Abstract
Allopolyploidy, or the combination of two or more distinct genomes in one nucleus, is usually accompanied by radical genomic changes involving transposable elements (TEs). The dynamics of TEs after an allopolyploidization event are poorly understood. In this study, we analyzed the methylation state and genetic rearrangements of a high copied, newly amplified terminal-repeat retrotransposon in miniature (TRIM) family in wheat termed Veju. We found that Veju insertion sites underwent massive methylation changes in the first four generations of a newly formed wheat allohexaploid. Hypomethylation or hypermethylation occurred in ∼43% of the tested insertion sites; while hypomethylation was significantly predominant in the first three generations of the newly formed allohexaploid, hypermethylation became predominant in the subsequent generation. In addition, we determined that the methylation state of Veju long terminal repeats (LTRs) might be correlated with the deletion and/or insertion of the TE. While most of the methylation changes and deletions of Veju occurred in the first generation of the newly formed allohexaploid, most Veju insertions were seen in the second generation. Finally, using quantitative PCR, we quantitatively assessed the genome composition of Veju in the newly formed allohexaploid and found that up to 50% of Veju LTRs were deleted in the first generation. Retrotransposition bursts in subsequent generations, however, led to increases in Veju elements. In light of these findings, the underlying mechanisms of TRIM rearrangements are discussed.
TRANSPOSABLE elements (TEs) are DNA sequences that range in size from several hundred base pairs to >15 kb and that have the ability to move to different locations within the genome. TE movement occurs through either a copy-and-paste mechanism involving RNA intermediates (class 1) or a cut-and-paste mechanism involving DNA intermediates (class 2). Class 1 elements are also called retrotransposons, or retroelements, and comprise two main types: (1) long terminal repeat (LTR) retrotransposons, flanked by LTRs, and (2) non-LTR elements (such as long interspersed nuclear elements and short interspersed nuclear elements).
LTR retrotransposons are the most abundant mobile elements in plant genomes (Feschotte et al. 2002), as the replicative mode of retroelement transposition enables the LTR retrotransposon to accrue in high copy number. Indeed, in some grasses, LTR retrotransposons represent up to 90% of the genome (Bennetzen and Kellogg 1997; Feschotte et al. 2002). As such, retrotransposon sequences function well as substrates for illegitimate and unequal recombinations that can lead to a variety of mutations, such as deletions, insertions, translocations, and others (Parisod et al. 2009).
The replicative nature of TEs seems to be stimulated by a variety of specific stress conditions (reviewed by Wessler 1996; Capy et al. 2000; Grandbastien et al. 2005), including challenges to the genome such as interspecific hybridization, an idea first proposed by Barbara McClintock 26 years ago (McClintock 1984). Accordingly, allopolyploidization is usually coupled with rapid and reproducible genomic changes, including the elimination of DNA sequences (Liu et al. 1998a,b; Ozkan et al. 2001; Shaked et al. 2001; Adams and Wendel 2005b; Skalicka et al. 2005), gene silencing (Chen and Pikaard 1997; Comai et al. 2000; Kashkush et al. 2002; Simons et al. 2006), alteration of cytosine methylation (Shaked et al. 2001; Madlung et al. 2002; Salmon et al. 2005; Beaulieu et al. 2009; Xu et al. 2009), activation of genes and retrotransposons (Kashkush et al. 2002, 2003; O'Neill et al. 2002), massively altered gene expression patterns (Kashkush et al. 2002; Wang et al. 2006), and organ-specific subfunctionalization, i.e., differential expression of homeoalleles in different tissues and at different developmental stages (Adams et al. 2003; Adams and Wendel 2004). These and other studies (Levy and Feldman 2002; Osborn et al. 2003; Adams and Wendel 2005a; Rapp and Wendel 2005; Chen and Ni 2006; Chen 2007) demonstrate the dynamic nature of allopolyploid plant genomes.
Although allopolyploidization has generally been assumed to induce large bursts of TE activity (Matzke and Matzke 1998), several studies that focused on different allopolyploid systems failed to provide any evidence for a transposition burst and offered only limited evidence for the transposition of specific TEs (Madlung et al. 2005; Ainouche et al. 2009; Beaulieu et al. 2009). In newly formed Arabidopsis allopolyploids, no evidence for transposition bursts was reported (Beaulieu et al. 2009), although limited evidence suggested that transposition events occurred in a specific TE called Sunfish (Madlung et al. 2005). Little evidence of TE transposition was found in a natural population of the 150-year-old allopolyploid, Spartina anglica (Ainouche et al. 2009), and no evidence of transposition of Wis 2-1A retrotransposons in a newly formed wheat allotetraploid was present (Kashkush et al. 2003). The results of these works and others indicate that, in the short term, TE proliferation after allopolyploidization may be restricted to a few specific TEs in particular allopolyploidy systems (Parisod et al. 2009).
This study entailed a detailed investigation of the methylation patterns and rearrangements of a one terminal-repeat retrotransposon in miniature (TRIM) family in wheat termed Veju. TRIM elements possess the classical structure of LTR retrotransposons, but they are distinguished by their small overall sizes (0.4 to ∼2.5 kb). A nonautonomous retrotransposon, Veju is 2520 bp long with 374 bp of identical LTRs, yet does not contain the proteins required for retrotransposition (Sanmiguel et al. 2002). However, because Veju elements contain polypurine tracts (PPTs) and primer binding sites (PBSs), they are capable of transposing if the retrotransposition proteins are available from another source. In addition, the identical sequences of the Veju 5′ and 3′ LTRs indicate that some members of the Veju family retain retrotransposition activity.
In silico analysis of Veju sequences revealed them to be one of the most active and most recently inserted sequences in the wheat genome (Sanmiguel et al. 2002; Sabot et al. 2005a). As such, we have determined and compared the methylation patterns of >880 Veju insertion sites in the first four generations of a newly formed wheat allohexaploid, as well as in the parental lines. We then tested the correlation between the cytosine methylation and genetic rearrangements (i.e., deletions and insertions) of Veju and addressed the precise developmental timing of these rearrangements. Finally, we successfully tested overall changes in the copy numbers of Veju in the newly formed allohexaploid using real-time quantitative PCR.
MATERIALS AND METHODS
Plant material:
The plant material used in this study comprised a newly formed allohexaploid (S1–S5 generations) and the parental lines Triticum turgidum ssp. durum (accession no. TTR19) and Aegilops tauschii (accession no. TQ27). The amphiploid was obtained through the spontaneous formation of unreduced gametes in F1 plants (Ozkan et al. 2001). Chromosome number was determined in newly formed allohexaploids, and only those having the expected euploid chromosome number were analyzed. Seed materials from the parental lines, S1, and S2 were kindly provided by Hakan Ozkan and Moshe Feldman.
DNA isolation:
DNA was isolated from young leaves (4 weeks post germination) using the CTAB technique (Kidwell and Osborn 1992) or the DNeasy plant kit (QIAGEN). In addition, DNA was isolated from various tissues of S1 and S2 plants (see details in results).
Transposon methylation display and transposon display:
DNA isolated from newly formed allohexaploids and the parental lines was subjected to transposon methylation display (TMD) according to a previously published protocol (Kashkush and Khasdan 2007). Veju-specific primers from the 5′ LTR (P2 in Table S1) and from the 3′ LTR (P3 in Table S1) were used in the TMD together with an adapter primer, +(TCAG) (Figure 1, P1) (see Kashkush and Khasdan 2007). Primer positions in the Veju sequence are displayed in Figure S1. The fluorescently labeled TMDs were analyzed by GeneMapper version 4 (see examples in Figure S2). Chimeric (Veju/host flanking sequence) TMD bands with evidence of alteration (loss/gain of bands) between newly formed allopolyploid generations and/or their parents were extracted from the polyacrylamide gel, reamplified (using the same PCR conditions as in the preamplification reaction for TMD), cloned, and sequenced.
Figure 1.—
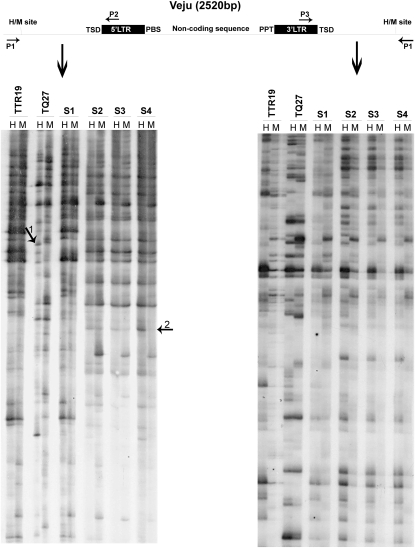
TMD patterns in T. turgidum ssp. durum (TTR19) and Ae. tauschii (TQ27) and in the first four generations (S1–S4) of the derived allohexaploid. Veju and its flanking sequences are shown (scheme at the top) together with HpaII (H) and MspI (M) cleavage sites and the positions of primers used in TMD reactions. Autoradiographs of TMD used P2 or P3 and the adapter primer (P1), +(TCAG). Primer sequences are listed in Table S1. Arrows 1 and 2 (left gel image) indicate representative TMD bands that are altered in the allohexaploid.
Transposon display (TD) was performed using a methylation-insensitive restriction enzyme, MseI with an adaptor primer, +(CAC, CTG, or CAT) (Table S1), together with a Veju-specific primer from the 5′ LTR (P2 in Table S1).
Site-specific PCR:
All PCR reactions were performed with 2 μl Taq DNA polymerase buffer 10× (Fisher Biotec), 2.0 μl of 25 mm MgCl2 (Fisher Biotec), 0.8 μl 2.5 mm dNTPs, 0.2–0.3 μl of Taq DNA polymerase (5000 units/ml, Fisher Biotec), 1 μl of forward primer (50 ng/μl), 1 μl of reverse primer (50 ng/μl), and 25–100 ng DNA template, to which ultra-pure, double-distilled water was added to achieve a final volume of 20 μl. PCR conditions were 94° for 3 min followed by a cycling stage of denaturation at 94° for 30 sec, annealing for 30 sec, and elongation at 72° for 30–70 sec, repeated for 29 cycles. Primers were designed using Primer 3 software version 0.4.0 (http://frodo.wi.mit.edu/primer3/input.htm). Ultra-pure double-distilled water served as template in all negative control PCR reactions. PCR products were visualized on 1.5–2% agarose gel using TAE buffer (40 mm Tris base, 0.1% HCl, and 1 mm EDTA, pH 8) under UV light after staining with ethidium bromide. The primer sequences used for genomic PCR amplifications are described in Table S1.
Quantitative PCR:
Primers for PCR were designed using the online program Primer 3 (version 0.4.0) and verified using Primer Express software version 3.0 (Applied Biosystems). Each quantitative PCR (qPCR) reaction was performed in a 15-μl reaction volume that consisted of 7.5 μl of Power SYBR Green PCR reaction mix 2× (Applied Biosystems), 5 μl of DNA template (0.5 ng/μl), 1 μl of forward primer (10 μm), 1 μl of reverse primer (10 μm), and 0.5 μl of ultra-pure water. The qPCR reactions were performed and analyzed using the 7500 Fast Real-Time PCR system and 7500 Software version 2.0.1 (Applied Biosystems), respectively. The thermal profile consisted of 2 min at 50°, 10 min at 95°, and 40 cycles of 4 sec at 95° and 30 sec at 60°. Amplification data were collected at the end of each extension step. Primer sequences used for qPCR amplifications are described in Table S2. The ratio of Veju LTRs to internal region sequences was calculated using the following formula: ratio = (the fold of template amplification at each cycle)−average ΔCt, where ΔCt (threshold cycle) = Ct(target) − Ct(reference control). Veju LTR was set as the target, and the internal region of Veju served as a reference.
A comparative 2−ΔΔCt method for determining a relative target quantity in samples was used in the normalization and analysis of the relative quantities of both the LTR and the internal Veju sequences. Using 7500 software version 2.0.1, we measured amplification of the target—either the Veju LTR or the internal region—and of the endogenous control of VRN1 in samples and in a reference sample (in this case the diploid parental line is TQ27). The same software was then used to determine the relative quantity of target in each sample by comparing the normalized target quantity in each sample to the normalized target quantity in the reference sample, TQ27, based on the following equation:
 |
Therefore, the relative quantity = (the fold of template amplification at each cycle)−ΔΔCt (Livak and Schmittgen 2001).
Reproducibility of the results was evaluated for each sample by running three technical replicates of each of the reactions. To ensure experimental reproducibility, three biological replicates were run for each genotype. To distinguish specific from nonspecific PCR products, a melting curve was generated immediately after amplification. It consisted of a 15-sec incubation at 95° and a 1-min incubation at 60°, after which the temperature was increased by increments of 0.1°/sec until 95° was reached. The same specific product was detected for either target or reference genes, while no amplification was detected in the no-template control wells.
PCR efficiencies of the target and reference genes were determined by generating standard curves, based on serial dilutions prepared from DNA templates. Fold amplification at each cycle was calculated according to PCR efficiency, which was deduced by the software from the slope of the regression line (y) according to the equation E = [(10−1/y) − 1] × 100. For primers with 100% efficiency, the fold equals 2. For other efficiencies, the software adjusts the fold accordingly (Table S2).
Computer-assisted analysis:
Sequence annotation relied on the BLAST 2.0 package from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/) and from the Institute for Genomic Research (http://tigrblast.tigr.org/tgi/).
RESULTS
Defining the methylation status of Veju LTR-flanking sequences in newly formed allohexaploids and in the parental lines:
The methylation status of Veju LTR-flanking CCGG sites in the two parental lines T. turgidum (TTR19) and A. tauschii (TQ27) and in the first four generations (S1–S4) of the derived allohexaploid (see materials and methods, Plant material) was investigated using transposon methylation display. TMD enables systematic, genome-wide analysis of the methylation status of CCGG sites in DNA that flanks high-copy-number TEs (Kashkush and Khasdan 2007). Gel-based analysis of the TMD products revealed that each TMD band contains a Veju LTR sequence at one end and an unmethylated HpaII (H) or MspI (M) site in the flanking sequences (Figure 1 and Figure S2). Note that because the two Veju LTRs are nearly identical, Veju internal sequences theoretically might also be amplified when a Veju LTR primer (P2) is used in a TMD reaction (see Figure S1), which thereby enables the analysis of the methylation status also in CCGG sites within Veju internal sequence. However, no such bands were identified in our analysis. The appearance of monomorphic bands (present in both the H and M lanes of the same individual) in the TMD gel indicates that no methylation was detected at the CCGG site flanking the LTR. However, the presence of polymorphic bands indicates the presence of methylation either at an internal cytosine residue (bands present in M lanes only) or at an external cytosine residue (bands present in H lanes only, reflecting hemi-methylation) of the CCGG site flanking the LTR. Using TMD, we analyzed 889 clear bands (Veju insertion sites) in the parental lines (559 and 330 bands in TTR19 and TQ27, respectively). Monomorphic bands in TMD gels in the two parental lines (which indicate similar insertions in the parents) were counted only once, and 207 such bands were counted. Of the 889 bands analyzed, 361 were polymorphic between the H and M lanes, indicating that 40.6% (361/889) of the CCGG sites flanking Veju LTRs were methylated in the DNA isolated from young leaves of the parental lines. The two parental lines, TTR19 and TQ27, showed similar levels of methylation, namely 41.3% and 40%, respectively (Table S3).
We expected all TMD bands to be inherited, reflecting additivity in the newly formed allohexaploid generations (i.e., the S generations) because both parents derived from inbred lines (Ozkan et al. 2001). Deviations from additivity (revealed as a loss or a gain of a band) in any of the S generations may be due to methylation alterations, deletions, and/or insertions. When the inheritance of the TMD bands (loci) was tested, 486 of the 889 (54.6%) TMD bands were altered in the first four generations (S1–S4). Hence, in summary, alterations of TMD patterns (Table 1) in the S generations can be classified into five groups (examples of TMD patterns representing the five groups are shown in Table S4):
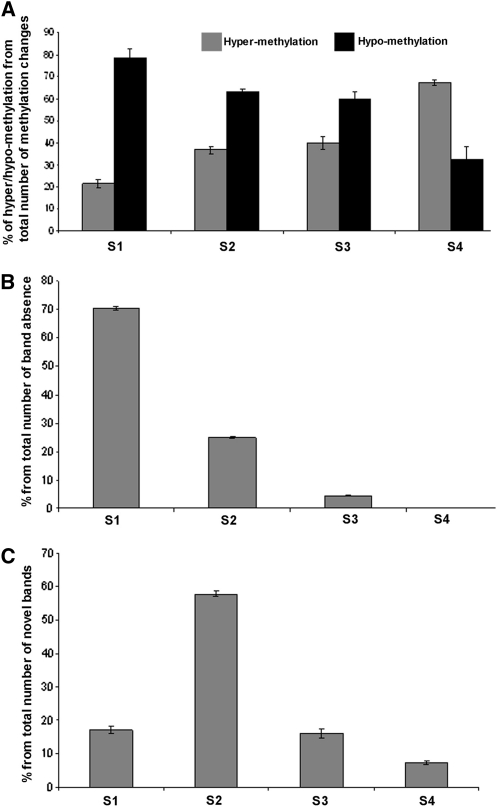
Clear, detectable alterations in the methylation of Veju LTR-flanking CCGG sites in S1–S4 accounted for 65.6% of the changes (319 bands, Table 1). Hypomethylation (release of methyl groups from one or both cytosines at CCGG sites that were methylated in one or both parental lines) or hypermethylation (gain of methyl groups on CCGG sites that were ummethylated at one or both cytosines in one or both parental lines) had occurred already in S1. Although hypomethylation occurred for ∼80% of the cases in S1, it decreased in subsequent generations to reach ∼30% in S4 (Figure 2A).
Absence of bands in S1 had already occurred for ∼6% (31 bands, Table 1) of the TMD changes.
Methylation alterations in either S1 or S2, followed by an absence of TMD bands in subsequent generations, occurred for ∼3% (13 bands, Table 1) of the TMD changes.
Appearance of novel unmethylated bands (bands present in both H and M lanes) in the amphiploid occurred for ∼5% (25 bands, Table 1) of the TMD changes.
Appearance of novel methylated bands (present in either H or M lanes) in the amphiploid occurred for ∼20% (98 bands, Table 1) of the TMD changes.
TABLE 1.
Classification of 486 TMD fragments that were altered in the amphiploid
| Type of alteration | No. of bands | % from total altered bands |
|---|---|---|
| Methylation alteration | 319 | 65.64 |
| Absence of bands already in S1 | 31 | 6.38 |
| Methylation alteration in S1 or S2 followed by absence of bands in subsequent generations | 13 | 2.68 |
| Appearance of novel unmethylated bands in S generations | 25 | 5.14 |
| Appearance of novel methylated bands in S generations | 98 | 20.16 |
| Total | 486 | 100 |
Figure 2.—
Analysis of changes in TMD patterns among the first four generations (S1–S4) of the newly formed allohexaploid. (A) Level of hypomethylation vs. hypermethylation from the total number of methylation changes in each generation. (B) Level of absence of TMD bands in each generation. (C) Level of appearance of novel bands (that were not seen in the parental lines) in each generation.
Figure 2B indicates that most of the band absence (∼70%) occurred in the S1 generations and decreased dramatically in the subsequent generations, while no band absence was detected in S4 (Figure 2B). On the other hand, appearance of novel bands in the amphiploid occurred in ∼57% of the cases in the S2 generation and were reduced in subsequent generations (Figure 2C).
A subset of 10 TMD bands was randomly chosen, extracted from the acrylamide gel, reamplified, and sequenced. Although all bands contained Veju LTR sequences at one end, their other ends contained LTR-flanking host DNA, two of which corresponded to wheat ESTs (accession nos. CD888659 and CJ569351) while the remainder were not found in wheat databases (see details in Table S5 and Table S6).
Validation of methylation changes by site-specific PCR:
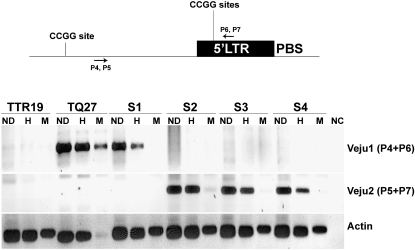
For validation of the methylation state, the 10 sequenced TMD bands mentioned above were further analyzed by site-specific PCR. While one band did not produce positive PCR products, the remaining 9 produced clear PCR products. To test the methylation status of CCGG sites in the Veju 5′ LTR by site-specific PCR, a primer was generated from the DNA sequence flanking the LTR (Figure 3, P4) of each one of the 10 sequences and used in PCR, in which undigested genomic DNA or DNA digested with either HpaII or MspI served as template. Specifically, the second primer was generated from a Veju 5′ LTR sequence upstream of the HpaII and MspI sites (five CCGG sites) (Figure 3, P6). The presence of a band in both the HpaII and the MspI lanes indicates that the LTR was methylated, the results of a fragment not being digested. In all 9 cases, the results of the site-specific PCR validated the TMD results (Figure 3 and Figure S3). For example, the analysis of two TMD fragments, termed Veju1 and Veju2 (Figure 3, Veju1 panel, and Figure 1, arrow 1), revealed that for Veju1, the insertion was unique to the TQ27 parental line and underwent demethylation (lack of PCR product in M lane, Figure 3) in the S1 generation. However, the band was absent in the subsequent generation as was seen also in TMD (Figure 1, arrow 1). For Veju2, the insertion was not seen in either the parental lines or the S1 generation; however, it was present in S2–S4 generations and was methylated (Figure 3, Veju2 panel, and Figure 1, arrow 2). In all other cases (Figure S3), the results of site-specific PCR were in full agreement with the TMD results.
Figure 3.—
Methylation of CCGG sites in the LTR. (Top) An LTR and flanking sequences shown together with the cleavage sites (CCGG) of the restriction enzymes HpaII and MspI (five CCGG sites upstream of P6 and P7 in the LTR) and the position of primers used in PCR analysis. (Bottom) PCR analysis (using primer pairs P4 and P6 for Veju1 or P5 and P7 for Veju2) of LTR methylation in leaves using undigested genomic DNA (ND) or DNA digested with either HpaII (H) or MspI (M) as the template (from parental lines and the first four generations of the derived allohexaploid). Primer sequences are listed in Table S1. Actin was used as a control for DNA quality, while no CCGG sites are present in the amplified region. As a negative control, water was used as template in the PCR reaction.
Correlation between cytosine methylation and Veju rearrangements in the newly formed allohexaploid:
The TMD analysis (Table 1) indicated that ∼35% of the changes in the amphiploid could be due to deletions or new insertions of Veju elements. In addition, the deletion (13 bands in Table 1) or possible new insertions (98 bands in Table 1) could indicate the possible correlation between methylation and rearrangements of Veju. To validate these claims, a TD was performed using a methylation-insensitive restriction enzyme, MseI, together with a Veju-specific primer (see materials and methods). Note that, in all cases where TMD bands disappeared in the S2 generation (Figure 2B), a methylation change (hypomethylation) occurred in the S1 generation. Similarly for newly appeared bands in the S2 generation (Figure 2C), the novel bands were accompanied by methylation.
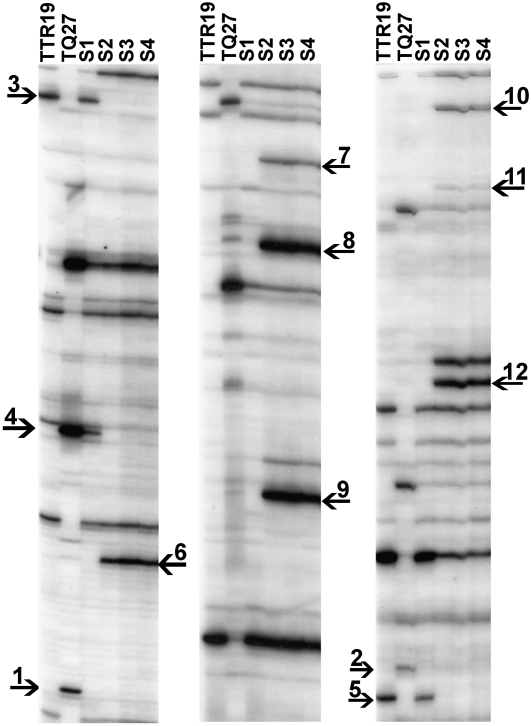
Figure 4 confirms that indeed deletion of Veju-containing sequences occurs already in the S1 generation (for example, bands 1 and 2, Figure 4), or it can occur in S2 (for example, bands 3–5, Figure 4). It is important to note that the ∼9% of the TMD changes that could be related to deletion of Veju-containing sequences (Table 1) could be underestimated because deletion of bands can be seen only among the polymorphic bands (in the TMD or TD gels) in the two parental lines. In addition, the TD analysis confirms the insertions of Veju sequences in the S2 generation (for example, bands 6–12, Figure 4). Extraction of four newly inserted bands and five deleted bands in the S2 generation from the TD gel, reamplification, and sequencing indicated that two newly inserted sequences and three deleted bands were already detected in the previous TMD experiments, whereas the newly inserted sequences in the S2 generation were methylated, and all three deleted bands in S2 were hypomethylated in S1. These data—together with the fact that all deleted bands in the S2 generation had changed their methylation pattern in S1 (hypomethylated), and all newly inserted bands in S2 were accompanied with methylation—indicate the direct link between methylation and rearrangements of Veju in the newly formed allohexaploid. Note that the flanking sequences of all nine chimeric isolated TD bands did not hit any known sequence in the database, except one newly inserted sequence that significantly hit a telomeric region (Hordeum vulgare telomeric chromosome 7H region).
Figure 4.—
Veju deletion and insertion as displayed by TD patterns in T. turgidum ssp. durum (TTR19) and Ae. tauschii (TQ27) and in the first four generations (S1–S4) of the derived allohexaploid. MseI, methylation-insensitive enzyme, was used for DNA cleavage. Autoradiographs of TD used P2 and the MseI-adapter primer. (Left) +(CAC). (Middle) +(CTG). (Right) +(CAT). Primer sequences are listed in Table S1. Arrows indicate deleted or newly inserted Veju elements in the newly formed allohexaploid (see text).
Additional support for the link between methylation and Veju rearrangements came from the site-specific PCR analysis (Figure 3, Veju1 and Veju2 panels, and Figure S3a). For Veju1 (Figure 3), it can be seen clearly that the demethylation of the element in the S1 generation was followed by its deletion in subsequent generations, while, for Veju2, a new insertion occurred in the second generations of the newly formed allohexaploid (Figure 3) that was accompanied by methylation of the new insertion. In another case (Figure S3a), a partially methylated Veju insertion in TTR19 parental lines was deleted in the S2 generation. Further validations of the deleted Veju1 and the newly inserted Veju2 were performed (see details in Figure S4 and Figure S5). Finally, the methylation of the new insertion (Veju 2) was further validated by bisulfite sequencing (Figure S6).
Precise developmental timing of Veju1 deletion and Veju2 insertion:
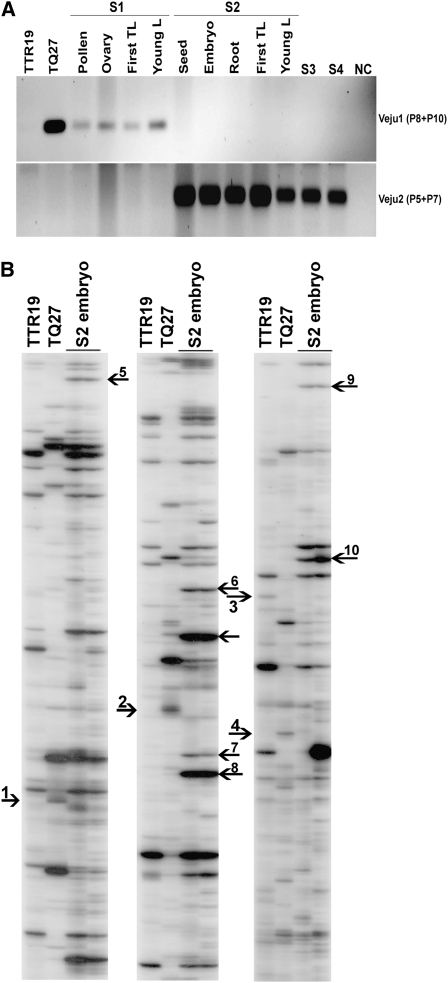
The developmental timing of the deletion or insertion of Veju sequences was tested by isolating DNA from (1) the pollen grains, ovaries, and first true leaves of S1 plants and (2) the immature seeds (i.e., 2 weeks after fertilization), mature embryos, first roots, and first leaves of S2 plants. The isolated DNA served as a template for PCR. For Veju1 (the deleted element in the S2 generation), PCR was performed using primers P8 and P10 (see Figure S5), designed to amplify a 193-bp fragment of the host flanking sequence. The band was detected in all S1 tissues but was absent from S2 tissues (Figure 5A, top), indicating that it was eliminated during the early stages of S2 embryo development. Similarly, PCR amplification of Veju2 (new insertion in S2 generation) was performed using primers P5 and P7 (see Figure 3) to amplify a 304-bp chimeric (Veju/host flanking) sequence. The band was absent in all S1 tissues yet present in S2 tissues (Figure 5A, bottom), indicating that Veju2 insertion occurred during the early stages of S2 embryo development. To validate these results, TD was performed in S2 embryos (Figure 5B), and it is clearly seen that deletion (for example, bands 1–3, Figure 5B) or insertion (for example, bands 4–10, Figure 5B) of Veju occurred in the embryo. These data indicate that rearrangement of Veju may occur somatically, transpiring as early as during zygote formation.
Figure 5.—
Developmental timing of deletion of Veju-containing fragments and new Veju insertions. (A) PCR analysis using primers P8 + P10 for Veju1 (top), and P5 + P7 for Veju2 (bottom) (see primer positions in Figure 3) on DNA templates of parental lines (TTR19 and TQ27); pollen, ovary, first true leaf (First TL), and young leaf of S1; seed, embryo, first roots, first true leaf, and young leaf of S2; S3; and S4. As negative control, water served as a template in a PCR reaction. (B) Veju deletion and insertion as displayed by TD patterns in T. turgidum ssp. durum (TTR19) and Ae. tauschii (TQ27) and in the S2 embryo. Primes used in TD reaction are the same as used in Figure 4. Arrows indicate deleted or newly inserted Veju elements in the embryos of the S2 generation (see text).
Quantitative evaluation of Veju composition in the newly formed allohexaploid and its parental lines:
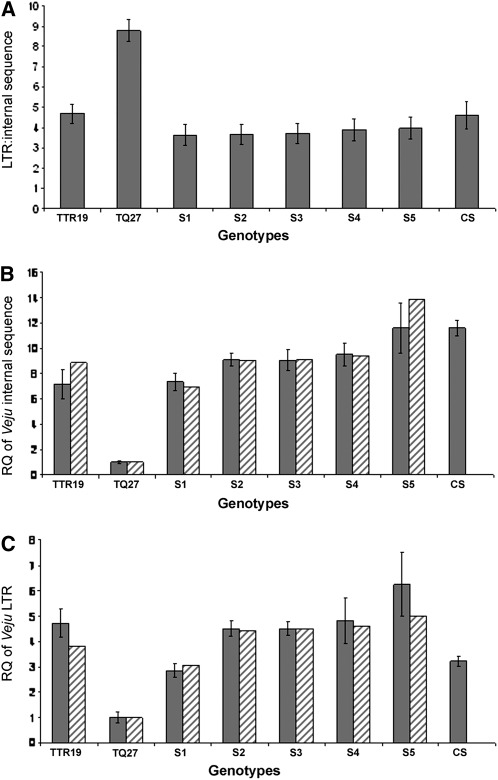
Our observation that the TMD patterns of Veju insertion sites changed dramatically (∼55%) in the newly formed allohexaploid, relative to its progenitors, and the fact that Veju methylation status can be correlated with Veju deletion and/or insertion facilitated the design of experiments to quantitatively test Veju composition using real-time PCR. Specifically, qPCR was used to calculate the ratio of Veju LTRs to Veju internal sequences to estimate the proportions of solo LTRs and intact elements. DNA serving as qPCR template was isolated from the parental lines and the S1 and S5 generations. Two primer pairs, one specific to the LTR sequence and the other designed from internal Veju sequences (Table S2), were employed. Figure 6A shows the relative quantities of Veju internal sequences and LTRs in the genomes of the parental lines, TTR19 and TQ27, and in those of the S1–S5 generations. The average internal sequence:LTR ratios were 1:4.7 in TTR19, 1:8.8 in TQ27, 1:3.7 in S1, 1:3.6 in S2, 1:37 in S3, 1:3:9 in S4, and 1:4 in S5. Most importantly, while the S1–S5 generations of the newly formed allohexaploid are each expected to possess an intermediate internal sequence:LTR ratio (∼1:6.75), relative to those of the two parental lines, in fact the offspring generations exhibited reductions of ∼43%, indicating that a massive number of LTRs may have been deleted in the newly formed allohexaploid. Interestingly, qPCR analysis of the newly formed allohexaploid displayed similar results as achieved with the natural hexaploid Triticum aestivum (cv. Chinese spring) (Figure 6A).
Figure 6.—
Quantitative PCR analysis of Veju retrotransposons in parental lines and in the S1–S5 generations of the derived allohexaploid. In addition, the natural hexaploid T. aestivum (cv. Chinese spring–CS) was used for comparison. (A) LTR:internal Veju sequence. (B) Relative quantity (RQ) of the Veju internal region (sequence). (C) RQ of the Veju LTR. Shaded bars represent the observed relative quantity (mean ± SE, n = 3), while hatched bars represent the expected relative quantity (see text for qPCR validation).
The observation that the number of Veju LTRs was significantly reduced in allohexaploid generations led to the design of experiments to test for changes in Veju element copy numbers in the newly formed allohexaploid. Relative Veju element quantities were measured in the parental lines and in the S1 and S5 generations using the single-copy vernalization gene 1 (VRN1) as a reference. Three orthologs of VRN1 that distinguish the AA, BB, and DD genomes of hexaploid wheat are correspondingly located in chromosomes 5A (Law et al. 1976; Dubcovsky et al. 1998), 5B (Barrett et al. 2002; Iwaki et al. 2002), and 5D (Law et al. 1976). Therefore, VRN1 exists in one copy in a diploid progenitor, in two copies in a tetraploid progenitor, and in three copies in the newly formed allohexaploid. To this end, a PCR reaction was performed as previously described ((Yan et al. 2004) to amplify the three VRN1 orthologs in the parental lines and in the S1 and S5 generations to ensure that VRN1 was not affected by the allopolyploidization process (Figure S7).
For the qPCR experiments, primers to VRN1 were designed on the basis of the multiple sequence alignment of previously published sequences from a conserved region of exon 4 (Figure S8). Our use of exon 4 enabled us to avoid amplifying a conserved MADS box encoded by the first exons (Yan et al. 2003) (Table S2 and Figure S8). Relative quantities of Veju internal sequences in the parental lines and in the S1–S5 genomes were measured using qPCR with primers specific for the Veju internal sequence (Figure 6B, shaded bars) (see Table S2). The copy number of Veju elements in the newly formed allohexaploid is expected to be the sum of the copy numbers of the two parental lines. While S1 displayed a reduction of ∼9.8% from this expected value, the subsequent generations displayed a >40% increase in Veju copy number relative to S1 (Figure 6B, shaded bars). These results indicate that a loss of Veju sequences in the first generation of the newly formed allohexaploid was followed by an accumulation of new Veju insertions in subsequent generations. Indeed, the copy number of Veju in the S5 generation was similar to the copy number of this sequence in the natural hexaploid (Figure 6B). Note that deletion of Veju-containing sequences might be completed in S4 (see Figure 2B), while Veju elements remain active (see Figure 2C), which may explain the relatively high quantities displayed in S5, even though the increase in S5 is not statistically significant compared to S4.
Relative Veju LTR quantities were also measured in the parental lines and in the S1 and S5 generations using LTR-specific primers (Figure 6C, shaded bars). As predicted from the previous experiment (Figure 6A), the S1 generation showed a reduction in LTR content of ∼50% from the number of Veju LTR copy numbers expected. The estimated quantity of Veju LTRs in S2–S5 indicated that these sequences were markedly reduced in the S1 generation and that a burst of insertions accrued in subsequent generations. However, as the Veju LTR content in the natural hexaploid was similar to that of S1, this might indicate that throughout evolution LTRs (or solo LTRs) are continuously removed from the genome.
All qPCR experiments had three biological replicates (see materials and methods, Quantitative PCR). Quality control for qPCR experiments to rule out possible competition effects in the PCR reactions using template mix was also performed (Figure S9). In addition, LTR:Veju internal sequence ratios were used to validate the observed relative quantities of Veju internal sequences and LTRs in the parental lines and in the S1 and S5 generations. Assuming that the observed ratios between the Veju elements are correct (Figure 6A), then the expected relative quantities of Veju LTRs (Figure 6C, hatched bars) should be calculable on the basis of the observed relative quantities of Veju internal sequences (Figure 6B, shaded bars). This can be realized by multiplying the observed relative quantities of Veju internal sequences by the observed LTR:internal sequence ratio (i.e., 4.7 in TTR19, 8.8 in TQ27, 3.7 in S1, and 3.4 in S5). For example, the expected relative quantity of Veju LTRs in the S1 generation will be the observed relative quantity of Veju internal sequence (Figure 6B, shaded bars) multiplied by 3.7. Similar calculations were also used to determine the expected relative quantities of Veju internal sequences, but in this case, the observed relative LTR quantities (Figure 6C, shaded bars) were divided, not multiplied, by the LTR:internal Veju sequence ratios. For example, the expected relative quantity of Veju internal sequences (Figure 6B, hatched bars) in the S1 generation can be attained by taking the observed relative quantity of Veju LTRs (Figure 6C, shaded bars) and dividing by 3.7. After such calculations, nearly complete agreement between the expected and the observed relative quantities was achieved for both Veju LTR and internal sequence experiments.
DISCUSSION
This study pinpointed changes in the methylation patterns of >880 Veju insertion sites using TMD. We found that ∼54% of the TMD patterns were altered in the first four generations of a newly formed allohexaploid. Using TMD and TD, we have observed deletions and/or new insertions of Veju elements in the newly formed allohexaploid, and in many cases we found that these alterations were correlated with changes in cytosine methylation. These observations led us to address the genomic compositions of Veju elements in S1–S5 generations of the newly formed allohexaploid. Surprisingly, we found that Veju elements, especially Veju LTRs, underwent massive deletions in the S1 generation and moderate insertions in subsequent generations. In addition, we noted that Veju rearrangements seemed to occur immediately after meiosis and formation of the zygote. This developmental timing might have important consequences as other work showed that TEs are epigenetically silenced in gametes, while they are active in the pollen vegetative nucleus (Slotkin et al. 2009).
Methylation of Veju elements:
On the basis of TMD analysis, we estimated that ∼41% of the CCGG sites flanking Veju elements could be cytosine-methylated in the young leaves of the parental lines T. turgidum ssp. durum (TTR19) and Ae. tauschii (TQ27). Over 54% of the Veju insertion sites in the parental plants showed altered TMD patterns in the first four generations (S1–S4) of the newly formed allohexaploid. In most cases, Veju sites were hypomethylated in the first generation (S1) of the newly formed allohexaploid, while hypermethylation was predominant in the S4 generation (Figure 2A). It is important to note that the analysis was performed qualitatively by assessing polymorphic vs. monomorphic bands. Furthermore, it is known that DNA methylation can vary from cell to cell in the same tissue, an outcome that can lead to different band intensities in a TMD gel.
The observed methylation and changes in TMD patterns of Veju insertion sites were significantly higher than one would expect from random methylation. Indeed, whole-genome methylation changes reported in newly formed allotetraploids are typically ∼13% (Shaked et al. 2001). Results similar to ours were reported for Spartina species, in which higher-than-random methylation was observed for the flanking sequences of three studied TEs (Parisod et al. 2009).
Our TMD analysis revealed that, in ∼23% of cases (Table 1), methylation alteration was accompanied by a deletion and/or insertion of Veju sequences. Most of the bands that were deleted in the newly formed allohexaploids were from the DD genome of TQ27, which could indicate that Veju loci inherited from the TQ27 parent in the newly formed allohexaploids may be more vigorously targeted for methylation/elimination than those inherited from the TTR19 parent. Accordingly, parent-dependent changes in TE methylation patterns were also reported in Spartina hybrids, in which the majority of the altered bands after hybridization, predominantly band losses, were of maternal origin (Parisod et al. 2009). However, in our case, TQ27 is the paternal line, so the phenomenon is more likely correlated with genome composition rather than with paternal origin (imprinting) because the same results were observed in reciprocal crosses. For example, Ozkan et al. (2001) and Shaked et al. (2001) reported the same pattern of DNA sequence elimination in reciprocal crosses of newly synthesized allopolyploids.
Our data indicate that deletion of Veju-containing sequences occurred in the S1–S3 generations of the amphiploid, while no deletion events were detected in the S4 (Figure 2B). This is in agreement with what was previously reported by Ozkan et al. (2001), who proposed that deletion of low-copy sequences in newly formed wheat allopolyploids was completed in the S3 generation.
TMD, TD analysis, and site-specific PCR experiments clearly indicated that a change in the methylation status (usually hypomethylation) in the S1 generation was followed by deletion in the S2 generation (Figures 1 and 3 and Figure S3). In addition, newly inserted Veju elements in the S2 generation exhibited LTR methylation. These data clearly show the correlation between methylation and post-allopolyploidization rearrangements that occur via a yet to be identified mechanism. In addition, small RNA corresponding to Veju elements might play a prominent role in Veju methylation in the newly formed allohexaploid (Avi Levy, personal communication).
Quantitative analysis of Veju by qPCR:
The existence of solo LTRs for nonautonomous elements and their evolutionary roles have been described for TRIM and LARD elements (Witte et al. 2001; Kalendar et al. 2004). However, detailed information about the extent and the timing of their formation in newly formed allopolyploids has yet to be studied. In this study, we performed a quantitative evaluation of the genomic composition of Veju elements using qPCR analysis.
The ratio of intact elements to solo LTRs fluctuates greatly between retrotransposons in different plant species—from ∼8:1 (eight intact elements to one solo LTR) in soybean (Wawrzynski et al. 2008), implying very slow rates of TE sequence removal, to ∼1:9 (one intact element to nine solo LTRs) for LARD retrotransposons in barley, probably due to abundant homologous recombination events (Kalendar et al. 2004). Our estimation of the genomic distribution of Veju elements showed that the genomes of the parental lines TTR19 and TQ27 possessed as many as 2.7 to 6.8 times more LTRs, respectively, than intact Veju elements.
The inherited Veju sequences exhibited clear losses of parental additivity. We observed an ∼44% reduction from the expected LTR:internal Veju ratios in newly formed allohexaploids, a finding that infers new genome reorganization following allopolyploidization and a massive loss of Veju LTRs. Moreover, we report a dramatic reduction in the relative quantities of Veju LTRs (∼40%) in the S1 generation (Figure 6, A and C). Apparently, the newly formed allopolyploid genome has attempted to reduce its burden of genetic “junk.” It is important to note that the qPCR data (Figure 6) and the TMD data (Figures 1 and 2) were in very good agreement, both indicating that most deletions of Veju sequences occurred in the S1 while most new insertions occurred in the S2 generation.
Species- and parent-specific elimination of high-copy genomic sequences has recently been reported in other plant allopolyploids. It was documented in Triticale (a synthetic allopolyploid hybrid produced by crossing wheat and rye) that allopolyploidy is associated with extensive, genome-wide genomic rearrangement. Prominent among these changes is the elimination of rye-specific fragments that often represent retrotransposons or their derivatives (Bento et al. 2008). In newly formed tobacco allopolyploids, parent-specific elimination of DNA repeats to a degree as high as 60% was also documented (Skalicka et al. 2005). In light of our observation that ∼68% of the TMD bands unaccounted for in the allopolyploids were from the DD genome, we propose that most TE elimination originated from the ancient DD genome while targeting solo LTRs or truncated elements for elimination.
Intriguingly, an increase in both Veju LTRs and internal regions was observed in the S5 generation. Considered together with the observed transcriptional activity of Veju, these results indicate that the immense loss of Veju sequences in the first generation after genome doubling is probably followed by retrotransposition in subsequent generations, a process that causes new insertions to accumulate in allohexaploids. Although some restructuring in the vicinity of TEs was reported in natural, 150-year-old Spartina anglica allopolyploids, no TE transposition was detected (Parisod et al. 2009). TE activation in newly formed allopolyploids, therefore, may be species-specific and limited to certain TEs.
A comparison of the genomic distribution of Veju in the S5 generation and in the ∼10,000-year-old natural hexaploid (Figure 6) revealed similar LTR:internal sequence ratios and similar quantities of Veju internal sequences. This might indicate that most rearrangements occur in the earliest generations of the nascent allopolyploid rather than on an evolutionary scale. This explains the data from Charles et al. (2008), according to which allopolyploidization neither enhanced nor repressed retrotranspositions when tested on an evolutionary timescale.
In natural populations, the abrupt proliferation of TEs could have important biological significance (Belyayev et al. 2009). First, differential TE insertion into the chromosomes of different parental genomes would reduce homologous recombination and promote disomic inheritance, thereby stabilizing the newly formed genome and increasing the fertility of the nascent allopolyploid species. Second, differential TE insertion contributes to genetic diversity and induces polymorphism in newly formed allopolyploids, a process that may increase allopolyploid fitness in different environments.
Underlying mechanisms for Veju elimination:
In plants, most of the full-length retrotransposons were estimated to be <5 million years old (Sanmiguel et al. 2002; Vitte and Panaud 2003; Wicker et al. 2003; Ma et al. 2004; Du et al. 2006; Wicker and Keller 2007; Charles et al. 2008), indicating that active deletion mechanisms that remove LTR retrotransposons from the genome exist. One molecular mechanism that may be responsible for massive LTR and internal Veju sequence loss in the first generation is unequal intrastrand homologous recombination. In rice, solo LTRs probably originated from intra-element recombination events (Vitte and Panaud 2003). In wheat allopolyploids, however, it is more likely that the observed rearrangements resulted from interelement recombination, which can affect more transposons than intra-element recombination and which can also eliminate flanking Veju sequences. In addition to unequal homologous recombination, illegitimate recombination is also a key process for retrotransposon deletion in rice (Ma et al. 2004). Moreover, illegitimate recombination was found to be the main cause of LTR-retrotransposon removal in Arabidopsis thaliana (Devos et al. 2002; Bennetzen et al. 2005) and the cause of LTR retroelement deletions in diploid and polyploid wheat and allopolyploid cotton. Thus, increased illegitimate recombination may be a general consequence of polyploidization (Wicker et al. 2003; Chantret et al. 2005; Grover et al. 2007).
Thus far, we do not have clear, definitive support for any of the mechanisms suggested as the driving force behind the observed high rates of genomic change between generations of allohexaploid wheat species. On the one hand, the finding that LTR levels were high in the parental lines, relative to the levels of intact elements, suggests that unequal homologous recombination is the more active and, therefore, more likely mechanism behind TE deletions (Ma et al. 2004). On the other hand, while unequal homologous recombination requires large (>50 bp) stretches of sequence homology, illegitimate recombination requires only a few base pairs of sequence identity, suggesting that illegitimate recombination is more widely applicable than unequal homologous recombination and, as such, is responsible for more deletions, including those of Veju-flanking sequences, than is unequal homologous recombination. Strikingly, a long form of the Veju retrotransposon was proposed to have originated from an illegitimate heterologous recombination (Sabot et al. 2005b), suggesting that this element can act as a “hot spot” that attracts illegitimate rearrangements. The molecular mechanism by which hypomethylated Veju elements undergo deletion remains unknown. Perhaps hypomethylation of Veju elements indicates an open chromatin that exposes these demethylated elements as targets for deletion by the host. As mentioned above, small RNAs might also have a major role in this process. On the other hand, methylation of new Veju insertions can be understood as a defensive mechanism of the host from the deleterious transposon insertions. Nevertheless, future studies should address these processes and their biological significance in nascent allopolyploids.
Acknowledgments
We thank Hakan Ozkan and Moshe Feldman for providing the amphiploid material and Avi Levy for helpful discussions. This work was supported by a grant from the Israel Science Foundation (grant no. 142/08) to K.K.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.120790/DC1.
References
- Adams, K. L., and J. F. Wendel, 2004. Exploring the genomic mysteries of polyploidy in cotton. Biol. J. Linn. Soc. 82 573–581. [Google Scholar]
- Adams, K. L., and J. F. Wendel, 2005. a Novel patterns of gene expression in polyploid plants. Trends Genet. 21 539–543. [DOI] [PubMed] [Google Scholar]
- Adams, K. L., and J. F. Wendel, 2005. b Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8 135–141. [DOI] [PubMed] [Google Scholar]
- Adams, K. L., R. Cronn, R. Percifield and J. F. Wendel, 2003. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl. Acad. Sci. USA 100 4649–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainouche, M. L., P. M. Fortune, A. Salmon, C. Parisod, M. A. Grandbastien et al., 2009. Hybridization, polyploidy and invasion: lessons from Spartina (Poaceae). Biol. Invasions 11 1159–1173. [Google Scholar]
- Barrett, B., M. Bayram and K. Kidwell, 2002. Identifying AFLP and microsatellite markers for vernalization response gene Vrn-B1 in hexaploid wheat using reciprocal mapping populations. Plant Breed. 121 400–406. [Google Scholar]
- Beaulieu, J., M. Jean and F. Belzile, 2009. The allotetraploid Arabidopsis thaliana-Arabidopsis lyrata subsp petraea as an alternative model system for the study of polyploidy in plants. Mol. Genet. Genomics 281 421–435. [DOI] [PubMed] [Google Scholar]
- Belyayev, A., R. Kalendar, L. Brodsky, E. Nevo, A. H. Schulman et al., 2009. Transposable elements in a marginal plant population: temporal fluctuations provide new insights into genome evolution of wild diploid wheat. Mobile DNA 1 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen, J. L., and E. A. Kellogg, 1997. Do plants have a one-way ticket to genomic obesity? Plant Cell 9 1509–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen, J. L., J. X. Ma and K. Devos, 2005. Mechanisms of recent genome size variation in flowering plants. Ann. Bot. 95 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento, M., H. S. Pereira, M. Rocheta, P. Gustafson, W. Viegas et al., 2008. Polyploidization as a retraction force in plant genome evolution: sequence rearrangements in Triticale. PLoS One 3 1402–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capy, P., G. Gasperi, C. Biemont and C. Bazin, 2000. Stress and transposable elements: Co-evolution or useful parasites? Heredity 85 101–106. [DOI] [PubMed] [Google Scholar]
- Chantret, N., J. Salse, F. Sabot, S. Rahman, A. Bellec et al., 2005. Molecular basis of evolutionary events that shaped the hardness locus in diploid and polyploid wheat species (Triticum and Aegilops). Plant Cell 17 1033–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles, M., H. Belcram, J. Just, C. Huneau, A. Viollet et al., 2008. Dynamics and differential proliferation of transposable elements during the evolution of the B and A genomes of wheat. Genetics 180 1071–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z. J., 2007. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant Biol. 58 377–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z. J., and Z. F. Ni, 2006. Mechanisms of genomic rearrangements and gene expression changes in plant polyploids. BioEssays 28 240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z. J., and C. S. Pikaard, 1997. Transcriptional analysis of nucleolar dominance in polyploid plants: biased expression/silencing of progenitor rRNA genes is developmentally regulated in Brassica. Proc. Natl. Acad. Sci. USA 94 3442–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai, L., A. P. Tyagi, K. Winter, R. Holmes-Davis, S. H. Reynolds et al., 2000. Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell 12 1551–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos, K. M., J. K. M. Brown and J. L. Bennetzen, 2002. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, C. G., Z. Swigonova and J. Messing, 2006. Retrotranspositions in orthologous regions of closely related grass species. BMC Evol. Biol. 6 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky, J., D. Lijavetzky, L. Appendino and G. Tranquilli, 1998. Comparative RFLP mapping of Triticum monococcum genes controlling vernalization requirement. Theor. Appl. Genet. 97 968–975. [Google Scholar]
- Feschotte, C., N. Jiang and S. R. Wessler, 2002. Plant transposable elements: where genetics meets genomics. Nat. Rev. Genet. 3 329–341. [DOI] [PubMed] [Google Scholar]
- Grandbastien, M., C. Audeon, E. Bonnivard, J. M. Casacuberta, B. Chalhoub et al., 2005. Stress activation and genomic impact of Tnt1 retrotransposons in Solanaceae. Cytogenet. Genome Res. 110 229–241. [DOI] [PubMed] [Google Scholar]
- Grover, C. E., H. Kim, R. A. Wing, A. H. Paterson and J. F. Wendel, 2007. Microcolinearity and genome evolution in the AdhA region of diploid and polyploid cotton (Gossypium). Plant J. 50 995–1006. [DOI] [PubMed] [Google Scholar]
- Iwaki, K., J. Nishida, T. Yanagisawa, H. Yoshida and K. Kato, 2002. Genetic analysis of Vrn-B1 for vernalization requirement by using linked dCAPS markers in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 104 571–576. [DOI] [PubMed] [Google Scholar]
- Kalendar, R., C. M. Vicient, O. Peleg, K. Anamthawat-Jonsson, A. Bolshoy et al., 2004. Large retrotransposon derivatives: abundant, conserved but nonautonomous retroelements of barley and related genomes. Genetics 166 1437–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush, K., and V. Khasdan, 2007. Large-scale survey of cytosine methylation of retrotransposons and the impact of readout transcription from long terminal repeats on expression of adjacent rice genes. Genetics 177 1975–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush, K., M. Feldman and A. A. Levy, 2002. Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics 160 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush, K., M. Feldman and A. A. Levy, 2003. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 33 102–106. [DOI] [PubMed] [Google Scholar]
- Kidwell, K. K., and T. C. Osborn, 1992. Simple plant DNA isolation procedures, pp. 1–13 in Plant Genomes: Methods for Genetic and Physical Mapping, edited by J. S. Beckmann and T. C. Osborn. Kluwer Academic Publishers. Dordrecht, The Netherlands.
- Law, C. N., A. J. Worland and B. Giorgi, 1976. Genetic control of ear emergence time by chromosomes 5a and chromosomes 5d of wheat. Heredity 36 49–58. [Google Scholar]
- Levy, A. A., and M. Feldman, 2002. The impact of polyploidy on grass genome evolution. Plant Physiol. 130 1587–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B., J. M. Vega and M. Feldman, 1998. a Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. II. Changes in low-copy coding DNA sequences. Genome 41 535–542. [DOI] [PubMed] [Google Scholar]
- Liu, B., J. M. Vega, G. Segal, S. Abbo, H. Rodova et al., 1998. b Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. I. Changes in low-copy noncoding DNA sequences. Genome 41 272–277. [DOI] [PubMed] [Google Scholar]
- Livak, K. J., and T. D. Schmittgen, 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25 402–408. [DOI] [PubMed] [Google Scholar]
- Ma, J. X., K. M. Devos and J. L. Bennetzen, 2004. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 14 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madlung, A., R. W. Masuelli, B. Watson, S. H. Reynolds, J. Davison et al., 2002. Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allotetraploids. Plant Physiol. 129 733–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madlung, A., A. P. Tyagi, B. Watson, H. M. Jiang, T. Kagochi et al., 2005. Genomic changes in synthetic Arabidopsis polyploids. Plant J. 41 221–230. [DOI] [PubMed] [Google Scholar]
- Matzke, M. A., and A. J. M. Matzke, 1998. Polyploidy and transposons. Trends Ecol. Evol. 13 241–241. [DOI] [PubMed] [Google Scholar]
- McClintock, B., 1984. The significance of responses of the genome to challenge. Science 226 792–801. [DOI] [PubMed] [Google Scholar]
- O'Neill, R. J. W., M. J. O'Neill and J. A. M. Graves, 1998. Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature 393 68–72. [DOI] [PubMed] [Google Scholar]
- Osborn, T. C., J. C. Pires, J. A. Birchler, D. L. Auger, Z. J. Chen et al., 2003. Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 19 141–147. [DOI] [PubMed] [Google Scholar]
- Ozkan, H., A. A. Levy and M. Feldman, 2001. Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 13 1735–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisod, C., A. Salmon, T. Zerjal, M. Tenaillon, M. A. Grandbastien et al., 2009. Rapid structural and epigenetic reorganization near transposable elements in hybrid and allopolyploid genomes in Spartina. New Phytol. 184 1003–1015. [DOI] [PubMed] [Google Scholar]
- Rapp, R. A., and J. F. Wendel, 2005. Epigenetics and plant evolution. New Phytol. 168 81–91. [DOI] [PubMed] [Google Scholar]
- Sabot, F., R. Guyot, T. Wicker, N. Chantret, B. Laubin et al., 2005. a Updating of transposable element annotations from large wheat genomic sequences reveals diverse activities and gene associations. Mol. Genet. Gen. 274 119–130. [DOI] [PubMed] [Google Scholar]
- Sabot, F., P. Sourdille and M. Bernard, 2005. b Advent of a new retrotransposon structure: the long form of the Veju elements. Genetica 125 325–332. [DOI] [PubMed] [Google Scholar]
- Salmon, A., M. L. Ainouche and J. F. Wendel, 2005. Genetic and epigenetic consequences of recent hybridization and polyploidy in Spartina (Poaceae). Mol. Ecol. 14 1163–1175. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P. J., W. Ramakrishna, J. L. Bennetzen, C. S. Busso and J. Dubcovsky, 2002. Transposable elements, genes and recombination in a 215-kb contig from wheat chromosome 5A m. Funct. Integr. Genomics 2 70–80. [DOI] [PubMed] [Google Scholar]
- Shaked, H., K. Kashkush, H. Ozkan, M. Feldman and A. A. Levy, 2001. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, K. J., J. P. Fellers, H. N. Trick, Z. C. Zhang, Y. S. Tai et al., 2006. Molecular characterization of the major wheat domestication gene Q. Genetics 172 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalicka, K., K. Y. Lim, R. Matyasek, M. Matzke, A. R. Leitch et al., 2005. Preferential elimination of repeated DNA sequences from the paternal, Nicotiana tomentosiformis genome donor of a synthetic, allotetraploid tobacco. New Phytol. 166 291–303. [DOI] [PubMed] [Google Scholar]
- Slotkin, R. K., M. Vaughn, F. Borges, M. Tanurdzic, J. D. Becker et al., 2009. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitte, C., and O. Panaud, 2003. Formation of solo-LTRs through unequal homologous recombination counterbalances amplifications of LTR retrotransposons in rice Oryza sativa L. Mol. Biol. Evol. 20 528–540. [DOI] [PubMed] [Google Scholar]
- Wang, J. L., L. Tian, H. S. Lee, N. E. Wei, H. M. Jiang et al., 2006. Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrzynski, A., T. Ashfield, N. W. G. Chen, J. Mammadov, A. Nguyen et al., 2008. Replication of nonautonomous retroelements in soybean appears to be both recent and common. Plant Physiol. 148 1760–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler, S. R., 1996. Plant retrotransposons: turned on by stress. Curr. Biol. 6 959–961. [DOI] [PubMed] [Google Scholar]
- Wicker, T., and B. Keller, 2007. Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families. Genome Res. 17 1072–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker, T., N. Yahiaoui, R. Guyot, E. Schlagenhauf, Z. D. Liu et al., 2003. Rapid genome divergence at orthologous low molecular weight glutenin loci of the A and A(m) genomes of wheat. Plant Cell 15 1186–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte, C. P., Q. H. Le, T. Bureau and A. Kumar, 2001. Terminal-repeat retrotransposons in miniature (TRIM) are involved in restructuring plant genomes. Proc. Natl. Acad. Sci. USA 98 13778–13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y. H., L. Zhong, X. M. Wu, X. P. Fang and J. B. Wang, 2009. Rapid alterations of gene expression and cytosine methylation in newly synthesized Brassica napus allopolyploids. Planta 229 471–483. [DOI] [PubMed] [Google Scholar]
- Yan, L., A. Loukoianov, G. Tranquilli, M. Helguera, T. Fahima et al., 2003. Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. USA 100 6263–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, L., M. Helguera, K. Kato, S. Fukuyama, J. Sherman et al., 2004. Allelic variation at the VRN-1 promoter region in polyploid wheat. Theor. Appl. Genet. 109 1677–1686. [DOI] [PubMed] [Google Scholar]