Ferree and Barbash 2009; Phadnis and Orr 2009; Tang and Presgraves 2009) have provided insights into the genetic bases of postzygotic reproductive isolation. Furthermore, these results are largely consistent with the notion that Dobzshanski–Muller incompatibility factors epistatically interact to promote hybrid inviability and sterility, helping to confirm a theory of how reproductive isolation can evolve (Dobzhansky 1937; Muller 1942). However, the Caenorhabditis system has made very few contributions to this issue (Baird et al. 1992; Baird and Yen 2001; Hill and L'Hernault 2001; Baird 2002; Seidel et al. 2008). This is somewhat surprising considering the breadth of subjects this system has been used to examine (e.g., de Bono and Bargmann 1998; Griffitts et al. 2001; Raizen et al. 2008). However, hybrid genetics has largely been impossible in this system due to the inability of any Caenorhabditis interspecies hybridization to successfully produce fertile hybrid progeny (Baird et al. 1992).
We have discovered a new gonochoristic Caenorhabditis species, provisionally named C. sp. 9, that is capable of producing fertile hybrids with the androdioecious C. briggsae. The existence of fertile hybrids between species of different reproductive mode opens up the possibility of using trait mapping approaches to examine the genetic basis of hermaphroditic spermatogenesis. Additionally, it allows the Caenorhabditis system to contribute to the study of the genetics of postzygotic reproductive isolation. Here, experiments conducted in this new hybrid system that pertain to both of these issues are described.
MATERIALS AND METHODS
Nomenclature:
To simplify discussion of the numerous hybrid crosses described herein, we developed a shorthand to denote specific hybrid generations (see Figure 2). The prefixes “F” and “B” are used for intercrosses and backcrosses, respectively, and are followed by standard size numbers denoting the nth generation since the pure species intercross. In addition, a subscript is used to specify the species identity of each sex in the crossing scheme that produced that generation. The sex of the animal corresponding to the subscript is assumed to be male unless otherwise denoted with an “f” for female or “h” for hermaphrodite. For instance, F1b denotes the generation resulting from the C. briggsae male × C. sp. 9 female parental hybrid cross (Figure 2A). Conversely, the generation resulting from the reciprocal parental cross would be the F19 (Figure 2B). A generation resulting from a scheme where the P0 father was C. briggsae and the hybrid female progeny were subsequently backcrossed to C. sp. 9 males would be the B2b,9 generation (Figure 2D). And, when a B2b,9 hybrid male is crossed to a C. briggsae hermaphrodite, the B3b,9,bh generation results (Figure 2G). When the directionality of a given cross is of no consequence, the subscript will be omitted.
Figure 2.—
Summary of various hybrid crosses. (A–L) Each panel represents a particular cross, with parents above and realized progeny below. Progeny are numbered via the scheme described in materials and methods. Numbers in parentheses represent the number of embryos and adult progeny scored to produce percentages of viable and female progeny, respectively. All hybrids derived using the C. briggsae strain AF16 and the C. sp. 9 strain JU1325, except in panels E and F, where C. sp. 9 JU1422 was used.
Maintenance and strains:
Animals were maintained according to standard C. elegans protocols (Wood 1988), with the exception of increased agar concentration in nematode growth medium (NGM) plates to 2.2%. Cultures were kept at 20° unless otherwise indicated. Inbred lines of C. sp. 9 were generated through 25 generations of full-sibling inbreeding. Strains used in this study include C. briggsae AF16 (sequenced reference strain), C. briggsae VT847 (mapping strain), C. briggsae HK104 (mapping strain), C. briggsae CP4 [dpy(nm4)II] C. briggsae CP99 [Cbr-unc-119 (nm67), courtesy of C. G. Thomas], C. briggsae CP116 (polyploid strain, this study), C. sp. 9 JU1325 (wild isolate from India), C. sp. 9 EG5268 (wild isolate from Congo, a gift of Michael Aillon), C. sp. 9 JU1422 (inbred derivative of JU1325), and C. sp. 9 JU1420 (inbred derivative of JU1325).
C. sp. 9 strain JU1325 was isolated by M.A.F. from rotting flowers and leaves sampled in the Zoo/Botanical Garden of Trivandrum, Kerala, India on December 21, 2007. The sample was kept in a plastic tube for 2 weeks. Nematodes were then isolated on agar plates seeded with Escherichia coli OP50 as described in Barrière and Félix (2006). Anatomical observation tentatively assigned JU1325 to the Elegans group of Caenorhabditis. Test crosses to C. remanei, C. briggsae, C. brenneri, C. sp. 5, and C. elegans indicated JU1325 was sufficiently reproductively isolated from other species of the Elegans group to consider it a new species of Caenorhabditis and that it was likely very closely related to C. briggsae. Sequence data are also consistent with this conclusion (Cutter et al. 2010).
Determination of viability, sex ratio, and brood size:
We define viability as the fraction of laid embryos that develop into adults. To measure viability, three females or hermaphrodites and five males were mated. C. sp 9 females were picked at the L4 stage to ensure virginity, and all L4 C. briggsae hermaphrodites used for hybrid crosses were purged of all self-sperm by daily plate transfers of solitary animals. After mating overnight, males were removed, the mothers were moved to a fresh plate, and the eggs on the previous plate were counted. This was repeated about every 12 hr until no more embryos were laid. The plates were scored for female and male adults 6 days after laying. The sex ratio is defined as the fraction of total adults that are female. Brood sizes, defined as the number of embryos laid by a given XX animal, were determined via a similar procedure, except matings with individual mothers were used. All brood size, viability, and sex ratio measurements were performed with C. sp. 9 JU1325 and C. briggsae AF16 unless otherwise indicated.
Fertility was measured on selected hybrid populations by single worm matings. For males, one male was placed with four wild-type C. sp. 9 females. If embryos were present on the plate the next day, the worm was marked as fertile. For females, the test was done with one virgin female and five C. sp. 9 males. The percentage of fertility is derived from the fraction of single worm matings that yield embryos. Plates where the individual worm being assayed had fled the plate were discarded. The extent of F1 male sterility was also evaluated by determining the fraction of males with abnormal gonad morphology under differential interference contrast (DIC) microscopy.
Determination of selfing and spermatogenesis incidence in hybrids:
We define “selfing” as the production of embryos in the absence of mating, which in all known Caenorhabditis species only occurs in the presence of XX spermatogenesis. To measure this in hybrids, XX L4 animals were removed from males and left overnight at 20°. Up to 50 L4 animals were picked to a single plate for the scoring of selfing. If embryos were observed on the plate, the plate was examined for the presence of an animal with embryos in its uterus. Typically, no more than one hybrid selfer was observed per plate. In addition, virgin young adult XX animals (produced as above) were scored for the presence of sperm-like or spermatocyte-like cells via DIC microscopy.
Immunoblotting:
Protein samples were prepared by transferring 100 worms into 30 μl of phosphate-buffered saline (PBS) (137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.76 mm KH2PO4), followed by addition of 30 μl 95% Laemmli sample buffer (Bio-Rad) + 5% β-mercaptoethanol. SDS polyacrylamide protein gels were run according to standard methods (Sambrook and Russell 2001). Both anti-MSP mouse monoclonal antibodies (provided by D. Greenstein; Kosinski et al. 2005) and anti-α-tubulin mouse antibodies (Sigma) were added to the blocking solution at dilutions of 1:1000. The secondary antibody, a horseradish peroxidase-conjugated anti-mouse conjugate (GE Healthcare), was used at a dilution of 1:1500 for 1.5 hr. Antibody-bound proteins were visualized using the SuperSignal Chemiluminescent substrate (Pierce Technology).
RT–PCR:
RNA preparations were made by transferring worms of the appropriate age and sex into RNA-ase free water at a concentration of 4 worms/μl, with ∼200 worms per sample. TRI reagent (Molecular Research Center) was added to each preparation, and the samples were frozen at −80°, thawed, pelleted in a microcentrifuge, and then lysed with a plastic pestle in a 1.5-ml microfuge tube. RNA was then purified via phenol/chloroform extraction and precipitation with isopropanol. RNA was reconstituted with RNA-ase free water, using 1 μl for every 4 worms in the initial preparation. Five microliters of an RNA prep was used in a RT–PCR reaction using the AccessQuick kit (Promega). Primers AD115 (5′-TCGACGACTTGGCTGTGCAAC-3′) and AD116 (5′-TTGACGAGCTGTTTGATGCCCACC-3′) were used to amplify a 245-bp fragment of the cb-fog-3 transcript, and primers EH37 and EH38 (Hill and Haag 2009) were used to amplify a 250-bp fragment of all C. briggsae actin paralogs. Reactions were then run on a 1% agarose ethidium bromide gel to visualize the amplicons.
Mutagenesis and screening:
Synchronized cultures of C. sp. 9 JU1422 L4 larvae were mutagenized for 4 hr using 50 mm ethyl methanesulfonate (EMS) according to standard methods (Brenner 1974). Animals were washed multiple times in M9 buffer and distributed onto seeded NGM plates. Approximately 10 such mutagenized virgin C. sp. 9 females were then mated with ∼20 C. briggsae AF16 males overnight, after which all parental males were removed. Plates were subsequently scored for the next 7 days for the presence of F2 embryos, which, given complete F1 male sterility, were likely to be due to XX self-fertility.
Construction of a polyploid C. briggsae strain:
Polyploid Caenorhabditis lines have been used to determine the chromosomal basis of sex determination in this genus (Nigon 1951; Madl and Herman 1979). A modified version of Madl and Herman's (1979) heat-shock protocol was used to generate a similar strain of C. briggsae. The wild-type C. briggsae AF16 and the dumpy C. briggsae CP4 (nm4) strains were shifted to 30° overnight. AF16 males were crossed with CP4 hermaphrodites, and 300 F1 L4 wild-type hermaphrodite progeny were singled to separate plates. The F2 self-progeny were scored for large animals, low brood size, and a high proportion of males, all of which are indicative of polyploids (Madl and Herman 1979). One such animal was found, and it was confirmed to sire polyploid progeny (likely 4A:3X, see Figure 8 below). This animal was used to generate the CP116 strain.
Figure 8.—
A polyploid strain of C. briggsae. (A) A wild-type C. briggsae AF16 hermaphrodite with 6 chromosomes. Bar, 50 μm. (B) A polyploid C. briggsae CP116 hermaphrodite with 11 chromosomes. Bar, 100 μm. chr, chromosome. Both of these images are focused upon oocytes arrested in diakinesis stage of meiotic prophase I, with DNA stained with Hoescht 33258.
DNA preparations for genotyping:
For DNA preparations, worms were picked into lysis buffer (50 mm KCl, 10 mm Tris pH 8.2, 2.5 mm MgCl2, 0.45% NP-40, 0.45% Tween 20, 0.01% gelatin) at a concentration of 2 worms/μl with ∼200 worms/prep. Proteinase K was added to a concentration of 100 μg/ml, and the preparation was frozen at −80° for at least 15 min. Samples were subsequently incubated for 1 hr at 65° and then at 95° for 30 min, after which they were used directly for PCR and genotyping. Control DNA samples for evaluating C. briggsae AF16/C. briggsae VT847 intrastrain segregation distortion were produced by using half AF16 worms and half VT847 worms. Samples representing the Mendelian expectations for B3b,9,bh animals were produced by mixing 50% C. sp. 9 JU1422 (not a productive template), 40% C. briggsae AF16, and 10% C. briggsae VT847. For X-linked markers, proportions of 25% C. sp. 9 JU1422, 50% C. briggsae AF16, and 25% C. briggsae VT847 were used for the Mendelian expectation control DNA preparation. Here, the expectation would be different from the autosomes because the hybrid X is donated by the male (Figure 9).
Figure 9.—
Scheme for genotyping hybrid animals with partially homozygosity for C. briggsae alleles. Existing markers for mapping C. briggsae mutations (Koboldt et al. 2010) were used to genotype B3b,9,bh hybrid animals. The C. briggsae strain VT847 was used for the parental cross, whereas a different C. briggsae strain, the AF16-derived Cbr-unc-119(nm67) strain CP99, was used for the final cross. The mapping generation was then heterozygous for AF16 and VT847 when homozygous for C. briggsae and was homozygous AF16 when heterozgygous C. briggsae/C. sp. 9. Following a Mendelian pattern of segregation, a given locus was expected to be heterozygous AF16/VT847 (or homozygous C. briggsae) 25% of the time.
Genotyping:
A total of 23 polymorphic molecular markers distinguishing C. briggsae AF16 and VT847 were used for genotyping, and all of these markers have been mapped to a physical position on the C. briggsae genome sequence (Koboldt et al. 2010). A total of 22 of these markers are single nucleotide polymorphisms (SNPs) and one an insertion/deletion (indel) of 18 bp. A total of 13 SNPs were genotyped via pyrosequencing technology and 9 via restriction fragment (“snip-SNP”) analysis, and the indel marker was assayed by agarose gel electrophoresis. All genotyping methods required a PCR amplification step. For the snip-SNP and indel assays, primers previously designed for interstrain C. briggsae mapping were used (Koboldt et al. 2010).
For pyrosequencing reactions, amplification and sequencing primers were designed around the SNPs of interest using software provided by the manufacturer (Qiagen, formerly Biotage; supporting information, Table S1). For the PCR step, 0.5 μl of a DNA preparation (as described above) was used with a 30-μl mixture containing: 0.5 μm of an untailed primer, 0.1 μm of a tailed primer, 0.4 μm of a universal biotinylated primer, 0.25 mm of each dNTP, 1X ThermoPol PCR buffer (New England Biolabs), 1.5 mm Mg2+, and 1 unit of Taq DNA polymerase per 10 μl. A single biotinylated primer was used for all pyrosequencing PCR reactions (Aydin et al. 2005). The cycling conditions were as follows: 95° denature (2 min), [95° denature (30 sec), 60° annealing (30 sec), 72° extension (30 sec)], 72° extension (5 min) with the bracketed subroutine repeated for 40 total cycles. The same conditions were used for each assay. Assays that only amplified C. briggsae DNA, and failed to amplify C. sp. 9 DNA, were used for genotyping. A total of 5 μl of all PCR reactions were visualized on an agarose gel to confirm amplification. Single-stranded PCR amplicons were purified with streptavidin sepharose beads (GE Healthcare) according to the manufacturer's instructions. Pyrosequencing reactions were performed using the PyroMarkTM Q96 ID machine (Qiagen) according to the manufacturer's instructions. Resulting data were analyzed using Allele Quantification software provided by the manufacturer. This software estimates allele frequencies of the polymorphic alleles through integration of the pyrogram peaks (Lavebratt et al. 2004).
For snip-SNP assays, the PCR conditions are as above, but with untailed, nonbiotinylated primers (0.5 μm each) designed by Koboldt et al. (2010), 25 μl reaction volumes, and a 1-min extension time. After amplification, 1.2 μl of the appropriate restriction endonuclease (NEB) was added to reaction and incubated at the appropriate temperature for 2 hr. To quantify the allele frequencies of the polymorphisms, a standard curve for every assay was generated by performing the assay on DNA preps of known AF16/VT847 allele frequencies of 0.5/0.5, 0.6/0.4, 0.7/0.3, 0.8/0.2, and 0.9/0.1. AF16, VT847, C. sp. 9 JU1422, and AF16/VT847 F2 controls were also run. All control and test DNA were run out on the same 1% ethidium bromide agarose gel to visualize the polymorphic bands. Band intensities were quantified using ImageJ software (Abramoff et al. 2004). The ratio of the VT847 and AF16 diagnostic band intensities for the control reactions were plotted against their known allele frequencies and a best-fit regression line was then used to estimate the allele frequencies of the test reactions. This same general process was used for the indel marker, but here no restriction digest step was necessary, and the samples were separated on a 2% agarose gel to resolve the bands differing in size by 18 bp. To facilitate the comparison between the Mendelian control and hybrid B3b,9,bh results, raw data were normalized by forcing the average allele frequency of each Mendelian expectation control to its known value.
RESULTS
C. briggsae and C. sp. 9 produce fertile hybrids:
A summary of the various hybrid crosses and backcrosses we have examined and the naming scheme used to describe them is presented in Figure 2. Prior to this study, the highest reported Caenorhabditis interspecies hybrid viability is 6% for crosses of C. remanei females to C. briggsae males (Baird et al. 1992). In contrast, one-third to one-half of hybrid F1 progeny from reciprocal crosses of C. briggsae and C. sp. 9 were viable, with the remainder arresting during embryonic development. This viability is dependent on the direction of the parental cross. When C. sp. 9 is the mother, the F1 have a viability of 45%, whereas when C. briggsae is the mother, the viability is 30% (Figure 2, A and B). This difference between these reciprocal crosses can be accounted for entirely by an extreme difference in male-specific viability. When C. sp. 9 is the mother, 34% of the progeny are male, whereas when C. briggsae is the mother, no viable F1 male progeny were observed. The average brood size (70 embryos laid, n = 5, SE = 11), sex ratio, and viability of the F1b are all significantly different (t-test P < 0.01) from the conspecific C. sp. 9 cross. Here the average brood size is 259 (n = 3, SE = 32), the sex ratio is 53% female (n = 1890), and the viability is 82% (n = 2312).
F1b males exhibit delayed development and most are atypically small. No cross ever performed with F1 males was successful, and all males examined under Nomarski microscopy (n = 94) had gonadal defects (Figure 3). All lacked obvious spermatocytes or sperm, and 37% had no gonad at all. In contrast, using single-pair mating tests (see materials and methods), the vast majority of F1 females successfully produce embryos when crossed with C. sp. 9 males (Table 1). The resulting B29 progeny are less viable than the F1 (Figure 2, D and F), but surviving females are comparable in fertility to F1 females (data not shown), and viable males, roughly one quarter of which are fertile, are also produced (Table 1 and Figure 2G).
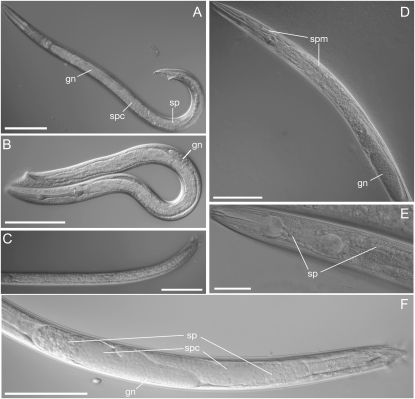
Figure 3.—
Hybrid males have abnormal gonads. (A) A wild-type C. sp. 9 JU1422 adult male, with a wild-type gonad (gn), spermatocytes (spc), and sperm (sp). (B) A hybrid F19 male with an underdeveloped gonad (gn) and no mature germ cells. (C) A hybrid F19 male with no discernible gonad. (D) A hybrid backcross (B2b,9) male with sperm (sp) located abnormally in the anterior. (E) A higher magnification image of the head of a hybrid backcross (B2b,9) male with mislocalized sperm. (F) B2b,9 male with developing spermatocytes (spc) and sperm (sp) oriented to both the anterior and posterior of the animal. Bars, 100 μm for all panels except for E, where it represents 50 μm. Hybrids shown in B and C were generated with the lines C. sp. 9 JU1325 and C. briggsae AF16, and the hybrids shown in C–F were generated with the lines C. sp. 9 1422 and C. briggsae AF16.
TABLE 1.
Incidence of fertility in hybrid animals
| Animal | Laid embryos % (n) |
|---|---|
| C. sp. 9 F | 93 (98) |
| C. sp. 9 M | 90 (100) |
| F1b F | 95 (99) |
| F19 F | 83 (101) |
| B2b,9 M | 24 (138) |
The incidence of fertility was determined through single worm mating tests. Animals were mated with C. sp. 9 males (M) or females (F). All hybrids were derived using the C. briggsae strain AF16 and the C. sp. 9 strain JU1422.
Multiple asymmetries in postzygotic isolation exist between C. briggsae and C. sp. 9:
In addition to the F1 sex ratio and viability asymmetries described above, other asymmetries were observed in later hybrid generations. F1 females in both reciprocal crosses are completely unable to produce viable progeny with C. briggsae males (Figure 2, C and E), with all progeny arresting during embryonic development. In contrast, they produce viable male and female progeny with C. sp. 9 males (Figure 2, D and F), with ∼24% of B2b,9 males being fertile (Table 1). Furthermore, the viability of B2 progeny depends on the identity of the P0 mother; the roughly twofold greater viability of B2b,9 progeny than B29,9 progeny (Figure 2, D vs. F) is highly significant (χ2 P = 0.0019, d.f. = 1). B2b,9 males exhibit a wider range of male germline phenotypes than F1 males (Figure 3, D–F). This included gonads with sperm oriented toward the anterior (Figure 3D), animals with sperm apparently localized outside of the gonad (Figure 3E), and gonads with female-like dual polarity (Figure 3F). A total of 34% (N = 167) of B2b,9 males have morphologically normal gonads. While fertile B2b,9 males can successfully mate with C. briggsae hermaphrodites to produce hybrid progeny (Figure 2G), their sisters can never produce viable hybrid progeny with C. briggsae males (Figure 2H). Hybrid females can only be backcrossed to C. briggsae males to produce viable hybrid progeny after being backcrossed with C. sp. 9 for two generations (Figure 2K).
In addition to the above asymmetries, pure species C. sp. 9 males greatly reduce the brood size of C. briggsae hermaphrodites and prevent them from laying self-progeny (Figure 4). About 50 hr after mating with a conspecific male, C. briggsae hermaphrodites are laying an average of 6 embryos per hour. However, after mating with C. sp. 9 males, C. briggsae hermaphrodites stop laying altogether by this time, despite the presence of both sperm and oocytes in the reproductive tract. C. sp. 9 females stop laying embryos ∼67 hr after mating (with either conspecifc or C. briggsae males). Examination of such postreproductive C. sp. 9 under DIC optics revealed that they had consistently run out of sperm.
Figure 4.—
C. sp. 9 males reduce the brood size of C. briggsae hermaphrodites. Conspecific crosses are solid lines, and hybrid crosses are dashed lines. Individual females or hermaphrodites were mated with four males overnight, after which the females were moved without the males to a new laying window twice a day. At least three replicates were performed for every cross. The error bars represent one standard error. The lines C. sp. 9 JU1325 and C. briggsae AF16 were used for all observations.
C. sp. 9 has a low viability at temperatures <20°:
It was noticed that C. sp. 9 strains grow poorly at 15°, so the viability of C. sp. 9 and hybrid F1 animals were examined at a range of temperatures (Figure 5). At 27°, the viability of C. briggsae is 95% and the viability of C. sp. 9 is 89%. However, at lower temperatures (i.e., <20°), C. briggsae has a much higher viability than does C. sp. 9 (87 vs. 6% at 15°, respectively). Hybrid F1 animals always displayed viability much lower than either C. sp. 9 or C. briggsae at all temperatures observed, but are similar to C. sp. 9 in performing particularly poorly below 20°.
Figure 5.—
Viabilities of C. briggsae, C. sp. 9, and F19 hybrids at different temperatures. Viability was measured as the number of adults resulting from the total number of embryos laid. All hybrid F19 were generated from crossing C. briggsae males to C. sp. 9 females. Error bars represent 95% confidence intervals. The lines C. sp. 9 JU1325 and C. briggsae AF16 were used for all observations. Sample sizes at temperatures 15°, 17°, 20°, 22.5°, 25°, and 27°, respectively: C. briggsae AF16 (342, 1883, 377, 867, 472, and 1800); C. sp. 9 JU1325 (136, 1078, 2312, 1396, 534, and 1616); and hybrid F1b (174, 195, 1004, 162, 510, and 383).
Hermaphroditism is rare in hybrid F1 XX animals:
Because C. sp. 9 and C. briggsae differ in reproductive mode, the germline sex of hybrid XX animals is of considerable interest. Three phenotypes were used to assess the presence of hermaphroditism among hybrid animals: the incidence of selfing (i.e., the fraction of apparent XX animals that laid embryos with eggshells in the absence of males), the incidence of spermatogenesis (the fraction of XX adults that appeared to have sperm-like cells under DIC microscopy), and the presence of sperm-specific molecular markers in XX hybrids.
Both selfing and spermatogenesis are rare in the F1 (Table 2). Overall, the incidence of any detectable selfing in the F1 is very low, and all selfed embryos observed died prior to hatching. However, the incidence of selfing varies with respect to the strain of C. sp. 9 that is used. When the wild isolates JU1325 and EG5862 are crossed with C. briggsae, selfing is observed in 1.2% and 0.3% of hybrid F1, respectively. If the inbred line JU1422 is used, then selfing is never seen. The use of other strains of C. briggsae, such as HK104 and VT847, does not reveal any significant increase in the incidence of selfing (Table 2). Progeny of the reciprocal parental crosses differ in their incidence of selfing, although not significantly so.
TABLE 2.
Number of hermaphrodites in hybrid generations
| Animal scored | C. briggsae strain(s) used | C. sp. 9 strain used | Selfers % (n) | Spermatogenic % (n) |
|---|---|---|---|---|
| P0 | NA | JU1422 | 0 (n > 100) | 0 (118) |
| P0 | NA | EG5268 | 0 (109) | — |
| F1b | AF16 | JU1422 | 0 (230) | 0 (92) |
| F1b | AF16 | JU1325 | 1.2 (494) | 0 (123) |
| F1b | AF16 | EG5268 | 0.3 (360) | 3.5 (114) |
| F19 | AF16 | JU1325 | 0 (106) | — |
| F1b | VT847 | JU1422 | 0 (34) | — |
| F1b | HK104 | JU1422 | 0 (29) | — |
| F1b | VT847 | JU1325 | 0 (32) | — |
| F1b | HK104 | JU1325 | 0 (68) | — |
| F1b | VT847 | EG5268 | 0 (26) | — |
| F1b | HK104 | EG5268 | 0 (31) | — |
| F1b | CP116 (polyploid AF16) | JU1325 | 0 (137) | 8.3 (24) |
| F1b | CP116 (polyploid AF16) | EG5268 | 2.3 (91) | 15 (177) |
| B3b,9,bh | AF16 | JU1422 | 0 (181) | 0 (65) |
| B3b,9,bh | AF16 | JU1325 | 0 (41) | — |
| B3b,9,bha | VT847 first AF16 second | JU1325 | 0 (128) | — |
| B4b,9,9,b | AF16 | JU1422 | 0 (202) | — |
| B4b,9,9,b | AF16 | JU1420 | 0 (396) | — |
| B4b,9,9,b | AF16 | JU1325 | 2.6 (117) | — |
Selfer, animal that lays embryos without mating; spermatogenic, female/hermaphrodite has sperm-like cells under DIC microscopy; —, not determined. See Figure 2 for how B3b,9,bh and B4b,9,9,b animals are constructed.
Because selfing requires more than just the generation of sperm and oocytes in a female soma (Baldi et al. 2009), XX hybrids were also observed under DIC to investigate the possibility that many hybrids made sperm (and were in fact hermaphrodite) but were unable to produce self-progeny. The incidence of spermatogenesis in young adult XX hybrid F1 animals is higher than that of the incidence of selfing itself, but never exceeds a few percent (Table 2). Also, in most F1 females germline development is delayed with respect to C. sp. 9 females. In C. sp. 9 females, typically at least one mature oocyte is fully developed by young adulthood (Figure 1A). However, in hybrid F1 animals, oftentimes no differentiated germ cells or incomplete oocytes (“ooids”) are seen in the proximal germline of young adult XX animals (Figure 6, A and B). However, the stacking oocyte phenotype characteristic of normal unmated females is seen in most older hybrid F1 animals (Figure 6C). This, in tandem with the result that most hybrid F1 females are fertile (above, Table 1), suggests that germline development is delayed but otherwise normal in F1 females. Indeed, young adult F1 females observed to have no differentiated germ cells under DIC microscopy were rescued, and all displayed the stacking oocyte phenotype after ∼12 hr at 20° (n = 10). Aside from rare hermaphrodites with clear populations of spermatocytes (Figure 5D), no hybrid F1 XX animals displayed sexually ambiguous populations of germ cells proximal to the oocytes.
Figure 6.—
Hybrid F1 female germlines. (A) A young hybrid F1b adult female displaying delayed germline development with no discernible differentiated germ cells despite having an adult vulva (v). The empty spermatheca (spth) is noted. (B) A young hybrid F1b adult female with small, proximal immature oocytes (imo). (C) A hybrid F1b adult female with stacking oocytes (oo) and an empty spermatheca (spth). (D) A rare hybrid F1b adult hermaphrodite with clear proximal spermatocytes (spc) and oocytes (oo). This animal was recovered and failed to lay any embryos. dgl, distal germline. Bar, 50 μm for A, B, and D; 100 μm for C. The lines C. sp. 9 JU1325 and C. briggsae AF16 were used for all panels, with the exception of D, where the C. sp. 9 line EG5468 was used.
The rarity of spermatogenesis and selfing in hybrid F1 XX animals does not entirely exclude the possibility that the hermaphroditism trait is codominant in this system. Despite the lack of morphologically sperm-like cells in the vast majority of F1 XX animals, their germlines could still possess cryptic male characteristics. Indeed, sex-specific germline molecular markers have been used in C. elegans to reveal the sexual identity of germ cells in the absence of morphological characteristics (Jones et al. 1996). Furthermore, the observed delayed oogenesis in young adult F1 XX animals is suggestive of codominant hermaphroditism. To investigate this possibility, the possible expression of male-specific molecular markers was examined in the F1b generation. Major sperm protein (MSP) is a crucial sperm cytoskeletal protein that is also implicated in oocyte maturation (Smith 2006). fog-3 is a TOB-domain protein that is necessary for spermatogenesis in C. elegans, C. briggsae, and C. remanei (Chen et al. 2001). Neither MSP nor fog-3 transcripts are detectably expressed in XX F1b L4 and young adult animals, though both are detectable in hermaphrodites and males (Figure 7, A and B). These results suggest that most hybrid F1 XX animals do not harbor germ cells with cryptic male character and that with only rare exceptions the female germline state is dominant in XX hybrid F1.
Figure 7.—
Sperm-specific molecular markers in hybrid F1 females. (A) RT–PCR for the sperm-specific transcript fog-3. Primers specific for an actin transcript were used as a positive control. All corresponding actin and fog-3 reactions used RNA from the same preparations in equal quantities. The asterisk denotes an uncharacterized amplicon that is not sex biased in C. sp. 9. H, hermaphrodite; A, adult; H F1, hybrid F1b. (B) Western blot for the sperm-specific protein major sperm protein (MSP). An anti-tubulin antibody was used as a positive control. Asterisks denote nonspecific proteins. All protein preparations were made with 100 worms. Blot was exposed overnight to ensure greatest possibility of protein detection in the hybrids.
Attempts to produce hybrid hermaphrodites via mutagenesis:
The genetic mechanism underlying female dominance may be due to a small number of hyperactive female-promoting genes in XX C. sp. 9 germ cells. If this were the case, then mutation of one of these genes could permit hermaphrodite-like levels of spermatogenesis in hybrid F1 XX animals and perhaps thereby provide enough viable F2 progeny to allow establishment of hybrid hermaphrodite lines. To test this possibility, mutagenized C. sp. 9 females were mated with C. briggsae males and their F1 progeny screened for the ability to produce viable self-progeny. No such F1 progeny were uncovered after screening ∼15,000 mutagenized haploid genomes. In C. elegans, this treatment would result in an average of 7.5 null mutations per gene (Brenner 1974).
Extent of F1 XX spermatogenesis is sensitive to the dosage of C. briggsae genetic material:
As an alternative to the oligogenic hypothesis investigated above, the genetic mechanism underlying the recessivity of hermaphroditism could be due to haploinsufficiency, perhaps at many loci. That is, the hermaphroditism trait may be rarely expressed in the F1 because there is only one copy of the C. briggsae genome in the F1 instead of the two copies that exist in the parent. To test this possibility, a tetraploid strain of C. briggsae, CP116, was created. Two lines of evidence suggest that hermaphrodites in this strain are 4A:3X tetraploids. First, there is a high incidence of male progeny arising from virgin CP116 hermaphrodites (39.7%, n adult progeny = 229). In addition, when CP116 diakinesis-stage oocytes were examined under fluorescent microscopy using Hoescht staining, a majority of them contained 11 or 12 chromosomes (70%, n animals = 40; Figure 8B). No such oocyte observed had <9 chromosomes. This is in contrast to wild-type C. briggsae oocytes, which all contain 6 chromosomes (Figure 8A).
Tetraploid (likely 4A:2X) C. briggsae CP116 males were crossed with diploid C. sp. 9 females to generate a triploid hybrid F1b with a 2:1 ratio of C. briggsae to C. sp. 9 genetic material. When triploid hybrid F1b are produced using wild isolates of C. sp. 9, the incidence of selfing increases to 2.3%, and the incidence of spermatogenesis increases significantly to 14% when compared to diploid hybrid F1 (χ2 P-value = 0.006, d.f. = 1; Table 2). Surprisingly, a small proportion of C. briggsae CP116/C. sp. 9 EG5268 self-progeny progress through embryonic development. Most undergo larval arrest, but one adult F2 was observed. It did not lay any embryos, but was observed to have both sperm and oocytes. No viable triploid hybrid F1 are produced when C. briggsae CP116 is crossed to the inbred C. sp. 9 strain JU1422. The great increase in both overt selfing and spermatogenesis in triploids with excess C. briggsae gene content suggests that hybrid F1 XX germline fate is at least somewhat sensitive to the dosage of “hermaphroditizing genes” and that haploinsufficiency can partly account for the dominance of the female germline state in the hybrid F1.
Partial homozygosity of C. briggsae loci in the hybrids does not reveal hermaphroditism:
To further investigate the potential of this hybrid system for understanding the genetic basis of hermaphroditism, possible segregation of the hermaphroditism trait was examined in recombinant hybrid generations. Segregation of the trait in the traditional F2 intercross and C. briggsae backcross populations cannot be examined due to the developmental and reproductive incompatibilities of the hybrid system and to the recessivity of the hermaphroditism (see above). For these reasons, more unconventional crossing designs were used to produce animals with substantial homozygosity for C. briggsae alleles, which allows potential segregation of the recessive hermaphroditism trait if it has a simple genetic architecture and key alleles are not linked to hybrid lethality and sterility factors.
Two generations were investigated for the segregation of the hermaphroditism trait. One is the progeny of B2b,9 males and C. briggsae hermaphrodites (B3b,9,bh animals; Figure 2G). The other results from crossing B3b,9,9 hybrid females with C. briggsae males to produce B4b,9,9,b animals (Figure 2K). These generations should be homozygous for C. briggsae alleles at a nonzero fraction of their genomes if they undergo a Mendelian pattern of segregation. Other crosses potentially yielding hybrids with homozygous C. briggsae regions were examined but either yielded no viable progeny (F1b female × B2b,9 male) or no hermaphrodites (B2b,9 female × B2b,9 male).
The incidence of selfing and spermatogenesis is 0% among B3b,9,bh XX animals (Table 2). In B4b,9,9,b XX animals, the selfing incidence varies, depending upon which C. sp. 9 strain is used. A total of 2.6% of XX B4b,9,9,b animals were selfers when the wild isolate strain of C. sp. 9, JU1325, was used, but none were seen with JU1420 and JU1422, the inbred strains derived from it (Table 2). This figure is not significantly different from that observed in hybrid F1 produced with JU1325 (1.2%; χ2 P = 0.5075, d.f. = 1). These results suggest that partial homozygosity for C. briggsae genes in hybrids does little or nothing to allow reemergence of the hermaphroditism trait.
Hybrid animals show segregation distortion:
It is possible that hermaphrodites are absent in the B3b,9,bh and B4b,9,9,b generations because multiple C. briggsae alleles must be homozygous for the trait to be observed. However, the patterns of hybrid viability suggest that there may be certain genotypes that promote hybrid lethality. If such hybrid lethal loci were linked to loci essential for hermaphrodite development, key genotypes would become inaccessible. To investigate this possibility, B3b,9,bh animals were genotyped at multiple loci to determine the extent of segregation distortion in this hybrid generation.
The crossing scheme in Figure 9 was used to allow the use of previously generated C. briggsae genetic markers (Koboldt et al. 2010). One strain of C. briggsae (VT847) was used as the P0 C. briggsae parent, and another strain of C. briggsae (the AF16-derived Cbr-unc-119(nm67) strain CP99) was used for the final backcross (Figure 9). This creates nonuncoordinated (non-Unc) B3b,9,bh hybrids that have one entire copy of the C. briggsae CP99 genome and one hybrid genome copy expected to contain ∼25% C. briggsae VT847 DNA. Such hybrids would thus be expected to be homozygous for C. briggsae at a given locus 25% of the time. Only assays that amplified C. briggsae DNA and failed to amplify C. sp. 9 DNA were utilized. Thus, homozygosity at a given C. briggsae locus would be revealed by presence of both polymorphic variants for the two C. briggsae strains (AF16/VT847; Figure 9). In contrast, heterozygosity at the species level (C. briggsae/C. sp. 9) would be revealed through hemizygosity for the C. briggsae strain AF16 (Figure 9).
A total of 23 markers were used with an average distance of 3.98 Mb (∼12.4 cM) (Hillier et al. 2007) between markers. Since the average size of the C. briggsae block of the recombinant hybrid chromosome was expected to be 12.5 cM, the assays provide reasonable power to detect segregation distortion if it were present. All 23 markers were confirmed to be dimorphic in C. briggsae AF16 and VT847, and no interstrain segregation distortion was seen (Figure S1; J. A. Ross, unpublished results). Control DNA preparation was made with proportions of C. briggsae AF16, C. briggsae VT847, and C. sp. 9 JU1422 animals equal to the proportions that would be expected in the hybrid B3b,9,bh generation if all of the genetic loci behaved in a Mendelian fashion. This facilitated the recognition of segregation distortion when compared with the hybrid B3b,9,bh generation genotypes.
Among the 23 markers genotyped, 13 displayed significant deviation (Mann–Whitney U test P-value <0.05) from the Mendelian expectation (Figure 10). Three markers showed no difference from expectation, and 7 markers displayed a nonstatistically significant deviation from the Mendelian expectation, all in the same direction. Strikingly, all 20 observed instances of substantial segregation distortion were underrepresentations of C. briggsae alleles, which is itself a highly significant deviation from random error model (binomial sign test, P < 0.0001). However, at no marker locus were C. briggsae alleles completely excluded. For certain regions of C. briggsae chromosomes II, III, IV, and X (Figure 10), it appears that either homozygosity in the B3b,9,bh generation or heterozygosity in the previous (B2) generation adversely affects hybrid fitness.
Figure 10.—
Patterns of segregation of loci in hybrids with homozygosity for some C. briggsae alleles. Animals in the hybrid B3b,9,bh generation (Figure 9), which represents the backcross with the highest potential to produce C. briggsae homozygosity, were used for genotyping. On the left are the physical positions of the markers used for the genotyping. On the right is displayed the percentage of B3b,9,bh homozygosity for C. briggsae at each locus. Pyrosequencing, RFLP, or indel analysis was used to estimate the frequencies of C. briggsae AF16 and C. briggsae VT847 alleles for each marker. The control (shaded bars) was a mixture of C. briggsae AF16, C. briggsae VT847, and C. sp. 9 JU1422 worms in the same proportions expected for hybrid alleles under Mendelian segregation. Note that C. sp. 9 DNA was included to mimic the hybrid genome composition, but does not support PCR amplification of polymorphic C. briggsae sites. The percentage of C. briggsae homozygosity measured for the Mendelian controls was normalized to 25% (50% for the X) to allow direct comparisons of the measurements of the hybrid markers. All DNA preparations used had at least 200 worms each. All tests had N ≥ 3. The error bars represent one standard error of the mean. *P < 0.05 for Mann–Whitney U test. †Genotyped using RFLP analyis. ‡Indel of 18 bp. All other markers were SNPs genotyped through pyrosequencing.
DISCUSSION
Reproductive isolation in Caenorhabditis:
The genetic basis of postzygotic isolation has been an intensely studied problem (Coyne and Orr 2004), and recent advances in fruit flies (Ferree and Barbash 2009), mice (Mihola et al. 2009), fish (Kitano et al. 2009), and plants (Bikard et al. 2009) have provided insights into the mechanisms that promote reproductive barriers. Recent work has also revealed genetic factors responsible for intraspecies postzygotic isolation in C. elegans (Seidel et al. 2008). However, little is known about the genetics of interspecies postzygotic isolation in Caenorhabditis due to the previous absence of fertile hybrids between Caenorhabditis species (Baird et al. 1992). The recently discovered C. sp. 9 promises to allow application of the genetic and genomic tools of the Caenorhabditis model genus to the problem.
The types of asymmetric patterns observed in the C. sp. 9/C. briggsae hybrid generations are not uncommon in hybrid systems. In many such systems, the heterogametic sex (XY/XO males or ZW/ZO females) is the disadvantaged sex with respect to hybrid viability and fertility (Presgraves 2008). This common phenomenon is referred to as Haldane's rule, and clearly applies to the C. sp. 9/C. briggsae hybrids: F1 males are either dead or sterile, depending upon cross direction, despite the presence of fertile F1 females in both directions. Many theories have been put forth to explain Haldane's rule, including the dominance theory, the “faster X” theory, and the “faster male” theory (Coyne and Orr 2004). Whichever applies, the leading candidate mechanism for incompatibility is the interaction of Dobzhansky–Muller incompatibility factors (Burke and Arnold 2001; Coyne and Orr 2004), and for the Caenorhabditis system several hypotheses can be framed with such factors in mind. The difference in the F1 sex ratio between reciprocal crosses (Figure 2, A and B) can be accounted for entirely by a difference in male-specific viability. This asymmetry in male viability could be explained by hemizygous X-linked C. briggsae factors that promote male inviability in the presence of C. sp. 9 autosomal factors. This explanation for this male-specific lethality is consistent with the dominance and faster-X theories of Haldane's rule.
In C. briggsae/C. remanei hybrid F1, an unusual Haldane's rule phenomenon is seen, in which XO male hybrids are transformed into females in a strain-dependent manner (Baird 2002). Although the possibility of male-to-female transformation was not specifically addressed here, the observation of hybrid males with bipolar gonads (Figure 3F) is suggestive that partial sexual transformation may occur in hybrids. Additionally, the segregation of both fertile and infertile males in the B2b,9 generation suggests that this system can be utilized to determine the genetic basis of hybrid male infertility. Indeed, the number of fertile males in this hybrid generation (24%; Table 1) suggests that as few as two loci may be needed to restore fertility in this generation (but see discussion of genotyping results below). This hybrid system will likely prove useful in teasing apart the basis of Haldane's rule in Caenorhabditis.
In addition to Haldane's rule, another common pattern of asymmetry is the strong dependence of hybrid F1 viability and fertility upon the directionality of the parental cross. This phenomenon has recently been dubbed “Darwin's corollary to Haldane's rule” (Turelli and Moyle 2007), and it is in effect here. If C. sp. 9 is the mother in the parental cross, viable male progeny are produced (Figure 2A), a larger percentage of F1 females are fertile (Table 1), and more viable B2 progeny are produced (Figure 2, D vs. F). F1 progeny exhibit lower fitness with respect to all of these categories when C. briggsae is the P0 mother. The older age of the C. briggsae mothers used in these experiments (see materials and methods) may be partly responsible for the poor performance of their progeny. However, although this is a possible explanation for the lowered viability and fertility of the F1 females, it is unlikely that moderate aging of the C. briggsae P0 mother would sex-specifically reduce F1 male viability to zero. Indeed, crosses using L4 C. briggsae AF16 hermaphrodites and C. sp. 9 JU1325 males also failed to produce males (data not shown). It is also unlikely it could explain the lower viability of B29,9 than B2b,9 embryos. This hybrid system then also has the potential to reveal potential causes of Darwin's corollary.
As an alternative to interactions between zygotic factors, there could be parental factors in the C. briggsae hermaphrodite or C. sp. 9 male gametes that adversely affect hybrid F1 fitness. The mutation rate of the C. briggsae mitochondrial genome has been reported to be much faster than that of other Caenorhabditis species (Howe and Denver 2008), and this difference could facilitate a nuclear–mitochondrial genome incompatibility between C. briggsae mitochondria and C. sp. 9 nuclear genes that is not reciprocal (Bolnick et al. 2008). This could provide a potential explanation for differences in F1 female fertility and backcross viability in reciprocal crosses. Alternatively, C. briggsae paternal effect factors may explain why hybrid progeny are inviable when F1 and backcross females are crossed to C. briggsae males (Figure 2, C, E, and H). Since F1 and backcross females are capable of producing viable hybrid progeny with C. sp. 9 males (Figure 2, D, F, and I), and because backcross males can produce viable progeny with C. briggsae hermaphrodites (Figure 2G), there may be a C. briggsae paternal effect factor that is incompatible with a hybrid background. As C. briggsae males can produce viable F1 hybrids, however, the hybrid's zygotic genotype may also have to have substantial C. briggsae homozygosity for this particular incompatibility to arise. The ability of C. briggsae males to produce viable progeny with B3b,9,9 females (Figure 2K and Table 2) is consistent with this.
The B3b,9,bh genotyping results also provide some insights into the genetic basis of hybrid male sterility in this system. That 24% of B2b,9 males are fertile is consistent with the existence of two unlinked C. sp. 9 loci that must be homozygous to allow male fertility. However, all loci genotyped were detectably homozygous for the C. briggsae allele in the B3b,9,bh generation, implying that no region of the C. briggsae genome was absolutely excluded from fertile B2b,9 males. Although clearly some genomic regions affect hybrid fitness more than others (particularly regions of chromosomes II, III, IV, and X; Figure 10), this suggests that there are multiple interactions between the C. briggsae and C. sp. 9 genomes that contribute to lowered hybrid fitness. Such polyfactorial interspecies incompatibility has been reported for both plants (e.g., Rieseberg et al. 1999; Jiang et al. 2000; Taylor et al. 2009) and other animals (e.g., Rogers and Bernatchez 2006; Good et al. 2008). In contrast, individual genes have been shown to play major roles in hybrid incompatibilities in Drosophila (Presgraves 2010), C. elegans (Seidel et al. 2008), and mice (Mihola et al. 2009).
Evolutionary implications for recessivity of the hermaphrodite germline:
The discovery of productive hybridization between the gonochoristic C. sp. 9 and the androdioecious C. briggsae suggested that these species could help reveal the genetic architecture of hermaphrodite development. That selfing is almost completely recessive is itself an important and surprising insight. In particular, this implies that no C. briggsae gene (or set of genes) in a single copy is sufficient for robust hermaphroditism in a gonochoristic background. This observation suggests that the female germline state in C. sp. 9 is extremely canalized in its female fate and thus highly resistant to the action of factors that promote XX spermatogenesis. In C. elegans and C. briggsae, loss-of-function mutations in these factors are recessive (Hodgkin 1986; Schedl and Kimble 1988; Zhang et al. 1997; Li et al. 2000; Guo et al. 2009; A.V. Doty, unpublished results), indicating they are not individually dose sensitive in present-day hermaphrodites. Therefore, perhaps an important part of the evolution of selfing C. briggsae was the weakening of female germline sexual canalization, so that the “sexual oscillator” that has been proposed to effect limited spermatogenesis (Haag 2009) can function.
We also note that the defining attributes of hermaphrodite germline development are not always congruent in their expression in hybrid generations. In a few cases, the incidence of spermatogenesis is higher than the incidence of overt selfing within a given hybrid generation. This suggests that in the hybrids selfing can become defective at multiple stages in the process. It would also suggest that it is more difficult to complete the self-fertilization and laying of embryos than it is to simply produce sperm. This is consistent with recent studies in C. remanei that suggest that multiple steps are necessary for hermaphroditism to evolve (Baldi et al. 2009).
Opportunities and limitations for the genetic investigation of hermaphroditism:
C. sp. 9 and C. briggsae open the possibility of mapping the historically crucial variants that led to C. briggsae hermaphroditism. However, the attempts to do this described here have so far been thwarted. One obvious problem is the extreme postzygotic isolation between these two species. Due to the inability to make self-fertile hybrid progeny, typical QTL designs based on recombinant inbred selfing lines are impossible (Doroszuk et al. 2009; Moyle and Payseur 2009). This necessitates the use of unconventional designs to tackle this problem in a genetic mapping context (Figure 9).
Another, larger problem is the low segregation of hermaphroditism in hybrid generations (Table 2), which falls to zero when inbred lines of C. sp. 9 are used. Further, in generations where hybrid hermaphrodites are observed, the incidence of hermaphroditism is no different from that seen in the F1 generation, indicating it is not due to rare combinations of homozygous C. briggsae alleles. The inability of C. sp. 9 inbred lines to yield any hybrid hermaphrodites suggests that either XX spermatogenesis in hybrids is especially sensitive to parental inbreeding depression or that there is standing genetic variation in C. sp. 9 for factors that specifically facilitate XX hybrid spermatogenesis. If the latter is true, then there should be heterogeneity among C. sp. 9 inbred lines with their ability to create hybrid hermaphrodites. This has not been observed, but only a small number of inbred lines have been investigated. However, even if such a polymorphic C. sp. 9 factor existed, its identity would not explain how C. briggsae became hermaphroditic. This would require the mapping of a C. briggsae hermaphroditizing factor that is not polymorphic.
Though it has proven difficult to use this system to map specific C. briggsae hermaphroditic factors, mechanisms patterning germline sex in this hybrid system can still be explored. One possibility is that this recessive trait has a complex (i.e., polygenic) genetic architecture. However, the presence of widespread segregation distortion biased against C. briggsae alleles in hybrids (Figure 10) prevents any such hypotheses about the nature of the genetic architecture of C. briggsae hermaphroditism from being rigorously evaluated. As an alternative approach, we screened 15,000 mutagenized haploid C. sp. 9 JU1422 genomes for a mutant that would facilitate the generation of hybrid F1 selfers. No such animal was observed. Given that loss-of-function alleles are generated for an average size gene about once in about every 2000 haploid mutagenized genomes (Anderson 1995), it is unlikely that there is any one C. sp. 9 feminizing factor that is responsible for the dominance of the female state in the hybrid germline. However, an increased dosage of C. briggsae factors to the hybrid background through the use of polyploids does elevate the incidence of hermaphroditism and in certain cases can almost triple it in the F1 (Table 2). This suggests that the haploinsufficiency of hermaphrodite-promoting C. briggsae alleles in the hybrid F1 is at least partly responsible for the recessivity of the hermaphrodite trait in this system.
Implications for the emergence of hermaphrodite lineages:
Phylogenetic studies suggest that hermaphroditism evolved multiple times in Caenorhabditis (Cho et al. 2004; Kiontke et al. 2004). If the ability of C. sp. 9 and C. briggsae to produce fertile hybrids (whereas all other Caenorhabditis pairwise crosses cannot) can be used to infer that these two species are recently diverged sister taxa (an inference further supported by Cutter et al. 2010; Raboin et al. 2010), then the most parsimonious scenario is one in which the last common ancestor of these two species had a female germline, similar to C. sp. 9 today. If that ancestral female was as resistant to the effects of hermaphroditizing factors as C. sp. 9, then mating with gonochoristic relatives would have completely destroyed the nascent trait. How, then, was a hermaphrodite lineage successfully established?
One simple scenario posits a single dominant factor sufficient for hermaphroditisim arising in a gonochoristic population, which then rapidly fixes due to the reproductive fitness benefits of selfing (Smith 1978). Alternatively, multiple hermaphrodite-promoting factors (with one or more being recessive) arise and accumulate within a gonochoristic population and are fixed in the population by physical or ecological isolation and resulting inbreeding. That our results support the latter, more convoluted process is unexpected for two reasons. First, developmental genetic studies (Schedl and Kimble 1988; Baldi et al. 2009) indicate a small number of genetic changes can interconvert females and hermaphrodites in the laboratory. Second, the multiple independent gains of selfing within Caenorhabditis might suggest that the process that leads to the evolution of hermaphroditism would be relatively simple. However, it is important to note that this is predicated upon the notion that the germline state of C. sp. 9 is a good proxy for the common ancestor of C. sp. 9 and C. briggsae. Given the degree of reproductive isolation demonstrated to exist between these two species, it is entirely possible that such an assumption is unsound. The strongly canalized C. sp. 9 female germline state may have evolved since the origin of C. briggsae by a number of possible mechanisms, including nonadaptive developmental system drift (True and Haag 2001), selection for greater robustness, or to resist introgression of the selfing trait from C. briggsae. Indeed, C. briggsae is cosmopolitan and has been so for a long period of time (Dolgin et al. 2008), and the high standing load of deleterious recessive mutations in gonochoristic Caenorhabditis creates strong inbreeding depression (Dolgin et al. 2007; Barriére et al. 2009). Further, selfing lineages appear to be relatively short-lived (Kiontke et al. 2004; Cutter et al. 2008), and pure selfing is predicted to quickly lead to extinction due to the accumulation of new deleterious mutations (Loewe and Cutter 2008). Gonochoristic lineages which can resist conversion to selfing may therefore be favored when such conversion is associated with low rates of outcrossing.
Other differences between C. sp. 9 and C. briggsae:
In addition to their reproductive barriers, other differences between C. briggsae and C. sp. 9 have been observed. One peculiar difference is that mated C. sp. 9 females stop laying embryos long before mated C. briggsae hermaphrodites do (Figure 4), even though the assays for the former initially placed multiple male mates with each female. The average C. sp. 9 total brood size of 259 is striking given that C. elegans hermaphrodites can lay >1000 embryos in a single mating assay (Hodgkin 1986). In the similarly gonochoristic C. remanei, outbred crosses of only 6-hr duration with single males (Dolgin et al. 2007) produce fecundities similar to those we report here for C. sp. 9, mated with multiple males for at least twice as long. This suggests that C. sp. 9 females cannot take full advantage of abundant sperm. This may be a general feature of the species, for example as a consequence of female spermatheca size, barriers to remating, male effects on female fecundity (Diaz et al. 2010), or as a consequence of unintended inbreeding in our laboratory stocks. Additionally, the reduction of C. briggsae hermaphrodite brood size by C. sp. 9 males may be in part due to a decrease in hermaphrodite survivorship after such matings (A. S. Chang, personal communication).
A more notable difference, however, is that C. sp. 9 animals have strikingly lower viabilities at low temperatures, whereas C. briggsae animals have relatively high viabilities at similar temperatures (Figure 5). Interestingly, this apparent pattern of C. sp. 9 as a high temperature specialist and C. briggsae as a temperature generalist correlates with these species' geographic distributions. C. briggsae has a relatively wide geographic range across at least five continents, and >100 isolates have been found in regions as disparate in temperature as Iceland and India (Cutter et al. 2010). Thus far, C. sp. 9 has only been found in tropical nations such as India and Congo, but this may change with more sampling. Despite the small sample size, the locations of its isolation and its low fitness at low temperatures are consistent with this species having a geographic range restricted to high-temperature environments. If this is the case, then C. sp. 9 and C. briggsae provide a new system within which to potentially understand the evolution of generalist and specialist modes of ecological adaptation. In tandem with the reproductive isolation between these species, these differences will likely prove the comparative C. sp. 9/C. briggsae system to be fruitful for future studies.
Future prospects:
We have described some attributes of the first hybrid genetic system in Caenorhabditis, formed between the gonochoristic species C. sp. 9 and the androdioecious species C. briggsae. Though mapping factors that distinguish hermaphrodites from true females has thus far been thwarted, a range of studies could be envisioned that would exploit this system's combination of speed and experimental resources to further studies of species formation, the evolution of mating systems, and any other phenotypic differences that may be discovered between the two species. Such experiments will be greatly aided by genomic tools for C. sp. 9. Indeed, a set of male- and female-specific transcriptome profiles (R. Jovelin and A. D. Cutter, personal communication) and a genome sequence (F. Piano, personal communication) of C. sp. 9 are currently being constructed. There is clearly great potential for this system that will begin to be realized in the near future.
Acknowledgments
We thank Michael Ailion for sharing the Congolese isolate of C. sp. 9 (EG5268) with us, Jiuzhou Song for assistance with pyrosequencing, and David Greenstein for the major sperm protein antibody. This work was supported by a fellowship from the University of Maryland Department of Biology (to G.W.), a grant from the National Institute of General Medical Sciences (to E.S.H.) (R01GM079414), and a research fellowship supplement for this grant (to O.E.). M.A.F. was supported by the Centre National de la Recherche Scientifique, France.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.120550/DC1.
References
- Abramoff, M. D., P. J. Magelhaes and S. J. Ram, 2004. Image processing with ImageJ. Biophotonics International 11 36–42. [Google Scholar]
- Anderson, P., 1995. Mutagenesis, pp. 31–58 in Caenorhabditis elegans: Modern Biological Analysis of an Organism (Methods in Cell Biology), Vol. 48, edited by H. F. Epstein and D. C. Shakes. Academic Press, San Diego.
- Aydin, A., M. R. Tollat, S. Bahring, C. Becker and P. Nurnberg, 2005. New universal primers facilitate pyrosequencing. Electrophoresis 27 394–397. [DOI] [PubMed] [Google Scholar]
- Baird, S. E., 2002. Haldane's rule by sexual transformation in Caenorhabditis. Genetics 161 1349–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird, S. E., M. E. Sutherlin and S. W. Emmons, 1992. Reproductive isolation in Rhabditidae (Nematoda: Secernentea); mechanisms that isolate six species of three genera. Evolution 46 585–594. [DOI] [PubMed] [Google Scholar]
- Baird, S. E., and W.-C. Yen, 2001. Reproductive isolation in Caenorhabditis: terminal phenotypes of hybrid embryos. Evol. Dev. 2 9–15. [DOI] [PubMed] [Google Scholar]
- Baldi, C., S. Cho and R. E. Ellis, 2009. Mutations in two independent pathways are sufficient to create hermaphroditic nematodes. Science 326 1002–1005. [DOI] [PubMed] [Google Scholar]
- Barrière, A., and M.A. Félix, 2006. Isolation of C. elegans and related nematodes (July 17, 2006) in WormBook. Edited by C. elegans Research Community, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Barriére, A., S. Wang, E. Pekarek, C. Thomas, E.S. Haag et al., 2009. Detecting heterozygosity in shotgun genome assemblies: lessons from obligately outcrossing nematodes. Genome Res. 19 470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard, D., D. Patel, C. Le Mette, V. Giorgi, C. Camilleri et al., 2009. Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science 323 623–626. [DOI] [PubMed] [Google Scholar]
- Bolnick, D. I., M. Turelli, H. Lopez-Fernandez, P. C. Wainwright and T. J. Near, 2008. Accelerated mitochondrial evolution and “Darwin's corollary”: asymmetric viability of reciprocal F1 hybrids in centrarchid fishes. Genetics 178 1037–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brideau, N. J., H. A. Flores, J. Wang, S. Maheshwari, X. Wang et al., 2006. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314 1292–1295. [DOI] [PubMed] [Google Scholar]
- Burke, J. M., and M. L. Arnold, 2001. Genetics and the fitness of hybrids. Annu. Rev. Genet. 35 31–52. [DOI] [PubMed] [Google Scholar]
- Chen, P.-J., S. Cho, S.-W. Jin and R. E. Ellis, 2001. Specification of germ cell fates by FOG-3 has been conserved during nematode evolution. Genetics 158 1513–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, S., S.-W. Jin, A. Cohen and R. E. Ellis, 2004. A phylogeny of caenorhabditis reveals frequent loss of introns during nematode evolution. Genome Res. 14 1207–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne, J. A., and H. A. Orr, 2004. Speciation. Sinauer, Sunderland, MA.
- Cutter, A. D., 2008. Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Mol. Biol. Evol. 25 778–786. [DOI] [PubMed] [Google Scholar]
- Cutter, A. D., J. D. Wasmuth and N. L. Washington, 2008. Patterns of molecular evolution in Caenorhabditis preclude ancient origins of selfing. Genetics 178 2093–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter, A. D., W. Yan, N. Tsvetkov, S. Sunil and M.-A. Felix, 2010. Molecular population genetics and phenotypic sensitivity to ethanol for a globally diverse sample of the nematode Caenorhabditis briggsae. Mol. Ecol. 19 798–809. [DOI] [PubMed] [Google Scholar]
- de Bono, M., and C. I. Bargmann, 1998. Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell 94 679–689. [DOI] [PubMed] [Google Scholar]
- Diaz, S. A., D. T. Haydon and J. Lindstrom, 2010. Sperm-limited fecundity and polyandry-induced mortality in female nematodes Caenorhabditis remanei. Biol. J. Linn. Soc. 99 362–369. [Google Scholar]
- Dobzhansky, T., 1937. Genetics and the Origin of Species. Columbia University Press, New York.
- Dolgin, E. S., B. Charlesworth, S. E. Baird and A. D. Cutter, 2007. Inbreeding and outbreeding depression in Caenorhabditis nematodes. Evolution 61 1339–1352. [DOI] [PubMed] [Google Scholar]
- Dolgin, E. S., M. A. Felix and A. D. Cutter, 2008. Hakuna Nematoda: genetic and phenotypic diversity in African isolates of Caenorhabditis elegans and C. briggsae. Heredity 100 304–315. [DOI] [PubMed] [Google Scholar]
- Doroszuk, A., L. B. Snoek, E. Fradin, J. Riksen and J. Kammenga, 2009. A genome-wide library of CB4856/N2 introgression lines of Caenorhabditis elegans. Nucleic Acids Res. 37 e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, R., and T. Schedl, 2006. Sex determination in the germ line, in WormBook. Edited by C. elegans Research Community, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Félix, M. A., 2007. Cryptic quantitative evolution of the vulva intercellular signaling network in Caenorhabditis. Curr. Biol. 17 103–114. [DOI] [PubMed] [Google Scholar]
- Ferree, P. M., and D. A. Barbash, 2009. Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol. 7 e1000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good, J. M., M. D. Dean and M. W. Nachman, 2008. A complex genetic basis to X-linked hybrid male sterility between two species of house mice. Genetics 179 2213–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffitts, J. S., J. L. Whitacre, D. E. Stevens and R. V. Aroian, 2001. Bt toxin resistance from loss of a putative carbohydrate-modifying enzyme. Science 293 860–864. [DOI] [PubMed] [Google Scholar]
- Guo, Y., S. Lang and R. E. Ellis, 2009. Independent recruitment of F box genes to regulate hermaphrodite development during nematode evolution. Curr. Biol. 19 1853–1860. [DOI] [PubMed] [Google Scholar]
- Haag, E. S., 2005. The evolution of nematode sex determination: C. elegans as a reference point for comparative biology, in WormBook. Edited by C. elegans Research Community, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Haag, E. S., 2009. Convergent evolution: regulatory lightning strikes twice. Curr. Biol. 19 R977–R979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag, E. S., and J. Kimble, 2000. Regulatory elements required for development of Caenorhabditis elegans hermaphrodites are conserved in the tra-2 homologue of C. remanei, a male/female sister species. Genetics 155 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag, E. S., S. Wang and J. Kimble, 2002. Rapid coevolution of the nematode sex-determining genes fem-3 and tra-2. Curr. Biol. 12 2035–2041. [DOI] [PubMed] [Google Scholar]
- Hill, K. L., and S. W. L'Hernault, 2001. Analyses of reproductive interactions that occur after heterospecific matings within the genus Caenorhabditis. Dev. Biol. 232 105–114. [DOI] [PubMed] [Google Scholar]
- Hill, R. C., C. E. de Carvalho, J. Salogiannis, B. Schlager, D. Pilgrim et al., 2006. Genetic flexibility in the convergent evolution of hermaphroditism in Caenorhabditis nematodes. Dev. Cell 10 531–538. [DOI] [PubMed] [Google Scholar]
- Hill, R. C., and E. S. Haag, 2009. A sensitized genetic background reveals evolution near the terminus of the Caenorhabditis germline sex determination pathway. Evol. Dev. 11 333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier, L. W., R. D. Miller, S. E. Baird, A. Chinwalla, L. A. Fulton et al., 2007. Comparison of C. elegans and C. briggsae genome sequences reveals extensive conservation of chromosome organization and synteny. PLoS Biol. 5 e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, J., 1986. Sex determination in the nematode C. elegans: analysis of tra-3 suppressors and characterization of fem genes. Genetics 114 15–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, D. K., and D. R. Denver, 2008. Muller's Ratchet and compensatory mutation in Caenorhabditis briggsae mitochondrial genome evolution. BMC Evol. Biol. 8 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, C., P. Chee, X. Draye, P. Morrell, C. Smith et al., 2000. Multilocus interactions restrict gene introgression in interspecific populations of polyploid Gossypium (cotton). Evolution 54 798–814. [DOI] [PubMed] [Google Scholar]
- Jones, A. R., R. Francis and T. Schedl, 1996. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev. Biol. 180 165–183. [DOI] [PubMed] [Google Scholar]
- Kiontke, K., N. P. Gavin, Y. Raynes, C. Roehrig, F. Piano et al., 2004. Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proc. Natl. Acad. Sci. USA 101 9003–9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano, J., J. A. Ross, S. Mori, M. Kume, F. C. Jones et al., 2009. A role for a neo-sex chromosome in stickleback speciation. Nature 461 1079–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt, D. C., J. Staisch, B. Thillainathan, K. Haines, S. E. Baird et al., 2010. A toolkit for rapid gene mapping in the nematode Caenorhabditis briggsae. BMC Genomics 11 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosinski, M., K. McDonald, J. Schwartz, I. Yamamoto and D. Greenstein, 2005. C. elegans sperm bud vesicles to deliver a meiotic maturation signal to distant oocytes. Development 132 3357–3369. [DOI] [PubMed] [Google Scholar]
- Lavebratt, C., S. Sengul, M. Jansson and M. Schalling, 2004. Pyrosequencing-based SNP allele frequency estimation in DNA pools. Hum. Mutat. 23 92–97. [DOI] [PubMed] [Google Scholar]
- Li, W., R. Boswell and W. B. Wood, 2000. mag-1, a homolog of Drosophila mago nashi, regulates hermaphrodite germ-line sex determination in Caenorhabditis elegans. Dev. Biol. 218 172–182. [DOI] [PubMed] [Google Scholar]
- Lin, K. T., G. Broitman-Maduro, W. W. Hung, S. Cervantes and M. F. Maduro, 2009. Knockdown of SKN-1 and the Wnt effector TCF/POP-1 reveals differences in endomesoderm specification in C. briggsae as compared with C. elegans. Dev. Biol. 325 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewe, L., and A. D. Cutter, 2008. On the potential for extinction by Muller's ratchet in Caenorhabditis elegans. BMC Evol. Biol. 8 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madl, J. E., and R. K. Herman, 1979. Polyploids and sex determination in Caenorhabditis elegans. Genetics 93 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihola, O., Z. Trachtulec, C. Vlcek, J. C. Schimenti and J. Forejt, 2009. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science 323 373–375. [DOI] [PubMed] [Google Scholar]
- Moyle, L. C., and B. A. Payseur, 2009. Reproductive isolation grows on trees. Trends Ecol. Evol. 24 591–598. [DOI] [PubMed] [Google Scholar]
- Muller, H., 1942. Isolating mechanisms, evolution, and temperature. Biol. Symp. 6 71–125. [Google Scholar]
- Nayak, S., J. Goree and T. Schedl, 2005. fog-2 and the evolution of self-fertile hermaphroditism in Caenorhabditis. PLoS Biol. 3 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigon, V., 1951. Experimental polyploidy in a free-living nematode, Rhabditis elegans Maupas. Bull. Biol. Fr. Belg. 85: 187–225 (in French).
- Orr, H. A., 2005. The genetic basis of reproductive isolation: insights from Drosophila. Proc. Natl. Acad. Sci. USA 102(Suppl 1): 6522–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadnis, N., and H. A. Orr, 2009. A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science 323 376–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves, D. C., 2008. Sex chromosomes and speciation in Drosophila. Trends Genet. 24 336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves, D. C., 2010. The molecular evolutionary basis of species formation. Nat. Rev. Genet. 11 175–180. [DOI] [PubMed] [Google Scholar]
- Presgraves, D. C., L. Balagopalan, S. M. Abmayr and H. A. Orr, 2003. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature 423 715–719. [DOI] [PubMed] [Google Scholar]
- Raboin, M., A. Timko, D. Howe, M. A. Félix and D. Denver, 2010. Evolution of Caenorhabditis mitochondrial genome pseudogenes and Caenorhabditis briggsae natural isolates. Mol. Biol. Evol. 27 1087–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizen, D. M., J. E. Zimmerman, M. H. Maycock, U. D. Ta, Y. J. You et al., 2008. Lethargus is a Caenorhabditis elegans sleep-like state. Nature 451 569–572. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L. H., J. Whitton and K. Gardner, 1999. Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics 152 713–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, S. M., and L. Bernatchez, 2006. The genetic basis of intrinsic and extrinsic post-zygotic reproductive isolation jointly promoting speciation in the lake whitefish species complex (Coregonus clupeaformis). J. Evol. Biol. 19 1979–1994. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and D. W. Russell, 2001. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schedl, T., and J. Kimble, 1988. fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis elegans. Genetics 119 43–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze, J., and E. Schierenberg, 2009. Embryogenesis of Romanomermis culicivorax: an alternative way to construct a nematode. Dev. Biol. 334 10–21. [DOI] [PubMed] [Google Scholar]
- Seidel, H. S., M. V. Rockman and L. Kruglyak, 2008. Widespread genetic incompatibility in C. elegans maintained by balancing selection. Science 319 589–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, H., 2006. Sperm motility and MSP, in WormBook. Edited by C. elegans Research Community, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Smith, J. M., 1978. The Evolution of Sex. Cambridge University Press, Oxford, UK.
- Tang, S., and D. C. Presgraves, 2009. Evolution of the Drosophila nuclear pore complex results in multiple hybrid incompatibilities. Science 323 779–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, S. J., M. Arnold and N. H. Martin, 2009. The genetic architecture of reproductive isolation in Louisiana irises: hybrid fitness in nature. Evolution 63 2581–2594. [DOI] [PubMed] [Google Scholar]
- True, J. R., and E. S. Haag, 2001. Developmental system drift and flexibility in evolutionary trajectories. Evol. Dev. 3 109–119. [DOI] [PubMed] [Google Scholar]
- Turelli, M., and L. C. Moyle, 2007. Asymmetric postmating isolation: Darwin's corollary to Haldane's rule. Genetics 176 1059–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, W. B. (Editor), 1988. The Nematode Caenorhabditis elegans. Cold Spring Laboratory Press, Cold Spring Harbor, NY.
- Zhang, B., M. Gallegos, A. Puoti, E. Durkin, S. Fields et al., 1997. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature 390 477–484. [DOI] [PubMed] [Google Scholar]