Abstract
Environment-specific quantitative trait loci (QTL) refer to QTL that express differently in different environments, a phenomenon called QTL-by-environment (Q × E) interaction. Q × E interaction is a difficult problem extended from traditional QTL mapping. The mixture model maximum-likelihood method is commonly adopted for interval mapping of QTL, but the method is not optimal in handling QTL interacting with environments. We partitioned QTL effects into main and interaction effects. The main effects are represented by the means of QTL effects in all environments and the interaction effects are represented by the variances of the QTL effects across environments. We used the Markov chain Monte Carlo (MCMC) implemented Bayesian method to estimate both the main and the interaction effects. The residual error covariance matrix was modeled using the factor analytic covariance structure. A simulation study showed that the factor analytic structure is robust and can handle other structures as special cases. The method was also applied to Q × E interaction mapping for the yield trait of barley. Eight markers showed significant main effects and 18 markers showed significant Q × E interaction. The 18 interacting markers were distributed across all seven chromosomes of the entire genome. Only 1 marker had both the main and the Q × E interaction effects. Each of the other markers had either a main effect or a Q × E interaction effect but not both.
GENOTYPE-BY-ENVIRONMENT (G × E) interaction is a very important phenomenon in quantitative genetics. With the advanced molecular technology and statistical methods for quantitative trait loci (QTL) mapping (Lander and Botstein 1989; Jansen 1993; Zeng 1994), G × E interaction analysis has shifted to QTL-by-environment (Q × E) interaction. In the early stage of QTL mapping, almost all statistical methods were developed in a single environment (Paterson et al. 1991; Stuber et al. 1992). Data from different environments were analyzed separately and the conclusions were drawn from the separate analyses of QTL across environments. These methods do not consider the correlation of data under different environments and thus may not extract maximum information from the data. Composite interval mapping for multiple traits can be used for Q × E interaction if different traits are treated as the same trait measured in different environments (Jiang and Zeng 1995). This multivariate composite interval mapping approach makes good use of all data simultaneously and increases statistical power of QTL detection and accuracy of the estimated QTL positions. However, the number of parameters of this method increases dramatically as the number of environments increases. Therefore, the method may not be applied when the number of environments is large. Several other models have been proposed to solve the problem of a large number of environments (Jansen et al. 1995; Beavis and Keim 1996; Romagosa et al. 1996). These methods were based on some special situations and assumptions. One typical assumption was independent errors or constant variances across environments. These assumptions are often violated in real QTL mapping experiments.
Earlier investigators realized the problem and adopted the mixed-model methodology to solve the problem (Piepho 2000; Boer et al. 2007). Under the mixed-model framework, people can choose which model effects are random and which are fixed. The mixed-model methodology is very flexible, leading to an easy way to model genetic and environmental correlation between environments using a suitable error structure. Piepho (2000) proposed a mixed model to detect QTL main effect across environments. Similar to the composite interval mapping analysis, his model incorporated one putative QTL and a few cofactors. The Q × E effects in the model were assumed to be random, which greatly reduced the number of estimated parameters. However, the fact that only one QTL is included in the model means that Piepho's (2000) model remains a single-QTL model rather than a multivariate model. Boer et al. (2007) proposed a step-by-step mixed-model approach to detecting QTL main effects, Q × E interaction effects, and QTL responses to specific environmental covariates. In the final step, Boer et al. (2007) rewrote the model to include all QTL in a multiple-QTL model and reestimated their effects.
In this study, we extended the Bayesian shrinkage method (Xu 2003) to map Q × E interaction effects of QTL. In the original study (Xu 2003), we treated each marker as a putative QTL and used the shrinkage method to simultaneously estimate marker effects of the entire genome. In the multiple-environment case, we can still use this approach to simultaneously evaluate marker effects under multiple environments but we can further partition the marker effects into main and Q × E interaction effects. For any particular marker, the mean of the marker effects represents the main effect and the variance of the marker effects represents the Q × E interaction effect for that marker. Under the Bayesian framework, we assigned a normal prior with zero mean and an unknown variance to each marker main effect and a multivariate normal prior with zero vector mean and homogeneous diagonal variance–covariance matrix to the Q × E interaction effects of each maker. In multiple environments, the structure of the error terms might be very complicated since we need to consider the correlation of the same genotype under different environments. In our analysis, we used different variance–covariance structures to model the error terms. The simplest case was the homogeneous diagonal matrix, and the most complex choice was an unstructured matrix. We also used a heterogeneous diagonal matrix whose parameters are somewhere between the two models. Finally, we considered several factor analytic models. The reason to use the factor analytic structure is that it can separate genetic effects into common effects and environment-specific effects. In addition, the factor analytic structure is parsimonious and thus can substantially reduce the computational burden of the mixed-model analyses.
THEORY AND METHOD
Hierarchical model:
Let 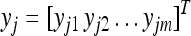 be an m × 1 column vector for the observed phenotypic values of individual j measured from m environments for
be an m × 1 column vector for the observed phenotypic values of individual j measured from m environments for 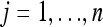 , where n is the sample size. Let q be the number of QTL included in the model. The multivariate linear model is
, where n is the sample size. Let q be the number of QTL included in the model. The multivariate linear model is
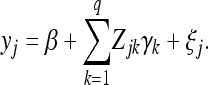 |
(1) |
In the above model, 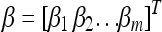 is an m × 1 vector for the intercepts. The dependent variable Zjk is a genotype indicator variable for individual j at marker k and it is defined as
is an m × 1 vector for the intercepts. The dependent variable Zjk is a genotype indicator variable for individual j at marker k and it is defined as 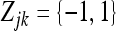 for the two genotypes of a backcross (BC) individual or
for the two genotypes of a backcross (BC) individual or 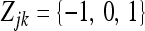 for the three genotypes of an
for the three genotypes of an 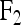 individual. The regression coefficient
individual. The regression coefficient 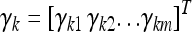 is an m × 1 vector of QTL effects for the m environments. Finally,
is an m × 1 vector of QTL effects for the m environments. Finally, 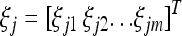 is an m × 1 vector for the residual errors. To model the Q × E interaction, we assume that γk follows a multivariate normal distribution,
is an m × 1 vector for the residual errors. To model the Q × E interaction, we assume that γk follows a multivariate normal distribution,
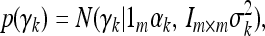 |
(2) |
where 1m is a unity vector with dimension m, Im×m is an m × m identity matrix, αk is the mean value representing the main QTL effect, and σk2 is the variance of γk representing the Q × E interaction. This type of model with further modeling on γk is called a hierarchical model. In the hierarchical model, the first moment parameter αk is the main effect and the second moment parameter σk2 represents the degree of Q × E interaction. The residual error vector ξj is assumed to be multivariate normal with density
 |
(3) |
where Θ is an m ×m variance–covariance matrix, which can be chosen from a class of available forms (to be discussed later). We have now defined the data and parameters. The nest step of the Bayesian analysis is to choose the prior distribution and infer the posterior distribution for each parameter.
Prior distribution:
We often have enough information from the data to estimate β and thus a flat (uninformative) prior was chosen for β; i.e., p(β)=1. The main effect for the kth QTL was assigned the following normal prior,
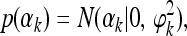 |
(4) |
where ϕ2k is the prior variance. A scaled inverse chi-square distribution was assigned to ϕ2k, which is
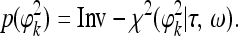 |
(5) |
A special case of this prior is τ=ω=0, leading to 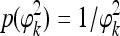 , called Jeffreys' prior. However, as mentioned by ter Braak et al. (2005), this prior is improper and leads to an improper posterior distribution. The revised prior is proposed by them and is claimed to lead to a proper posterior distribution. The revised prior is
, called Jeffreys' prior. However, as mentioned by ter Braak et al. (2005), this prior is improper and leads to an improper posterior distribution. The revised prior is proposed by them and is claimed to lead to a proper posterior distribution. The revised prior is
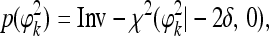 |
(6) |
where 0<δ≤0.5. In this study we used the proper prior to avoid any potential problems caused by the improper posterior distribution. The same scaled inverse chi-square distribution was also assigned to σk2,
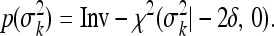 |
(7) |
Finally, we assumed 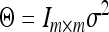 , where
, where 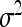 is a common residual variance and was assigned the same scaled inverse prior,
is a common residual variance and was assigned the same scaled inverse prior,
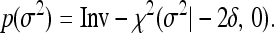 |
(8) |
Other structures of Θ are considered and described in a later section.
Posterior distribution:
The Markov chain Monte Carlo (MCMC) algorithm was used to implement the Bayesian shrinkage analysis. In the MCMC sampling, we need to derive only the fully conditional posterior distribution for each parameter. For example, the fully conditional posterior distribution for β is denoted by p(β | …), where the dots after the vertical bar represent the data and all other parameters. Except for the prior of β, all other priors we chose in the previous section are conjugate. Therefore, the fully conditional posterior has the same form as the prior distribution. Derivation of the posterior distribution was not given and we simply provided the parameters of the fully conditional posterior distribution for each variable.
The posterior distribution for β is multivariate normal
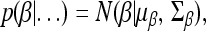 |
(9) |
where
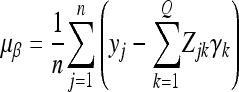 |
(10) |
and
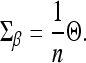 |
(11) |
The posterior for γk is also multivariate normal,
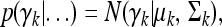 |
(12) |
where
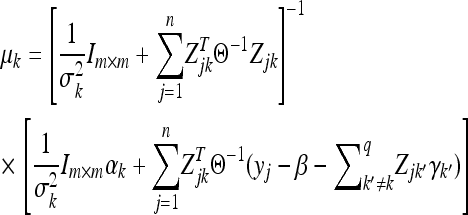 |
(13) |
and
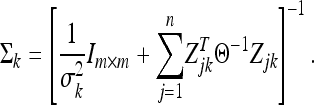 |
(14) |
The posterior distribution for αk is normal,
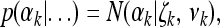 |
(15) |
where
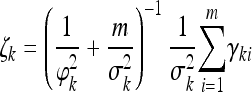 |
(16) |
and
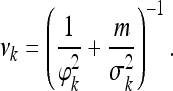 |
(17) |
We now discuss the posterior distributions for all the variance components, σ2, σ2k, and ϕ2k for k = 1,…, q. All of them are scaled inverse chi squares as given below,
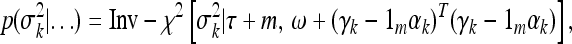 |
(18) |
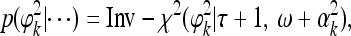 |
(19) |
and
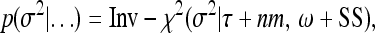 |
(20) |
where
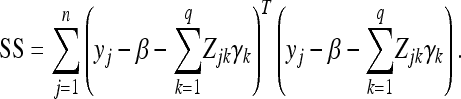 |
(21) |
MCMC sampling:
Since all the fully conditional posterior distributions have closed-form distributions, either a normal or a scaled inverse chi-square, Gibbs sampler was used for sampling all the variables, which is summarized below:
Initialize all variables by sampling the values from their prior distributions.
Sample the parameters sequentially from their corresponding posterior distributions.
Repeat the sampling cycle until the chain reaches a desired length.
The posterior sample contains all the observations after burn-in deletion and chain thinning. Post-MCMC analysis was performed on the posterior sample. We often ran multiple chains and took the average posterior statistics across the chains as the Bayesian estimates of the parameters.
Covariance structure:
We now introduce several alternative covariance structures for the residual errors.
Identity matrix:
The simple structure described earlier, Θ=Imσ2, is called the scaled identity matrix structure. This assumption is oversimplified and should be relaxed in real data analysis.
Diagonal matrix:
The covariance matrix is defined as
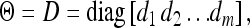 |
(22) |
which represents uncorrelated residual errors but has taken into account nonhomogenous residual variances for different environments. This assumption may hold in most situations. Each d was assigned a scaled inverse chi-square distribution and the fully conditional posterior distribution for di is
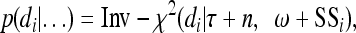 |
(23) |
where
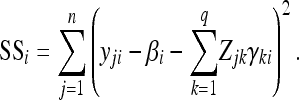 |
(24) |
Unstructured matrix:
The unstructured covariance matrix has been used before by Jiang and Zeng (1995) for multivariate QTL mapping. The only restriction for matrix Θ is positive definite. We assigned an inverse Wishart prior distribution to Θ. This prior is the multivariate version of the scaled inverse chi-square distribution,
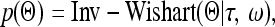 |
(25) |
where τ > m − 1 is the prior degree of belief and ω>0 is a positive definite matrix with the same dimension as matrix Θ. The posterior distribution remains inverse Wishart and thus
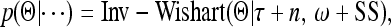 |
(26) |
where
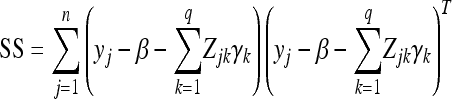 |
(27) |
is an m×m sum of squares matrix, different from the SS defined in Equation 20.
Factor analytic structured matrix:
The covariance matrix has the following structure,
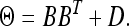 |
(28) |
It is called factor analytic structure because this structure has been used in factor analysis. This factor analytic structure was derived on the basis of the following latent variable linear model for the residual errors,
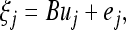 |
(29) |
where uj is an r × 1 latent factor (r < m) with a
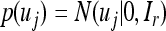 |
(30) |
distribution, B is an m× r matrix called factor loading, and 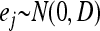 is a vector of independent errors and
is a vector of independent errors and 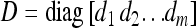 is a diagonal matrix for the independent error variances.
is a diagonal matrix for the independent error variances.
Under the factor analytic structure, the MCMC algorithm requires sampling B and uj for j=1,…, n, in addition to other parameters. We now describe the prior and posterior of these new variables. The prior for uj is standardized multivariate normal given in Equation 30. The fully conditional posterior distribution remains multivariate normal,
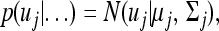 |
(31) |
where
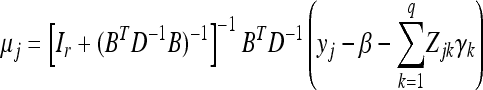 |
(32) |
and
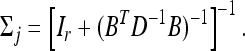 |
(33) |
The factor loadings are represented by an m × r matrix B. Let 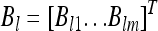 be the lth column of matrix B for l = 1,…, r. We now rewrite Equation 29 as
be the lth column of matrix B for l = 1,…, r. We now rewrite Equation 29 as
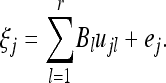 |
(34) |
Given 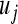 and knowing that
and knowing that
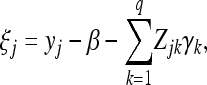 |
(35) |
Equation 34 is a typical multivariate regression problem. The fully conditional posterior distribution of Bl is multivariate normal,
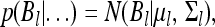 |
(36) |
where
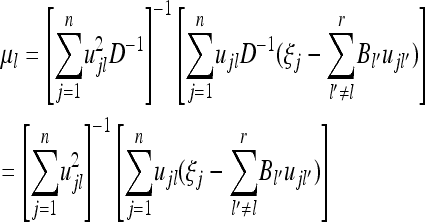 |
(37) |
and
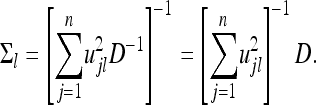 |
(38) |
Having provided the fully conditional posterior distribution for every variable, we are now ready to conduct the Gibbs sampler to infer the empirical posterior distribution for each variable.
APPLICATIONS
Barley data analysis:
We used barley data obtained from the North American Barley Genome Mapping Project (Tinker et al. 1996) to demonstrate the application of the new method. In the barley QTL mapping project, there were 127 mapped markers covering 1500 cM of the barley genome. Seven traits were investigated in the project. In this study, we used the yield trait analysis for the demonstration. The doubled haploid (DH) population was initiated from the cross between Harrington and TR306. The DH population consisted of 145 lines, each grown in 28 different environments. The data set was updated after it was first published in 1996, but the difference between the original and the updated data was minor so that we could still compare the current result with that from the original study.
We used six different covariance structures to analyze the data, which were (1) the homogeneous (constant) variance 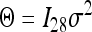 , (2) the heterogeneous variances
, (2) the heterogeneous variances 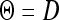 , (3) unstructured matrix Θ (positive definite), (4) the first-order factor analytic structure
, (3) unstructured matrix Θ (positive definite), (4) the first-order factor analytic structure 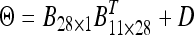 , (5) the second-order factor analytic structure
, (5) the second-order factor analytic structure 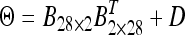 , and (6) the third-order factor analytic structure
, and (6) the third-order factor analytic structure 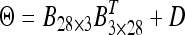 . The length of the Markov chain consisted of 200,000 sweeps. The first 100,000 sweeps were deleted as burn-in and thereafter the chain was thinned by keeping 1 observation in every 100 sweeps, producing 1000 observations in the collected posterior sample for post-MCMC analysis.
. The length of the Markov chain consisted of 200,000 sweeps. The first 100,000 sweeps were deleted as burn-in and thereafter the chain was thinned by keeping 1 observation in every 100 sweeps, producing 1000 observations in the collected posterior sample for post-MCMC analysis.
To test the significance of the QTL effects, we conducted a permutation test to generate the null distribution of each main effect and each Q × E interaction effect. In the permutation analysis, we repeated the MCMC sampling method as described before but reshuffled the phenotypic values. The permutation analysis was proposed by Che and Xu (2010), who called it permutation inside the Markov chain. In the permutation analysis, the length of the Markov chain was 200,000 sweeps. The first 100,000 sweeps were deleted as burn-in and the chain thinning rate was 1/25. The posterior sample contained 4000 observations. From the null distribution, we drew a confidence interval for each estimated effect. An effect was claimed to be significant if the estimated value fell outside of the 99% confidence interval of the null distribution.
Among the six covariance structures, the second structure Θ=D (a diagonal matrix) detected the maximum number of QTL (main and Q × E interaction effects). Although more QTL does not mean better, it is hard to use cross-validation to evaluate different structures under the MCMC implemented Bayesian analysis. Bayes factors are often used to evaluate different models. However, the complexity of our proposed model makes the calculation of the Bayes factors difficult. Therefore, we used the Bayesian information criteria (BIC) to evaluate the performance of the six different models. The BIC score was calculated using
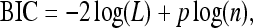 |
(39) |
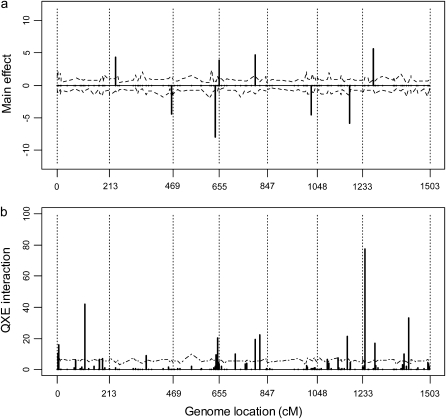
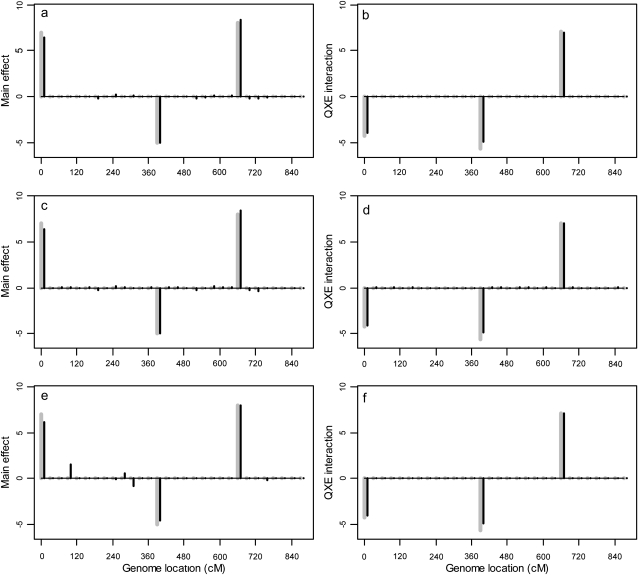
where L is the likelihood function evaluated at the estimated parameters, p is the number of parameters, and n is the sample size. The Bayesian estimates of the parameters in L are the posterior means of β, γ, and Θ. The BIC scores are shown in Table 1, which indicates that the second (heterogeneous residual variance) model performed better than all other models. The first-order factor analytic model was the second best model with a BIC score slightly larger than that of the best model. The result of the best model is depicted in Figure 1, where the posterior means of the main effects and the Q × E interaction effects are plotted against the genome locations of the markers. Figure 1 also gives the 99% confidence intervals for the main and Q × E interaction effects. Eight markers showed significant main effects and 18 markers showed significant Q × E interaction. The 18 interacting markers were distributed across all seven chromosomes of the entire genome. Only 1 marker had both the main and the Q × E interaction effects. Each of the other markers had either a main effect or a Q × E interaction effect but not both. The estimated main and Q × E interaction effects for the markers are given in Table 2.
TABLE 1.
BIC scores of the six variance–covariance structures for the barley data analysis
| Structure | Log likelihood | p | BIC |
|---|---|---|---|
| Homogeneous | −18,055.18 | 1 | 36,118.67 |
| Heterogeneous | −17,239.85 | 28 | 34,712.35 |
| Unstructured | −17,234.20 | 406 | 37,841.83 |
| First-order factor | −17,171.71 | 56 | 34,808.72 |
| Second-order factor | −17,155.33 | 84 | 35,008.60 |
| Third-order factor | −17,143.79 | 118 | 35,218.19 |
The number of parameters is denoted by p.
Figure 1.—
The estimated main and Q × E interaction effects for markers of the entire barley genome. (a) Main effects under the heterogeneous residual variance model. (b) Q × E interaction effects under the heterogeneous residual variance model. Chromosomes are separated by the dotted vertical reference lines. The dashed curves represent the 99% confidence intervals generated from the permutation analysis.
TABLE 2.
Estimated main and Q × E interaction effects and their 99% confidence intervals of the null distributions for the barley yield data
| Marker no. (position, chromosome) | Main effect |
Q × E interaction |
||
|---|---|---|---|---|
| Effect | 99% C.I. | Effect | 99% C.I. | |
| 2 (3.516, 1) | — | — | 10.240 | (0, 7.719) |
| 3 (7.892, 1) | — | — | 15.805 | (0, 8.066) |
| 13 (111.273, 1) | — | — | 42.046 | (0, 6.797) |
| 18 (182.978, 1) | — | — | 7.070 | (0, 6.356) |
| 23 (24.59, 2) | 4.379 | (−0.904, 1.063) | — | — |
| 32 (145.863, 2) | — | — | 8.974 | (0, 5.420) |
| 38 (247.586, 2) | −4.447 | (−1.868, 1.340) | — | — |
| 50 (168.297, 3) | −7.927 | (−0.739, 0.725) | — | — |
| 51 (172.134, 3) | — | — | 9.226 | (0, 6.239) |
| 52 (178.493, 3) | — | — | 20.160 | (0, 4.541) |
| 53 (182.222, 3) | — | — | 4.557 | (0, 4.214) |
| 54 (185.101, 3) | 3.875 | (−0.975, 1.018) | — | — |
| 59 (63.732, 4) | — | — | 9.724 | (0, 5.383) |
| 64 (145.469, 4) | 4.711 | (−1.230, 1.541) | 19.317 | (0, 5.378) |
| 65 (163.074, 4) | — | — | 22.422 | (0, 5.337) |
| 77 (178.118, 5) | −4.542 | (−1.256, 0.836) | — | — |
| 86 (41.568, 6) | — | — | 6.603 | (0, 4.198) |
| 89 (85.438, 6) | — | — | 7.548 | (0, 7.476) |
| 95 (122.839, 6) | — | — | 21.108 | (0, 7.386) |
| 96 (131.476, 6) | −5.819 | (−1.539, 0.884) | — | — |
| 102 (7.247, 7) | — | — | 77.311 | (0, 5.606) |
| 105 (43.111, 7) | 5.605 | (−0.704, 0.958) | — | — |
| 106 (47.487, 7) | — | — | 16.614 | (0, 6.140) |
| 116 (164.789, 7) | — | — | 9.899 | (0, 5.959) |
| 118 (173.542, 7) | — | — | 33.210 | (0, 3.860) |
All the estimated effects are outside of the 99% confidence intervals.
We also performed an individual marker analysis to compare the result with that of the Bayesian analysis. For the individual marker analysis, QTL mapping was conducted separately for each environment. The average estimated effect for each marker across the 28 environments represented the main effect while the variance of the estimated effects across the environments represented the Q × E interaction effect. The estimated main and Q × E interaction effects of the single-marker analysis are shown in Figure 2. We can see that Figure 2 is quite similar to Figure 1 in the Bayesian analysis. The main difference between the two figures is the different sharpness of the marker effects. The Bayesian analysis generated very clean (sharp) signals of the plots.
Figure 2.—
Estimated main and Q × E interaction effects using single-marker analysis. (a) Main effects. (b) Q × E interaction effects. Chromosomes are separated by the dotted vertical reference lines.
Arabidopsis data analysis:
The barley data contain many environments, which is hard to find in most studies. So we also applied our model to recombinant inbred line data of Arabidopsis (Loudet et al. 2002), where two parents initiating the line cross were Bay-0 and Shahdara, with Bay-0 as the female parent. Flowering time was recorded for each line in two environments: long day (16-hr photoperiod) and short day (8-hr photoperiod). The population contained 420 lines. A total of 38 microsatellite markers were used for QTL mapping. We inserted a pseudomarker in every 2 cM of the genome and had a total of 200 markers (38 true markers plus 162 pseudomarkers) in our analysis.
The variance of Q × E interaction 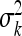 may not be estimated accurately due to small environments. So in small environments the variance would then simply serve as a tool to shrink the environment-specific QTL effects. The bias of
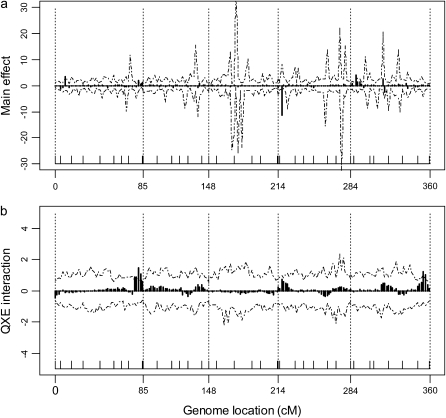
may not be estimated accurately due to small environments. So in small environments the variance would then simply serve as a tool to shrink the environment-specific QTL effects. The bias of 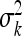 would lead to biased estimation of main effect as well. Although the MCMC algorithm remains the same as before, we need to revise our post-MCMC procedure. We use vector γk to estimate Q × E interaction effects of the kth marker. The differences between vector γk and its mean represent Q × E interaction effects. In the two-environments case, we can just use the differences between the two components of γk as the Q × E interaction effects because vector γk is a 2 × 1 vector. Since there are only two environments, we did not use the factor analytic model to analyze the data. The BIC scores for the three models (homogeneous, heterogeneous, and unstructured covariance matrices) are 3798.49, 3775.64, and 3645.55. Figure 3 shows the main and Q × E interaction effects of the Arabidopsis data under the unstructured covariance model. The 95% confidence intervals for the main and Q × E interaction effects are also given. Four markers showed significant main effects and six markers showed significant Q × E interaction effects.
would lead to biased estimation of main effect as well. Although the MCMC algorithm remains the same as before, we need to revise our post-MCMC procedure. We use vector γk to estimate Q × E interaction effects of the kth marker. The differences between vector γk and its mean represent Q × E interaction effects. In the two-environments case, we can just use the differences between the two components of γk as the Q × E interaction effects because vector γk is a 2 × 1 vector. Since there are only two environments, we did not use the factor analytic model to analyze the data. The BIC scores for the three models (homogeneous, heterogeneous, and unstructured covariance matrices) are 3798.49, 3775.64, and 3645.55. Figure 3 shows the main and Q × E interaction effects of the Arabidopsis data under the unstructured covariance model. The 95% confidence intervals for the main and Q × E interaction effects are also given. Four markers showed significant main effects and six markers showed significant Q × E interaction effects.
Figure 3.—
Estimated main and Q × E interaction effects for markers of the entire Arabidopsis genome. (a) Main effects under the unstructured model. (b) Q × E interaction effects (expressed as differences between the QTL effects in the two environments) under the unstructured model. Chromosomes are separated by the dotted vertical reference lines. The marker positions are represented by the ticks on the horizontal line. The dashed curves represent the 95% confidence intervals generated from the permutation analysis.
Simulation study:
The barley data analysis did not show the advantage of fitting appropriate covariance structures over the simple diagonal covariance matrix because the 28 different environments did not seem to be correlated. Therefore, we conducted two simulation experiments (simulations 1 and 2) in this section to demonstrate the importance of covariance structure to the Bayesian analysis of Q × E interaction. We also did a two-environment simulation (simulation 3) to demonstrate the fitness of our model for small environments.
In simulation 1, we used the real marker information from the North American Barley Genome Mapping Project (Tinker et al. 1996) to simulate the genome. We simulated 127 markers from seven chromosomes with marker distances exactly the same as the real data. We simulated 145 DH lines in 28 environments. The intercept β was given values ranging from 200 to 605 for the 28 different environments. We assumed that 10 of the 127 markers had main effects and also Q × E interaction effects in the 28 environments. In the simulation experiment, we chose the factor analytic covariance structure 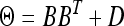 with B defined as a 28×3 matrix, indicating that correlations had occurred between different environments. The true values of β, the QTL effects, the B matrix, and the D matrix are given in Tables 4 and 5. The simulated data were analyzed using the six different covariance structures described earlier in the barley data analysis. We expected that the first three structures (homogeneous variance, heterogeneous variance, and unstructured matrices) would perform poorly but the last three structures (first-order, second-order, and third-order factor analytic structures) would perform better, especially the third-order factor analytic structure.
with B defined as a 28×3 matrix, indicating that correlations had occurred between different environments. The true values of β, the QTL effects, the B matrix, and the D matrix are given in Tables 4 and 5. The simulated data were analyzed using the six different covariance structures described earlier in the barley data analysis. We expected that the first three structures (homogeneous variance, heterogeneous variance, and unstructured matrices) would perform poorly but the last three structures (first-order, second-order, and third-order factor analytic structures) would perform better, especially the third-order factor analytic structure.
TABLE 4.
True and estimated QTL main effects and Q × E interaction effects from 20 replicated simulation experiments under the second-order factor analytic covariance structure
| Marker no. (position, chromosome) | Main effect |
Q × E |
||
|---|---|---|---|---|
| True | Estimated | True | Estimated | |
| 1 (0, 1) | 5 | 1.14 | 36 | 41.33 |
| 14 (127.208, 1) | −7 | −6.51 | 0.25 | 0.20 |
| 32 (145.863, 2) | −1 | −0.01 | 16 | 19.76 |
| 48 (153.931, 3) | 10 | 8.08 | 4 | 2.38 |
| 52 (178.493, 3) | −4 | −0.75 | 25 | 22.45 |
| 65 (163.074, 4) | −8 | −8.14 | 49 | 45.03 |
| 71 (109.389, 5) | 15 | 15.52 | 9 | 7.66 |
| 84 (10.533, 6) | −1 | 0.00 | 1 | 0.25 |
| 96 (131.476, 6) | 0 | 0.00 | 64 | 65.64 |
| 110 (122.584, 7) | 3 | 0.01 | 100 | 123.19 |
TABLE 5.
The true and estimated intercepts, the B and D matrices used in simulation 1
| E | Intercept |
B matrix: |
D matrix |
||||
|---|---|---|---|---|---|---|---|
| True | Estimated | true | True | Estimated | |||
| 1 | 200 | 199.59 | 50 | 0 | 0 | 7569 | 7354 |
| 2 | 215 | 215.11 | 50 | 0 | 0 | 6561 | 6698 |
| 3 | 230 | 230.28 | 50 | 0 | 0 | 1849 | 1790 |
| 4 | 245 | 243.15 | 50 | 0 | 0 | 2116 | 2166 |
| 5 | 260 | 259.01 | 50 | 0 | 0 | 784 | 758 |
| 6 | 275 | 276.03 | 50 | 0 | 0 | 729 | 758 |
| 7 | 290 | 290.02 | 50 | 0 | 0 | 1296 | 1324 |
| 8 | 305 | 305.01 | 50 | 0 | 0 | 1296 | 1239 |
| 9 | 320 | 320.02 | 50 | 0 | 0 | 5184 | 5285 |
| 10 | 335 | 335.68 | 0 | 30 | 0 | 4225 | 4207 |
| 11 | 350 | 350.78 | 0 | 30 | 0 | 121 | 138 |
| 12 | 365 | 365.58 | 0 | 30 | 0 | 1444 | 1454 |
| 13 | 380 | 381.38 | 0 | 30 | 0 | 529 | 516 |
| 14 | 395 | 397.56 | 0 | 30 | 0 | 1296 | 1250 |
| 15 | 410 | 411.56 | 0 | 30 | 0 | 1521 | 1503 |
| 16 | 425 | 425.82 | 0 | 30 | 0 | 529 | 529 |
| 17 | 440 | 441.16 | 0 | 30 | 0 | 2809 | 2697 |
| 18 | 455 | 456.33 | 0 | 30 | 0 | 529 | 514 |
| 19 | 470 | 469.16 | 0 | 0 | 10 | 2116 | 2250 |
| 20 | 485 | 484.17 | 0 | 0 | 10 | 2304 | 2359 |
| 21 | 500 | 498.4 | 0 | 0 | 10 | 1156 | 1227 |
| 22 | 515 | 515.16 | 0 | 0 | 10 | 729 | 865 |
| 23 | 530 | 530.17 | 0 | 0 | 10 | 1369 | 1393 |
| 24 | 545 | 544.75 | 0 | 0 | 10 | 400 | 496 |
| 25 | 560 | 559.78 | 0 | 0 | 10 | 1369 | 1476 |
| 26 | 575 | 575.4 | 0 | 0 | 10 | 625 | 705 |
| 27 | 590 | 589.9 | 0 | 0 | 10 | 2401 | 2629 |
| 28 | 605 | 604.43 | 0 | 0 | 10 | 841 | 933 |
The estimated parameters are from the second-order factor analytic model. The estimated B is not given (see details in the simulation study). E stands for environment.
In the MCMC-implemented Bayesian analysis, the length of the Markov chain was 50,000 sweeps. The first 25,000 sweeps (burn-in period) were deleted. The chain thinning rate was 1 in 50. The empirical posterior sample contained 500 observations for the post-MCMC analysis. The MCMC experiment with the same simulated data was repeated a few times to make sure that the chain had converged to the stationary distribution.
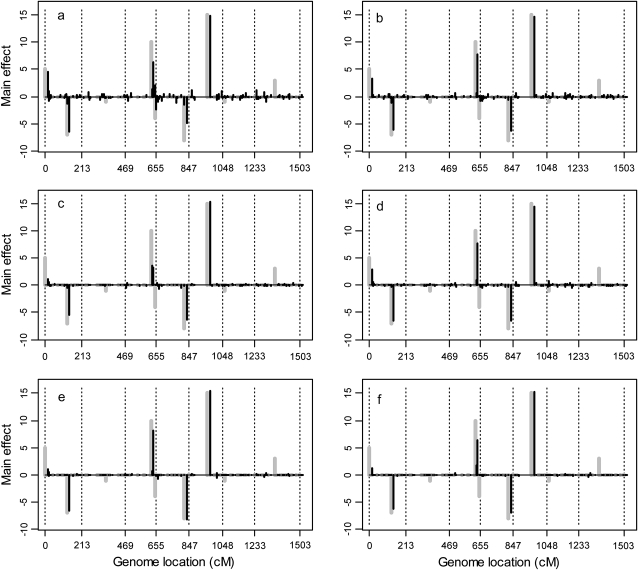
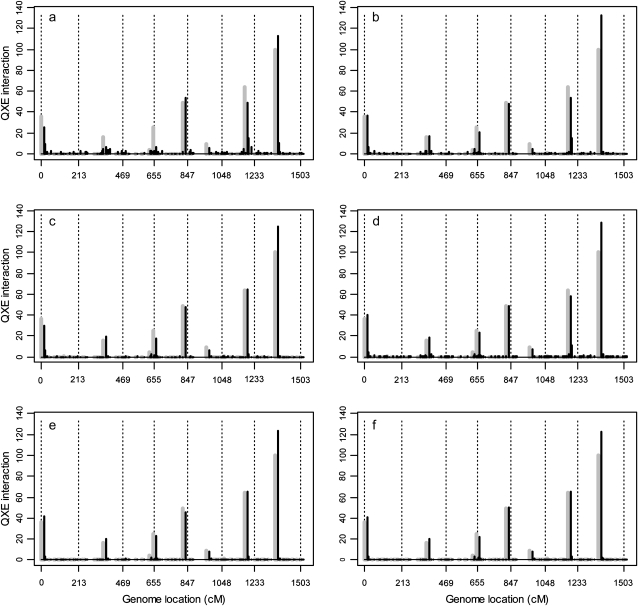
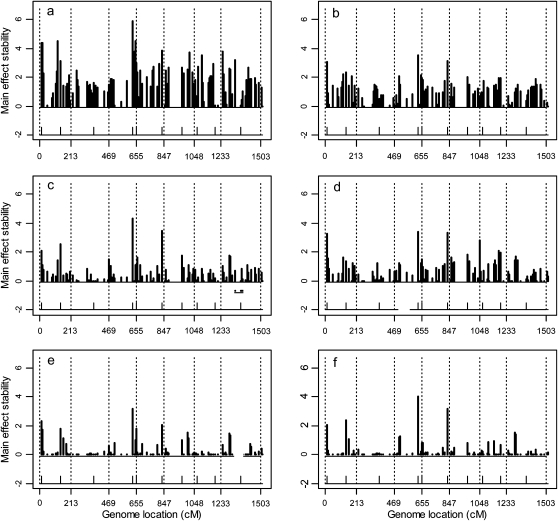
The results of the simulation studies are depicted in Figure 4 for the average main effects of 20 replicates and in Figure 5 for the average Q × E interaction effects of 20 replicates. These two figures show that the estimated QTL effects agreed well with the true effects. Figures 4 and 5 also show that the first four covariance structures (homogeneous residual, heterogeneous residual, unstructured covariance, and first-order factor analytic structure) have some notable background noise, indicating some false positives had occurred. However, the last two factor analytic structures have very little background noise. From the two figures, the background noise of the first four covariance structures may not be very clear. So we calculated standard deviations of each marker's main effect among the 20 replicates and plotted them in Figure 6, from which we can see the difference among the six models. The performance of the last two factor analytic models is very stable for the majority of the markers without main effects. While the first four structures, especially the first two, cannot achieve such a nice performance, which means that among the 20 replicates these models generated some false main effects. Table 3 shows the average BIC scores for the six different covariance structures. We see that the factor analytic structure outperformed the other three models. This is consistent with our expectation. The lowest BIC occurred in the second-order factor analytic structure. However, the third-order factor analytic structure (the true model) was just slightly higher in value than the second-order structure. The log-likelihood value of the third-order factor was higher than that of the second-order factor. Table 4 gives the average estimated main and Q × E interaction effects obtained from the 20 replicates based on the second-order factor analytic model. When we compared the estimated main effects and the true effects, we noted that large main effects were estimated quite accurately but small effects were shrunken to zero. The Q × E interaction effects were always detectable regardless of the sizes of the effects. Table 5 gives the true intercepts and the residual error variances (the D matrix) along with their estimated values. The true B matrix is also given in Table 5. The estimated B was not given here because the columns of B are independent and thus are exchangeable. This does not affect the estimate of the covariance structure. We checked the average estimated variance–covariance matrix and did observe three separate environment groups.
Figure 4.—
The average main effects across 20 replicated simulation experiments for the entire genome in the first simulation experiment. (a) Homogeneous residual variance (scaled identity matrix); (b) heterogeneous residual variances (diagonal matrix); (c) unstructured covariance matrix; (d) first-order factor analytic structure; (e) second-order factor analytic structure; (f) third-order factor analytic structure. The dotted vertical reference lines divide the genome into seven chromosomes. The black solid needles are the average marker main effects. The true effects of the markers are indicated by the shaded needles.
Figure 5.—
(a–f) The average Q × E interaction effects across 20 replicated simulation experiments for the entire genome in the first simulation experiment. Chromosomes are separated by the dotted vertical reference lines. The black solid needles are the average Q × E interaction effects. The true effects of Q × E interaction are indicated by the shaded needles.
Figure 6.—
(a–f) The standard deviations (stabilities) of estimated main effects across 20 replicated simulation experiments for the entire genome in the first simulation experiment. The positions of markers with (nonzero) simulated effects are indicated by the ticks on the horizontal lines at a value of −2.
TABLE 3.
Average BIC score for six different variance–covariance structures in the simulation study
| Covariance structure | Log likelihood | p | BIC |
|---|---|---|---|
| Homogeneous | −18,096.83 | 1 | 36,201.97 |
| Heterogeneous | −17,564.80 | 28 | 35,362.26 |
| Unstructured | −16,815.18 | 406 | 37,003.79 |
| First-order factor | −17,176.53 | 56 | 34,818.36 |
| Second-order factor | −16,887.74 | 84 | 34,473.44 |
| Third-order factor | −16,858.56 | 118 | 34,647.73 |
The number of parameters is denoted by p.
Although the stability test and BIC scores showed the advantage of the factor analytic model, the differences of marker main effects for the six models are not very obvious in Figure 4. So we performed the second simulation experiment (simulation 2) to further demonstrate the advantage of the factor analytic model. We focused mainly on comparison of heterogeneous diagonal structure and three factor analytic models. In this simulation, 100 DH lines in eight environments were generated with 30 markers. The distance between two nearby markers was 30 cM. The intercept β was given values ranging from 200 to 305 for the eight environments. We assumed that 3 of the 30 markers had main effects and also Q × E interaction effects in the eight environments. We also chose the factor analytic covariance structure 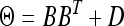 with B defined as an 8×2 matrix. The first column of B had values of 20, 10, 10, 5, 0, 0, 0, 0. The second column had values of 0, 0, 0, 0, 15, 15, 10, 2. Matrix D was an identity matrix. Figure 7 shows the estimated main effects of the four models. We can clearly see many false positive main effects in the heterogeneous diagonal structure. The BIC scores for the four models are 4264, 3362, 2495, and 2558, which also are in favor of the factor analytic models.
with B defined as an 8×2 matrix. The first column of B had values of 20, 10, 10, 5, 0, 0, 0, 0. The second column had values of 0, 0, 0, 0, 15, 15, 10, 2. Matrix D was an identity matrix. Figure 7 shows the estimated main effects of the four models. We can clearly see many false positive main effects in the heterogeneous diagonal structure. The BIC scores for the four models are 4264, 3362, 2495, and 2558, which also are in favor of the factor analytic models.
Figure 7.—
The marker main effects in the second simulation experiment. (a) Heterogeneous residual variances (diagonal matrix); (b) first-order factor analytic structure; (c) second-order factor analytic structure; (d) third-order factor analytic structure. The black solid needles are the estimated marker main effects. The shaded needles are the estimated main effects.
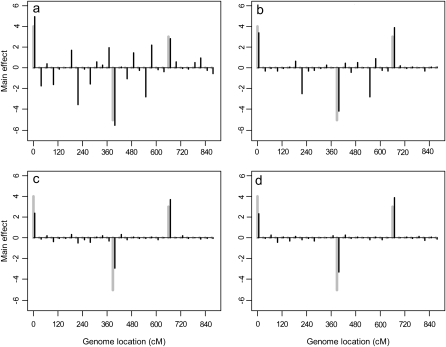
In simulation 3, we also generated 100 DH lines using the same marker information given by simulation 2, but this time we simulated only two environments. The intercepts were 200 and 215 for the two environments. The factor analytic covariance structure was used with B defined as a 2×2 matrix. The first column of B had values of 1 and 2. The second column had values of 2 and 1. Matrix D was an identity matrix. The MCMC and post-MCMC analyses of these data used the same setup as Arabidopsis data analysis. Figure 8 gives the comparison of the true and the estimated main and Q × E interaction effects. From Figure 8, the true and the estimated marker effects are very close to each other for all three models. The promising results also demonstrate that our proposed method is a good choice to handle data with small environments.
Figure 8.—
The marker main effects and Q × E interaction effects in the third simulation experiment. (a and b) Homogeneous residual variance (scaled identity matrix); (c and d) heterogeneous residual variances (diagonal matrix); (e and f) unstructured covariance matrix. The black solid needles are the estimated effects (main and interaction). The shaded needles are the true values (main and interaction).
DISCUSSION
The importance of this study is reflected by two major contributions to Q × E study, the multiple-QTL model and the factor analytic covariance structure. The multiple-QTL model for Q × E is an extension of the Bayesian shrinkage analysis for mapping QTL in a single environment (Xu 2003). The factor analytic covariance structure is available in the literature but has never been applied to QTL mapping. Other covariance structures may be considered in future studies, e.g., the autoregressive model of order 1 [AR(1)] and compound symmetry (CS) covariance structures. These alternative structures can be used to fit models when the environments represent temporal or spatial variation. The 28 environments in the barley experiments represent 28 different locations (spatial variation). However, the information about the location was not available to us. We believe that the factor analytic structure is robust and can be fit to a wide variety of covariance structures, ranging from the simplest diagonal matrix to the most complicated unstructured matrix, by choosing different orders of the factors. This has been demonstrated by the similarity of the diagonal matrix and the first-order factor analytic model in our data analyses. The factor analytic model is also easy to fit under the general linear model framework. Both the factor loadings and the factors themselves have normal posterior distributions and can be sampled using the Gibbs sampler approach.
The most significant contribution of this study was to use the variance of QTL effects across environments to measure the size of the Q × E interaction for a particular QTL. This has significantly simplified the Q × E study. If the number of environments were small, however, the variance would not be accurately estimated. In this case, one should use some kind of linear contrast of the environment-specific effects as a measure of the Q × E interaction. Arabidopsis data and simulation 3 are two examples of such a treatment. The variance would then simply serve as a tool to shrink the environment-specific QTL effects. The MCMC sampling procedure remains the same, but the post-MCMC analysis needs to be modified.
The method developed in the current study applies only to plants where the same genotype can be replicated in multiple environments. In animals where the same genotype cannot be replicated (except identical twins), some modification is required. For example, if an F2 family is raised in three environments, each animal may have a different genotype from other animals. This argument also applies to QTL-by-sex interaction, where the same individual cannot be split into male and female. The modification is not trivial and thus deserves further study.
Although the environment-specific QTL effects, denoted by vector 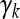 for the kth marker, are used only to draw the posterior distributions for the main and Q × E interaction effects, they may be interesting parameters in their own rights. The posterior mean of each
for the kth marker, are used only to draw the posterior distributions for the main and Q × E interaction effects, they may be interesting parameters in their own rights. The posterior mean of each 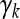 can be used to predict the molecular breeding value of each line in a particular environment. This information may facilitate marker-assisted selection (using a few markers) or genome selection (using all markers of the entire genome). Genome selection has been an important strategy for animal (Meuwissen et al. 2001) and plant breeding (Xu 2003).
can be used to predict the molecular breeding value of each line in a particular environment. This information may facilitate marker-assisted selection (using a few markers) or genome selection (using all markers of the entire genome). Genome selection has been an important strategy for animal (Meuwissen et al. 2001) and plant breeding (Xu 2003).
The Bayesian method presented here applies only to multiple-marker analysis; i.e., each marker is treated as a putative QTL. If the markers are not evenly placed in the genome, one may insert some pseudomarkers in regions not well covered by markers. In the regions with saturated markers, one may use only a few selected markers to avoid a potential multicollinearity problem. With the current molecular technology, genomes of most species of agricultural importance may be saturated very soon with high-density markers. Pseudomarker insertion will no longer be necessary, but marker selection will become important. One strategy for marker selection is to include one marker in every d cM for the Bayesian model. The optimal strategy may be the moving interval approach proposed by Wang et al. (2005), in which a fixed number of putative QTL were included in the model for each chromosome and the position of the putative QTL can move (jump) among a few neighboring markers. This approach may be adopted in the second stage of mapping, i.e., fine mapping after the important QTL regions have been identified.
One drawback of the MCMC-implemented Bayesian method is the slow computation process due to the large number of environments and the high dimensionality of the model. A quick method may be the posterior mode estimation in which only the conditional posterior modes are presented as the Bayesian estimates for the parameters of interest. Although the estimates are no longer Bayesian estimates, the results may be comparable. This quick posterior mode estimation may provide preliminary results to be used for further analysis using the fully Bayesian analysis.
Finally, the entire data analyses were conducted using a program developed in R. Interested readers may visit our website (www.statgen.ucr.edu) to download the program and the sample data to test the method and analyze their own data.
Acknowledgments
This project was supported by the National Plant Genome Initiative of the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service grant 2007-02784 (to S.X.).
References
- Beavis, W. D., and P. Keim, 1996. Identification of quantitative trait loci that are affected by environment, pp. 123–150 in Genotype-by-Environment Interaction, edited by M. S. Kang and H. G. Gauch. CRC Press, Boca Raton, FL.
- Boer, M. P., D. Wright, L. Feng, D. W. Podlich, L. Luo et al., 2007. A mixed-model quantitative trait loci (QTL) analysis for multiple-environment trial data using environmental covariables for QTL-by-environment interactions, with an example in maize. Genetics 177 1801–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che, X. and S. Xu, 2010. Significance test and genome selection in Bayesian shrinkage analysis. Int. J. Plant Genomics doi:10.1155/2010/893206 [DOI] [PMC free article] [PubMed]
- Jansen, R. C., 1993. Interval mapping of multiple quantitative trait loci. Genetics 135 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, R. C., J. W. Ooijen, P. Stam, C. Lister and C. Dean, 1995. Genotype-by-environment interaction in genetic mapping of multiple quantitative trait loci. Theor. Appl. Genet. 91 33–37. [DOI] [PubMed] [Google Scholar]
- Jiang, C., and Z. B. Zeng, 1995. Multiple trait analysis of genetic mapping for quantitative trait loci. Genetics 140 1111–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E. S., and D. Botstein, 1989. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet, O., S. Chaillou, C. Camilleri, D. Bouchez and F. Daniel-Vedele, 2002. Bay-0 × Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor. Appl. Genet. 104 1173–1184. [DOI] [PubMed] [Google Scholar]
- Meuwissen, T. H., B. J. Hayes and M. E. Goddard, 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157 1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, A. H., S. Damon, J. D. Hewitt, D. Zamir, H. D. Rabinowitch et al., 1991. Mendelian factors underlying quantitative traits in tomato: comparison across species, generations, and environments. Genetics 127 181–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepho, H. P., 2000. A mixed-model approach to mapping quantitative trait loci in barley on the basis of multiple environment data. Genetics 156 2043–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romagosa, I., S. E. Ullrich, F. Han and P. M. Hayes, 1996. Use of the additive main effects and multiplicative interaction model in QTL mapping for adaptation in barley. Theor. Appl. Genet. 93 30–37. [DOI] [PubMed] [Google Scholar]
- Stuber, C. W., S. E. Lincoln, D. W. Wolff, T. Helentjaris and E. S. Lander, 1992. Identification of genetic factors contributing to heterosis in a hybrid from two elite maize inbred lines using molecular markers. Genetics 132 823–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Braak, C. J. F., M. P. Boer and M. Bink, 2005. Extending Xu's Bayesian model for estimating polygenic effects using markers of the entire genome. Genetics 170 1435–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinker, N. A., D. E. Mather, B. G. Rossnagel, K. J. Kasha, A. Kleinhofs et al., 1996. Regions of the genome that affect agronomic performance in two-row barley. Crop Sci. 36 1053–1062. [Google Scholar]
- Wang, H., Y. M. Zhang, X. Li, G. L. Masinde, S. Mohan et al., 2005. Bayesian shrinkage estimation of quantitative trait loci parameters. Genetics 170 465–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, S., 2003. Estimating polygenic effects using markers of the entire genome. Genetics 163 789–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Z. B., 1994. Precision mapping of quantitative trait loci. Genetics 136 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]