Abstract
Cre/loxP recombination enables cellular specificity and, in the case of inducible systems, temporal control of genomic deletions. Here we used a SM22α tamoxifen-inducible Cre line to inactivate β1 integrin in adult smooth muscle. Interestingly, analysis of two distinct β1 loxP transgenic mice revealed vastly different outcomes after β1 integrin deletion. Lethality occurred 4 weeks postinduction in one Cre/loxP line, while no apparent phenotype was seen in the other line. Genetic analysis revealed appropriate DNA excision in both cases; however, differences were found in the degree of protein loss with absolutely no change in protein levels in the model that lacked a phenotype. Seeking to understand protein persistence despite appropriate recombination, we first validated the flox allele using a constitutive Cre line and demonstrated its ability to mediate effective protein inactivation. We then examined the possibility of heterozygous cell selection, protein turnover, and deletion efficiency with no success for explaining the phenotype. Finally, we documented the presence of the Cre-recombination episomal product, which persisted in tissue samples with no protein loss. The product was only noted in cells with low proliferative capacity. These findings highlight the potential for protein expression from the products of Cre-recombinase excised genes, particularly when deletion occurs in low turnover populations.
INTEGRINS are a group of transmembrane, extracellular matrix receptors composed of heterodimeric pairs of α- and β-subunits that have been shown to function in cell survival, migration, and mechanotransduction (Schmidt et al. 1993; Wang et al. 1993; Chen et al. 1997; Hynes 2002; Stupack and Cheresh 2002; Martinez-Lemus et al. 2005; Abraham et al. 2008). To study the function of β1 integrin in vivo, knockout mice were generated by homologous recombination and found to be embryonic lethal prior to implantation (Fassler and Meyer 1995; Stephens et al. 1995). Due to this early lethality, Cre/loxP technology has been employed to dissect the contribution of β1 integrin in specific cell types (Raghavan et al. 2000; Pietri et al. 2004; Jones et al. 2006; Lei et al. 2008).
As with all targeted recombination technologies, the Cre/loxP system has advantages and some known limitations (Branda and Dymecki 2004). In the case of loxP sites, location is critical. LoxP sites can flank miRNAs resulting in their inadvertent deletion (Kuhnert et al. 2008) and insertion of a floxed neo cassette in the intron of a gene has been shown to unintentionally disrupt gene splicing (Wassarman et al. 1997; Meyers et al. 1998). Furthermore, unidentified alternative transcriptional start sites (Han and Zhang 2002) may circumvent attempts to inactivate a gene by loxP flanking a single exon. Given these caveats, it is important to use multiple controls to facilitate data interpretation and, whenever possible, validate the phenotype with an additional floxed allele having loxP sites located in different regions of the gene.
Four β1 integrin loxP constructs have been developed (Potocnik et al. 2000; Raghavan et al. 2000; Graus-Porta et al. 2001; Keller et al. 2001). Each of these transgenic alleles varies in their placement of loxP sites; however, they all have been shown to successfully eliminate β1 integrin protein when used with constitutive Cre-expressing systems (Li et al. 2005; Benninger et al. 2006; Jones et al. 2006; Lei et al. 2008). Our goal was to examine β1 integrin gene deletion in smooth muscle of adult mice. For this, we decided to use two distinct loxP transgenes designed to inactivate β1 integrin: an allele with loxP sites flanking exon 3 (β1e3) (Raghavan et al. 2000) and another allele with loxP sites spanning the region between exon 2 and the 3′-UTR (β1fl) (Potocnik et al. 2000). Both transgenic mice were crossed to an inducible SM-22α Cre-recombinase mouse (Kuhbandner et al. 2000) to temporally restrict deletion in adult smooth muscle cells.
To our surprise, the double transgenic lines provided different phenotypes despite clear verification of genomic recombination. Assessment of protein levels showed gradual protein loss in the β1e3-Cre mouse, but no loss of protein was seen in the β1fl-Cre mouse. This study communicates our path to reconcile the conundrum of having genomic recombination and yet no loss of protein, and in the process we provide evidence for episomal protein expression.
MATERIALS AND METHODS
Mice:
β1fl/fl mice (Potocnik et al. 2000) were a gift from Reinhard Fassler (Max Planck Institute of Biochemistry, Germany). The β1e3/e3 (Raghavan et al. 2000), SM22αCre (Holtwick et al. 2002), R26R (Soriano 1999), and the β1+/− (Stephens et al. 1995) mice were purchased from Jackson Laboratories (Bar Harbor, ME). The SM22aCre-ERT2 mice (Kuhbandner et al. 2000) were generously provided by Robert Feil (Interfakultäres Institut für Biochemie, Universität Tübingen, Germany). SM22α protein binds actin and contributes to smooth muscle contractility (Je and Sohn 2007; Assinder et al. 2009) and is expressed specifically in adult smooth muscle cells of the vasculature and viscera. Genotyping primers used are listed in supporting information,Table S1.
Tamoxifen (MP Biomedical) was prepared at a concentration of 20 mg/ml in 10% ethanol and 90% sunflower seed oil (Sigma) according to previous reports (Monvoisin et al. 2006). Animals were allowed to mature (5–6 weeks old) and then received 1 mg of tamoxifen at 24-hr intervals for the specified number of injections. Mice receiving the 28-day injection regimen were aged an additional 2 weeks prior to evaluation.
Tissue to be immunostained was fixed in 2% PFA overnight. Samples were histologically sectioned and stained according to the manufacturer's protocol for phosphorylated histone 3 (Cell Signaling; 1:100). FITC-conjugated α-sm actin (Sigma, 1:100) was used for staining the bladder tissue.
β-Gal staining:
To facilitate penetration of the reagents, the mucosa was dissociated from the bladder under a dissecting microscope. Embryos were stained whole mount. Tissue was then fixed at room temperature for 15 min in glutaraldhyde (0.2% glutaraldehyde, 5 mm EGTA pH 8, 2 mm MgCl, 1× PBS, pH 7.3). This was followed by three rinses of 30 min in detergent wash solution (2 mm MgCl, 0.01% DOC, 0.02% NP40, 1× PBS, pH 7.3). Tissues were stained overnight on an incubated shaker at 37° in detergent rinse with 5 mm ferrocyanide (II), 5 mm ferricyanate (III), 20 mm Tris, pH 7.3, and 1 mg/ml X-gal, pH 7.3. After staining overnight, samples were washed several times in 1× PBS at room temperature. Then they were fixed in 2% PFA overnight at 4°.
Western blot analysis:
After the mucosa was removed, a portion of the bladder was placed in RIPA buffer (Lee et al. 2006). The Western blot antibodies used were anti-β1 integrin (Millipore, 1:750), anti-α-tubulin (Sigma, 1:5000) and GAPDH (Millipore, 1:5000).
Cell culture:
Mouse embryonic fibroblasts (MEFs) were isolated from E13.5–E16.5 embryos according to Nagy et al. (2003). Mitomycin C (Calbiochem) was prepared at 200 mg/ml in HBSS and then filtered at 0.2 μm. Cells were incubated in 15 μg/ml mitomycin C in media for 3 hr at 37°. Cells were then rinsed two times with 1× HBSS and fresh media were added. Cells were allowed to recover overnight before replating. Induced quiescence was confirmed by a 6-hr incubation in 100 μm BrdU followed by fixation in 2% PFA for 30 min. Cells were then stained for BrdU incorporation using an anti-BrdU antibody (Thermo Scientific) and analyzed via confocal microscopy. Adeno-cre and adeno-gfp control viruses were applied to cells at an MOI of 30. DNA and protein were isolated from each treatment group, which included mock-infected controls.
Quantitative PCR primer design:
Unique quantitative PCR (qPCR) primers were designed to facilitate a quantitative analysis of the rate of β1 loxP recombination and the rate of maintenance of the loxP-flanked DNA sequence (see Table S1 for primer sequences). The first primer set, “total-DNA,” was designed to quantify the total number of chromosomal copies in each DNA sample from a template. This value was used to normalize all subsequent quantitative PCR values.
To determine the rate of recombination, a pair of primers, “unrecombined,” was designed to anneal to the sequences on either side of a loxP site in the unrecombined β1 integrin locus. Following Cre-mediated excision of the loxP-flanked sequence, these primers anneal to distinctly separate fragments of DNA and will not generate an amplicon. Thus, the difference in the value from this primer set relative to the value from the total-DNA primer set indicates the degree to which the β1 integrin gene has been recombined and excised.
The total number of copies of β1-flanked DNA, both unrecombined (chromosomal) and recombined (episomal circles), were determined by the “flanked DNA” primer set. This primer set amplifies a segment just upstream of exon 3 that lies within the loxP-flanked region in both β1fl and β1e3 cells. Comparison of the value from this primer set relative to the value from the total-genomic primer set indicates the degree of loss of loxP-flanked sequence.
Validation experiments with these primers confirmed that each pair gave a unique, correctly sized product and that this was quantitative, relative to the amount of template present. The exception to this was the unrecombined-β1fl primer set, which produced a marginal amount of an incorrectly sized product. To eliminate this from the analysis, a TaqMan probe was designed for this template to ensure that only the correct product was quantified.
Determination of recombination and floxed DNA persistence:
DNA was extracted from bladder tissue, cardiac tissue, or MEFs using Phase Lock gel tubes (5 PRIME) according to the manufacturer's instructions. DNA was then resuspended in water and the concentration was normalized prior to PCR. Primer sequences are included in Table S1.
For qPCR, DNA was resuspended in TE (10 mm Tris, 1 mm EDTA) and subjected to qPCR with the following conditions: Each DNA sample was assayed in triplicate reactions in optical-grade, 96-well plates (Applied Biosystems, Foster City, CA). In the cases of total DNA, flanked DNA, and unrecombined-β1e3, each 25-μl reaction contained 12.5 μl of Power SYBR Green Universal Master Mix (Applied Biosystems), 5 μl of purified DNA template, and primers (900 nm each final concentration, IDTDNA). Unrecombined β1fl was performed in 25 μl total reaction containing 12.5 μl of TaqMan Universal Master Mix (Applied Biosystems), 5 μl of purified DNA template, primers (900 nm each final concentration), and a FAM-conjugated TaqMan probe (250 nm final concentration; Operon, Huntsville, AL). All PCR runs were performed on an ABI 7500 Real Time Thermocycler using SDS software v.1.3 (Applied Biosystems). Reaction conditions were as follows: 10 min at 94° followed by 40 cycles of 10 sec at 94° and 30 sec at 64°. The starting template copy number of each reaction was determined by comparison to a standard curve, which was generated by the serial dilution of DNA from uninfected β1fl or β1e3 MEFs.
RESULTS
Induced β1 integrin deletion in vascular smooth muscle:
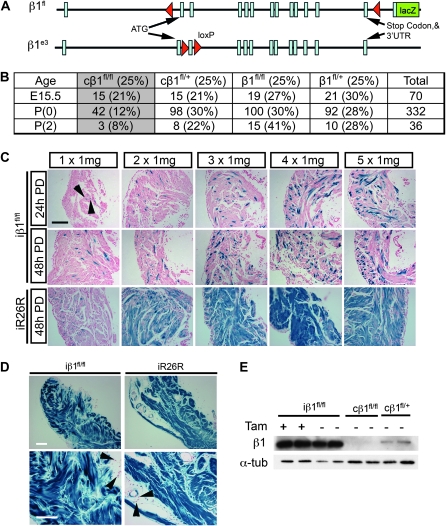
Two floxed mice were used for deletion of β1 integrin in smooth muscle cells: β1fl and β1e3 (Figure 1A; Potocnik et al. 2000; Raghavan et al. 2000). The β1fl allele contains loxP sites flanking the entire protein-coding region followed by a lacZ gene that functions as a reporter for the β1 promoter in cells with at least one recombined allele (Figure 1A; Potocnik et al. 2000). Expression of the lacZ reporter gene occurs upon excision of the floxed portion of the β1 integrin gene and subsequent splicing from exon 1 to a short segment of exon 2 placed in front of the lacZ gene. To examine the role of β1 integrin in smooth muscle in vivo, we crossed the β1fl mouse to the SM22αCre (identified by “c” to indicate constitutive Cre) mouse, which constitutively expresses Cre-recombinase in smooth muscle and cardiomyocytes during development (Holtwick et al. 2002; Figure S2). We chose the SM22αCre because, in adult tissues, SM22α (transgelin) expression is restricted to smooth muscle cells (Duband et al. 1993; Kuhbandner et al. 2000). Deletion of β1 integrin using this Cre line resulted in lethality with ∼50% of the progeny dying between E15.5 and P(0) (Figure 1B). The remaining progeny died prior to weaning. Due to their early lethality, the cβ1fl/fl mice could not be used to study β1 integrin deletion in the adult. To bypass this lethality, the β1fl allele was crossed to the inducible SM22αCre-ERT2 (identified by “i” to indicate inducible Cre) transgenic mouse (Kuhbandner et al. 2000).
Figure 1.—
Inducing β1 integrin deletion in smooth muscle cells. (A) Schematic representation of the location of the loxP sites in the β1fl and β1e3 transgenic mice (Potocnik et al. 2000; Raghavan et al. 2000). (B) Distribution of genotypes and viability of genetic crosses between cβ1+/fl mice and β1fl/fl mice. (C) Histology of β-gal–stained bladders from iβ1fl/fl and iR26R either 24 hr or 48 hr postdose (PD) at indicated doses. Arrows denoted β-gal positive cells. (D) Histology of β-gal–stained bladders from iβ1fl/fl and iR26R mice exposed to 1 mg tamoxifen daily for 28 days. Arrows point to cells negative for β-gal. (E) Western blot from bladder lysates of 28 × 1 mg iβ1fl/fl, oil-injected iβ1fl/fl, with cβ1fl/fl as a positive control and cβ1+/fl as a negative control for protein loss and α-tubulin as a loading control.
To identify an appropriate induction protocol for deletion of β1 integrin in adult smooth muscle, a regimen of five injections of 1 mg tamoxifen was tested (Figure 1C). Bladder smooth muscle was collected and stained for β-galactosidase (β-gal) at either 24 hr or 48 hr postfinal injection. Each tamoxifen injection was additive to total recombination, as indicated by β-gal positive cells (Figure 1C and Figure S1); however, a significant portion of cells remained negative for β-gal. To determine whether this poor degree of recombination was due to heterogeneous Cre-recombinase expression, the SM22αCre-ERT2 mouse was also crossed to the ROSA26R reporter (R26R) mouse (Soriano 1999). As seen previously (Kuhbandner et al. 2000), a sequence of five daily injections induced recombination in most bladder smooth muscle cells (Figure 1C), indicating that Cre was expressed and active in the large majority of these cells. To determine whether the low level of β-gal positive cells in the iβ1fl/fl mouse was associated with β1 integrin promoter activity in adult smooth muscle cells, bladders from the constitutive cβ1fl/+ and the cR26R were harvested and stained for lacZ expression (Figure S1). In the constitutive mouse, most bladder smooth muscle cells were positive for β-gal, indicating that the β1 integrin promoter was indeed active in these cells. From this we concluded that the loxP sites flanking the β1fl allele did not recombine as efficiently as the R26R construct, most likely due to the distance between loxP sites (Zheng et al. 2000; Coppoolse et al. 2005; Wang et al. 2009) which is ∼28 kb in β1fl (Potocnik et al. 2000) vs. <2 kb in R26R (Soriano 1999). Therefore, to improve excision, additional tamoxifen injections were used.
Extending the induction protocol to 28 days increased the number of β-gal positive cells (Figure 1D). β1 integrin deletion was further tested in bladder protein lysates by Western blot analysis (Figure 1E). Surprisingly, a Western blot for β1 integrin revealed no detectable change in protein levels in the iβ1fl/fl when compared to oil-injected controls despite the clear abundance of β-gal positive cells. This was in sharp contrast to protein isolated from the hearts of perinatal cβ1fl/fl and cβ1fl/+ animals (Figure 1E).
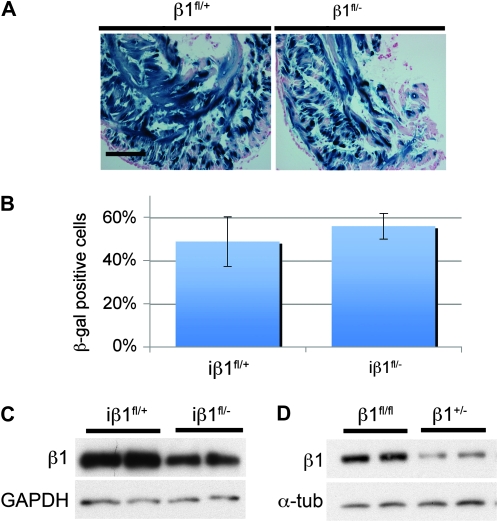
Findings from the cβ1fl/fl mouse demonstrated that recombination of the β1fl allele could indeed reduce β1 integrin protein levels. To determine whether the persistence of protein in the iβ1fl/fl mouse was due to a possible selection of heterozygous recombined cells (monoallelic recombination), we took advantage of the null (β1−) allele (Stephens et al. 1995) and generated iβ1fl/− and iβ1fl/+ animals. Histological analysis of β-gal–stained bladders revealed a high proportion of recombination in both the iβ1fl/− and in the iβ1fl/+ (Figure 2, A and B). Both iβ1fl/− and iβ1fl/+ animals had approximately equivalent β-gal positive cells; therefore, heterozygous cells were not being preferentially selected to survive in the iβ1fll− bladders. Further, a Western blot showed reduced β1 integrin protein in iβ1fl/− bladders compared to the iβ1fl/+ (Figure 2C), but approximately equivalent reduction was observed in β1+/− and the β1fl/fl (Figure 2D). These data demonstrate that persistence of the β1 integrin protein in the iβ1fl/fl bladders was not the result of selection of heterozygous cells and further showed that loss of a single β1 allele was detectable by Western blot.
Figure 2.—
Heterozygous and homozygous gene deletion. (A) Histology of β-gal–stained bladders from iβ1+/fl and iβ1fl/− mice that received 1 mg tamoxifen daily for 28 days. (B) Quantification of β-gal positive cells from iβ1+/fl and iβ1fl/− mice exposed to 1 mg tamoxifen daily for 28 days. (C) Western blot for β1 integrin on bladder lysates from iβ1fl/− and iβ1+/fl mice. (D) Western blot for β1 integrin bladder lysates from β1+/− and β1fl/fl mice.
Cre-induced recombination products:
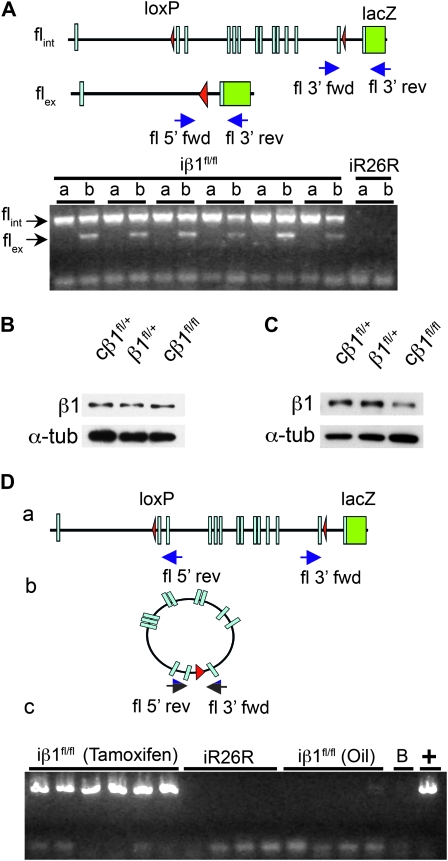
The persistence of β1 integrin protein in the iβ1fl/fl bladders could suggest that the flox sites in the gene were not being recombined. This is unlikely as β-gal staining indicated recombination of the β1 integrin locus. Nonetheless, we verified genetic excision by PCR analysis using primers that flanked the 5′ end in conjunction with the 3′ reverse primer (Figure 3A schematic). For the PCR, bladders from six iβ1fl/fl mice were used to examine recombination in smooth muscle (Figure 3A, b). We used abdominal muscle (Figure 3A, a), which is skeletal muscle as a negative control for recombination. The results confirmed the recombination (Figure 3A) and supported findings from the lacZ assay (Figure 1, C and D).
Figure 3.—
Recombination and the generation of a β1fl episome. (A) Schematic representation of excision product and primer locations (blue arrows). A representative PCR gel shows recombined (flex) and intact (flint) alleles from DNA of abdominal muscle (a) and bladder (b) of iβ1fl/fl and iR26R mice. (B and C) Western blot analysis of bladder and heart protein lysates, respectively, from early postnatal cβ1fl/fl, β1+/fl, and cβ1+/fl progeny. (D) Schematic representation of circular product and primer (blue arrows) with representative PCR results of DNA from bladder of iβ1fl/fl and iR26R and oil-injected iβ1fl/fl mice. B in panel c denotes a water blank and (+) is the cβ1+/fl bladder positive control.
To reconcile the lack of protein loss after confirmed recombination, we examined the constitutive SM22αCre mice and assessed levels of β1 integrin protein in the heart and bladder (Figure 3, B and C). While the bladder of the knockout had similar β1 integrin protein levels as controls (Figure 3B), the heart, with higher cell proliferation rate at time of recombination, showed a significant decrease in β1 integrin protein over controls (Figure 3C and Figure S2). These results provide evidence that the β1fl allele was appropriately targeted and excised during development, yet inactivation of the gene product was not accomplished in the adult bladder, despite clear evidence of recombination.
Recombination at loxP sites provides two outcomes: (a) elimination of chromosomal DNA flanking the loxP sites and (b) formation of an extrachromosomal circular product generated by the excised DNA (Sternberg and Hamilton 1981; Hoess et al. 1985; Metzger and Feil 1999). Since excision in the inducible model was validated by PCR, we next sought the fate of the excised extrachromosomal DNA product. In the case of the β1fl allele, the entire coding region, from the ATG translation initiation site in exon 2 to the stop codon and 3′-UTR, is flanked by loxP sites and forms a circle following recombination, which we refer to as an episome. Thus, we considered that the persistent β1 integrin protein in recombined cells might result from the activity of this extrachromosomal DNA. Next, we asked whether the circular excision product was present in recombined cells. PCR analysis using primers from opposite ends of the loxP flanked DNA showed that the episomal DNA was indeed present in the bladders of both the six iβ1fl/fl mice exposed to tamoxifen and in the adult constitutive cβ1fl/+ bladders, but not in the four iR26R mice, which lack the allele, or in the four oil-injected iβ1fl/fl mice (Figure 3D).
Generation and retention of episomes from different β1 integrin flox alleles:
The generation of an episomal product by Cre-recombinase has been examined at length (Sternberg and Hamilton 1981; Hoess et al. 1985; Sternberg 1990; Bigger et al. 2001). In fact, persistence of the episomal Cre-recombinase product was documented in plants (Srivastava and Ow 2003) and was also noted to be retained after viral Cre infection in mice (Dorigo et al. 2004; Gallaher et al. 2009). Yet, the potential expression from an intended gene deletion product has not been previously described in mammals. If the β1fl episome was the reason for the persistence of β1 integrin protein in the recombined adult tissue, then utilization of a second loxP construct that disrupted the β1 integrin coding region should address protein persistence.
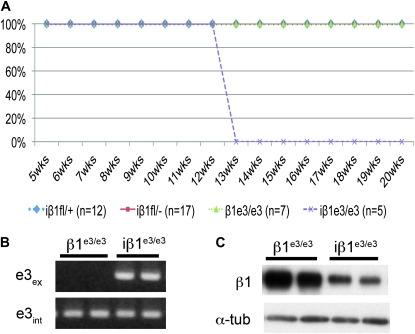
The β1e3/e3 mouse has loxP sites that flank exon 3 and generate a frameshift and early termination mutation upon recombination (Raghavan et al. 2000). We crossed the β1e3 mouse to the SM22αCre-ERT2 mouse. Upon treatment with tamoxifen, the iβ1e3/e3 mice die 4 weeks after the final injection with 100% penetration (Figure 4A), whereas tamoxifen-treated controls that included β1e3/e3 and iβ1fl/+ as well as the iβ1fl/− mice remained viable throughout the experiment. PCR analysis confirmed genetic recombination (Figure 4B). Comparison of β1 integrin protein from bladders of iβ1e3/e3 and β1e3/e3 mice showed the iβ1e3/e3 had a significant reduction in β1 integrin protein (Figure 4C). These results eliminated the possibility of an extended protein half-life. The findings further support the possibility that in the adult β1fl mice, the persistence of protein was due to the excised β1fl episome.
Figure 4.—
Deletion of β1 integrin in β1e3 mice. (A) Survival curve for iβ1fl/−, iβ1+/fl, iβ1e3/e3, and β1e3/e3 mice from the first injection through 3 months after final injection. (B) PCR of recombination (e3ex) and intact allele (e3int) in the iβ1e3/e3 and β1e3/e3 mouse bladders. (C) Western blot analysis of bladder from iβ1e3/e3 and β1e3/e3 adult mice exposed to 1 mg tamoxifen daily for 28 days.
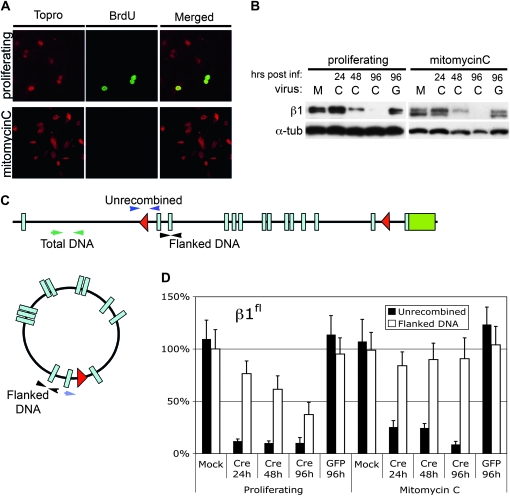
To evaluate episomal retention and expression in a more controlled environment, primary MEFs were isolated from β1fl/fl animals. Cells were expanded and proliferative quiescence was induced by mitomycin C treatment in half of the β1fl/fl MEFs. These lines were then subjected to either mock infection or infected with adeno-Cre virus or an adeno-gfp control virus. Mock-infected β1fl/fl quiescent and proliferating MEFs were harvested following similar treatments to that of the infected cells, but lacking virus. Adeno-cre–infected cells were harvested for protein and DNA at 24, 48, and 96 hr postinfection. Adeno-gfp control cells were harvested at 96 hr postinfection. BrdU was used to evaluate proliferation vs. quiescence (Figure 5A). Indeed, the quiescent MEFs were negative for BrdU. Western blots from the β1fl/fl MEFs indicate loss of protein at approximately the same rate as the β1e3/e3 MEFs regardless of their proliferative status (Figure 5B and Figure S3). Quantitative PCR was performed to evaluate recombination and retention of the episome (Figure 5, C and D). The qPCR findings demonstrated high levels of recombination in all cells exposed to adeno-cre (Figure 5D). Further, β1fl/fl cells retained the episome in the quiescent state and lost it from proliferating cells. In contrast to the findings in the bladder, the data demonstrate that the β1fl/fl-generated episome was not active in MEFs in culture (Figure 5B). Clearly the findings in vitro did not recapitulate either the rate of protein loss in vivo nor the expression from episomal recombined DNA, but demonstrated that the episome is retained in quiescent cells, while it is lost in proliferating cells.
Figure 5.—
Episome kinetics in β1fl/fl MEFs. (A) BrdU staining in untreated and mitomycin C treated β1fl/fl MEFs (BrdU-green, Topro-red). (B) Western blot showing a time course of β1 integrin levels in mock (M), adeno-Cre (C), and adeno-GFP (G) infected cells up to 96 hr postinfection. (C) Schematic showing the location of the primers used in qPCR for detection of the flanked (episome) and unrecombined DNA. (D) Quantitative PCR of flanked and unrecombined DNA for the proliferating and mitomycin C-treated β1fl/fl MEFs.
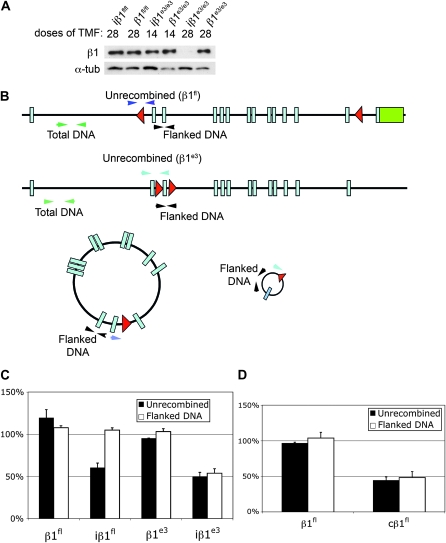
To evaluate the episome and protein levels in vivo, protein and DNA was isolated from iβ1fl/fl, iβ1e3/e3, β1fl/fl, and β1e3/e3 mice after 28 doses of tamoxifen. Western blot analysis showed a precipitous loss of protein at 28 doses of tamoxifen in the iβ1e3/e3, while the iβ1fl/fl mouse receiving 28 doses of tamoxifen showed no loss of protein (Figure 6A).
Figure 6.—
Episome kinetics in β1fl/fl and β1e3/e3 in vivo. (A) Western blot of iβ1fl/fl and iβ1e3/e3 with β1fl/fl and β1e3/e3 controls for tamoxifen dose indicated. The 14-dose bladders were collected 48 hr after the final injection while the 28-dose samples were collected 2 weeks after the last injection. (B) Schematic of the qPCR primers for detection of β1fl/fl and β1e3/e3 flanked and unrecombined DNA. (C) Quantitative PCR for flanked and unrecombined DNA in iβ1fl/fl, β1fl/fl, iβ1e3/e3, and β1e3/e3 bladders of animals receiving 28 doses of tamoxifen. (D) Quantitative PCR of flanked and unrecombined DNA in the β1fl/fl and cβ1fl/fl hearts.
Quantitative PCR for recombination and presence of the episome in all four groups: iβ1fl/fl, β1fl/fl, iβ1e3/e3, and β1e3/e3, showed approximately equivalent total recombination in both Cre positive groups, but marked retention of episomal DNA with poor protein loss in the iβ1fl/fl group only (Figure 6C). In contrast, DNA analysis from cβ1fl/fl and β1fl/fl hearts showed that the episome was eliminated in the cβ1fl/fl hearts, where protein also decreased significantly (Figures 1E, 3C, and 6D). A central difference between the cβ1fl/fl hearts and the iβ1fl/fl bladders is its proliferation status at times of recombination that was verified by the proliferative marker phosphorylated histone 3 (pHis3, Figure S2).
DISCUSSION
Here we have shown that recombination of the β1fl and the β1e3 alleles in smooth muscle cells yield different outcomes. Our results indicate that this difference in phenotype can be attributed to the persistence of protein in the β1fl allele. We addressed five hypotheses regarding the persistence of β1 integrin protein following recombination: (1) appropriate targeting, (2) heterozygous selection, (3) protein half-life, (4) recombination efficiency, and (5) episomal expression.
Testing the first three hypotheses was straightforward. Through PCR and DNA sequencing we confirmed that both loxP alleles were appropriately targeted within the β1 integrin gene. Further, work by us, as well as others, has demonstrated that both alleles were capable of reducing β1 integrin protein levels in vivo, in the presence of Cre-recombinase when constitutive systems were used (Li et al. 2005; Benninger et al. 2006; Jones et al. 2006; Lei et al. 2008; Zovein et al. 2010). β1 integrin had been shown to be important in cell survival (Stupack and Cheresh 2002; Manohar et al. 2004; Pinkse et al. 2004). We showed by analysis of β-gal–stained bladders from iβ1fl/+ and iβ1fl/− mice that heterozygous selection was unlikely. Finally, while the protein half-life of β1 integrin in cultured cells is 6–8 hr (De Strooper et al. 1991), there is no information as to β1 integrin turnover in vivo. Nonetheless, the iβ1e3/e3 findings confirmed that an extended protein half-life was not causing protein persistence. Thus, having eliminated the first three hypotheses, we turned our attention to recombination efficiency.
It has been shown that even small changes in the distance between loxP sites can affect recombination efficiency (Zheng et al. 2000; Coppoolse et al. 2005; Wang et al. 2009). The distance between loxP sites in the β1fl allele is ∼28 kb (Potocnik et al. 2000), while the loxP sites for the β1e3 allele (Raghavan et al. 2000) are <2 kb apart. The 28-day protocol was developed to compensate for the inefficiency of the β1fl construct and, indeed, we observed a significant increase in the number of β-gal positive cells when compared to the 5-day regimen. Surprisingly, there was no change in protein levels. Moreover, Western blot analysis of the β1+/− and iβ1fll− bladder tissue when compared to β1fl/fl showed that loss of a single allele was significant enough to reduce β1 integrin protein. This finding indicates that β1 integrin does not change promoter activity to compensate for lower total β1 integrin protein. Furthermore, it demonstrates that recombination efficiency of the β1fl allele is lower than the β1e3, but this low efficiency is an unlikely explanation for the persistence of protein. At this point we began to consider the episomal hypothesis.
We found the episome generated by Cre-recombinase persisted in vivo, particularly with the β1fl transgene. Interestingly, unlike the β1e3 mice, the β1fl recombination product included the ATG site at exon 2 through the final exon containing the stop codon. It is assumed that the lack of exon 1 and, therefore the promoter, would make this product transcriptionally irrelevant. However, recent analysis of the mouse and human genomes revealed that >50% of genes have alternative promoters and transcriptional start sites (Carninci et al. 2006). Of the genes with alternative transcriptional start sites, 97% generate identical protein products (Carninci et al. 2006). The persistence of the episome in the bladder of iβ1fl/fl mice and the apparent loss of the episome and subsequent protein depletion in the heart of cβ1fl/fl mice suggest potential expression in vivo. Consistent with this possibility Cre-recombinase–generated episomes have been shown to be competent for transcription in mammalian cells in culture, as well as in vivo (Bigger et al. 2001; Dorigo et al. 2004; Gallaher et al. 2009).
We attempted to recapitulate our in vivo data using MEFs in vitro. While we were able to generate quiescent β1fl/fl MEFs, which retained the episome upon adeno-cre–mediated recombination, β1 integrin protein loss was equivalent between proliferating and quiescent β1fl/fl MEFs. The discrepancy between the in vivo and in vitro experiments is difficult to interpret. It is possible that the promoter active in smooth muscle cells in vivo is not active in MEFs. Given the in vivo and in vitro qPCR data combined and the Western blots, it is difficult to resolve the difference in protein persistence by efficiency alone. We achieve nearly comparable recombination levels in the iβ1fl/fl and iβ1e3/e3 bladders in vivo, yet changes in total protein do not equate and, more importantly, the biological outcome of the deletion in each model is distinct. While we find in vitro analyses are valuable, it may be a less optimal approach when exploring questions of gene regulation in vivo.
In summary, our data illustrate the value of analyzing a phenotype using multiple loxP alleles and reinforce the importance of verifying protein loss, not just genetic recombination in Cre/loxP experiments. Further, protein depletion needs to be validated in vivo for every novel Cre/loxP combination. Finally, this work illustrates a new approach to the Cre/loxP system in vivo. Our data together with previous work (Dorigo et al. 2004; Gallaher et al. 2009) indicate the potential for a new method for noninvasive gene expression that would be permanent in quiescent cells and transitory in proliferating cells. The technique may eventually enable novel and temporary gene therapies.
Acknowledgments
We thank Arnold Berk (Department of Microbiology, Immunology, and Molecular Genetics, UCLA) for helpful discussion of the data. We also thank Reid Johnson (Department of Biological Chemistry, UCLA) for comments on the manuscript. We appreciate the β1fl mouse provided by Reinhard Fassler (Max Planck Institute, Germany) and the SM22α-CreERT2 mouse provided by Robert Feil (Interfakultäres Institut für Biochemie, Universität Tübingen, Germany). Also, we thank the Translational Pathology Core Lab for the sectioning of histological samples. The work was supported by a grant from the National Institutes of Health to M.L.I.A. (RO1CA126935).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.121608/DC1.
References
- Abraham, S., N. Kogata, R. Fassler and R. H. Adams, 2008. Integrin beta1 subunit controls mural cell adhesion, spreading, and blood vessel wall stability. Circ. Res. 102 562–570. [DOI] [PubMed] [Google Scholar]
- Assinder, S. J., J. A. Stanton and P. D. Prasad, 2009. Transgelin: an actin-binding protein and tumour suppressor. Int. J. Biochem. Cell. Biol. 41 482–486. [DOI] [PubMed] [Google Scholar]
- Benninger, Y., H. Colognato, T. Thurnherr, R. J. Franklin, D. P. Leone et al., 2006. Beta1-integrin signaling mediates premyelinating oligodendrocyte survival but is not required for CNS myelination and remyelination. J. Neurosci. 26 7665–7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigger, B. W., O. Tolmachov, J. M. Collombet, M. Fragkos, I. Palaszewski et al., 2001. An araC-controlled bacterial cre expression system to produce DNA minicircle vectors for nuclear and mitochondrial gene therapy. J. Biol. Chem. 276 23018–23027. [DOI] [PubMed] [Google Scholar]
- Branda, C. S., and S. M. Dymecki, 2004. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev. Cell 6 7–28. [DOI] [PubMed] [Google Scholar]
- Carninci, P., A. Sandelin, B. Lenhard, S. Katayama, K. Shimokawa et al., 2006. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 38 626–635. [DOI] [PubMed] [Google Scholar]
- Chen, C. S., M. Mrksich, S. Huang, G. M. Whitesides and D. E. Ingber, 1997. Geometric control of cell life and death. Science 276 1425–1428. [DOI] [PubMed] [Google Scholar]
- Coppoolse, E. R., M. J. de Vroomen, F. van Gennip, B. J. Hersmus and M. J. van Haaren, 2005. Size does matter: cre-mediated somatic deletion efficiency depends on the distance between the target lox-sites. Plant Mol. Biol. 58 687–698. [DOI] [PubMed] [Google Scholar]
- De Strooper, B., F. Van Leuven, G. Carmeliet, H. Van Den Berghe and J. J. Cassiman, 1991. Cultured human fibroblasts contain a large pool of precursor beta 1-integrin but lack an intracellular pool of mature subunit. Eur. J. Biochem. 199 25–33. [DOI] [PubMed] [Google Scholar]
- Dorigo, O., J. S. Gil, S. D. Gallaher, B. T. Tan, M. G. Castro et al., 2004. Development of a novel helper-dependent adenovirus-Epstein-Barr virus hybrid system for the stable transformation of mammalian cells. J. Virol. 78 6556–6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duband, J. L., M. Gimona, M. Scatena, S. Sartore and J. V. Small, 1993. Calponin and SM 22 as differentiation markers of smooth muscle: spatiotemporal distribution during avian embryonic development. Differentiation 55 1–11. [DOI] [PubMed] [Google Scholar]
- Fassler, R., and M. Meyer, 1995. Consequences of lack of beta 1 integrin gene expression in mice. Genes Dev. 9 1896–1908. [DOI] [PubMed] [Google Scholar]
- Gallaher, S. D., J. S. Gil, O. Dorigo and A. J. Berk, 2009. Robust in vivo transduction of a genetically stable Epstein-Barr virus episome to hepatocytes in mice by a hybrid viral vector. J. Virol. 83 3249–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graus-Porta, D., S. Blaess, M. Senften, A. Littlewood-Evans, C. Damsky et al., 2001. Beta1-class integrins regulate the development of laminae and folia in the cerebral and cerebellar cortex. Neuron 31 367–379. [DOI] [PubMed] [Google Scholar]
- Han, B., and J. T. Zhang, 2002. Regulation of gene expression by internal ribosome entry sites or cryptic promoters: the eIF4G story. Mol. Cell. Biol. 22 7372–7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoess, R., A. Wierzbicki and K. Abremski, 1985. Formation of small circular DNA molecules via an in vitro site-specific recombination system. Gene 40 325–329. [DOI] [PubMed] [Google Scholar]
- Holtwick, R., M. Gotthardt, B. Skryabin, M. Steinmetz, R. Potthast et al., 2002. Smooth muscle-selective deletion of guanylyl cyclase-A prevents the acute but not chronic effects of ANP on blood pressure. Proc. Natl. Acad. Sci. USA 99 7142–7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes, R. O., 2002. Integrins: bidirectional, allosteric signaling machines. Cell 110 673–687. [DOI] [PubMed] [Google Scholar]
- Je, H. D., and U. D. Sohn, 2007. SM22alpha is required for agonist-induced regulation of contractility: evidence from SM22alpha knockout mice. Mol. Cells 23 175–181. [PubMed] [Google Scholar]
- Jones, R. G., X. Li, P. D. Gray, J. Kuang, F. Clayton et al., 2006. Conditional deletion of beta1 integrins in the intestinal epithelium causes a loss of Hedgehog expression, intestinal hyperplasia, and early postnatal lethality. J. Cell Biol. 175 505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, R. S., S. Y. Shai, C. J. Babbitt, C. G. Pham, R. J. Solaro et al., 2001. Disruption of integrin function in the murine myocardium leads to perinatal lethality, fibrosis, and abnormal cardiac performance. Am. J. Pathol. 158 1079–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhbandner, S., S. Brummer, D. Metzger, P. Chambon, F. Hofmann et al., 2000. Temporally controlled somatic mutagenesis in smooth muscle. Genesis 28 15–22. [DOI] [PubMed] [Google Scholar]
- Kuhnert, F., M. R. Mancuso, J. Hampton, K. Stankunas, T. Asano et al., 2008. Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development 135 3989–3993. [DOI] [PubMed] [Google Scholar]
- Lee, N. V., M. Sato, D. S. Annis, J. A. Loo, L. Wu et al., 2006. ADAMTS1 mediates the release of antiangiogenic polypeptides from TSP1 and 2. EMBO J. 25 5270–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, L., D. Liu, Y. Huang, I. Jovin, S. Y. Shai et al., 2008. Endothelial expression of beta1 integrin is required for embryonic vascular patterning and postnatal vascular remodeling. Mol. Cell. Biol. 28 794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, N., Y. Zhang, M. J. Naylor, F. Schatzmann, F. Maurer et al., 2005. Beta1 integrins regulate mammary gland proliferation and maintain the integrity of mammary alveoli. EMBO J. 24 1942–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manohar, A., S. G. Shome, J. Lamar, L. Stirling, V. Iyer et al., 2004. Alpha 3 beta 1 integrin promotes keratinocyte cell survival through activation of a MEK/ERK signaling pathway. J. Cell. Sci. 117 4043–4054. [DOI] [PubMed] [Google Scholar]
- Martinez-Lemus, L. A., Z. Sun, A. Trache, J. P. Trzciakowski and G. A. Meininger, 2005. Integrins and regulation of the microcirculation: from arterioles to molecular studies using atomic force microscopy. Microcirculation 12 99–112. [DOI] [PubMed] [Google Scholar]
- Metzger, D., and R. Feil, 1999. Engineering the mouse genome by site-specific recombination. Curr. Opin. Biotechnol. 10 470–476. [DOI] [PubMed] [Google Scholar]
- Meyers, E. N., M. Lewandoski and G. R. Martin, 1998. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat. Genet. 18 136–141. [DOI] [PubMed] [Google Scholar]
- Monvoisin, A., J. A. Alva, J. J. Hofmann, A. C. Zovein, T. F. Lane et al., 2006. VE-cadherin-CreERT2 transgenic mouse: a model for inducible recombination in the endothelium. Dev. Dyn. 235 3413–3422. [DOI] [PubMed] [Google Scholar]
- Nagy, A., M. Gertenstein, K. Vintersten and R. Behringer, 2003. Preparing mouse embryo fibroblasts, pp. 371–373 in Manipulating the Mouse Embryo. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Pietri, T., O. Eder, M. A. Breau, P. Topilko, M. Blanche et al., 2004. Conditional beta1-integrin gene deletion in neural crest cells causes severe developmental alterations of the peripheral nervous system. Development 131 3871–3883. [DOI] [PubMed] [Google Scholar]
- Pinkse, G. G., M. P. Voorhoeve, M. Noteborn, O. T. Terpstra, J. A. Bruijn et al., 2004. Hepatocyte survival depends on beta1-integrin-mediated attachment of hepatocytes to hepatic extracellular matrix. Liver Int. 24 218–226. [DOI] [PubMed] [Google Scholar]
- Potocnik, A. J., C. Brakebusch and R. Fassler, 2000. Fetal and adult hematopoietic stem cells require beta1 integrin function for colonizing fetal liver, spleen, and bone marrow. Immunity 12 653–663. [DOI] [PubMed] [Google Scholar]
- Raghavan, S., C. Bauer, G. Mundschau, Q. Li and E. Fuchs, 2000. Conditional ablation of beta1 integrin in skin. Severe defects in epidermal proliferation, basement membrane formation, and hair follicle invagination. J. Cell Biol. 150 1149–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, C. E., A. F. Horwitz, D. A. Lauffenburger and M. P. Sheetz, 1993. Integrin-cytoskeletal interactions in migrating fibroblasts are dynamic, asymmetric, and regulated. J. Cell Biol. 123 977–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano, P., 1999. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21 70–71. [DOI] [PubMed] [Google Scholar]
- Srivastava, V., and D. W. Ow, 2003. Rare instances of Cre-mediated deletion product maintained in transgenic wheat. Plant Mol. Biol. 52 661–668. [DOI] [PubMed] [Google Scholar]
- Stephens, L. E., A. E. Sutherland, I. V. Klimanskaya, A. Andrieux, J. Meneses et al., 1995. Deletion of beta 1 integrins in mice results in inner cell mass failure and peri-implantation lethality. Genes Dev. 9 1883–1895. [DOI] [PubMed] [Google Scholar]
- Sternberg, N., 1990. Bacteriophage P1 cloning system for the isolation, amplification, and recovery of DNA fragments as large as 100 kilobase pairs. Proc. Natl. Acad. Sci. USA 87 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg, N., and D. Hamilton, 1981. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. J. Mol. Biol. 150 467–486. [DOI] [PubMed] [Google Scholar]
- Stupack, D. G., and D. A. Cheresh, 2002. Get a ligand, get a life: integrins, signaling and cell survival. J. Cell Sci. 115 3729–3738. [DOI] [PubMed] [Google Scholar]
- Wang, N., J. P. Butler and D. E. Ingber, 1993. Mechanotransduction across the cell surface and through the cytoskeleton. Science 260 1124–1127. [DOI] [PubMed] [Google Scholar]
- Wang, S. Z., B. H. Liu, H. W. Tao, K. Xia and L. I. Zhang, 2009. A genetic strategy for stochastic gene activation with regulated sparseness (STARS). PLoS ONE 4 e4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman, K. M., M. Lewandoski, K. Campbell, A. L. Joyner, J. L. Rubenstein et al., 1997. Specification of the anterior hindbrain and establishment of a normal mid/hindbrain organizer is dependent on Gbx2 gene function. Development 124 2923–2934. [DOI] [PubMed] [Google Scholar]
- Zheng, B., M. Sage, E. A. Sheppeard, V. Jurecic and A. Bradley, 2000. Engineering mouse chromosomes with Cre-loxP: range, efficiency, and somatic applications. Mol. Cell. Biol. 20 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovein, A. C., A. Luque, K. A. Turlo, J. J. Hofmann, K. M. Yee et al., 2010. Beta1 integrin establishes endothelial cell polarity and arteriolar lumen formation via a Par3-dependent mechanism. Dev. Cell 18 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]