Abstract
Transcriptional regulation is a key mechanism that controls the fate and response of cells to diverse signals. Therefore, the identification of the DNA-binding proteins, which mediate these signals, is a crucial step in elucidating how cell fate is regulated. In this report, we applied both bioinformatics and functional genomic approaches to scrutinize the unusually large promoter of the IME1 gene in budding yeast. Using a recently described fluorescent protein-based reporter screen, reporter-synthetic genetic array (R-SGA), we assessed the effect of viable deletion mutants on transcription of various IME1 promoter–reporter genes. We discovered potential transcription factors, many of which have no perfect consensus site within the IME1 promoter. Moreover, most of the cis-regulatory sequences with perfect homology to known transcription factor (TF) consensus were found to be nonfunctional in the R-SGA analysis. In addition, our results suggest that lack of conservation may not discriminate against a TF regulatory role at a specific promoter. We demonstrate that Sum1 and Sok2, which regulate IME1, bind to nonperfect consensuses within nonconserved regions in the sensu stricto Saccharomyces strains. Our analysis supports the view that although comparative analysis can provide a useful guide, functional assays are required for accurate identification of TF-binding site interactions in complex promoters.
TRANSCRIPTIONAL regulation is the key mechanism that controls cell fate in both prokaryotes and eukaryotes. DNA-binding proteins, and proteins recruited by them, modulate the transcription pattern of genes in response to changing signals. Thus, the identification of DNA-binding proteins and the specific promoter regulatory roles they play are vital for elucidating how cell fate is determined in response to changing signals. The transcription of master regulators of developmental pathways is in many cases controlled by large and complex promoters that are subject to multiple and diverse signals that act through specific cis-regulatory sequences. Although each promoter element in isolation often has a small impact on transcriptional output, the sum of all regulatory signals determines proper transcriptional control (Davidson et al. 2002). Given this complexity, it remains a challenge to: (1) identify the individual trans-acting regulatory components of promoters and (2) identify regions in promoters bound by these proteins.
Several approaches can be used for the identification of specific DNA-binding proteins that regulate the transcription of genes, which include combining sensitive genetic and biochemical assays. In recent work, an approach called reporter-synthetic genetic array (R-SGA) analysis was developed to carry out reverse genetic promoter–reporter screens genome-wide (Kainth et al. 2009). These screens allow measurement of a test promoter–GFP reporter gene as well as a control promoter–red fluorescent protein (RFP) reporter gene in an array of yeast deletion mutants and provide quantitative measures of reporter gene activity in each mutant background.
Combinations of in vivo approaches and bioinformatic analysis of sequence features within promoters are commonly used to match consensus transcription factor (TF) binding sites to DNA-binding proteins. TF-binding sites are generally mapped by employing the following methodologies: (i) direct mutational analysis of sites bound by a specific TF (see for instance Shimizu et al. 1998), (ii) identification of common promoter sequence motifs in groups of coregulated genes using gene expression microarray analysis in mutant backgrounds of specific TFs or in strains where each TF is overexpressed (Roth et al. 1998; Chua et al. 2006), (iii) comparative analysis of all the sites bound by a specific TF following genome-wide location analysis (ChIP–chip) (Harbison et al. 2004), and (iv) systematic exploration of sequence motifs recognized by DNA-binding proteins using high-resolution protein binding microarrays (PBMs) (Berger and Bulyk 2006). However, these methods alone may be insufficient since single TFs interact with a range of related sequences (Lapidot et al. 2008). To better predict sequences likely influencing transcriptional output, comparative DNA sequence analysis between sensu stricto Saccharomyces species is useful. This analysis is based on the assumption that important cis-regulatory motifs in promoter regions are conserved throughout evolution, unlike other intergenic regions of DNA (Cliften et al. 2001, 2003).
In this report, we examined the feasibility of relying primarily on consensus TF-binding sites to faithfully identify true TF–promoter regulatory links in Saccharomyces cerevisiae by studying the IME1 promoter. In budding yeast, the regulated transcription of most genes is mediated through short upstream regulatory sequences, (∼437 bp, Saccharomyces Genome Database at http://www.yeastgenome.org/), residing 100–200 bp upstream of the start codon (Tirosh et al. 2007). This means that most genes are not suitable for such an analysis. However, complex regulation by an exceptionally large region can be found at the IME1 gene (>2 kb) (Granot et al. 1989; Sagee et al. 1998). IME1 encodes a transcriptional activator that serves as the master regulator of meiosis in budding yeast. Ime1 is required to initiate a transcriptional cascade that consists of a network of meiosis-specific genes and all the signals that regulate meiosis converge at IME1, regulating its transcription, translation, and activity (Kassir et al. 2003). Thus, IME1 may serve as an important tool to compare and integrate both bioinformatic and experimental approaches as described above. At least three general signals, carbon source, nitrogen depletion, and mating type, regulate the transcription of IME1. In the presence of glucose IME1 transcription is repressed, while in the presence of a nonfermentable carbon source such as acetate, a basal level of IME1 mRNA is detected. Upon nitrogen depletion, the transcription of IME1 is transiently induced but only in MATa/MATα diploids (Kassir et al. 1988). Deletion analysis of the 5′ regulatory region of the IME1 gene revealed that it is regulated by at least 10 distinct elements, 6 of which confer negative regulation, while 4 confer positive regulation (Figure 1; Kassir et al. 2003). However, knowledge of the DNA-binding proteins that directly affect transcription of the IME1 promoter remains incomplete with only a few known regulators (Msn2/4, Rme1, Sok2, and Yhp1) (Sagee et al. 1998; Shimizu et al. 1998; Kunoh et al. 2000; Shenhar and Kassir 2001).
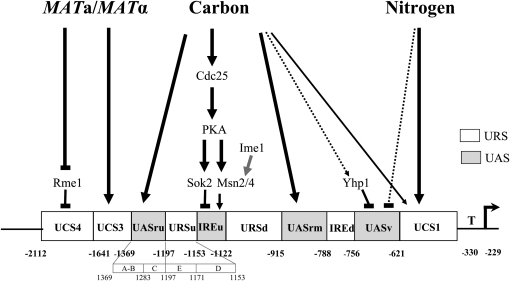
Figure 1.—
A schematic structure of IME1 5′ untranslated region. The MAT signal mediates repression activity of two elements UCS3 and UCS4. The carbon source signal is transmitted to UCS1, UASru, and IREu, which function as repression elements in the presence of glucose. In addition, UASru IREu and UASrm function as activation elements in the absence of glucose and the presence of acetate as the sole carbon source. Preliminary data suggest that the activity of UASv requires the absence of glucose as well as nitrogen. UCS1 also functions as a negative element in the presence of nitrogen. Solid boxes, elements required for transcriptional activation; open boxes, elements required for transcriptional repression. A positive role is marked with an arrow, a negative role by a line. Larger effects are denoted by thick lines while lesser effects are denoted by thin lines. Putative regulation is marked with a dashed line. Known regulators of IME1 transcription are shown.
In this report we combine bioinformatic and functional genomic approaches to discover regulators of IME1 transcription. The simple presence of a consensus binding site was generally a poor predictor of direct binding by a specific TF, since most of the sites appeared nonfunctional; rather, a combination of in vivo tests and the presence of a consensus site was a more powerful predictor of a true regulator. While most of the functional TF sites we identified resided in conserved regions, detailed analysis of one element, IREu, suggests that this element, and the TFs that bind and regulate its activity, are functional but not conserved. Similarly, Sum1 was identified and verified in our screens as a functional TF that binds to a nonconserved region.
MATERIALS AND METHODS
Bioinformatic approaches:
Consensus sequences for known TFs were taken from SGD (http://www.yeastgenome.org/), Saccharomyces Cerevisiae Promoter Database (SCPD) (http://rulai.cshl.edu/SCPD/), and YEASTRACT (http://www.yeastract.com/), as well as from reports based on ChIP–chip (Harbison et al. 2004; MacIsaac et al. 2006). In addition, several consensus sites were taken from specific articles (details are given in supporting information, Table S1). Location analysis was based on the http://fraenkel.mit.edu/yeast_map_2006/. Conservation analysis was based on S. cerevisiae genome browser (http://genome.ucsc.edu/cgi-bin/hgGateway?hgsid=101879728&clade=other&org=0&db=0).
High-throughput functional assay:
We screened the viable deletion array of ∼4500 genes for mutants that affect the transcription of GFP reporter genes whose expression was controlled by distinct upstream activation sequence (UAS) elements as previously described (Kainth et al. 2009). Four positive elements from the IME1 promoter, namely, UASru, UASrm, UASv, and UASru(AB) (which carries only the 5′ half of UASru) were fused to his4TATA–GFP, which by itself is not expressed. The use of UASru(AB) allowed us, as expected, to identify TFs, for instance, Swi4, whose function was masked in the UASru element. The negative elements, UCS3, URSuE, URSd, and UCS1 were fused to his4UAS–his4TATA–GFP. These constructs were used to transform strain BY4256 carrying the RPL39pr–tdTomato reporter (RFP). The constructs were introduced into the deletion array and MATa haploids carrying the GFP and RFP reporters, as well as the deletion alleles were selected as previously described (Kainth et al. 2009). The activity of the UCS3 element was screened in a diploid array homozygous for the deletions. To generate a diploid deletion array, we introduced a URA3 marker on a 2μ vector to the deletion array [following transformation of Y8205 with pRS426 (Sikorski and Hieter 1989), mating, and sporulation], and MATα haploids carrying the deletion alleles were selected. Following mating between the two arrays, diploids carrying URA3, GFP, and RFP were selected. Colony fluorescence was assayed following 2 and 6 days of incubation on minimal glucose and SPO plates, respectively, using Typhoon Trio variable mode imager (GE Healthcare). The log2 GFP/RFP ratio from each colony on the array was calculated as described (Kainth et al. 2009).
Plasmids and yeast strains:
Table 1 lists the plasmids used in this study. Detailed description on how these plasmids were constructed is available in File S1. Table 2 lists the genotype of the strains used in this study. The genotype of the constructed strains, including the copy number of inserted genes, was verified by PCR and quantitative PCR, respectively. A precise deletion of IREu in the genomic locus was constructed in two steps, essentially as described (Gray et al. 2004). First, strains Y1065 and Y1064, respectively, were transformed with a 2.9-kb SpeI fragment from YIp2887 to construct a large deletion of IME1 marked with URA3. Deletion of IME1 was confirmed by lack of sporulation when mated to a known ime1Δ strain. The resulting strains were cotransformed with a BamHI–XhoI fragment from p2980 and pRS423 (Sikorski and Hieter 1989), selecting for HIS+ colonies. Colonies that were −Ura were identified by plating on 5-FOA–containing medium. Y1837 carries SUM1–6HA–kiTRP1–sum1. It was constructed by transforming Y1214 with YIp3131 cut EcoNI.
TABLE 1.
List of plasmids
| Plasmid number | Description | Source |
|---|---|---|
| YIp1090 | LEU2, IME1(−1318 to +202)-lacZ | Kassir lab |
| YIp2102 | IME1UASru-HIS4TATA-LacZ, LEU2 | This work |
| YEp2536 | TRP1, 2μ, pCDC28-GST-MSN2 | Shenhar and Kassir (2001) |
| YEp2562 | 2μ, URA3, pCDC28-3xHA-sok2(1–32, 766–2721) | This work |
| YEp2780 | TRP1, 2μ, pADH1-GAL4(bd)-ime1(270–360) | Rubinstein et al. (2007) |
| YIp2887 | ime1(−3762 to −1118)-URA3-ime1(+945 to +2100) | This work |
| YIp2895 | sum1-URA3-sum1(Sum1 is from −49 to −1 and from +3189 to +3244, complete deletion of ORF) | This work |
| P2980 | IME1w/o IREu (IME1 is from −3.7 to +2.1) | This work |
| YCp3010 | IME1(−1283 to −1370)UASruab-GFP-ADH1t, pTEF-natR-tTEF, CEN6, ARSH4 | This work |
| YCp3021 | IME1(−1203 to −1370)UASru-GFP-ADH1t, pTEF-natR-tTEF, CEN6, ARSH4 | This work |
| YCp3022 | IME1(−789 to −915)UASrm-GFP-ADH1t, pTEF-natR-tTEF, CEN6, ARSH4 | This work |
| YCp3023 | IME1(−623 to −757)UASv-GFP-ADH1t, pTEF-natR-tTEF, CEN6, ARSH4 | This work |
| YCp3031 | UAShis4-IME1(−916 to −1122)URSd-GFP-ADH1t, pTEF-natR-tTEF, CEN6, ARSH4 | This work |
| YCp3032 | UAShis4-IME1(−1370 to −1643)UCS3-GFP-ADH1t, pTEF-natR-tTEF, CEN6, ARSH4 | This work |
| YCp3039 | UAShis4-IME1(−345 to −622)UCS1-GFP-ADH1t, pTEF-natR-tTEF, CEN6, ARSH4 | This work |
| YCp3040 | UAShis4-2xIME1(−1171 to −1197)URSuE-GFP-ADH1t, pTEF-natR-tTEF, CEN6, ARSH4 | This work |
| YIp3085 | gzf3Δ∷hisG-URA3-hisG | This work |
| YIp3101 | yox1Δ∷hisG-URA3-hisG | This work |
| YIp3107 | rim101∷hisG-URA3-hisG | This work |
| YIp3125 | IME1(Sc)(−3.8 to +2.1), URA3 | This work |
| YIp3129 | IME1(Sp −2175 to −1, Sc −33 to +2100), URA3 | This work |
| YIp3130 | IME1(Sb −2119 to −1, Sc −33 to +2100), URA3 | This work |
| YIp3131 | sum1(+1782 to 3168)-6HA, k1TRP1 | This work |
| YIp3153 | sum1Δ∷URA3 | This work |
| YIp3198 | upc2Δ∷hisG-URA3-hisG | This work |
| YIp3199 | spt23Δ∷hisG-URA3-hisG | This work |
Sc, Saccharomyces cerevisiae; Sp, Saccharomyces paradoxus; Sb, Saccharomyces bayanus.
TABLE 2.
Yeast strains
| Strain | Relevant genotype | Source |
|---|---|---|
| BY4256 | MATα, hoΔ∷RPL39pr-tdTomato-hph, can1Δ∷STE2pr-SpHIS5, lyp1Δ, his3Δ1, leu2Δ0, ura3Δ0, met15Δ0 | Kainth et al. (2009) |
| Y8205 | MATα, hoΔ∷RPL39pr-tdTomato-hph, can1Δ∷STE2pr-SpHIS5, leu2Δ∷STE3pr-LEU2, lyp1Δ, his3Δ1, ura3Δ0, met15Δ0 | Kainth et al. (2009) |
| Y422 | MATa/MATα, ura3-52/ura3-52, trp1Δ/trp1Δ, leu2-3,112/leu2-3,112, ade2-1/ade2-R8 | Sagee et al. (1998) |
| Y1061 | MATα, ura3-52, trp1Δ, leu2-3,112, his3∷hisG-URA3-hisG, ade2-R8, gal80∷hisG | Parent of Y1065 |
| Y1064 | MATa, ura3-52, leu2,3-112, trp1Δ, his3∷hisG, ade2-1, gal80∷hisG, gal4∷hisG | Shenhar and Kassir (2001) |
| Y1065 | MATα, ura3-52, trp1Δ, leu2-3,112, his3∷hisG, ade2-R8, gal80∷hisG gal4∷hisG | Shenhar and Kassir (2001) |
| Y1075 | MATa, ura3-52, leu2,3-112, trp1Δ, his3∷hisG, ime1∷hisG | (Shenhar and Kassir 2001) Isogenic to Y1064 |
| Y1076 | MATα, ura3-52, leu2-3,112, trp1Δ, his3∷hisG, ime1∷hisG | (Shenhar and Kassir 2001) Isogenic to Y1065 |
| Y1162 | MATa, ura3-52, sok2Δ∷TRP1,trp1Δ, his3∷hisG | (Shenhar and Kassir 2001) Isogenic to Y1064 |
| Y1171 | MATa/MATα, ura3-52/ura3-52, trp1Δ/trp1Δ, his3∷hisG/his3∷hisG, msn2∷HIS3/msn2∷HIS3, msn4∷URA3/msn4∷URA3 | HO induced diploidization of Y1132 (Shenhar and Kassir 2001) |
| Y1214 | MATa, ura3-52, leu2,3-112∷LEU2-UASru-his4-lacZ, trp1Δ, his3∷hisG | Y1064 transformed with YIp2102 cut PpuMI |
| Y1631 | MATa/MATα, IME1/IME1 | Y1064 × Y1065 |
| Y1699 | MATα, IME1-ΔIREu | Isogenic to Y1065 |
| Y1721 | MATa/MATα, ura3-52/ura3-52, trp1Δ/trp1Δ, leu2-3,112/leu2,3-112∷LEU2-UASru-his4-lacZ, his3∷hisG/his3∷hisG-URA3-hisG, ade2-R8/ade2-1, gal80∷hisG/gal80∷hisG, gal4∷hisG/GAL4, met/MET | Y1214 × Y1061 |
| Y1794 | MATa, IME1-ΔIREu | Isogenic to Y1064 |
| Y1795 | MATa/MATα IME1-ΔIREu/IME1-ΔIREu | Y1794 × Y1699 |
| Y1802 | MATa, ura3-52∷URA3- IME1(−3.8 to +2.1), leu2,3-112, trp1Δ, his3∷hisG, ime1∷hisG | Y1075 transformed with YIp3125 cut StuI |
| Y1803 | MATα, ura3-52∷URA3- IME1(−3.8 to +2.1), trp1Δ, leu2-3,112, his3∷hisG, ime1∷hisG | Y1076 transformed with YIp3125 cut StuI |
| Y1804 | MATa, ura3-52∷URA3-IME1p-SP-Ime1-SC, leu2,3-112, trp1Δ, his3∷hisG, ime1∷hisG | Y1075 transformed with YIp3129 cut StuI |
| Y1805 | MATα, ura3-52∷URA3-IME1p-SP-Ime1-SC, trp1Δ, leu2-3,112, his3∷hisG, ime1∷hisG | Y1076 transformed with YIp3129 cut StuI |
| Y1806 | MATa, ura3-52∷URA3-IME1p-SB-Ime1-SC, leu2,3-112, trp1Δ, his3∷hisG, ime1∷hisG | Y1075 transformed with YIp3130 cut StuI |
| Y1807 | MATα, ura3-52∷URA3-IME1p-SB-Ime1-SC, trp1Δ, leu2-3,112, his3∷hisG,, ime1∷hisG | Y1076 transformed with YIp3130 cut StuI |
| Y1808 | MATa/MATα, ura3-52∷URA3- IME1(-3.8 to +2.1)/ura3-52∷URA3- IME1(-3.8 to +2.1), ime1∷hisG/ime1∷hisG | Y1802 × Y1803 |
| Y1809 | MATa/MATα, ura3-52∷URA3- IME1p-SP-Ime1-SC/ura3-52∷URA3-IME1p-SP-Ime1-SC, ime1∷hisG/ime1∷hisG | Y1804 × Y1805 |
| Y1810 | MATa/MATα, ura3-52∷URA3-IME1p-SB-Ime1-SC/ura3-52∷URA3-IME1p-SB-Ime1-SC, ime1∷hisG/ime1∷hisG | Y1806 × Y1807 |
| Y1824 | MATa, ura3-52, leu2,3-112∷LEU2-UASru-his4-lacZ, yox1Δ∷hisG-URA3-hisG, trp1Δ, his3∷hisG | Y1214 transformed with a 6.7-kb SacII fragment from YIp3101 |
| Y1825 | MATa, ura3-52, leu2,3-112∷LEU2-IME1-lacZ, trp1Δ, his3∷hisG, ade2-1, gal80∷hisG, gal4∷hisG | Y1064 transformed with YIp1090 cut PpuMI |
| Y1827 | MATa, sum1Δ, ura3-52, leu2,3-112∷LEU2-UASru-his4-lacZ, trp1Δ, his3∷hisG | Y1214 transformed with 1.6-kb EcoRI fragment from YIp3153 |
| Y1836 | MATa, ura3-52, leu2,3-112∷LEU2-IME1-lacZ, trp1Δ, gzf3Δ∷hisG-URA3-hisG, his3∷hisG, ade2-1, gal80∷hisG, gal4∷hisG | Y1825 transformed with 7.7-kb SpeI–XhoI fragment from YIp3085 |
| Y1837 | MATa, SUM1-6HA-kiTRP1-sum1, ura3-52, leu2,3-112∷LEU2-UASru-his4-lacZ, trp1Δ, his3∷hisG | Y1214 transformed with YIp3131 cut EcoNI |
| Y1854 | MATa, rim101Δ∷hisG-URA3-hisG, ura3-52, trp1Δ, leu2,3-112∷LEU2-UASru-his4-lacZ, his3∷hisG | Y1214 transformed with 4.8-kb NotI fragment from YIp3107 |
| Y1855 | MATa, swi4∷URA3, ura3-52, trp1Δ, his3∷hisG, leu2,3-112∷LEU2-UASru-his4-lacZ | Y1214 transformed with 1636 (from K. Nasmyth) cut SacI–SalI |
| Y1858 | MATa, tec1∷HIS3, ura3-52, trp1Δ, his3∷hisG, leu2,3-112∷LEU2-UASru-his4-lacZ | Y1214 transformed with p55217 (G. Fink) cut SacI–EcoRI |
| Y1859 | MATa, upc2∷ hisG-URA3-hisG, ura3-52, trp1Δ, his3∷hisG, leu2,3-112∷LEU2-UASru-his4-lacZ | Y1214 transformed with 6.0-kb SacI–SacII fragment from YIp3198 |
| Y1860 | MATa, spt23Δ∷hisG-URA3-hisG, ura3-52, trp1Δ, leu2,3-112∷LEU2-UASru-his4-lacZ, his3∷hisG | Y1214 transformed with 6.0-kb SacI–SacII fragment from YIp3107 |
| Y1877 | MATα, sum1Δ∷URA3, ura3-52, trp1Δ, leu2-3,112, his3∷hisG, ade2-R8, gal80∷hisG, gal4∷hisG | Y1065 transformed with 1.6-kb EcoRI fragment from YIp3153 |
| Y1891 | MATa/MATα, sum1Δ∷URA3/sum1Δ | Y1827 × Y1877 |
Media and growth conditions:
SD, minimal glucose (synthetic glucose medium with glutamic acid as a nitrogen source), PSP2 (SA), SPM, and SPO media were prepared as previously reported (Kassir and Simchen 1991; Tong et al. 2001). Meiosis was induced as follows: cells were grown in PSP2 supplemented with the required amino acids to early exponential stage (0.8–1.2 × 107 cells/ml), washed once with water, and resuspended in SPM.
β-Galactosidase activity was assayed as described previously (Miller 1972).
Quantitative analysis of RNA level:
RNA was extracted from 108 cells by the hot acidic phenol method. One microgram of total RNA was used for a reverse transcription reaction (total 20 μl) with random hexamer primers and SuperScript Reverse-iT transcriptase. A total of 100 ng of the resulting cDNA was then used in real-time PCR analysis (qPCR) according to the manufacturer's instructions (ABGene, Surrey, UK).
ChIP:
The chromatin immunoprecipitation assay was done essentially as described (Pnueli et al. 2004). Msn2, Sok2, and Ime1 were tagged with GST, 3xHA, and Gal4(1–147), and placed on a 2μ vector. The expression of SOK2 in SA media is substantially reduced in comparison to SD media (Shenhar and Kassir 2001). Therefore, to determine the effect of the carbon source on binding of Sok2 to its target, we expressed Sok2, and as a control also Msn2, from the CDC28 promoter. The transcription of IME1 is under glucose repression (Kassir et al. 1988). Therefore, to detect Ime1 in SD media, we expressed it from the ADH1 promoter. As can be observed by the results (Figure 3), overexpression of these proteins did not affect their regulation, validating the use of a 2μ plasmid. Finally, Sum1 tagged with 6xHA was expressed from its own promoter and present in the genome. Following IP, qPCR on 100 ng genomic DNA was performed.
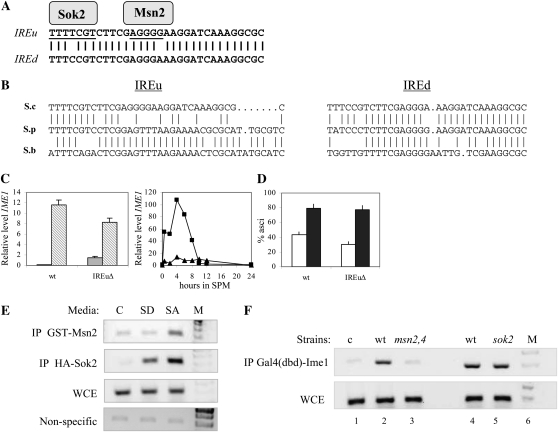
Figure 3.—
IREu functions as a positive element in IME1 promoter. (A) Alignment of IREu and IREd elements and the putative Msn2 and Sok2 binding sites. (B) Alignment of IREu and IREd in the three Saccharomyces sensu stricto strains. (C) RNA was isolated from 1 × 107 cells/ml grown in either SD (shaded column) or SA (striped column) media (left panel). In addition, cells grown in SA to 1 × 107 cells/ml were shifted to SPM, and at the indicated hours samples were taken to isolate RNA (right panel). Level of RNA was determined by qPCR. The relative level of IME1 RNA in comparison to RNA levels of SUM1 is given. The isogenic strains used were Y1631 (wild type, squares) and Y1795 (IME1–ΔIREu/IME1–ΔIREu, triangle). (D) Following 24 (open column) and 48 (solid column) hr of incubation in SPM, the percentage of asci was determined. (E) Samples for ChIP were taken from cells grown in either SD or SA media to 1 × 107 cells/ml. Strain used was Y422 (wild-type diploid) carrying either pCDC28–3xHA–sok2 (YEp2562) or pCDC28–GST–MSN2 (YEp2536). The PCRs amplified IME1 IREu region or the nonspecific ARS305 loci. Detection of ARS305 DNA required a 10-fold increase in the level of the input DNA. C, control (a strain without tagged proteins); WCE, whole-cell extract (input); M, marker. (F) Samples for ChIP were taken from cells grown in SA media to 1 × 107 cells/ml. Strains used were Y422 (wild-type diploid, lanes 1 and 2), Y1064 (wild-type haploid, lane 4), Y1171 (msn2Δ msn4Δ/msn2Δ msn4Δ diploid, lane 3), and Y1162 (sok2Δ haploid, lane 5). These strains carried on 2μ plasmid pADH1–GAL4(bd)–ime1(id) (YEp2780).
Oligonucleotides:
Primers used for qPCR:
IME1, CAGCTGCAGAACTTGGTTCA and GTGGAACGTAGATGCGGATT; ACT1, ATCACCGCTTTGGCTCCAT and CCAATCCAGACGGAGTACTTTCTT; and SUM1, TCTACGACCTCTGCGACAAT and CCGTCATCAAGGAAGTCAAA.
Primers used for ChIP–PCR:
IME1–IREu, GACCCAAGAAGCCACCATGA and CGGTGTTATAGCAGCCGCAA; ARS305, CTATCTAAACTGGCTTTC and GAGAGAAACGCAACTACC; IME1–UASru, CGTTGATGTCATCCGCTATT and ACAGCGCAGTTGTGCTATG; and TEL1, GCGTAACAAAGCCATAATGCCTCC and CTCGTTAGGATCACGTTCGAATCC.
RESULTS
Computational assignment of TFs to the IME1 promoter:
S. cerevisiae has ∼150 DNA-binding proteins that affect transcription, whose consensus binding site(s) was reported, although 218 DNA-binding proteins are known (Badis et al. 2008). We used the dataset of known consensus binding sites to predict the TFs that may bind to the 2117-bp regulatory region of the IME1 gene (Sagee et al. 1998). We discovered 75 putative TFs, present on 346 sites, which showed a perfect match to the reported consensus sites. Table S1 gives the complete list of TFs and sites. For statistical analysis, TFs that form a complex were counted as a single TF (see details in Table 3 legend). This approach, which led to the identification of a surprisingly large number of putative TFs, was unlikely to indicate “true” TF-promoter regulators in yeast. To refine our list, we examined a ChIP–chip dataset that defined the genome-wide location of 203 DNA-binding transcription regulators. This analysis identified nine TFs spanning 55 sites within the IME1 promoter (Table S1). Only 16 (29%) of the identified sites showed a perfect match to the consensus sequences, while the rest had a variation in at least one nucleotide (Table S1). We note that this assay was done in haploid cells grown in glucose-containing medium (Harbison et al. 2004). It is possible that true TFs that affect the transcription of IME1 under different conditions were not represented. The large list of putative TFs and “perfect” consensus sites that we generated, together with the information from the ChIP–chip dataset, suggest that a bioinformatic approach relying solely on perfect consensus site identification may not identify the majority of true TFs that regulate a given promoter.
TABLE 3.
Summary of TFs identified using different approaches
| Effect |
No effect |
Not determined |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Maintained site |
Maintained site |
Maintained site |
|||||||||||||
| Approach | No site | Conserved | Preserved | Relocated | Total | No site | Conserved | Preserved | Relocated | Total | No site | Conserved | Preserved | Relocated | Total |
| Consensus | 16 | 3 | 1 | 1 | 21 | 91 | 20 | 2 | 20 | 133 | 133 | 9 | 4 | 25 | 171 |
| ChIP cons. | 5 | 2 | 0 | 0 | 7 | 5 | 2 | 1 | 1 | 9 | 4 | 1 | 0 | 0 | 5 |
| Variation | 16 | 4 | 5 | 18 | 43 | — | — | — | — | — | — | — | — | — | — |
| ChIP var. | 1 | 0 | 0 | 1 | 2 | 9 | 0 | 1 | 9 | 19 | 7 | 2 | 2 | 7 | 18 |
| Total | 38 | 9 | 6 | 20 | 73 | 105 | 22 | 4 | 30 | 161 | 144 | 12 | 6 | 32 | 194 |
Approach: Consensus, site identified by the presence of a perfect match to a reported consensus; ChIP cons, site identified by the presence of a perfect match to a reported consensus as well as by ChIP–chip data; variation, TF identified by the R-SGA assay with an alteration from the known consensus; ChIP var., site identified by ChIP–chip data with an alteration from the known consensus; effect, effect was observed in either the R-SGA assay or reported data (four TFs spanning five sites were added to ChIP cons.); no effect, no effect was found when TF was deleted in the R-SGA assay; not determined, gene not present or not grown in the array. Proteins that bind as a complex were referred to as a single TF. These are: Gcr1-2, Hap2-5, Ino2-4, Rtg1-3, and Swi4-6. Maintained sites, the presence of TF-binding sites was examined in S. cerevisiae, S. bayanus, and S. paradoxus and characterized as follows: conserved, identical sequence in all strains; preserved, sequence was not identical, but consensus was present; relocated, sequence was found at an adjacent site, up to 200 bp; no site, TF-binding site was not found in the three examined strains. Results calculated from Table S1. Additional information is given in Fig. S1 and Table S1.
Analyzing sequence conservation to identify true TFs:
Cliften et al. (2001) suggested that within the promoter regions, conserved sequences between closely related species of Saccharomyces may reveal true sites that bind specific TFs. The promoter region of IME1 from five sensu stricto Saccharomyces strains was aligned using a genome browser. We searched within the promoter of IME1 for conservation between three sensu stricto Saccharomyces strains: S. cerevisiae, its closest relative S. paradoxus, and its distant relative S. bayanus (Figure S1). Because the smallest consensus site for TF binding is 5 bp, a minimum of 5 bp was taken as a measure for complete conservation. Overall, the promoter of IME1 from that of S. cerevisiae and S. paradoxus showed 62% conservation; that of S. cerevisiae and S. bayanus showed, as expected, lower conservation, 22%; and the three sensu stricto strains showed 19% conservation. Within the putative DNA-binding sites that we identified in our computational survey (29 TFs) 11% (37/346) resided in conserved regions. These results imply that regions that may not serve as TF-binding sites were also conserved. This analysis may provide an underestimate of the number of TFs present in conserved regions. This may result from the facts that TFs bind to more than one specific sequence, and lack of conservation may not reflect the absence of a TF-binding site (designated as preserved sites). Moreover, it is also possible that the DNA-binding site exists within the promoter, but at an adjacent location (designated as relocation). Therefore, we also searched the IME1 promoter sequences from S. paradoxus and S. bayanus for the presence of sequences that might bind the putative TFs, which are present in nonconserved regions. By allowing either preservation or relocation of sites, we found that the number of putative sites increased to 92 (27%) and the number of potential TFs to 35 (Table S1).
The transcription of IME1 from three sensu stricto Saccharomyces strains responds similarly to meiotic signals:
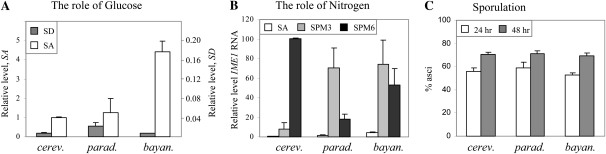
The assumption that conserved sequences are true TF-binding sites predicts that replacing the IME1 promoter of S. cerevisiae with that of S. paradoxus or S. bayanus, both the closest and distant relatives, respectively, would not affect the pattern and/or level of IME1 transcription. We tested this hypothesis by constructing three isogenic S. cerevisiae strains with the endogenous IME1 gene deleted and the IME1 ORF expressed from either S. cerevisiae, S. paradoxus, or S. bayanus IME1 promoters, integrated at the URA3 locus. We discovered that the pattern of IME1 transcription in the three constructed strains was similar (Figure 2, A and B); transcription was repressed under vegetative growth conditions with glucose as the sole carbon source and was induced when cells were grown with acetate as the sole carbon source (Figure 2A). However, the relative level of expression in SA media was about fourfold higher in the strain carrying the S. bayanus promoter in comparison to the S. cerevisiae and S. paradoxus promoters (Figures 2A). This result implies that the promoter of IME1 from either S. cerevisiae or its closest relative S. paradoxus carries additional upstream repression sequence (URS) elements, or that additional UAS elements that respond to the carbon source are present in S. bayanus. It is also possible that the effect resulted from reduced affinity of the TFs for the binding sites due to sequence differences. In S. cerevisiae nitrogen depletion leads to a transient induction in the transcription of IME1 (peak between 6 and 8 hr in SPM) (Kassir et al. 1988). The IME1 promoters of both S. bayanus and S. paradoxus support a transient transcription, albeit the increase as well as decline in transcription is faster (Figure 2B). The different timing in expression did not affect the efficiency of sporulation (Figure 2C). Thus, the similar pattern of expression, namely, glucose repression and a transient increase upon nitrogen depletion, is in agreement with the suggestion that essential positive (UAS) and negative (URS) elements are conserved between the three sensu stricto strains. Nonetheless, the differences in the level of expression in SA media and the kinetic of expression in SPM (Figure 2B) suggest that some URS and/or UAS elements are not conserved.
Figure 2.—
The transcription of IME1 is regulated in a similar manner by IME1 promoters isolated from S. cerevisiae, S. paradoxus, or S. bayanus. (A) RNA was isolated from 1 × 107 cells/ml grown in either SD (shaded column) or SA media (open column). (B) Cells grown in SA to 1 × 107 cells/ml (open column) were shifted to SPM, and at 3 (shaded column) and 6 (solid column) hr, samples were taken to isolate RNA. Level of RNA was determined by qPCR. The relative level of IME1 RNA in comparison to RNA levels of ACT1 is given. (C) Following 24 (open column) and 48 (shaded column) hr of incubation in SPM, the percentage of asci was determined. The results are the average of at least three colonies and standard deviation is given. The isogenic strains used were: Y1808 (pIME1Sc–IME1Sc/pIME1Sc–IME1Sc), Y1809 (pIME1Sp–IME1sc/pIME1Sp–IME1sc), and Y1810 (pIME1Sb–IME1sc/pIME1Sb–IME1sc). cerev., S. cerevisiae; parad., S. paradoxus; bayan., S. bayanus.
IREu is an important but nonconserved element in the IME1 promoter:
The S. cerevisiae IME1 promoter carries two short repeats designated IREu and IREd in which 30 of the 32 bp are identical (Sagee et al. 1998) (Figure 3A). The IREd repeat is conserved in S. paradoxus and S. bayanus, but the IREu repeat is absent (Figure 3B). This is surprising because IREu is an active UAS element in the promoter of IME1, whereas IREd shows only a weak UAS activity (Sagee et al. 1998; Shenhar and Kassir 2001). To verify the function of IREu in the transcription of IME1, we constructed a diploid strain with a precise deletion of the IREu element. We compared the level and pattern of IME1 transcription in this strain relative to the isogenic wild-type strain. Under vegetative growth conditions with glucose as the sole carbon source, the mutant strain showed a 20-fold increase in the level of transcription in comparison to the wild-type strain (Figure 3C). In the presence of acetate as the sole carbon source, a 0.4-fold reduction was observed (Figure 3C). Moreover, under meiotic conditions the level of transcription of IME1 was significantly reduced (Figure 3C). These results reinforce the conclusion that IREu serves as a negative control element in the presence of glucose as the sole carbon source and as a positive element in the presence of acetate, with or without a nitrogen source. The reduced level of IME1 mRNA had no effect on the efficiency of asci formation (Figure 3D), in agreement with our recent report that modulating the levels of IME1 RNA and protein has no effect on the level of sporulation (Gurevich 2010).
The activity of the IREu element in the presence of glucose is regulated by Sok2, while its UAS activity in the presence of acetate as the sole carbon source is regulated by Msn2/4 and Ime1. Genetic analysis suggests that Sok2 binds the TTTTCGTC site (Shenhar and Kassir 2001), while Msn2/4 binds to the STRE element—AGGGG (Sagee et al. 1998) (Figure 3A). We used ChIP assays (Figure 3E) to demonstrate that Sok2 binds to a region encompassing its predicted binding site and to confirm the localization of Msn2 to this region. Consistent with previous findings showing that the presence of glucose excludes Msn2 from the nucleus (Gorner et al. 1998), we found that Msn2 bound IREu only in the absence of glucose (Figure 3E). Sok2 was present on the promoter regardless of the carbon source (Figure 3D), confirming genetic analysis suggesting that relief of Sok2 repression in the absence of glucose is not mediated by its sequestering from the promoter (Shenhar and Kassir 2001). The Sok2-binding site does not correspond to the reported consensus site for Sok2; rather, Sok2 binds to an Swi4,6-dependent cell cycle box (SCB)-like element in the IME1 promoter (Figure 3A), probably reflecting its extensive homology to the DNA-binding domain of Swi4 (Ward et al. 1995), which binds SCB elements. Ime1 was present on the promoter in wild-type cells, but not in the msn2Δ msn4Δ double mutant (Figure 3F), suggesting that Msn2 and/or Msn4 recruits Ime1 to the IREu element. In summary, the IREu element is an essential, but nonconserved element whose activity is directly regulated by Sok2, Msn2/4, and Ime1. These data suggest that discrimination of putative TFs due to lack of conservation between species may be misleading.
High-throughput functional screen for genes affecting transcription of IME1:
The promoter of IME1 consists of distinct elements that are regulated by the same signal through specific TFs. For instance, the glucose signal is transmitted through at least four elements. Consequently, deletion of a single TF may have only a minor effect on the transcription of IME1. Therefore, reporter gene screens are useful because fragments of the IME1 promoter consisting of each individual regulatory element can be screened in isolation to identify specific mutants causing a defect in the ability of that particular element to drive reporter gene expression. We used the R-SGA approach (Kainth et al. 2009) to screen the viable deletion array of ∼4500 genes for mutants that affect the transcription of IME1UAS–HIS4TATA–GFP and HIS4UAS–IME1URS–HIS4TATA–GFP reporter genes. The expression of these reporter genes was controlled by seven distinct UAS and URS elements from the IME1 promoter. The R-SGA assay was performed under two conditions: (1) SD media, which includes glucose as the sole carbon source as well as a nitrogen source, nutrients that repress the transcription of IME1 (Kassir et al. 1988) and (2) SPO media, which induces the transcription of IME1 and meiosis (Kassir et al. 1988). In addition, since UCS3 functions as a repression element in the absence of the Mata1/Matα2 complex (Sagee et al. 1998), the expression of the UCS3–GFP reporter was examined in a homozygous MATa/MATα diploid array. The normalized log2 GFP:RFP ratio was calculated as described (Kainth et al. 2009) (Figure S2 show representative results obtained for two reporter genes). We transformed the above ratios to Z-scores, and P-values were assigned on the basis of a normal distribution (Kainth et al. 2009). A cutoff of <10% was used to identify putative regulators (Table S1). We used the SGA annotation to identify putative TFs, whose direct effect on IME1 transcription was inferred by identifying the TF consensus within the element, allowing imperfect homology (up to three alterations) (Table S1). If there were more than one putative imperfect site to which our potential TF could bind, to calculate the percentage of sites, only one site per element was taken (Table S1).
Our screens identified 41 TFs spanning 68 sites (Table S1). Using a 1% cutoff identified 31 TFs spanning 43 sites. Table 3 and Table S1 also include 4 additional TFs, namely Msn2, Msn4, Rme1, and Sok2 spanning 5 sites, which were identified by a functional assay of additional elements (not tested in this study) and verified by binding assays (Covitz and Mitchell 1993; Sagee et al. 1998; Shenhar and Kassir 2001). Interestingly, only 34% (23/68) (30% for a 1% cutoff) of the identified sites (without the above-mentioned 5 sites) showed a perfect match to the reported consensus. Moreover, 50% (34/68) (65% for a 1% cutoff) of the affected sites were in conserved, preserved, or relocated regions (Table 3, data rows 1, 2, and 5 in data column 5 and Table S1).
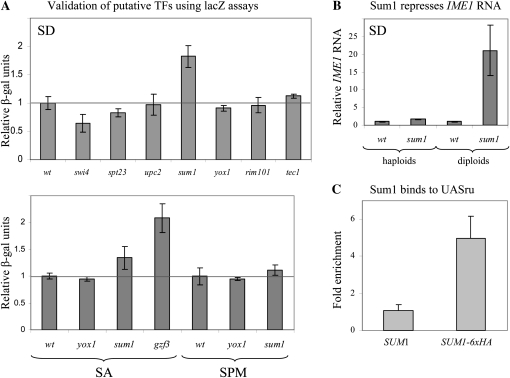
To validate the function of putative TFs identified by the functional genomics screen, we constructed their complete deletion in a different strain background, and determined the level of expression of a lacZ reporter gene. We focused on TFs that affect UASru activity, because this is an important element in IME1 promoter. It functions as a negative element in the presence of glucose and as an essential UAS element in the absence of glucose (Kassir et al. 2003). Eight TFs affecting UASru or UASru(AB) activity with the highest GFP/RFP score were examined (see Figure S2). Deletion of SPT23, SUM1, or SWI4 resulted in a significant effect on the expression of the reporter gene UASru–lacZ (Figure 4A). On the other hand, deletions of UPC2, YOX1, and RIM101 had no effect (Figure 4A). Gzf3 was identified by the R-SGA assay as a regulator of several UAS and URS elements in IME1 promoter (Table S1). We examined therefore its effect on an IME1–lacZ construct, showing that when deleted the level of expression of IME1–lacZ was induced (Figure 4A) (the effect was examined in SA media since in SD IME1 is under repression from several elements). Thus, four of the eight TFs tested showed an effect, suggesting that the genome-wide screen gave ∼50% false positives. Reducing the cutoff to 1%, cast three TFs, namely, Sum1, Yox1, and Rim101, reducing the false positive to only 40%. Nevertheless, this low cutoff could discard true TFs, for instance, Sum1. To further validate the effect of Sum1, we examined the level of IME1 RNA in wild-type and sum1Δ isogenic haploid and diploid strains, using qPCR. Figure 4B shows that IME1 RNA level is increased 1.6- and 21-fold in sum1Δ haploids and diploids, respectively. To determine that the effect of Sum1 is direct, we tested the location of Sum1 on UASru using ChIP-enriched DNA (qChIP). The level of bound Sum1 was calculated relative to the level of binding to a nonspecific locus (TEL1). Figure 4C shows that Sum1 bound specifically to UASru. Thus, our analysis verified the suggestion that Sum1 directly represses the activity of UASru. We note that one of the reported binding sites for Sum1 is GNCRCAAAW, whereas the identified site in UASru is GCCGCAAAG, which has a single alteration from the consensus site.
Figure 4.—
Validation of putative TFs identified by the R-SGA screen. (A) Samples were taken to extract proteins and measure lacZ levels following growth in either SD or SA media to 107 cells/ml. In addition, proteins were extracted from cells grown in SA media to 1 × 107 cells/ml and shifted to SPM for 6 hr. The relative level of β-gal is given. Strains used are wild-type (Y1214) and its isogenic swi4Δ (Y1855), spt23Δ (Y1860), upc2Δ (Y1859), sum1Δ (Y1827), yox1Δ (Y1824), rim101 (Y1854), and tec1Δ (Y1858) strains. These strains carry IME1UASru–HIS4TATA–lacZ integrated in the genomic LEU2 gene. Additional strains were the wild-type Y1825 and its isogenic gzf3Δ (Y1836). These strains carry IME1–lacZ integrated in the genomic LEU2 gene. The results are the averages of at least three independent colonies, and standard deviation is given. (B) RNA was isolated from cells grown in SD media to 1 × 107 cells/ml. Level of RNA was determined by qPCR. The relative level of IME1 RNA in comparison to RNA levels of ACT1 is given. Strains used are: Y1214 and Y1721, the wild-type haploid and diploid strains, respectively, and their isogenic sum1Δ (Y1827) and sum1Δ/sum1Δ (Y1891) strains. The results are the averages of at least three independent colonies. (C) Samples for ChIP assay were taken from 3 × 108 logarithmic cells grown in SD. Real-time PCR was used to amplify the specific UASru element present on IME1UASru–HIS4TATA–lacZ and the nonspecific TEL1. Fold enrichment of the specific vs. the nonspecific PCR products for IP without antibody (w/o α) or with antibody directed against the HA epitope (α HA) are given. The results are the averages of three independent colonies, and standard deviation is given. Strain used is Y1837.
The high-throughput functional assay allowed us to determine whether the putative TFs, which were assigned to the IME1 promoter using bioinformatic or genome-wide location analysis, had any effect on the transcription of IME1. As stated above, the functional analysis was done for only 7 of the 10 designated UAS and URS elements; therefore, the function of many sites was not determined (ND in Table 3 and Table S1). Moreover, some of the TFs were not present on the deletion array, and thus the function of additional sites (total of 194) was not determined. Nonetheless, our in vivo screens showed that most of the TFs (86%, 142/165, for a 10% cutoff; Table 3, data rows 1 and 2 in data columns 5 and 10, and 92%, 152/165, for a 1% cutoff; Table S1) identified by the presence of a perfect site through our computational analysis, showed no effect when analyzed in the deletion strains under the conditions tested.
DISCUSSION
We used IME1 as a paradigm to explore different approaches to identify the TFs that bind to and regulate the transcription of any gene of interest, specifically master regulators of developmental pathways. Moreover, only five TFs that bind to the IME1 promoter and regulate its function were known and we reasoned that further scrutiny of IME1 transcription would enrich our knowledge of TFs that regulate meiosis. The decoration of IME1 promoter with the TFs identified by the different approaches is summarized in Figure S1, which also shows the sequence alignment of the three sensu stricto strains.
The bioinformatic approach:
Consensus sequences have been reported for most predicted TFs in yeast. Therefore, our first approach involved searching of the IME1 sequence for these reported consensus sites. In this way, we identified a large number of putative TF sites (346) and distinct TFs (75), which probably reflects the observation that single TFs can interact with a range of related sequences (Lapidot et al. 2008). We explored the functionality of the putative TFs and binding sites in the IME1 intergenic region by screening various IME1–GFP reporter genes for their sensitivity to gene deletion by using the array of viable yeast deletion mutants. We examined 170 sites (including the ones reported previously), only 28 of which (16%) had an effect (only 18 sites when a 1% cutoff was used), namely, deletion of the TFs with a consensus binding site, did not impair the expression of the specific reporter gene (Figure 5; Table 3, data rows 1 and 2 in data columns 5 and 10; and Table S1). Thus, simply scanning the promoter region for consensus TF-binding sites results in a significant number of false positives sites. There are several possible explanations for the high level of false positives obtained by compiling lists of consensus sites. First, the complex structure of IME1 promoter may mean that each TF may have only a small effect when tested individually. Consequently, our assay may be lacking the sensitivity required to detect a transcriptional defect upon deletion of these TFs. Second, we examined reporter gene expression in two physiological relevant conditions: (1) vegetative growth conditions with glucose as the sole carbon source and (2) sporulation conditions. Therefore, effects of other TFs might be observed in different, unstudied conditions. For instance, Yhp1 has been reported to bind UASv and repress transcription of a UASv–PHO84p–PHO5 reporter gene (Kunoh et al. 2000). In our R-SGA assay, the UASv–GFP reporter lacking a heterologous UAS element could not reveal a repressive activity for Yhp1 (Table S1). These results suggest that some of the false negatives may in fact be true TFs that regulate IME1 transcription. Third, chromatin architecture may mask the ability of a TF to bind to a putative consensus sequence, and therefore such a site might not affect transcription in a reporter assay (Yuan et al. 2005). Finally, most consensus sites were revealed by comparing sequences from genes whose expression depends on a specific TF, or ones that bind a specific TF. However, direct mutational analysis has been performed for only a few TFs. Therefore, some of the reported consensus sequences may not be accurate, which may explain our failure to discover a perfect match to reported consensus sites for most of the TFs identified by the functional analysis (62%, 45/73; Table 3, data rows 3 and 4 in data column 5). This hypothesis is supported by the observation that Sum1 binds to and represses the activity of IME1 UASru element (Figure 4), while a perfect match to any of the reported consensus binding sites for Sum1 is not present. We suggest that Sum1 binds to the sequence GCCGCAAAG, which deviates from the GNCRCAAAW consensus (YEASTRACT) by a single alteration.
Figure 5.—
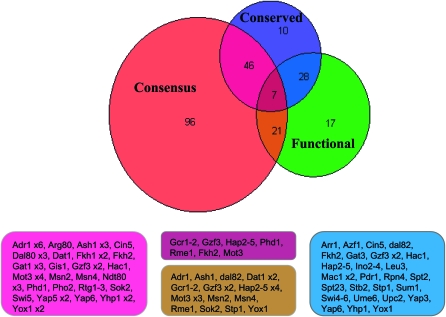
Venn diagram based on Table 3. The TFs belonging to the intersecting groups are given in boxes. Most functional TFs are not present in conserved regions nor in sites that match the reported consensus.
Conservation:
We examined the feasibility of using conservation to predict true regulation by TFs from false positives; 48% (35/73) of the functional sites were maintained in the sensu stricto strains (Table 3, data row 5 in data columns 2–5, and Figure 5). A similar percentage, namely 35% (56/161) of the nonfunctional sites were maintained (Table 3, data row 5 in data columns 7–10). Moreover, within the sites identified by the presence of consensus sequence, conservation did not discriminate between functional and nonfunctional sites (4 vs. 27%; Table 3, data rows 1 and 2 compare data columns 5 and 10). These results do not support the hypothesis that functional sites are maintained during evolution. We directly examined this hypothesis by exchanging the promoter of IME1 from S. cerevisiae with that from S. paradoxus or S. bayanus. Swapping of the promoter had no effect on the pattern of transcription of IME1 (Figure 2), suggesting that in these three strains, the pattern of transcription of IME1 is similar and that the binding sites for the essential TFs are conserved. However, we reached an opposite conclusion when we examined the conservation of two repeated elements in IME1 promoter, IREu, which serves as an essential element, and IREd (Figure 3). The IREd element differs from IREu at two positions, corresponding to the binding sites for Sok2 and Msn2/4 (Figure 3A), explaining why IREd serves as a weak UAS (Kassir et al. 2003). Therefore, we expected that the IREu element would be conserved in the sensu stricto strains. To our surprise the IREd element could be identified in these strains, whereas IREu showed no conservation (Figure 3B and Figure S1). This result implies that binding sites for critical transcription factors may not be conserved in evolution, suggesting that conservation cannot be used to discriminate between true and false positive TFs. This lack of conservation may be due to the growth of these yeast strains in different ecological niches. Therefore, the transcription of IME1 and consequently meiosis, may be regulated by different promoter elements. Nonetheless, IME1 may present an unusual example as meiosis is a robust process, which is neither sensitive to the levels of IME1 mRNA nor to its time of expression (Gurevich 2010). Thus, the shuffling of the IME1 promoter in S. cerevisiae with that of S. paradoxus and S. bayanus, which affected the time of transcription of IME1 or creating an IME1 allele with a deletion of IREu, which resulted in drastic reduction in the levels of IME1 RNA, had no effect on the level of asci formation (Figures 2, B and C, and 3). Since evolution will select for spore formation rather than the level of expression of IME1, the DNA-binding sites, even for important TFs, were not conserved. Moreover, multiple elements, each with a small impact, regulate the transcription of IME1. For this reason the elimination of a single binding site will have a weak rather than a strong and critical effect on the transcription of IME1.
The functional assay:
In this report we used a functional, high-throughput assay to identify genes that regulate the transcription of IME1. Previously, this approach was successfully used to identify chromatin-associated proteins that modulate the transcription of histone genes (Fillingham et al. 2009). A revised approach was devised for screening the IME1 promoter. First, the transcription of IME1 is regulated by multiple and distinct elements, and thus a mutation in a single TF may have only a minor effect. Therefore, we did not fuse the entire IME1 promoter to the GFP reporter; rather, distinct elements were used. The activity of these elements in the GFP reporter genes was identical to their function when fused to lacZ reporters (data not shown). Second, to identify TFs that affect the function of a URS element, we inserted these elements between HIS4UAS and the HIS4 TATA box of a HIS4UAS–HIS4TATA–GFP. Our ability to identify genes, whose deletions affect the activity of the specific URS element, suggests that this modification of the R-SGA method is useful. Third, the assay was performed under both vegetative and meiotic conditions. This screen enabled the identification of many genes, including the expected chromatin remodeling factors, mediators, as well as various TFs.
We considered a TF with a direct effect if a perfect or nonperfect match to the reported consensus was found. However, since for most (62%; Figure 5) of the TFs that were identified as affecting IME1 a perfect consensus site was not found, this approach cannot be used to rule out a direct effect. Thus, it is possible that the list lacks TFs that did not pass the threshold for defining regulators in our screen (FDR of 10%) or ones whose binding sites were not reported. Moreover, it is possible that the list of the identified TFs (Table S1) contains false positives, and therefore, their direct effect needs to be validated. Indeed, we reexamined the effect of eight TFs identified by our functional assay, and only four of them were validated (Figure 4). Thus, using a cutoff of 10% gave ∼50% false positives.
In summary, in this report we applied both bioinformatics and functional genomic approaches to scrutinize the unusually large promoter of the IME1 gene, which encodes the master transcriptional activator of a developmental pathway, meiosis in budding yeast. The functional R-SGA analysis identified new TFs, whose function was validated. The simple approach, namely, the presence of a consensus site, gave rise to too many transcription factors, most of which were nonfunctional. We also showed that conservation between the sensu stricto Saccharomyces strains can be misleading, as in many cases, functional TFs were present within a nonconserved region. We conclude, therefore, that there is no “easy” bioinformatic method to predict the function of a TF, and functional tests are still critical.
The role of the putative TFs in the transcriptional regulation of IME1:
The R-SGA screen identified 69 new putative TFs that affect the transcription of IME1. We reexamined the function of 8 putative TFs, demonstrating that the function of only 4 (Sum1, Swi4, Gzf3, and Spt23) was verified (Figure 4). This suggests that the screen could identify ∼30 new putative TFs. The direct function of one gene, SUM1, was determined by qChIP analysis (Figure 4C). Previous reports showed that Sum1 is a negative regulator of NDT80 and the middle meiosis-specific genes, with no apparent effect on the transcription of an early meiosis-specific gene, HOP1 (Xie et al. 1999; Lindgren et al. 2000). Because the activity of Ime1 is regulated by glucose (Rubin-Bejerano et al. 2004), it is not surprising that the effect of Sum1 on the transcription of IME1 did not affect the transcription of HOP1. GZF3 encodes a zinc finger protein that negatively regulates nitrogen catabolic gene expression (Soussi-Boudekou et al. 1997). Since the transcription of IME1 is negatively regulated by nitrogen (Kassir et al. 1988), it is not surprising that deletion of this gene affected the UCS1 activity (Table S1), a negative element in the IME1 promoter whose repression activity depends on the presence of nitrogen (Kassir et al. 2003). Moreover, UASv is a UAS element whose activity is induced upon nitrogen depletion (V. Gurevich and Y. Kassir, unpublished results). Gzf3 also affected the activity of URSuE, URSd, and UASrm (Table S1). Further work is required to verify whether the activity of these elements is directly regulated by nitrogen and Gzf3. Swi4 is a TF required for entry into the cell cycle (Breeden and Nasmyth 1987; Andrews and Herskowitz 1989), and Spt23 is required for the transcription of OLE1 (Chellappa et al. 2001), a gene involved with lipid biosynthesis. Further work is required to elucidate whether these TFs have a direct effect on the transcription of IME1.
Acknowledgments
We thank Lourdes Peña-Castillo for the statistical analysis of the raw data and Nir Kahana for help in the use of computational tools to analyze the data. This work was supported by grants from the Israel Science Foundation (to Y.K.), Canadian Institutes for Health Research (to B.A.), an European Molecular Biology Organization (EMBO) short-term fellowship (to S.K.), and an Ontario graduate scholarship (to P.K.).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.122200/DC1.
References
- Andrews, B. J., and I. Herskowitz, 1989. Identification of a DNA binding factor involved in cell-cycle control of the yeast HO gene. Cell 57 21–29. [DOI] [PubMed] [Google Scholar]
- Badis, G., E. T. Chan, H. van Bakel, L. Pena-Castillo, D. Tillo et al., 2008. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell 32 878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, M. F., and M. L. Bulyk, 2006. Protein binding microarrays (PBMs) for rapid, high-throughput characterization of the sequence specificities of DNA binding proteins. Methods Mol. Biol. 338 245–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeden, L., and K. Nasmyth, 1987. Cell cycle control of the yeast HO gene: cis- and trans-acting regulators. Cell 48 389–397. [DOI] [PubMed] [Google Scholar]
- Chellappa, R., P. Kandasamy, C. S. Oh, Y. Jiang, M. Vemula et al., 2001. The membrane proteins, Spt23p and Mga2p, play distinct roles in the activation of Saccharomyces cerevisiae OLE1 gene expression. Fatty acid-mediated regulation of Mga2p activity is independent of its proteolytic processing into a soluble transcription activator. J. Biol. Chem. 276 43548–43556. [DOI] [PubMed] [Google Scholar]
- Chua, G., Q. D. Morris, R. Sopko, M. D. Robinson, O. Ryan et al., 2006. Identifying transcription factor functions and targets by phenotypic activation. Proc. Natl. Acad. Sci. USA 103 12045–12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliften, P. F., L. W. Hillier, L. Fulton, T. Graves, T. Miner et al., 2001. Surveying Saccharomyces genomes to identify functional elements by comparative DNA sequence analysis. Genome Res. 11 1175–1186. [DOI] [PubMed] [Google Scholar]
- Cliften, P., P. Sudarsanam, A. Desikan, L. Fulton, B. Fulton et al., 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301 71–76. [DOI] [PubMed] [Google Scholar]
- Covitz, P. A., and A. P. Mitchell, 1993. Repression by the yeast meiotic inhibitor RME1. Genes Dev. 7 1598–1608. [DOI] [PubMed] [Google Scholar]
- Davidson, E. H., J. P. Rast, P. Oliveri, A. Ransick, C. Calestani et al., 2002. A genomic regulatory network for development. Science 295 1669–1678. [DOI] [PubMed] [Google Scholar]
- Fillingham, J., P. Kainth, J. P. Lambert, H. van Bakel, K. Tsui et al., 2009. Two-color cell array screen reveals interdependent roles for histone chaperones and a chromatin boundary regulator in histone gene repression. Mol. Cell 35 340–351. [DOI] [PubMed] [Google Scholar]
- Gorner, W., E. Durchschlag, M. T. Martinez-Pastor, F. Estruch, G. Ammerer et al., 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12 586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granot, D., J. P. Margolskee and G. Simchen, 1989. A long region upstream of the IME1 gene regulates meiosis in yeast. Mol. Gen. Genet. 218 308–314. [DOI] [PubMed] [Google Scholar]
- Gray, M., M. Kupiec and S. M. Honigberg, 2004. Site-specific genomic (SSG) and random domain-localized (RDL) mutagenesis in yeast. BMC Biotechnol. 4 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich, V., Y. Kassir, 2010. A switch from a gradient to a threshold mode in the regulation of a transcriptional cascade promotes robust execution of meiosis in budding yeast. PLoS ONE 5 e11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison, C. T., D. B. Gordon, T. I. Lee, N. J. Rinaldi, K. D. Macisaac et al., 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kainth, P., H. E. Sassi, L. Pena-Castillo, G. Chua, T. R. Hughes et al., 2009. Comprehensive genetic analysis of transcription factor pathways using a dual reporter gene system in budding yeast. Methods 48 258–264. [DOI] [PubMed] [Google Scholar]
- Kassir, Y., and G. Simchen, 1991. Monitoring meiosis and sporulation in Saccharomyces cerevisiae. Methods Enzymol. 194 94–110. [DOI] [PubMed] [Google Scholar]
- Kassir, Y., D. Granot and G. Simchen, 1988. IME1, a positive regulator gene of meiosis in S. cerevisiae. Cell 52 853–862. [DOI] [PubMed] [Google Scholar]
- Kassir, Y., N. Adir, E. Boger-Nadja, N. Guttmann-Raviv, I. Rubin-Bejerano et al., 2003. Transcriptional regulation of meiosis in budding yeast. Int. Rev. Cytol. 224 111–171. [DOI] [PubMed]
- Kunoh, T., Y. Kaneko and S. Harashima, 2000. YHP1 encodes a new homeoprotein that binds to the IME1 promoter in Saccharomyces cerevisiae. Yeast 16 439–449. [DOI] [PubMed] [Google Scholar]
- Lapidot, M., O. Mizrahi-Man and Y. Pilpel, 2008. Functional characterization of variations on regulatory motifs. PLoS Genet. 4 e1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren, A., D. Bungard, M. Pierce, J. Xie, A. Vershon et al., 2000. The pachytene checkpoint in Saccharomyces cerevisiae requires the Sum1 transcriptional repressor. EMBO J. 19 6489–6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIsaac, K. D., T. Wang, D. B. Gordon, D. K. Gifford, G. D. Stormo et al., 2006. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics 7 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J., 1972. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Pnueli, L., I. Edry, M. Cohen and Y. Kassir, 2004. Glucose and nitrogen regulate the switch from histone deacetylation to acetylation for expression of early meiosis-specific genes in budding yeast. Mol. Cell. Biol. 24 5197–5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, F. P., J. D. Hughes, P. W. Estep and G. M. Church, 1998. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat. Biotechnol. 16 939–945. [DOI] [PubMed] [Google Scholar]
- Rubin-Bejerano, I., S. Sagee, O. Friedman, L. Pnueli and Y. Kassir, 2004. The in vivo activity of Ime1, the key transcriptional activator of meiosis-specific genes in Saccharomyces cerevisiae, is inhibited by the cyclic AMP/protein kinase A signal pathway through the glycogen synthase kinase 3-beta homolog Rim11. Mol. Cell. Biol. 24 6967–6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein, A., V. Gurevich, Z. Kasulin-Boneh, L. Pnueli, Y. Kassir et al., 2007. Faithful modeling of transient expression and its application to elucidating negative feedback regulation. Proc. Natl. Acad. Sci. USA 104 6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagee, S., A. Sherman, G. Shenhar, K. Robzyk, N. Ben-Doy et al., 1998. Multiple and distinct activation and repression sequences mediate the regulated transcription of IME1, a transcriptional activator of meiosis-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 18 1985–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenhar, G., and Y. Kassir, 2001. A positive regulator of mitosis, Sok2, functions as a negative regulator of meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 21 1603–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, M., W. Li, P. A. Covitz, M. Hara, H. Shindo et al., 1998. Genomic footprinting of the yeast zinc finger protein Rme1p and its roles in repression of the meiotic activator IME1. Nucleic Acids Res. 26 2329–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R. S., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soussi-Boudekou, S., S. Vissers, A. Urrestarazu, J. C. Jauniaux and B. Andre, 1997. Gzf3p, a fourth GATA factor involved in nitrogen-regulated transcription in Saccharomyces cerevisiae. Mol. Microbiol. 23 1157–1168. [DOI] [PubMed] [Google Scholar]
- Tirosh, I., J. Berman and N. Barkai, 2007. The pattern and evolution of yeast promoter bendability. Trends Genet. 23 318–321. [DOI] [PubMed] [Google Scholar]
- Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader et al., 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294 2364–2368. [DOI] [PubMed] [Google Scholar]
- Ward, M. P., C. J. Gimeno, G. R. Fink and S. Garrett, 1995. SOK2 may regulate cyclic AMP-dependent protein kinase-stimulated growth and pseudohyphal development by repressing transcription. Mol. Cell. Biol. 15 6854–6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, J., M. Pierce, V. Gailus-Durner, M. Wagner, E. Winter et al., 1999. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 18 6448–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, G. C., Y. J. Liu, M. F. Dion, M. D. Slack, L. F. Wu et al., 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309 626–630. [DOI] [PubMed] [Google Scholar]