Abstract
Robust mechanisms for tissue repair are critical for survival of multicellular organisms. Efficient cutaneous wound repair requires the migration of cells at the wound edge and farther back within the epidermal sheet, but the genes that control and coordinate these migrations remain obscure. This is in part because a systematic screening approach for in vivo identification and classification of postembryonic wound closure genes has yet to be developed. Here, we performed a proof-of-principle reporter-based in vivo RNAi screen in the Drosophila melanogaster larval epidermis to identify genes required for normal wound closure. Among the candidate genes tested were kinases and transcriptional mediators of the Jun N-terminal kinase (JNK) signaling pathway shown to be required for epithelial sheet migration during development. Also targeted were genes involved in actin cytoskeletal remodeling. Importantly, RNAi knockdown of both canonical and noncanonical members of the JNK pathway caused open wounds, as did several genes involved in actin cytoskeletal remodeling. Our analysis of JNK pathway components reveals redundancy among the upstream activating kinases and distinct roles for the downstream transcription factors DJun and DFos. Quantitative and qualitative morphological classification of the open wound phenotypes and evaluation of JNK activation suggest that multiple cellular processes are required in the migrating epidermal cells, including functions specific to cells at the wound edge and others specific to cells farther back within the epidermal sheet. Together, our results identify a new set of conserved wound closure genes, determine putative functional roles for these genes within the migrating epidermal sheet, and provide a template for a broader in vivo RNAi screen to discover the full complement of genes required for wound closure during larval epidermal wound healing.
CELL migration is a critical feature of wound healing responses (Martin 1997; Singer and Clark 1999). During postembryonic cutaneous repair in humans, rodents, and Drosophila larvae, highly differentiated barrier epidermal cells assume a polarized and motile morphology (Odland and Ross 1968; Clark et al. 1982; Galko and Krasnow 2004; Wu et al. 2009). This change to a migratory phenotype is essential for efficient repair. The identification of genes involved in repair and assignment of specific cellular functions to these genes in vertebrate models have been hindered by the redundancy and complexity of the vertebrate tissue repair response (Martin 1997; Grose and Werner 2004). Cell culture-based assays (Simpson et al. 2008; Vitorino and Meyer 2008) have allowed high-throughput identification of genes required for closure of simple epithelial scratch wounds. Further, a genetic screen for genes required for the mechanistically distinct process of embryonic wound closure was recently reported (Campos et al. 2009). However, the tissue repair field still lacks a method for systematic in vivo identification and functional classification of genes required for postembryonic wound closure.
One pathway implicated in postembryonic repair in both vertebrates and Drosophila is the Jun N-terminal kinase (JNK) signaling pathway (Ramet et al. 2002; Li et al. 2003; Galko and Krasnow 2004), which often controls epithelial migrations (Xia and Karin 2004). In Drosophila, JNK has been implicated in developmentally programmed epithelial migrations, including dorsal closure (DC) (Riesgo-Escovar et al. 1996; Sluss et al. 1996), thorax closure (Zeitlinger and Bohmann 1999), and border cell migration (Llense and Martin-Blanco 2008). Of these responses, JNK signaling during DC is the most extensively studied. In this canonical context the intracellular signaling relay has been defined as follows: the JNK kinase kinase kinase (Jun4K) Misshapen (Treisman et al. 1997; Su et al. 1998), the JNK kinase kinase (Jun3K) Slipper (Stronach and Perrimon 2002), the JNK kinase (Jun2K) Hemipterous (Glise et al. 1995), and the JNK Basket (Riesgo-Escovar et al. 1996; Sluss et al. 1996). Phosphorylated Basket activates the transcription factors DJun and DFos (Riesgo-Escovar and Hafen 1997; Kockel et al. 2001) encoded by the genes Jun-related antigen (Jra) and kayak (kay), respectively. Despite intensive study, the signal(s) that activate this pathway during both DC and larval wound closure remain unidentified. In larval wound healing, the only JNK pathway component so far shown to be required for healing is Basket (Galko and Krasnow 2004), but the architecture of this signaling pathway both upstream and downstream of JNK activation remains unclear. Similarly unclear is the relationship between JNK activation and the actin cytoskeletal dynamics that likely drive epidermal cell migration across the wound gap, although an increase of actin at the wound edge in larvae lacking JNK within the epidermal sheet was recently shown (Wu et al. 2009).
Localized actin polymerization is thought to drive the protrusive cell behavior observed during wound healing and other instances of cell migration (Pollard and Borisy 2003). Biochemical and cell culture approaches have identified many regulators of actin cytoskeletal dynamics including the Rho GTPases (Nobes and Hall 1995) and the Arp2/3 complex (Welch et al. 1997) and its activators like SCAR, all of which can stimulate polymerization of new or rearrangement of existing actin filaments. SCAR, for instance, is clearly required for polymerization of new actin filaments that branch from the shafts of preexisting filaments (Goley and Welch 2006), a prerequisite for lamellipodial protrusion in cell culture (Kiger et al. 2003). SCAR is also required for certain cell migrations in the developing Drosophila embryo (Zallen et al. 2002). However, the in vivo requirement of most such factors during physiologically induced cell migrations such as wound healing remains unclear.
Here, by combining an epidermal reporter of cell morphology, a larval wound healing assay, and UAS-RNAi gene knockdown technology (Kennerdell and Carthew 2000; Dietzl et al. 2007) we performed a proof-of-principle genetic screen targeting primarily putative JNK pathway kinases, transcription factors, substrates (Otto et al. 2000), and selected regulators of actin cytoskeletal dynamics. Although all members of the canonical JNK pathway defined in DC are also required for normal wound closure, the architecture of the signaling pathway during larval wound healing differs in important ways. Further, we established functional roles for several regulators of the actin cytoskeleton. Some of the genes appeared to function primarily in leading edge cells, while others acted farther back in the epidermal sheet or in both groups of cells. Thus, our screen is capable of identifying new wound closure genes and subsequent morphological analysis can assign putative functional roles to these genes. Since UAS-RNAi transgene libraries now cover nearly the whole genome (Dietzl et al. 2007) and functional annotation of these lines is rapidly proceeding (Mummery-Widmer et al. 2009), the methodology described here is highly scalable. Thus, we anticipate eventually identifying and dissecting the specific roles of the full complement of genes required within the epidermis for proper wound healing.
MATERIALS AND METHODS
Fly strains and genetics:
Drosophila melanogaster strains were reared at 25° on standard cornmeal media. We used the GAL4/UAS system (Brand and Perrimon 1993) for tissue-specific expression of transgenes: A58-Gal4 (Galko and Krasnow 2004) and UAS-Dcr-2;A58-Gal4 (see below) express Gal4 predominantly in the larval epidermis, while e22c-Gal4 (Lawrence et al. 1995) expresses Gal4 in the embryonic and larval epidermis. We constructed three “wound reporter” lines on the basis of these epidermal drivers. All involved recombining the driver with a UAS-src-GFP transgene to label cell membranes and a UAS-DsRed2-Nuc transgene to label cell nuclei. w1118;;UAS-src-GFP,UAS-DsRed2-Nuc,A58-Gal4/TM6B (A58) was built as follows: DsRed2-Nuc was PCR amplified from pDsRed2-Nuc (BD Biosciences Clontech) using a 5′ primer with an EcoRI restriction site (5′-GAAGGAATTCATGGCCTCCTCCGAGAACG-3′) and a 3′ primer with an EagI restriction site (5′-GAAGCGGCCGTTATCTAGATCCGGTGG-3′), digested with EcoRI and EagI, and ligated into EcoRI/EagI-digested pUAST. Transgenic flies were generated by standard procedures and UAS-DsRed2-Nuc inserts on the third chromosome were recombined with A58-Gal4 and a third chromosome UAS-src-GFP transgene. In parallel, this A58 reporter was combined with a second chromosome UAS-Dicer-2 (Dcr-2) transgene (Dietzl et al. 2007) to enhance RNAi potency (Dcr-2;A58). The “e22c reporter” w1118;e22c-Gal4,UAS-src-GFP,UAS-DsRed2-Nuc/CyO (e22c) was constructed similarly. For detailed morphological analysis (see below) we used w1118;e22c-Gal4,UAS-DsRed2-Nuc/CyO hs-hid and crossed it to the respective RNAi lines. UAS-basketRNAi or UAS-basketDN served as a positive control for wound closure defects, while progeny of w1118 crossed with the respective reporter served as a negative control. We refer to the parental line bearing the UAS-RNAi transgene as the “RNAi line”. When an RNAi line is crossed with one of the Gal4 wound reporters, we refer to the progeny as “geneX RNAi”.
All 190 UAS-RNAi lines used in our pilot screen (supporting information, Table S1) were obtained from NIG-Fly (http://www.shigen.nig.ac.jp/fly/nigfly/index.jsp) or from the Vienna Drosophila RNAi Center (VDRC). For optimization of screening the following RNAi lines targeting the following genes were selected: 1388R-1 and 1388R-2 (TGF- β activated kinase 1, Tak1); 2190R-2 and 4353R-2 (hemipterous, hep); 2248R-1 (Rac1); 2272R-1, 2272R-2, 2272 #33516*, and 2272 #33518 (slipper, slpr); 2275R-2 (Jun-related antigen, DJun/Jra); 4636R-1 (SCAR); 4720 #34891 and 4720 #34892* (Protein kinase at 92B, Pk92B); 4803 #34898 (Tak1-like 2, Takl2); 5336R-2 and 5336R-3* (Ced-12); 5680R-1 and 5680R-2 (basket, bsk); 7717 #25528 (Mekk1); 8261R-1 (G protein γ 1, Gγ1); 9738R-1 and 9738R-3* (MAP kinase kinase 4, Mkk4); 9901R-2 (Actin-related protein 14D, Arp14D); 10076R-1 (spire, spir); 10379R-1 (myoblast city, mbc); 12235R-1* and 12235R-3 (Arp11); 12530R-2 and 12530R-3 (Cdc42); 15509R-2 (kayak, DFos/kay); 16973R-1 and 16973R-2 (misshapen, msn); 31421 #25760 (Tak1-like 1, Takl1); and DPXN IR N1 and DPXN IR N3 (Paxillin, Pax). Lines indicated above with an asterisk are the ones plotted in Figure 3, B and C. Lines with two UAS-RNAi inserts targeting the same gene are denoted by x2 in Figure 3, B and C Tak1x2 for 1388R-1,1388R-2: hepx2 for 2190R-2,4353R-2; bskx2 for 5680R-1,5680R-2; Cdc42x2 for 12530R-2,12530R-3; msnx2 for 16973R-2;16973R-1; and Paxx2 for DPXN IR N1,DPXN IR N3.
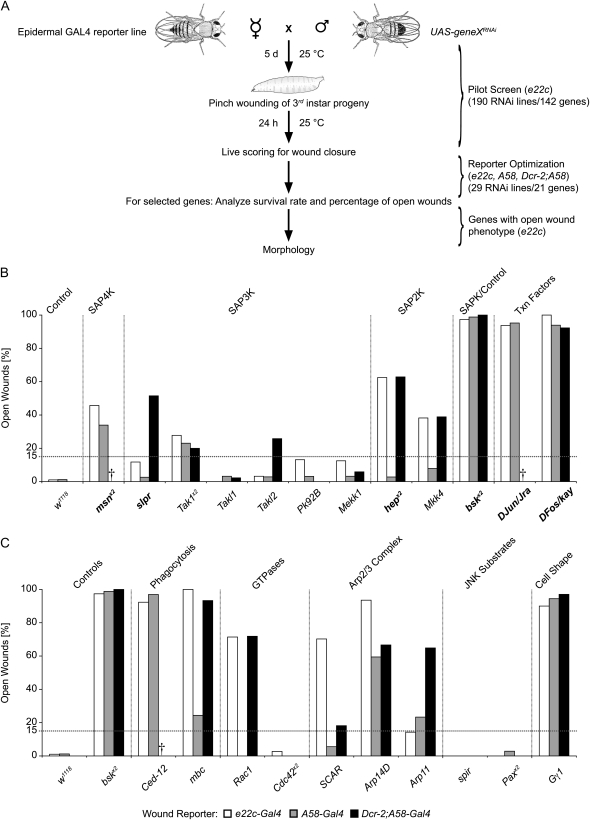
Figure 3.—
Quantification of wound closure upon epidermal expression of UAS-RNAi transgenes. (A) Workflow of the UAS-RNAi–based screen for wound closure genes. (B and C) Percentage of open wounds upon expression of UAS-RNAi transgenes targeting indicated genes of the JNK pathway and other SAPK signaling components (B) and genes involved in actin cytoskeletal remodeling (C). Open bars, e22c reporter; shaded bars, A58 reporter; solid bars, Dcr-2;A58 reporter. w1118 crossed to the respective reporter was used as a negative control and bskRNAi as a positive control. Dashed line, arbitrary 15% cutoff for detailed morphological analysis. Daggers indicate insufficient L3 larvae for testing. Lines with two UAS-RNAi inserts targeting the same gene are denoted by x2. Genes in boldface type in B indicate members of the canonical JNK pathway. The numbers of scored larvae for each RNAi knockdown using A58 or Dcr-2;A58 in B and C were as follows: n(A58-Gal4) = 82 (w1118), 56 (msnx2), 39 (slpr), 39 (Tak1x2), 31 (Takl1), 35 (Takl2), 32 (Pk92B), 31 (Mekk1), 36 (hepx2), 38 (Mkk4), 84 (bskx2), 42 (DJun/Jra), 33 (DFos/kay), 33 (Ced-12), 41 (mbc), 31 (Rac1), 36 (Cdc42x2), 36 (SCAR), 37 (Arp14D), 30 (Arp11), 46 (spir), 35 (Paxx2), and 36 (Gγ1); and n(Dcr-2;A58-Gal4) = 72 (w1118), 33 (slpr), 35 (Tak1x2), 44 (Takl1), 31 (Takl2), 33 (Pk92B), 34 (Mekk1), 35 (hepx2), 36 (Mkk4), 69 (bskx2), 65 (DFos/kay), 30 (mbc), 35 (Rac1), 40 (Cdc42x2), 33 (SCAR), 33 (Arp14D), 37 (Arp11), 46 (spir), 34 (Paxx2), and 34 (Gγ1); see also Table S1 for n(e22c).
We constructed two RNAi lines, each targeting two different genes: 4353R-2,9738R-1 for hep,Mkk4 and 1388R-1,1388R-2;2272 #33516 for Tak1x2;slpr. UAS-Rac1-N17DN and UAS-cdc42-N17DN (Luo et al. 1994) were used to inhibit Rac1 and Cdc42 function, and viable alleles of Tak1 (Tak12527) and slipper (slprBS06) (Polaski et al. 2006) were used to test their functions in larval wound healing.
To confirm the open wound phenotypes listed in Figure 3 we tested additional overlapping and nonoverlapping RNAi lines (the NIG-Fly and VDRC identifiers for these strains are listed in Table S2, columns 3 and 4).
Wounding assays:
Pinch and puncture wounds were performed as described (Galko and Krasnow 2004). Briefly, early third instar (L3) Drosophila larvae were pinched with blunted forceps on the dorsal aspect of a single abdominal segment, usually A4, -5, or -6. Larvae were maintained at 25° with exception of wounding and imaging, which were performed at room temperature.
Pilot screen for wound closure genes:
Females bearing one of the epidermal reporters were crossed to males harboring UAS-RNAi inserts to block target gene expression in the larval epidermis. These sexes were reversed for UAS-RNAi transgenes on the X chromosome. Early L3 progeny larvae harboring both the reporter and the UAS-RNAi transgene were etherized, pinch wounded, and allowed to recover on fly food for 24 hr, a time at which control wounds were almost always closed. For visualization, etherized larvae were mounted on double-sided tape, flattened with coverslips, and viewed dorsal side up with a GFP2 filter on a Leica MZ16FA stereomicroscope using a Planapo 1.6× objective. Images were captured using a color digital camera (Leica DFC300 FX) and Image-Pro AMS v5.1 software (Media Cybernetics). Wounds were scored as “open” if a dark gap free of nuclei remained in the epidermal sheet.
Statistical analysis of the survival rate:
A minimum of 30 unwounded and wounded larvae of each genotype were analyzed for survival rates 24 hr post wounding or post mock treatment. We performed pairwise comparisons (A58 vs. Dcr-2;A58, e22c vs. A58, and e22c vs. Dcr-2;A58) of unwounded (Table S3) and wounded (Table S4) larval survival rates for each UAS-RNAi transgene using chi-square tests and Fisher's exact tests, when appropriate. In addition, we compared the survival rate of unwounded vs. wounded UAS-RNAi transgenes for a given reporter strain (Table S5). To account for the inflated type 1 error rate, a Bonferroni correction was applied (P-value for significant difference was set at 0.0001792 instead of 0.05).
Whole mount immunofluorescence and histochemistry:
Dissection and immunostaining of larval epidermal whole mounts were performed as described except that 4% paraformaldehyde was used as fixative for some antigens (Galko and Krasnow 2004). Primary antibodies were anti-Fasciclin III diluted 1:50 (Patel et al. 1987), anti-phospho-Histone H3 (Ser10) (Upstate Cell Signaling) diluted 1:500, anti-DFos (Zeitlinger et al. 1997) diluted 1:1000, and anti-DJun (Bohmann et al. 1994) diluted 1:2000. All primary and secondary antibodies were diluted in phosphate-buffered saline, 1% heat-inactivated goat serum, and 0.3% Triton X-100 (PHT) buffer. Secondary antibodies were FITC-conjugated goat anti-mouse IgG (H+L) (1:200; Jackson ImmunoResearch), Alexa Fluor 488-conjugated goat anti-mouse IgG2a (1:200; Invitrogen Molecular Probes), and Cy3-conjugated goat anti-rabbit IgG (H+L) (1:300; Jackson ImmunoResearch).
For analysis of JNK activation, RNAi lines were crossed to either w1118;e22c-Gal4/CyO Act∷GFP;msn-lacZ/TM6B or w1118;;msn-lacZ,A58-Gal4/TM6B and progeny larvae carrying msn-lacZ, the Gal4 driver, and the UAS-RNAi transgene were dissected 6 hr post wounding or post mock wounding. Histochemistry was performed as described (Galko and Krasnow 2004) with the following modifications: Samples were fixed for 20 min in cold 2% glutaraldehyde at room temperature, rinsed with phosphate-buffered saline, and stained at 37° for 20 min. Samples were imaged under a 10×/0.40 HC PLAN APO objective with a digital camera (Leica DFC300 FX) mounted on a Leica DM5500 B stereomicroscope. Eighteen to 52 Z-stacks of 2-μm depth were collected and the most in-focus information was extracted using the extended-depth of field algorithm of Image-Pro MDA v6.1 software (Media Cybernetics).
Quantitative analysis of morphological features:
Twenty-four hours post wounding, various wound features were measured. For measurement of wound area (Figure 5B) the wound gap was defined as an area free of epidermal cells. Outlining the wound perimeter using Adobe Photoshop CS3 allowed measurement of this area. To quantify clustering of wound-edge nuclei (Figure 5C) we defined “front line” nuclei as those closest to the wound gap that were >20 μm2 and located <40 μm back from the wound margin. The center-to-center distance between all adjacent front line nuclei was measured. When this distance was <17.8 μm, we considered these nuclei a cluster. Clusters of five or more adjacent nuclei were then counted for open wounds resulting from targeting of genes in classes I, III, and IV. To quantify nuclear crowding farther from the wound edge (Figure 5D) the area occupied by nuclei in a 50-μm strip extending two to three cell rows from the wound margin was measured using Image-Pro version 6.2. To normalize the effect of wound size, we divided this nuclear area by the wound gap area for each wound. Figure 5A provides a schematic overview of the parameters (described above) that we quantified for open wounds. We used the nonparametric Mann–Whitney test to assess statistical significance (Figure 5, B–D). To quantify epidermal syncytium formation in closed wounds (Figure 5E) we counted the number of nuclei in each syncytial cell near three wounds of control larvae (w1118) and of msnRNAi x2 -expressing larvae.
Figure 5.—
Quantification of wound closure classes identified by qualitative morphological features. (A) Schematic of parameters quantified for classes of genes showing open wounds at 24 hr post wounding. Hexagons, epidermal cells; solid circles, epidermal nuclei; b, wound area; c, clustering of front line nuclei; d, crowding of nuclei near the wound margin. The lightly shaded area indicates the region where the area occupied by nuclei was measured to assess nuclear crowding. For details of quantification procedures see materials and methods. (B) Quantification of wound area in select classes. n = 35, 10, and 9 for classes I, II, and V, respectively. ***P < 0.001, Mann–Whitney test. (C) Quantification of clusters of front line nuclei in select classes. n = 35, 32, and 10 for classes I, III, and IV, respectively. ***P < 0.001, Mann–Whitney test. For nonsignificant comparison (NS) P = 0.725. (D) Quantification of nuclear area within 50 μm of the wound edge. n = 15, 15, and 10 for classes I, III, and IV, respectively. ***P < 0.001, Mann–Whitney test. For nonsignificant comparison (NS) P = 0.467. (E) Quantification of epidermal syncytium formation in closed wounds of msnRNAi-expressing larvae. The number of nuclei in each syncytial cell near three wounds of control and msnRNAi x2 was measured. The diameter of the bubbles reflects the number of cells with that number of nuclei. Control wounds have on average more syncytial cells with small numbers of nuclei (2–12) while wounds in msnRNAi-expressing larvae have fewer syncytia with greater numbers of nuclei.
RESULTS
Normal progression of wound closure:
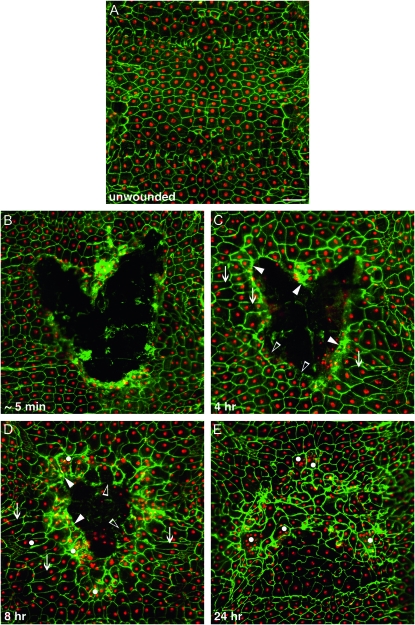
To provide context for analysis of our UAS-RNAi screen, we first observed normal pinch wound closure in control larvae. The unwounded epidermis of these larvae consisted of a regularly patterned monolayer of primarily mononucleate and polygonal cells (Figure 1A). Immediately after wounding, a gap of ∼0.2 mm2 in the epidermal sheet was apparent (Figure 1B). Cells at the wound margin were partially destroyed and intact cells farther away from the wound gap maintained their regular shape. Four hours later, some cells in the first two rows surrounding the wound elongated toward the gap (arrows in Figure 1C). In addition, some wound margin cells showed intense Fasciclin III staining (solid arrowheads), while others lacked Fasciclin III staining at the wound-facing side of the cell (open arrowheads). By 8 hr migrating cells covered much of the original wound gap and cells behind these ones also appeared elongated (arrows in Figure 1D). Further, cells with multiple nuclei (white dots) were visible. By 24 hr almost all wounds were closed and possessed irregularly shaped epidermal cells at the former wound site including some syncytia containing 2–12 nuclei (Figure 1E). In summary, during normal wound closure cells adjacent to the wound change their shape. These cells also begin to migrate to close the wound gap while epidermal cells farther away elongate toward the wound. Moreover, formation of small syncytia at the wound site is a normal event during wound closure. Because immunostaining of dissected larvae is laborious, we next sought to develop reporters that would permit rapid live scoring of wound closure.
Figure 1.—
Temporal progression of wound closure in control larvae. (A–E) Dissected whole mounts of larvae heterozygous for the e22c-Gal4 driver and UAS-DsRed2-Nuc. Nuclei expressing DsRed2-Nuc are red and membranes immunostained for Fasciclin III are green. (A) Unwounded epidermal sheet. (B–E) Epidermal sheet after wounding: (B) ∼5 min post wounding, (C) 4 hr post wounding, (D) 8 hr post wounding, and (E) 24 hr post wounding. Note that the epidermal sheet is closed. Solid arrowheads, wound-edge cells that retain Fasciclin III staining facing the wound gap; open arrowheads, wound-edge cells that lack Fasciclin III staining facing the wound gap; arrows, elongated epidermal cells; dots, multinucleate epidermal cells. Bar, 100 μm.
Epidermal reporters allow live assessment of wound closure:
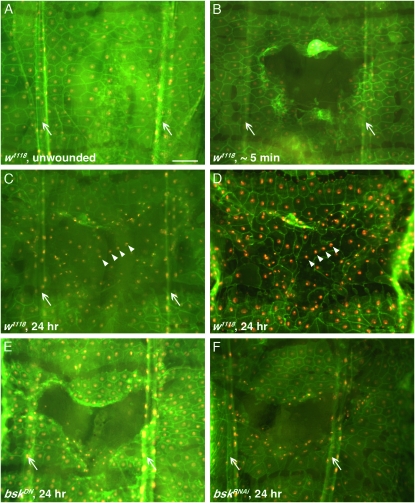
We built wound reporters that consist of a Gal4 driver with either embryonic (e22c) or larval (A58 or Dcr-2;A58) onset of epidermal expression recombined with transgenes to label epidermal cell membranes (UAS-src-GFP) and nuclei (UAS-DsRed2-Nuc). UAS-Dicer-2 is a transgene reported to enhance RNAi potency (Dietzl et al. 2007). We first established that the wound reporters could identify wound closure defects when viewed live. Unwounded A58 reporter-bearing larvae possessed largely polygonal mononuclear epidermal cells of highly uniform size and shape (Figure 2A), similar to those observed in immunostained whole mounts (Figure 1A and Galko and Krasnow 2004). Immediately after wounding, the wound gap was apparent (Figure 2B). Twenty-four hours post wounding, when wound closure was complete in control larvae, irregularly spaced nuclei and a haze of green fluorescence overlaid the previous wound gap (Figure 2C). Although cellular membranes retaining GFP within the former wound gap were not as bright viewed live as in dissected samples (compare Figure 2C to 2D), the src-GFP and nuclear RFP signals were sufficient to unambiguously score defective wound closure (Figure 2, E and F). Similar results were obtained with the e22c and the Dcr2;A58 reporters (data not shown). In conclusion, the three epidermal wound reporters are useful tools for rapid live scoring of wound closure defects. We next used these reporters to establish a viable method for identification of wound closure genes.
Figure 2.—
An epidermal wound reporter allows live scoring of wound closure. (A–C and E–F) Live larvae bearing one copy of the A58 epidermal wound reporter. (A–D) Control (crossed to w1118). (A) Mock wounded. (B–F) Pinch wounded. (B) Immediately post wounding. (C) Twenty-four hours post wounding. Not all cell membranes are visible, but nuclei are present in the former wound gap. (D) Dissected whole mount of the same larva as in C, immunostained for Fasciclin III (green). The arrowheads mark identical positions in C and D. Faint membranes difficult to see in live larvae are now apparent. (E) Larva expressing a UAS-bskDN transgene. (F) Larva expressing a UAS-bskRNAi transgene exhibits a wound closure defect similar to UAS-bskDN. Arrows, tracheal dorsal trunks. Bar, 100 μm.
Screening strategy:
To test the efficacy of RNAi for screening, we first asked whether a tissue-specific UAS-RNAi transgene could phenocopy the wound closure defect observed upon expression of a dominant-negative version of basket (UAS-basketDN), a known wound closure gene (Galko and Krasnow 2004). Reporter-bearing larvae expressing a UAS-bskRNAi transgene phenocopied the UAS-bskDN wound closure defect (Figure 2, E and F), confirming that RNAi-mediated gene knockdown, at least for this gene, could lead to a wound closure defect. Assessment of the RNAi potency of the three reporters using a UAS-GFPRNAi transgene (Levi et al. 2006) to target UAS-src-GFP revealed for the e22c and Dcr-2;A58 reporters strong and approximately equal knockdown that was greater than that achieved with the A58 reporter (Figure S1).
Using the e22c wound reporter, which retained src-GFP membrane fluorescence in wound-proximal cells better than the other reporters (data not shown), we screened 190 UAS-RNAi lines targeting 142 genes (see Table S1 and general scheme in Figure 3A). Fifteen UAS-RNAi transgenes showed an open wound phenotype and 4 transgenes did not produce a substantial population of testable larvae (Table S1). We wished to determine whether onset of expression (embryonic or larval) or RNAi potency would be the determining factor in the yield of the screen. To assess this, we retested a subset of UAS-RNAi transgenes with all three reporters. For this optimization, we surveyed lethality, the percentage of open wounds, and the morphology of the wounded and unwounded epidermis. Our test set included 29 UAS-RNAi lines targeting 21 candidate wound closure genes, most of which scored positive in the first round of screening. Some surprising negatives were also included to give a preliminary assessment of whether there were false negatives in the initial screen. For some of the candidates (slpr, Pk92B, Mkk4, Ced-12, and Arp11) we tested different independent RNAi lines but we present here only the UAS-RNAi transgene that showed the highest percentage of open wounds (Figure 3, B and C). The candidate genes fell into two categories. The first comprised genes of stress-activated protein kinase (SAPK) signaling pathways such as the canonical Jun kinase relay and associated transcription factors (in total 17 UAS-RNAi transgenes targeting 11 genes). The second contained genes involved in actin cytoskeletal remodeling, including Rho-like GTPases and genes involved in phagocytosis (in total 12 UAS-RNAi transgenes targeting 10 genes).
For most of the hits in either category we were able to confirm an open wound phenotype with RNAi lines that target nonoverlapping sequences within the gene (nine of nine lines), alternative RNAi lines that target completely or partially overlapping sequences (five of six lines), dominant-negative transgenes (two of two transgenes), or larval viable mutant alleles (two of two mutants) (Figure S2 and Table S2). For two proteins for which good antibodies for immunostaining in larval whole mounts exist (DFos and DJun), we were also able to verify on-target knockdown of the targeted protein (Figure S3). Together these results suggest that the wound closure phenotype of most of the hits is highly unlikely to be due to off-target RNAi effects.
Canonical and noncanonical JNK pathway components are involved in wound closure:
Genes of the first test category (Figure 3B) encoded canonical JNK pathway and other SAPK signaling components. The highest percentage of open wounds was obtained upon targeting bsk, DJun/Jra, and DFos/kay (92–100% open wounds). These were followed by transgenes targeting the Jun4 kinase Msn (46%), the Jun/SAP2 kinases Hep and Mkk4 (38–63%), and the Jun/SAP3 kinases (0–52%). Interestingly, expression of UAS-RNAi transgenes targeting members of the canonical JNK signaling pathway involved in DC showed a higher percentage of open wounds compared to other kinases at the same level of the kinase cascade. For example, the Jun2K Hep (required for DC) showed 63% open wounds while the SAP2K Mkk4 (not required for DC) showed 38% open wounds using the e22c reporter. Among the six SAP3 kinases we tested, only transgenes targeting slpr, Tak1, and Takl2 displayed open wounds above an arbitrary 15% threshold with the Jun3K Slpr (required for DC) showing the highest percentage (52% with Dcr-2;A58) within the group. In short, downstream members of the JNK/SAPK pathways showed a higher percentage of open wounds than the upstream SAP kinases. It is likely that Hemipterous/Mkk4 at the Jun2K level and Slipper/Tak1 at the Jun3K level are redundant with each other since coexpression of RNAi transgenes targeting both Jun2 kinases or both Jun3 kinases led to a nearly fully penetrant wound closure defect (Figure S4). In conclusion, all genes of the canonical JNK pathway and three noncanonical components (Tak1, Takl2, and Mkk4) showed an open wound phenotype with at least one of the three reporters.
Actin cytoskeletal remodeling is required for normal wound closure:
The second test set comprised 10 genes involved in actin cytoskeletal remodeling (Figure 3C). Four of the UAS-RNAi transgenes exhibited an open wound phenotype similar to the positive control of bskRNAi and three showed >50% open wounds with at least one of the three wound reporters. The most penetrant open wound phenotype (92–100%) was achieved upon targeting Gγ1, Ced-12, Arp14D, and mbc, followed by Rac1, SCAR, and Arp11 (65–72%) in combination with at least one of the reporters. Three UAS-RNAi transgenes (spir, Cdc42, and Pax) showed no open wounds. This latter result could be due to inefficient knockdown of the targeted gene or could indicate that these genes play no role in wound closure (see discussion below).
Comparison of the wound reporters:
In total, an open wound phenotype was scored for 9 UAS-RNAi transgenes using A58 and for 12 UAS-RNAi transgenes using e22c or Dcr-2;A58 (Figure 3, B and C). However, the latter reporter was lethal when crossed to transgenes targeting msn, DJun/Jra, and Ced-12. In addition, we observed that the larval epidermis was more fragile upon expression of Dcr-2 (data not shown). When the percentage of open wounds was high (90–100%), we obtained similar results with all three reporters. In contrast, the choice of the reporter was critical for RNAi lines with a medium to low percentage of open wounds. Where there were differences between the reporters, RNAi potency was the major determinant of the strength of the wound closure phenotype for most of these differences. For instance, targeting of hep, Mkk4, mbc, and Rac1 showed more open wounds with the stronger e22c and Dcr-2;A58 reporters. Only SCAR and Arp14D showed more open wounds with the e22c reporter than with Dcr-2;A58, suggesting that for these genes the early onset of the RNAi expression was the major determinant. For nonlethal crosses, statistical comparison of survival rates between UAS-RNAi transgenes in unwounded (Table S3) and wounded (Table S4) larvae showed that there were only sporadic cases where the choice of reporter significantly influenced subsequent survival. Further, comparison of unwounded vs. wounded survival for a given UAS-RNAi transgene with a particular reporter (Table S5) showed that there was only one case where survival after wounding was significantly different using the e22c-Gal4 reporter. In sum, the e22c reporter has the advantages of both early onset of expression and high RNAi potency (Figure S1) without the lethality and epidermal fragility complications resulting from Dicer-2 expression. Given that e22c also affords a better visualization of wound morphology, we used the e22c reporter for the detailed analysis of wound morphology presented below.
Different classes of wound closure genes can be identified by quantitative and qualitative assessment of wound morphology:
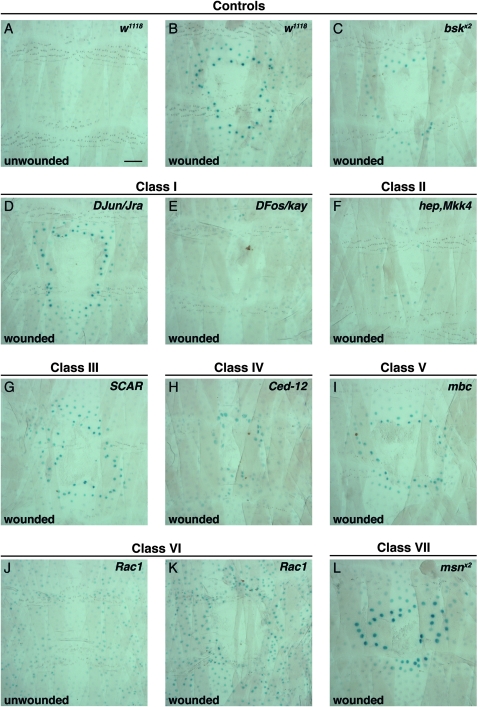
We next aimed to classify the wound closure genes according to the morphology of epidermal cells surrounding the wound. For optimal morphological analysis we used an e22c reporter labeling only the epidermal nuclei in red and then stained the membranes in dissected whole mounts with anti-Fasciclin III. To limit the scope of the analysis of wound morphology, we compared only genes whose open-wound phenotype exceeded an arbitrary 15% cutoff with the e22c reporter (see Figure 3, B and C). Differences in epidermal organization were apparent and could be grouped into seven classes (detailed below) on the basis of qualitative (Figure 4) and quantitative (Figure 5) criteria.
Figure 4.—
UAS-RNAi transgenes affecting wound closure show distinct wound morphologies. (A–I) Dissected whole mounts of larvae heterozygous for e22c-Gal4,UAS-DsRed2-Nuc (red) and the indicated UAS-RNAi transgene were immunostained for Fasciclin III (green) 24 hr post wounding. UAS-RNAi transgenes were grouped into seven classes on the basis of wound morphology. (A) Class I, bskx2; (B) class I, DFos/kay; (C) class I, DJun/Jra; (D) class II, hepx2; (E) class III, SCAR; (F) class IV, Ced-12; (G) class V, mbc; (H) class VI, Rac1; (I) class VII, msnx2. See results for classification criteria. Stars indicate disorganized epidermal sheet; arrowheads indicate smooth wound edge with pronounced fluorescence; brackets span zones of nuclear crowding. Lines with two UAS-RNAi inserts targeting the same gene are denoted by x2. Bar, 100 μm.
Class I (bsk, DFos/kay, and DJun/Jra):
The distinguishing feature of class I genes was that the cells surrounding the wound gap and the cells in the epidermal sheet farther away from the wound failed to elongate and remained similar in shape and size to unwounded epidermal cells (Figure 4, A–C). At least upon targeting of DFos/kay and DJun/Jra, defects in the progress of wound closure, including a failure of leading edge and more distal cells to elongate toward the wound edge, were apparent at earlier time points (Figure S5).
Class II (hep, Tak1, and Mkk4):
The distinguishing feature of class II genes was the smaller size of the open wounds (Figures 4D and 5B). This phenotype resembled closing wounds of control larvae at 8 hr post wounding (Figure 1D) and was exacerbated by coexpression of RNAi transgenes targeting either the two Jun2 kinases or the two Jun3 kinases (Figure S4, C and E), again suggesting redundancy at these levels of the JNK pathway.
Class III (SCAR, Arp14D, and Gγ1):
The distinguishing feature of class III genes was that leading-edge epidermal nuclei appeared in “pearl necklace”-like clusters at the wound margin (Figure 4E). This could be demonstrated quantitatively by measuring the presence of clusters containing five or more front line nuclei that were within a certain minimal distance (see materials and methods) of each other (Figure 5C). This analysis revealed that both class III and class IV genes shared this property of front line nuclear clustering.
Class IV (Ced-12):
The distinguishing feature of the class IV gene was stronger nuclear crowding within a much broader band that extended farther back from the wound margin compared to class III genes. Smaller epidermal cells in the region near the wound edge accompanied this crowding (Figure 4F). When we quantified nuclear area within a 50-μm distance of the wound edge (Figure 5D), this measure of nuclear crowding was significantly higher only with the class IV gene Ced-12 but not with class I or class III genes.
Class V (mbc):
The distinguishing features of the class V gene were a pronounced accumulation of green fluorescence at the wound margin, smoother wound edges (Figure 4G), and a larger average wound size (Figure 5B).
Class VI (Rac1):
The distinguishing feature of the class VI gene was the disorganization of the epidermal cells surrounding the open wound (Figure 4H). This was not surprising since the unwounded epidermal sheet in Rac1RNAi-expressing larvae is already highly disorganized (see Figure S6 for unwounded larvae expressing the UAS-RNAi transgenes shown in Figure 4).
Class VII (msn):
Open wounds of larvae expressing msnRNAi were similar to the ones of Rac1RNAi larvae in class VI (data not shown). However, closed wounds of msnRNAi larvae exhibited a distinct morphology (Figure 4I), in which the original wound area (Figure S7A) became a single syncytial epidermal cell containing tens of nuclei (Figure 5E). To accommodate these differences, msn was grouped in a separate class, class VII. This distinct morphology did not appear to be the result of nuclear division in the absence of cytokinesis since there was no anti-phospho-Histone H3 staining observed in msnRNAi larvae, similar to controls and to knockdown of the class I genes DJun/Jra and DFos/kay (Figure S8). Nor did it appear to be due to misregulated apoptosis, which is not observed around control or msnRNAi wounds (data not shown).
Finally, targeting of at least some of the genes described here with nonoverlapping RNAi transgenes revealed that their distinctive morphologies were consistent and not a property of the specific RNAi transgenes first used in the screen [examples for misshapen (class VII) and Arp14D (class III) are shown in Figure S7].
JNK activation on expression of UAS-RNAi transgenes affecting wound closure:
As a final mode of classifying the genes identified in our screen, we used the JNK reporter msn-lacZ (Spradling et al. 1999; Galko and Krasnow 2004) to assess the extent of JNK pathway activation during expression of UAS-RNAi transgenes that block or alter wound closure. In unwounded control larvae, msn-lacZ is expressed only at very low levels (Figure 6A), whereas 6 hr after wounding it is induced in a graded fashion in several cell rows surrounding the wound (Figure 6B). Expression of UAS-bskRNAi reduces activation of msn-lacZ (Figure 6C), although not to the extent observed with UAS-bskDN (data not shown and Wu et al. 2009). Targeting putative JNK pathway components blocked msn-lacZ activation except for two notable surprises. The first was that targeting the transcription factors DJun and DFos, which are often thought to work in tandem and exhibit a similar wound closure morphology (Figure 4, B and C), did not affect msn-lacZ activation equally. Expression of DJun/JraRNAi had only a slight effect on msn-lacZ activation (Figure 6D), while expression of DFos/kayRNAi led to a complete block (Figure 6E), similar to that observed with coexpression of UAS-RNAi transgenes targeting the upstream SAP2 kinases Hep and Mkk4 (Figure 6F) or the SAP3 kinases Slpr and Tak1 (data not shown). The second surprise was that expression of msnRNAi targeting the Jun4K required for DC did not block msn-lacZ activation and may indeed enhance it slightly (Figure 6L).
Figure 6.—
JNK activation in larvae expressing UAS-RNAi transgenes affecting wound closure. (A–L) Dissected epidermal whole mounts of unwounded (A and J) or pinch wounded (B–I and K and L) larvae heterozygous for the e22c-Gal4 driver (A–K) or the A58-Gal4 driver (L), the JNK activity reporter msn-lacZ (A–L), and the indicated UAS-RNAi transgene. All whole mounts were stained 6 hr post wounding or post mock wounding with X-Gal to detect β-galactosidase reporter activity (blue). (A) w1118, unwounded; (B) w1118, wounded; (C) bskx2; (D) DJun/Jra; (E) DFos/kay; (F) hep,Mkk4; (G) SCAR; (H) Ced-12; (I) mbc; (J) Rac1, unwounded; (K) Rac1, wounded; (L) msnx2. Morphological classes are indicated above the panels. Bar, 100 μm.
Notably, most of the other classes of genes that included actin cytoskeletal modulators, classes III and V, had little or no effect on msn-lacZ expression (Figure 6, G and I). The only exception to this was the class IV gene, Ced-12 (Figure 6H), whose targeting led to a decrease in JNK activation similar to that observed upon expression of bskRNAi (Figure 6C). Only Rac1RNAi affected JNK activation in the unwounded state (Figure 6J and data not shown), showing a higher level of msn-lacZ activation than in unwounded controls (Figure 6A). We found that upon expression of Rac1RNAi (Figure 6K) or Rac1DN (data not shown) msn-lacZ was activated even further following wounding, although curiously this activation was uniform and not obviously graded around the wound site. Taken together, these results suggest that either an increase or a decrease of Rac1 signaling can somehow activate the JNK pathway. In conclusion, the msn-lacZ reporter serves as a useful tool to further classify the genes identified and originally grouped by their effects on epidermal morphology.
DISCUSSION
Morphological classification of wound closure genes suggests multiple cellular processes are required for normal closure:
We found that the genes identified in our screen could be grouped into seven classes, each possessing a distinct morphological feature or defect. Here, we use these classes to infer some of the biological functions that occur within the epidermal sheet to bring about proper closure. These inferences are also illustrated graphically in Figure 7.
Figure 7.—
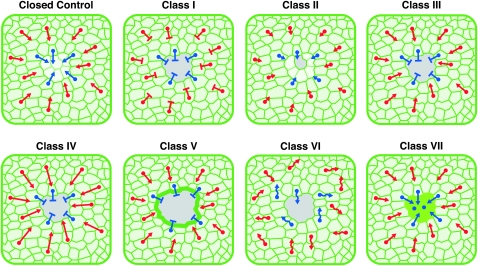
Model of epidermal cell behaviors in normal and perturbed wound healing. Cell behaviors during wound healing are indicated on a schematic template. On these templates, select cells contacting the wound edge are marked with blue nuclei, whereas select cells within the sheet in close vicinity to the wound are marked with red nuclei. Arrows indicate the direction of cellular migration toward the wound gap. The length of the arrow symbolizes the speed of the migrating cells (longer, fast; shorter, slow). A T-bar indicates that migration is blocked. Wavy arrows show improper directionality of migration. The bright green wound margin of class V indicates pronounced membrane fluorescence. The green central wound area of class VII indicates a large syncytial cell. See text for model details.
Targeting of class I genes (bsk, DJun/Jra, and DFos/kay) led to open wounds where epidermal cells surrounding the wound appeared to largely maintain their original shapes and distribution. This is in contrast to class III genes (SCAR, Arp14D, and Gγ1), whose targeting led to a pearl necklace-like clustering of epidermal nuclei along the wound edge, a more exaggerated version of what is seen in control wounds that are actively closing (8-hr control in Figure 1D). Comparing classes I and III, we hypothesize that at least two processes are required for closure. One is the ability of leading-edge cells to move into the wound gap. The second is a genetically separable ability of cells farther back from the wound edge to move within the sheet toward the gap. We speculate that class I genes are defective in both of these processes while class III genes are defective only in the ability of the cells to move into the gap.
Class II genes (hep, Mkk4, and Tak1) are distinguished mostly by the smaller area of their open wounds that suggests a delay in wound closure. Since there is no pronounced nuclear crowding when these genes are targeted, we hypothesize that they, like class I genes (but likely to a lesser extent due to their redundancy), are partially deficient in both movement into the gap and movement within the sheet.
The four remaining classes are to date defined by single genes. The sole class IV gene, Ced-12, is characterized by extreme cell and nuclear crowding behind the wound edge. In contrast to class III genes, this crowding extends farther behind the wound edge and is in excess of that seen during normal closure. This phenotype suggests that targeting Ced-12 may actually lead to a hyperactivation of directional cell migration within the wounded sheet (but not into the wound gap). Ced-12 was initially identified as a gene required for apoptotic corpse clearance through phagocytosis in Caenorhabditis elegans (Gumienny et al. 2001). Migrating epidermal cells following wounding in Drosophila larvae extend impressive phagocytic processes (Galko and Krasnow 2004). We speculate that this phagocytic activity may actually be positively required for movement into the gap as well as an inhibitor of movement within the sheet.
The only class V gene (myoblast city) leads to a distinct phenotype of larger wounds with smooth wound edges that exhibit pronounced green fluorescence. This suggests that migrating epidermal cells must properly organize wound edge membrane dynamics to form a normal wound edge and migrate into the wound gap. Why are mbcRNAi open wounds larger? One possibility is that the wound closure defect of mbcRNAi is more severe than that of genes in classes I, III, and IV. A second possibility is that mbcRNAi wounds gape to some extent. This latter possibility is supported by an apparently weak wound edge that may retract or fold over from the original border (data not shown). Such folding may cause the increased green fluorescence observed at the edges of these wounds.
Targeting Rac1 (class VI) leads to a disorganized epidermis even within the unwounded sheet and also to constitutive activation of JNK signaling. The odd spacing and irregular shapes of the epidermal cells in both the unwounded and the wounded epidermal sheet suggest that cells lacking Rac1 may be constitutively motile within the sheet but that this motility may lack the directionality that usually leads to successful wound closure. An alternative interpretation is that loss of Rac1 somehow affects the adhesive interactions between epidermal cells so that they can no longer maintain their normal shapes. There is considerable evidence in other systems for Rac having effects on both migration (Monypenny et al. 2009; Wang et al. 2010) and adhesion of epithelial cells (Eaton et al. 1995; Chihara et al. 2003) so these possibilities are not mutually exclusive.
The sole class VII gene, misshapen, shows the strangest phenotype to come from our pilot screen. Most wounds in msnRNAi-expressing larvae closed, albeit aberrantly. These closed wounds exhibit an abnormal morphology, in which much of the initial wound gap becomes a single syncytial cell that can contain >60 nuclei. Although syncytium formation occurs normally during larval wound closure, it usually involves relatively small numbers (10–12) of cells. The msnRNAi phenotype suggests that syncytium formation is a tightly regulated process during normal closure but also that hyperactivation of syncytium formation does not preclude closure of the wound.
In summary, our classification scheme suggests that there are at least five different processes that are important for normal wound closure: migration into the wound gap, migration within the epidermal sheet near the wound, directionality of these migrations, organization of the wound edge, and regulation of syncytium formation. A recent scratch-wound study employing adherent endothelial cells that migrate as a sheet reached similar conclusions about some of the cellular processes required for closure (Vitorino and Meyer 2008). Our scheme, which is certainly incomplete at the level of the genes contained within each class, and is likely also incomplete in terms of the number of distinct classes, will hopefully serve as a useful framework for analyzing genes that emerge from further reporter-based screening or candidate gene analysis (Kwon et al. 2010).
Architecture of the JNK signaling pathway in larval wound closure:
Despite intensive study, the signal(s) that activate this pathway during both DC and larval wound closure remain unidentified. Our results suggest that the architecture of the JNK signaling pathway in wound healing differs from that in DC, as it does in other JNK-dependent cellular processes such as innate immunity (Boutros et al. 2002; Silverman et al. 2003; Geuking et al. 2009) and cell death (Igaki 2009). Given the unique morphological phenotype and the persistence of JNK reporter activity upon expression of msnRNAi (see above), it is unlikely that this kinase acts linearly upstream of the rest of the kinase relay. Like Misshapen, Rac has also been reported to be upstream of JNK activation in DC (Woolner et al. 2005) and was recently reported to be upstream of JNK activation in the larval epidermis where expression of an activated form of Rac1 led to activation of JNK signaling (Baek et al. 2010). The notion that Rac1 is linearly upstream of JNK in the larval epidermis is likely to be an oversimplification since we find here that loss of Rac1 function also leads to JNK activation. One gene that is partially required upstream of JNK activation in wound closure is Ced-12, which performs a similar role in thorax closure (Ishimaru et al. 2004).
Unlike in DC, where one Jun3K and one Jun2K are exclusively required for morphogenesis, in wound closure these positions appear to be redundant. Two SAP3 kinases, Slipper and Tak1, and two SAP2 kinases, Hemipterous and Mkk4, give a moderate wound closure defect when individually targeted. Redundancy is suggested by the more severe open-wound phenotype in double-RNAi experiments and double-mutant analysis for Tak1 and slipper (C. Lesch, J. Jo and Y. Wu, unpublished observations). JNK reporter results that show complete absence of JNK activation upon hepRNAi-Mkk4RNAi expression also support redundancy.
Finally, our results, as do other recent studies (Campos et al. 2009; Pearson et al. 2009), indicate the importance of DJun- and DFos-mediated transcription following wounding. In our study, differences in morphology, msn-lacZ activation, survival, and fragility of the cuticle (not shown) also suggest that DJun and DFos may not act together but rather serve distinct functions in wound closure.
Importance of actin cytoskeletal remodeling in wound closure:
Unlike DC and embryonic wound closure, which depend at least in part on actin cable formation and contraction (Kiehart et al. 2000; Jacinto et al. 2002; Wood et al. 2002), larval wound closure proceeds primarily through a process of directed cell migration (Wu et al. 2009). Although actin accumulates at the larval wound margin (Wu et al. 2009; Kwon et al. 2010), this accumulation is discontinuous and filopodial and lamellipodial process extension is apparent even during the relatively early stages of closure (Wu et al. 2009). Given these differences in cytoskeletal dynamics between DC and larval and embryonic wound closure, one might expect differences in the set of genes required to regulate actin polymerization. Indeed, this is what we observe. At the level of Rho-GTPases, both DC and embryonic wound closure require Rac1, RhoA, and Cdc42 (Harden et al. 1999; Stramer and Martin 2005), whereas larval wound closure requires Rac1 and Cdc42 (Figure 3C and Figure S2) but not RhoA (data not shown).
Although Arc-p20, an Arp2/3 complex component, has recently been shown to be required for embryonic wound closure (Campos et al. 2009), the Arp2/3 complex has not been extensively analyzed to date in DC or embryonic wound closure. Here, we find that at least one component of this complex (Arp 14D) is required for larval wound closure, as is SCAR, an activator of the complex. Both of these genes fall morphologically into class III, which suggests that they act together to positively regulate epidermal cell movement into the wound gap but not movement within the epidermal sheet. A third gene in this class, previously implicated in control of cell morphology in an in vitro RNAi screen (Kiger et al. 2003), is Gγ1, which suggests a possible connection between G-protein–coupled receptor signaling and regulation of cytoskeletal dynamics in epithelial cells.
Future screening prospects:
A systematic approach for identifying genes required for postembryonic wound closure has long been lacking in the field of tissue repair. This is partly due to the complexities of the wound healing response in vertebrates and the time and cost large-scale screens in vertebrate models would entail. RNAi-based in vitro scratch wounds have partially filled this gap in the field (Simpson et al. 2008; Vitorino and Meyer 2008) although it is unlikely that these assays recapitulate the full complexity of wound healing as it occurs in a living organism. Here, we have developed a reasonably rapid, medium-throughput methodology for identifying wound closure genes in vivo in Drosophila larvae. The success and future scalability of the screen depend critically on two elements: the reporter that allows live visualization of closure and the recent development of near whole-genome UAS-RNAi libraries (Dietzl et al. 2007). Because it is RNAi based, the screen is likely to show a false-negative rate (29.4%) similar to that of other screens (Mummery-Widmer et al. 2009) where a larger selection of positive control genes was available for analysis. Like other screens involving readouts to physiological challenge (Kambris et al. 2006; Brandt et al. 2008; Campos et al. 2009), the screen is labor intensive. However, because this is the first systematic in vivo approach for screening for postembryonic wound closure defects, it holds great promise for identifying the set of evolutionarily conserved genes required tissue autonomously for wound closure. This is especially the case given the numerous recent studies (Li et al. 2003; Galko and Krasnow 2004; Mace et al. 2005; Pujol et al. 2008; Tong et al. 2009; Wang et al. 2009; Wu et al. 2009) that point to a deep evolutionary conservation in epidermal wound healing responses.
Acknowledgments
We thank Georg Halder, Randy Johnson, Andreas Bergmann, and members of the Galko laboratory for comments; Shana Palla for assistance with statistical analysis; Beth Stronach for mutant strains; Amin Ghabrial for UAS-GFPRNAi; Leisa McCord for graphical help; LaGina Nosavanh and Denise Weathersby for help with screening; and the Developmental Studies Hybridoma Bank and Dirk Bohmann for antibodies. UAS-RNAi lines were generously provided in advance of public distribution by Ulrich Theopold and Ryu Ueda. This work was supported by a University of Texas MD Anderson Cancer Center institutional research grant and by National Institutes of Health grants 1 R01GM083031 and R01GM083031-02 S1 (American Recovery and Reinvestment Act supplement to the preceding grant) to M.J.G.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.121822/DC1.
References
- Baek, S. H., Y. C. Kwon, H. Lee and K. M. Choe, 2010. Rho-family small GTPases are required for cell polarization and directional sensing in Drosophila wound healing. Biochem. Biophys. Res. Commun. 394 488–492. [DOI] [PubMed] [Google Scholar]
- Bohmann, D., M. C. Ellis, L. M. Staszewski and M. Mlodzik, 1994. Drosophila Jun mediates Ras-dependent photoreceptor determination. Cell 78 973–986. [DOI] [PubMed] [Google Scholar]
- Boutros, M., H. Agaisse and N. Perrimon, 2002. Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev. Cell 3 711–722. [DOI] [PubMed] [Google Scholar]
- Brand, A. H., and N. Perrimon, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118 401–415. [DOI] [PubMed] [Google Scholar]
- Brandt, S. M., G. Jaramillo-Gutierrez, S. Kumar, C. Barillas-Mury and D. S. Schneider, 2008. Use of a Drosophila model to identify genes regulating Plasmodium growth in the mosquito. Genetics 180 1671–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos, I., J. A. Geiger, A. C. Santos, V. Carlos and A. Jacinto, 2009. Genetic screen in Drosophila melanogaster uncovers a novel set of genes required for embryonic epithelial repair. Genetics 184 129–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chihara, T., K. Kato, M. Taniguchi, J. Ng and S. Hayashi, 2003. Rac promotes epithelial cell rearrangement during tracheal tubulogenesis in Drosophila. Development 130 1419–1428. [DOI] [PubMed] [Google Scholar]
- Clark, R. A., J. M. Lanigan, P. DellaPelle, E. Manseau, H. F. Dvorak et al., 1982. Fibronectin and fibrin provide a provisional matrix for epidermal cell migration during wound reepithelialization. J. Invest. Dermatol. 79 264–269. [DOI] [PubMed] [Google Scholar]
- Dietzl, G., D. Chen, F. Schnorrer, K. C. Su, Y. Barinova et al., 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448 151–156. [DOI] [PubMed] [Google Scholar]
- Eaton, S., P. Auvinen, L. Luo, Y. N. Jan and K. Simons, 1995. CDC42 and Rac1 control different actin-dependent processes in the Drosophila wing disc epithelium. J. Cell Biol. 131 151–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galko, M. J., and M. A. Krasnow, 2004. Cellular and genetic analysis of wound healing in Drosophila larvae. PLoS Biol. 2 E239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuking, P., R. Narasimamurthy, B. Lemaitre, K. Basler and F. Leulier, 2009. A non-redundant role for Drosophila Mkk4 and hemipterous/Mkk7 in TAK1-mediated activation of JNK. PLoS ONE 4 e7709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glise, B., H. Bourbon and S. Noselli, 1995. hemipterous encodes a novel Drosophila MAP kinase kinase, required for epithelial cell sheet movement. Cell 83 451–461. [DOI] [PubMed] [Google Scholar]
- Goley, E. D., and M. D. Welch, 2006. The ARP2/3 complex: an actin nucleator comes of age. Nat. Rev. Mol. Cell. Biol. 7 713–726. [DOI] [PubMed] [Google Scholar]
- Grose, R., and S. Werner, 2004. Wound-healing studies in transgenic and knockout mice. Mol. Biotechnol. 28 147–166. [DOI] [PubMed] [Google Scholar]
- Gumienny, T. L., E. Brugnera, A. C. Tosello-Trampont, J. M. Kinchen, L. B. Haney et al., 2001. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell 107 27–41. [DOI] [PubMed] [Google Scholar]
- Harden, N., M. Ricos, Y. M. Ong, W. Chia and L. Lim, 1999. Participation of small GTPases in dorsal closure of the Drosophila embryo: distinct roles for Rho subfamily proteins in epithelial morphogenesis. J. Cell Sci. 112(Pt. 3) 273–284. [DOI] [PubMed] [Google Scholar]
- Igaki, T., 2009. Correcting developmental errors by apoptosis: lessons from Drosophila JNK signaling. Apoptosis 14 1021–1028. [DOI] [PubMed] [Google Scholar]
- Ishimaru, S., R. Ueda, Y. Hinohara, M. Ohtani and H. Hanafusa, 2004. PVR plays a critical role via JNK activation in thorax closure during Drosophila metamorphosis. EMBO J. 23 3984–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto, A., W. Wood, S. Woolner, C. Hiley, L. Turner et al., 2002. Dynamic analysis of actin cable function during Drosophila dorsal closure. Curr. Biol. 12 1245–1250. [DOI] [PubMed] [Google Scholar]
- Kambris, Z., S. Brun, I. H. Jang, H. J. Nam, Y. Romeo et al., 2006. Drosophila immunity: a large-scale in vivo RNAi screen identifies five serine proteases required for Toll activation. Curr. Biol. 16 808–813. [DOI] [PubMed] [Google Scholar]
- Kennerdell, J. R., and R. W. Carthew, 2000. Heritable gene silencing in Drosophila using double-stranded RNA. Nat. Biotechnol. 18 896–898. [DOI] [PubMed] [Google Scholar]
- Kiehart, D. P., C. G. Galbraith, K. A. Edwards, W. L. Rickoll and R. A. Montague, 2000. Multiple forces contribute to cell sheet morphogenesis for dorsal closure in Drosophila. J. Cell Biol. 149 471–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiger, A. A., B. Baum, S. Jones, M. R. Jones, A. Coulson et al., 2003. A functional genomic analysis of cell morphology using RNA interference. J. Biol. 2 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kockel, L., J. G. Homsy and D. Bohmann, 2001. Drosophila AP-1: lessons from an invertebrate. Oncogene 20 2347–2364. [DOI] [PubMed] [Google Scholar]
- Kwon, Y. C., S. H. Baek, H. Lee and K. M. Choe, 2010. Nonmuscle myosin II localization is regulated by JNK during Drosophila larval wound healing. Biochem. Biophys. Res. Commun. 393 656–661. [DOI] [PubMed] [Google Scholar]
- Lawrence, P. A., R. Bodmer and J. P. Vincent, 1995. Segmental patterning of heart precursors in Drosophila. Development 121 4303–4308. [DOI] [PubMed] [Google Scholar]
- Levi, B. P., A. S. Ghabrial and M. A. Krasnow, 2006. Drosophila talin and integrin genes are required for maintenance of tracheal terminal branches and luminal organization. Development 133 2383–2393. [DOI] [PubMed] [Google Scholar]
- Li, G., C. Gustafson-Brown, S. K. Hanks, K. Nason, J. M. Arbeit et al., 2003. c-Jun is essential for organization of the epidermal leading edge. Dev. Cell 4 865–877. [DOI] [PubMed] [Google Scholar]
- Llense, F., and E. Martin-Blanco, 2008. JNK signaling controls border cell cluster integrity and collective cell migration. Curr. Biol. 18 538–544. [DOI] [PubMed] [Google Scholar]
- Luo, L., Y. J. Liao, L. Y. Jan and Y. N. Jan, 1994. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 8 1787–1802. [DOI] [PubMed] [Google Scholar]
- Mace, K. A., J. C. Pearson and W. McGinnis, 2005. An epidermal barrier wound repair pathway in Drosophila is mediated by grainy head. Science 308 381–385. [DOI] [PubMed] [Google Scholar]
- Martin, P., 1997. Wound healing–aiming for perfect skin regeneration. Science 276 75–81. [DOI] [PubMed] [Google Scholar]
- Monypenny, J., D. Zicha, C. Higashida, F. Oceguera-Yanez, S. Narumiya et al., 2009. Cdc42 and Rac family GTPases regulate mode and speed but not direction of primary fibroblast migration during platelet-derived growth factor-dependent chemotaxis. Mol. Cell. Biol. 29 2730–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummery-Widmer, J. L., M. Yamazaki, T. Stoeger, M. Novatchkova, S. Bhalerao et al., 2009. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature 458 987–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobes, C. D., and A. Hall, 1995. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81 53–62. [DOI] [PubMed] [Google Scholar]
- Odland, G., and R. Ross, 1968. Human wound repair. I. Epidermal regeneration. J. Cell Biol. 39 135–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto, I. M., T. Raabe, U. E. Rennefahrt, P. Bork, U. R. Rapp et al., 2000. The p150-Spir protein provides a link between c-Jun N-terminal kinase function and actin reorganization. Curr. Biol. 10 345–348. [DOI] [PubMed] [Google Scholar]
- Patel, N. H., P. M. Snow and C. S. Goodman, 1987. Characterization and cloning of fasciclin III: a glycoprotein expressed on a subset of neurons and axon pathways in Drosophila. Cell 48 975–988. [DOI] [PubMed] [Google Scholar]
- Pearson, J. C., M. T. Juarez, M. Kim and W. McGinnis, 2009. Multiple transcription factor codes activate epidermal wound-response genes in Drosophila. Proc. Natl. Acad. Sci. USA 106 2224–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polaski, S., L. Whitney, B. W. Barker and B. Stronach, 2006. Genetic analysis of slipper/mixed lineage kinase reveals requirements in multiple Jun-N-terminal kinase-dependent morphogenetic events during Drosophila development. Genetics 174 719–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, T. D., and G. G. Borisy, 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell 112 453–465. [DOI] [PubMed] [Google Scholar]
- Pujol, N., S. Cypowyj, K. Ziegler, A. Millet, A. Astrain et al., 2008. Distinct innate immune responses to infection and wounding in the C. elegans epidermis. Curr. Biol. 18 481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramet, M., R. Lanot, D. Zachary and P. Manfruelli, 2002. JNK signaling pathway is required for efficient wound healing in Drosophila. Dev. Biol. 241 145–156. [DOI] [PubMed] [Google Scholar]
- Riesgo-Escovar, J. R., and E. Hafen, 1997. Common and distinct roles of DFos and DJun during Drosophila development. Science 278 669–672. [DOI] [PubMed] [Google Scholar]
- Riesgo-Escovar, J. R., M. Jenni, A. Fritz and E. Hafen, 1996. The Drosophila Jun-N-terminal kinase is required for cell morphogenesis but not for DJun-dependent cell fate specification in the eye. Genes Dev. 10 2759–2768. [DOI] [PubMed] [Google Scholar]
- Silverman, N., R. Zhou, R. L. Erlich, M. Hunter, E. Bernstein et al., 2003. Immune activation of NF-kappaB and JNK requires Drosophila TAK1. J. Biol. Chem. 278 48928–48934. [DOI] [PubMed] [Google Scholar]
- Simpson, K. J., L. M. Selfors, J. Bui, A. Reynolds, D. Leake et al., 2008. Identification of genes that regulate epithelial cell migration using an siRNA screening approach. Nat. Cell Biol. 10 1027–1038. [DOI] [PubMed] [Google Scholar]
- Singer, A. J., and R. A. Clark, 1999. Cutaneous wound healing. N. Engl. J. Med. 341 738–746. [DOI] [PubMed] [Google Scholar]
- Sluss, H. K., Z. Han, T. Barrett, R. J. Davis and Y. T. Ip, 1996. A JNK signal transduction pathway that mediates morphogenesis and an immune response in Drosophila. Genes Dev. 10 2745–2758. [DOI] [PubMed] [Google Scholar]
- Spradling, A. C., D. Stern, A. Beaton, E. J. Rhem, T. Laverty et al., 1999. The Berkeley Drosophila Genome Project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics 153 135–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stramer, B., and P. Martin, 2005. Cell biology: master regulators of sealing and healing. Curr. Biol. 15 R425–R427. [DOI] [PubMed] [Google Scholar]
- Stronach, B., and N. Perrimon, 2002. Activation of the JNK pathway during dorsal closure in Drosophila requires the mixed lineage kinase, slipper. Genes Dev. 16 377–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, Y. C., J. E. Treisman and E. Y. Skolnik, 1998. The Drosophila Ste20-related kinase misshapen is required for embryonic dorsal closure and acts through a JNK MAPK module on an evolutionarily conserved signaling pathway. Genes Dev. 12 2371–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, A., G. Lynn, V. Ngo, D. Wong, S. L. Moseley et al., 2009. Negative regulation of Caenorhabditis elegans epidermal damage responses by death-associated protein kinase. Proc. Natl. Acad. Sci. USA 106 1457–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman, J. E., N. Ito and G. M. Rubin, 1997. misshapen encodes a protein kinase involved in cell shape control in Drosophila. Gene 186 119–125. [DOI] [PubMed] [Google Scholar]
- Vitorino, P., and T. Meyer, 2008. Modular control of endothelial sheet migration. Genes Dev. 22 3268–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., V. Tsarouhas, N. Xylourgidis, N. Sabri, K. Tiklova et al., 2009. The tyrosine kinase Stitcher activates Grainy head and epidermal wound healing in Drosophila. Nat. Cell Biol. 11 890–895. [DOI] [PubMed] [Google Scholar]
- Wang, X., L. He, Y. I. Wu, K. M. Hahn and D. J. Montell, 2010. Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat. Cell Biol. 12 591–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch, M. D., A. Iwamatsu and T. J. Mitchison, 1997. Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature 385 265–269. [DOI] [PubMed] [Google Scholar]
- Wood, W., A. Jacinto, R. Grose, S. Woolner, J. Gale et al., 2002. Wound healing recapitulates morphogenesis in Drosophila embryos. Nat. Cell Biol. 4 907–912. [DOI] [PubMed] [Google Scholar]
- Woolner, S., A. Jacinto and P. Martin, 2005. The small GTPase Rac plays multiple roles in epithelial sheet fusion–dynamic studies of Drosophila dorsal closure. Dev. Biol. 282 163–173. [DOI] [PubMed] [Google Scholar]
- Wu, Y., A. R. Brock, Y. Wang, K. Fujitani, R. Ueda et al., 2009. A blood-borne PDGF/VEGF-like ligand initiates wound-induced epidermal cell migration in Drosophila larvae. Curr. Biol. 19 1473–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, Y., and M. Karin, 2004. The control of cell motility and epithelial morphogenesis by Jun kinases. Trends Cell Biol. 14 94–101. [DOI] [PubMed] [Google Scholar]
- Zallen, J. A., Y. Cohen, A. M. Hudson, L. Cooley, E. Wieschaus et al., 2002. SCAR is a primary regulator of Arp2/3-dependent morphological events in Drosophila. J. Cell Biol. 156 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger, J., and D. Bohmann, 1999. Thorax closure in Drosophila: involvement of Fos and the JNK pathway. Development 126 3947–3956. [DOI] [PubMed] [Google Scholar]
- Zeitlinger, J., L. Kockel, F. A. Peverali, D. B. Jackson, M. Mlodzik et al., 1997. Defective dorsal closure and loss of epidermal decapentaplegic expression in Drosophila fos mutants. EMBO J. 16 7393–7401. [DOI] [PMC free article] [PubMed] [Google Scholar]