Abstract
Social interactions among individuals are abundant both in natural and domestic populations. Such social interactions cause phenotypes of individuals to depend on genes carried by other individuals, a phenomenon known as indirect genetic effects (IGE). Because IGEs have drastic effects on the rate and direction of response to selection, knowledge of their magnitude and relationship to direct genetic effects (DGE) is indispensable for understanding response to selection. Very little is known, however, of statistical power and optimum experimental designs for estimating IGEs. This work, therefore, presents expressions for the standard errors of the estimated (co)variances of DGEs and IGEs and identifies optimum experimental designs for their estimation. It also provides an expression for optimum family size and a numerical investigation of optimum group size. Designs with groups composed of two families were optimal and substantially better than designs with groups composed at random with respect to family. Results suggest that IGEs can be detected with ∼1000–2000 individuals and/or ∼250–500 groups when using optimum designs. Those values appear feasible for agriculture and aquaculture and for the smaller laboratory species. In summary, this work provides the tools to optimize and quantify the required size of experiments aiming to identify IGEs. An R-package SE.IGE is available, which predicts SEs and identifies optimum family and group sizes.
SOCIAL interactions among individuals, such as competition and cooperation, are fundamental to evolution by natural selection (Darwin 1859; Hamilton 1964; Wilson 1975; Frank 1998; Keller 1999; Clutton-Brock 2002). Most research on social interactions among individuals has focused on the fitness cost and benefit of such interactions. This focus on fitness effects seems to originate from an interest in the factors determining evolutionary success of populations and from the observation of altruistic and cooperative behaviors, which seem to contradict the intuitive notion of “survival of the fittest.”
Effects of social interactions, however, are not limited to fitness. There is increasing evidence that not only fitness, but also trait values of individuals are affected by genes in other individuals (reviewed in Wolf et al. 1998; Mcglothlin and Brodie 2009). Such effects are known as indirect genetic effects (IGE; Cheverud and Moore 1994; Moore et al. 1997) or associative effects (Griffing 1967; Muir 2005). An IGE is a heritable effect of one individual on the trait value of another individual. A well-known example in mammals is the maternal-genetic effect of a mother on preweaning growth rate of her offspring (Willham 1963; Mousseau and Fox 1998). The IGE modeling approach has its roots in quantitative genetic models for maternally affected traits, which are widely used in agriculture (Dickerson 1947; Willham 1963; Cheverud 1984; Kirkpatrick and Lande 1989; Lynch and Walsh 1998), but is increasingly applied to other traits, both in animals and plants (e.g., Muir 2005; Petfield et al. 2005; Mutic and Wolf 2007; Bergsma et al. 2008; Cappa and Cantet 2008; Bleakley and Brodie 2009; Chenoweth et al. 2010).
IGEs may have drastic effects on the rate and direction of response to natural or artificial selection (Griffing 1967, 1976, 1981a,b; Moore et al. 1997; Wolf et al. 1998; Agrawal et al. 2001; Bijma and Wade 2008; Mcglothlin et al. 2010). Griffing (1967), for example, showed that IGEs may cause traits to respond in the direction opposite to selection. This theoretical expectation has been confirmed empirically in selection experiments (Muir 1996, 2005). Bijma et al. (2007a) showed theoretically that IGEs may substantially increase the heritable variance determining a population's potential to respond to selection. Moreover, IGEs alter the impacts of multilevel selection and relatedness among individuals on response to selection (Griffing 1976, 1981a,b; Agrawal et al. 2001; Bijma and Wade 2008; Mcglothlin et al. 2010). The above clearly illustrates that knowledge of IGEs and their relationship to direct genetic effects (DGEs) is indispensable for understanding response to natural or artificial selection.
The magnitude of IGEs and their contribution to heritable variance can be estimated empirically using mixed models that are common in the field of animal breeding (Muir and Schinckel 2002; Van Vleck and Cassady 2004, 2005; Muir 2005; Arango et al. 2005; Bijma et al. 2007b; Chen et al. 2008; Mcglothlin and Brodie 2009). Such models are empirically powerful, because they allow estimating IGEs without knowledge of the underlying mechanisms and without the need to observe the interactions per se. Mixed models including IGEs can be fitted to field data, using restricted maximum likelihood estimation (Patterson and Thompson 1971; Kruuk 2004; Muir 2005; Bijma et al. 2007b). Using field data on domestic pigs, for example, Bergsma et al. (2008) found that IGEs contributed more than half of heritable variance in growth rate.
At present, knowledge of the magnitude of IGEs and their relationship to DGEs is limited. More knowledge is required to better understand the relevance of IGEs for evolution by natural selection and for genetic improvement in livestock, aquaculture, and plants. Very little is known, however, of the factors determining the standard errors (SE) of the estimated genetic parameters for IGEs, which hampers the optimization of experimental designs. Although the variance of IGEs can be estimated when groups of interacting individuals are composed at random with respect to family, it is unknown whether such designs are efficient. Moreover, it has been shown that genetic covariance components are not identifiable when groups consist of entire families, when families are allocated to groups in a systematic manner (Bijma et al. 2007b), or when fixed group effects are fitted (Cantet and Cappa 2008).
Here I present equations for the SE of the estimated variances, covariance, and correlation of DGEs and IGEs. Accuracy of the prediction equations is evaluated using stochastic simulations. Subsequently, the prediction equations are used to identify optimum experimental designs for estimating IGEs.
BACKGROUND
This section briefly summarizes the quantitative genetic theory of IGEs and may be skipped by readers familiar with this theory. An IGE is a heritable effect of one individual on the trait value of an other. IGEs can be modeled in two alternative ways (reviewed in Mcglothlin and Brodie 2009): first, using a trait-based model, where an individual's IGE is a direct consequence of its trait value (Falconer 1965; Moore et al. 1997) and second, using a variance-component model, where the genetic variance in trait value is partitioned into a direct component due to the focal individual's genotype, and an indirect component due to the genotypes of its interactants (Willham 1963; Griffing 1967). Here I use the variance-component model, because it allows identification of the full variance due to IGEs without requiring knowledge of the trait(s) or mechanisms underlying them.
Consider a population consisting of groups of nw members each, where social interactions occur among the members of a group. Then the trait value of focal individual i may be modeled as the sum of a direct effect rooted in the focal individual itself, 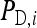 , and the sum of the indirect effects,
, and the sum of the indirect effects, 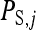 , of each of its nw − 1 group mates,
, of each of its nw − 1 group mates,
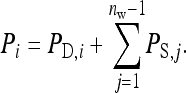 |
(1) |
Hence, i denotes the focal individual, j one of its nw − 1 group mates, D direct effects, S indirect effects, and 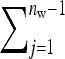 the sum taken over the nw−1 group mates of focal individual i. (S, suggesting “social,” is used as subscript for indirect effects, since I may be confused with i or 1. See Table 1 for a notation key.)
the sum taken over the nw−1 group mates of focal individual i. (S, suggesting “social,” is used as subscript for indirect effects, since I may be confused with i or 1. See Table 1 for a notation key.)
TABLE 1.
Notation key
| Symbol | Meaning |
|---|---|
| DGE, IGE | Direct genetic effect, indirect genetic effect |
| T, N, n | Total number of individuals, number of families, family size, T = Nn |
| i, j | Focal individual, group mate of focal individual |
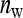 , ,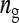 , ,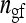
|
Group size, number of groups, number of groups per family, 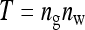
|
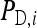 , , 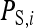
|
Direct effect of i, indirect effect of i |
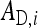 , , 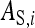
|
Direct genetic effect of i, indirect genetic effect of i |
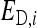 , , 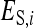
|
Direct nongenetic effect of i, indirect nongenetic effect of i |
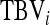 , , 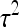
|
Total breeding value of i (Equation 4), relative heritable variance (Equation 5) |
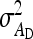 , , 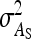
|
Variance of DGE among individuals, variance of IGE among individuals |
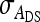 , , 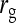
|
Covariance between DGEs and IGEs, correlation between DGEs and IGEs |
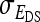 , , 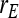
|
Covariance and correlation between nongenetic direct and indirect effects |
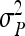 , , 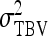
|
Phenotypic variance among individuals, variance of TBVs among individuals |
| k, l, m | Focal family, index for records within family, effective no. records per family |
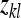 |
lth effective record on family k for use in ANOVA (Equation 6) |
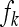 , , 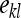
|
Effect of kth family, residual of lth record on family k, 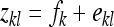 . . |
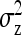 , , 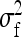 , , 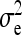
|
Full variance of records, between-family variance, residual variance |
 |
Optimum family size for parameter x |
SE, 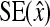
|
Standard error, standard error of estimate of x |
| d | Degree of dilution of indirect effects with group size (Equation 18) |
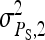 |
Variance of indirect effects in groups of two individuals |
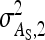 |
Variance of IGEs in groups of two individuals |
Throughout, hats (^) denote estimates, whereas symbols without hats refer to true values.
Both direct and indirect effects may be decomposed into an additive genetic (i.e., heritable) component, A, and a remaining nonheritable component, E (Griffing 1967),
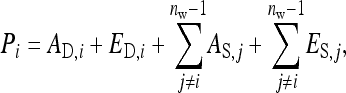 |
(2) |
where 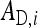 is the DGE of the focal individual,
is the DGE of the focal individual, 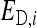 the corresponding nonheritable direct effect,
the corresponding nonheritable direct effect, 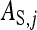 the IGE of group mate j, and
the IGE of group mate j, and 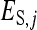 the corresponding nonheritable indirect effect.
the corresponding nonheritable indirect effect.
On the population level, the magnitude of DGEs and IGEs is measured by their (co)variances. Hence, interest is in the variance of DGE,  , the variance of IGE,
, the variance of IGE, 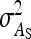 , and their covariance,
, and their covariance, 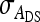 . Moreover, interest is in the total heritable variance arising from the joint effect of DGEs and IGEs. The total heritable impact of an individual's genes on the mean trait value of the population is given by the individual's total breeding value (TBV; Bijma et al. 2007a)
. Moreover, interest is in the total heritable variance arising from the joint effect of DGEs and IGEs. The total heritable impact of an individual's genes on the mean trait value of the population is given by the individual's total breeding value (TBV; Bijma et al. 2007a)
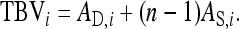 |
(3) |
Note that, in contrast to the trait value, the TBV in Equation 3 is entirely a heritable property of the focal individual. It is a generalization of the classical breeding value and is the heritable property relevant for response to selection in traits affected by IGEs. The total heritable variance in the trait due to both DGEs and IGEs, i.e., the magnitude of the heritable differences among individuals determining the potential of the population to respond to selection, equals the variance in TBVs among individuals (Bijma et al. 2007a),
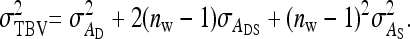 |
(4) |
The 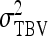 expresses heritable variance in absolute units. Interpretation is often facilitated by expressing heritable variance relative to phenotypic variance. In classical theory, heritability measures heritable variance relative to phenotypic variance,
expresses heritable variance in absolute units. Interpretation is often facilitated by expressing heritable variance relative to phenotypic variance. In classical theory, heritability measures heritable variance relative to phenotypic variance, 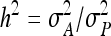 (Falconer and Mackay 1996). By analogy, in case of IGEs we may define the ratio of total heritable variance over phenotypic variance (Bergsma et al. 2008),
(Falconer and Mackay 1996). By analogy, in case of IGEs we may define the ratio of total heritable variance over phenotypic variance (Bergsma et al. 2008),
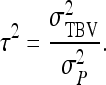 |
(5) |
A comparison of h2 and 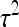 reveals the proportional contribution of IGEs to heritable variance.
reveals the proportional contribution of IGEs to heritable variance.
PREDICTION OF STANDARD ERRORS
This section presents expressions for the SE of estimated values of 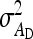 ,
, 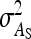 ,
,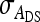 ,
, 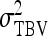 , and
, and 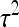 , for two experimental designs.
, for two experimental designs.
Experimental designs:
Two experimental designs were investigated; groups were either composed at random with respect to family, or groups were composed of members of precisely two families. For both designs there was a one-way classification of families, where N unrelated families each contribute n individuals, yielding a total of T = Nn individuals with records. Genetic relatedness within family, r, is the same for all families. The T individuals are allocated to ng groups, each of size nw, so that T = ngnw.
Random group composition:
In this design, the individuals making up a group are sampled at random with respect to family. Hence, there is no relatedness among group mates, other than by chance, so that genotypes of group mates are independent.
Two families per group:
In this design, each group is composed of members of two distinct families, each family contributing 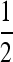 nw individuals (Table 2). Hence, group size is an even number, and each family contributes to ngf = n/(
nw individuals (Table 2). Hence, group size is an even number, and each family contributes to ngf = n/(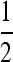 nw) groups. Thus each family is combined with ngf distinct other families. This design leads to a block structure, with ngf + 1 families per block. Each of the ngf + 1 families within a block is combined with each of the other ngf families in that block, each combination occurring precisely once. In the stochastic simulations, design parameters were chosen such that the number of blocks was an integer number.
nw) groups. Thus each family is combined with ngf distinct other families. This design leads to a block structure, with ngf + 1 families per block. Each of the ngf + 1 families within a block is combined with each of the other ngf families in that block, each combination occurring precisely once. In the stochastic simulations, design parameters were chosen such that the number of blocks was an integer number.
TABLE 2.
Example of the design with groups composed of two families
| Family B |
|||||
|---|---|---|---|---|---|
| Family A | 2 | 3 | 4 | 5 | 6 |
| 1 | 2/2 | 2/2 | 2/2 | 2/2 | 2/2 |
| 2 | 2/2 | 2/2 | 2/2 | 2/2 | |
| 3 | 2/2 | 2/2 | 2/2 | ||
| 4 | 2/2 | 2/2 | |||
| 5 | 2/2 | ||||
Group size equals 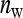 = 4. Each group consists of members of two families, each family contributing 2 individuals. Family size equals n = 10, so that each family is combined with ngf = 10/(½ × 4) = 5 other families. Thus a block consists of 6 families, each being combined with each of the 5 other families in that block. Hence, there are ngb = ngf (ngf + 1)/2 = 5 × 6/2 = 15 groups per block, ngb × nw = 15 × 4 = 60 individuals per block, and nb = T/60 blocks.
= 4. Each group consists of members of two families, each family contributing 2 individuals. Family size equals n = 10, so that each family is combined with ngf = 10/(½ × 4) = 5 other families. Thus a block consists of 6 families, each being combined with each of the 5 other families in that block. Hence, there are ngb = ngf (ngf + 1)/2 = 5 × 6/2 = 15 groups per block, ngb × nw = 15 × 4 = 60 individuals per block, and nb = T/60 blocks.
Expressions for SEs:
Prediction equations for the SE of estimated genetic parameters are based on one-way ANOVA, partitioning the full variance into a between- and within-family component. Thus records were modeled as
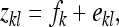 |
(6) |
where 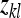 is the lth effective record on family k,
is the lth effective record on family k, 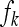 is the effect of the kth family, and
is the effect of the kth family, and 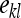 is the residual. The definition of an effective record
is the residual. The definition of an effective record 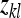 will depend on the experimental design and on the genetic parameter of interest. In general, the
will depend on the experimental design and on the genetic parameter of interest. In general, the 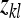 refers to the variable that provides information on the genetic parameter of interest in the experimental design considered (Appendix B). There are m effective records per family and Nm effective records in total.
refers to the variable that provides information on the genetic parameter of interest in the experimental design considered (Appendix B). There are m effective records per family and Nm effective records in total.
Additive genetic variances:
From one-way ANOVA, the SE of an estimated genetic variance equals (Appendix A)
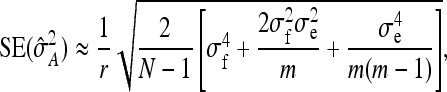 |
(7) |
where 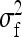 denotes the between-family variance and
denotes the between-family variance and 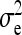 the residual variance,
the residual variance,
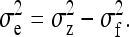 |
(8) |
Equations 7 and 8 are used to predict  ,
, 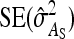 , and
, and 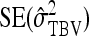 . Application of those equations requires knowledge of
. Application of those equations requires knowledge of 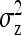 and
and 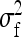 , which depend on the parameter and experimental design of interest, and are given in Tables 3 and 4.
, which depend on the parameter and experimental design of interest, and are given in Tables 3 and 4.
TABLE 3.
Components of SEs with random group composition
| Parameter | Expression | |
|---|---|---|
| All VC | m = n | |
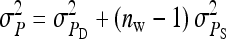 |
||
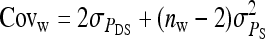 |
||
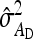 |
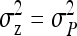 |
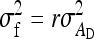 |
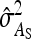 |
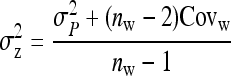 |
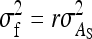 |
 |
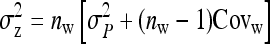 |
 |
SEs for the estimated genetic variance of interest follow from substituting 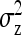 and
and 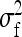 for that parameter into Equations 7 and 8. See also Equations 19 and 20, and see Appendix B for a derivation.
for that parameter into Equations 7 and 8. See also Equations 19 and 20, and see Appendix B for a derivation.
TABLE 4.
Components of SEs with group composed of two families
| Parameter | Expression | |
|---|---|---|
| All VC | 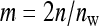 |
|
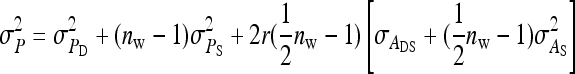 |
||
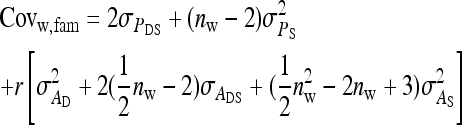 |
||
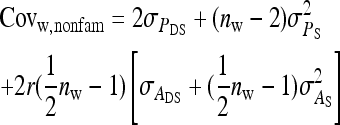 |
||
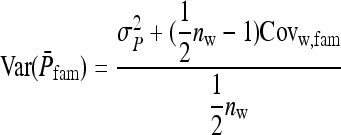 |
||
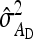 |
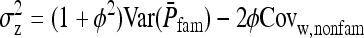 |
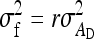 |
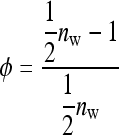 |
||
 |
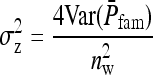 |
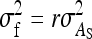 |
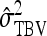 |
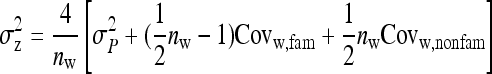 |
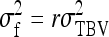 |
SEs for the estimated genetic variance of interest follow from substituting 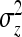 and
and 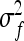 for that parameter into Equations 7 and 8. See also Equations 19 and 20, and see Appendix B for a derivation.
for that parameter into Equations 7 and 8. See also Equations 19 and 20, and see Appendix B for a derivation.
Direct–indirect additive genetic covariance:
The SE of the estimated genetic covariance, 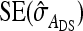 , was obtained from a general expression for the variance of an estimated covariance (Appendix A),
, was obtained from a general expression for the variance of an estimated covariance (Appendix A),
 |
(9) |
The values of 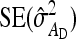 and
and 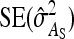 required for Equation 9 follow from Equations 7 and 8.
required for Equation 9 follow from Equations 7 and 8.
Relative genetic variance:
The SE of the ratio of total heritable variance over phenotypic variance is given by
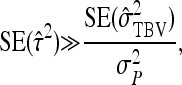 |
(10) |
where 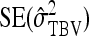 follows from Equations 7 and 8. This approximation assumes that
follows from Equations 7 and 8. This approximation assumes that 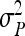 is accurately known, or equivalently that
is accurately known, or equivalently that 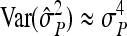 and
and 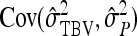 ≈ 0.
≈ 0.
Additive genetic correlation:
The SE of the estimated correlation between DGEs and IGEs,
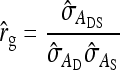 |
(11) |
was approximated by using a Taylor-series expansion (Appendix A), yielding
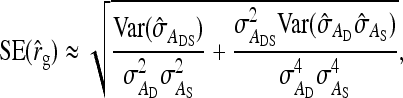 |
(12) |
where 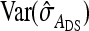 is given by the square of Equation 9. Equation 12 ignores covariances among the components of
is given by the square of Equation 9. Equation 12 ignores covariances among the components of 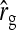 . The
. The 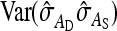 , required for Equation 12, was calculated as (Appendix A)
, required for Equation 12, was calculated as (Appendix A)
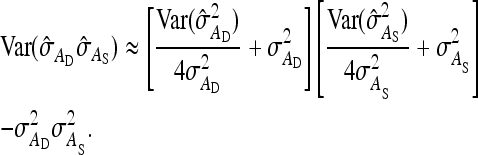 |
(13) |
The values of 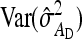 and
and 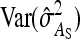 , required for Equation 13, follow from the square of Equation 7.
, required for Equation 13, follow from the square of Equation 7.
Accuracy of predicted SEs:
To evaluate the precision of predicted SEs, experimental populations were stochastically simulated, variance components were estimated using ASReml (Gilmour et al. 2006), and realized SEs of estimates were calculated from the variance among the estimates of 1000 replicates (Appendix C). Accuracies of predicted SEs were evaluated for a range of input parameters (Table 5). A basic scheme with 50 families of 100 members each was used as default. In the basic scheme, direct and indirect effects contributed equally to phenotypic variance, heritabilities of direct and indirect effects were 0.3, and total heritable variance was 60% of phenotypic variance (T2 = 0.6).
TABLE 5.
Schemes used for validation of prediction equations
| Scheme | Deviation from basic schemea |
|---|---|
| Alt. 1 |
 = = 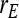 = 0.7 = 0.7 |
| Alt. 2 |
 = = 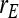 = −0.7 = −0.7 |
| Alt. 3 |
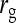 = 0.7, = 0.7, 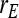 = −0.7 = −0.7 |
| Alt. 4 |
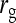 = −0.7, = −0.7, 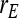 = 0.7 = 0.7 |
| Alt. 5 |
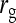 = 0.7 = 0.7 |
| Alt. 6 |
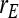 = 0.7 = 0.7 |
| Alt. 7 |
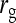 = −0.7 = −0.7 |
| Alt. 8 |
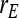 = −0.7 = −0.7 |
| Alt. 9 |
nw = 2, ng = 2,500, 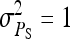 , T = 2450 , T = 2450 |
| Alt. 10 |
nw = 10, ng = 500, 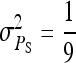
|
| Alt. 11 |
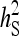 = 0.1 = 0.1 |
| Alt. 12 |
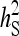 = 0.5 = 0.5 |
| Alt. 13 | 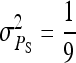 |
| Alt. 14 |
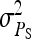 = 1 = 1 |
| Alt. 15 | N = 20, n = 250 |
| Alt. 16 | N = 100, n = 50 |
| Alt. 17 | n = 40, T = 2,000 |
| Alt. 18 | n = 200, T = 10,000 |
| Alt. 19 | N = 100, n = 10, T = 1,000 |
| Alt. 20 | N = 200, n = 10, T = 2,000 |
Basic scheme: N = 50 families, n = 100 members per family, so that T = 5000; nw = 4 individuals per group, so that ng = 1250 groups; 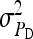 = 1,
= 1, 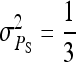 ,
, 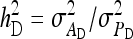 = 0.3,
= 0.3, 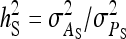 = 0.3,
= 0.3, 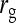 =
= 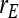 = 0. For schemes with groups composed of two families, T was adjusted when needed to obtain an integer number of blocks.
= 0. For schemes with groups composed of two families, T was adjusted when needed to obtain an integer number of blocks.
Results show that predicted SEs are accurate, both for groups composed at random (Table 6) and for groups composed of two families (Table 7), except for the genetic correlation between DGEs and IGEs. When the true 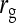 deviated considerably from zero (Alt. 1 through 5, and 7),
deviated considerably from zero (Alt. 1 through 5, and 7), 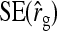 was severely overpredicted. Errors in the predicted
was severely overpredicted. Errors in the predicted 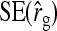 originated from correlations between the components of
originated from correlations between the components of 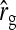 (Equation 11), which were ignored in the derivation (Appendix A). Hence, SEs of
(Equation 11), which were ignored in the derivation (Appendix A). Hence, SEs of 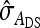 ,
, 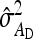 , and
, and 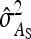 were predicted accurately, but
were predicted accurately, but 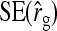 was inaccurate due to correlations among
was inaccurate due to correlations among 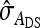 ,
, 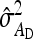 , and
, and 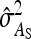 . Moreover, the approximate SEs reported by ASReml (Gilmour et al. 2006) were generally in good agreement with the empirical SEs from the simulations. In conclusion, prediction equations presented above are accurate, except for
. Moreover, the approximate SEs reported by ASReml (Gilmour et al. 2006) were generally in good agreement with the empirical SEs from the simulations. In conclusion, prediction equations presented above are accurate, except for 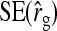 in cases the true
in cases the true 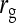 deviates considerably from zero.
deviates considerably from zero.
TABLE 6.
Precision of predicted SEs for groups composed at random
| Error% |
||||||
|---|---|---|---|---|---|---|
| Scheme | 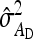 |
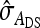 |
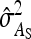 |
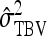 |
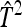 |
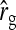 |
| Basic | 2 | 0 | −1 | 1 | 5 | 1 |
| Alt. 1 | 1 | −11 | −3 | −2 | 3 | 146 |
| Alt. 2 | 3 | 3 | 1 | 1 | 2 | 129 |
| Alt. 3 | 4 | 0 | 0 | 2 | 9 | 121 |
| Alt. 4 | 0 | 8 | −1 | −1 | −1 | 47 |
| Alt. 5 | 3 | −6 | −1 | 0 | 7 | 134 |
| Alt. 6 | 1 | −4 | −1 | −2 | 1 | −5 |
| Alt. 7 | 2 | 7 | −1 | 0 | 1 | 85 |
| Alt. 8 | 3 | 1 | 0 | 2 | 6 | 2 |
| Alt. 9 | −1 | 2 | 2 | 3 | 5 | −2 |
| Alt. 10 | 2 | −1 | 0 | 3 | 5 | −10 |
| Alt. 11 | 2 | 0 | −3 | 0 | 2 | −7 |
| Alt. 12 | 2 | 1 | 0 | 1 | 7 | 2 |
| Alt. 13 | 2 | 0 | −1 | −1 | 2 | −1 |
| Alt. 14 | 2 | 2 | 0 | 2 | 7 | 0 |
| Alt. 15 | −2 | 1 | −1 | −2 | 3 | 0 |
| Alt. 16 | 2 | −3 | 4 | 2 | 5 | −6 |
| Alt. 17 | −1 | −1 | 4 | 2 | 5 | −10 |
| Alt. 18 | 3 | −4 | 1 | 0 | 3 | −5 |
| Alt. 19 | 1 | −1 | 4 | 4 | 6 | −38 |
| Alt. 20 | 0 | −6 | −1 | 1 | 2 | −33 |
See Table 5 for description of schemes. Error% = 100% × (predicted − empirical)/empirical. Empirical SEs were based on stochastic simulations with 1000 replicates. With 1000 replicates, the relative SE of the empirical SE equals 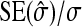 ≈
≈ 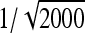 ≈ 0.022 (Stuart and Ord 1994). Therefore, when predictions equal the true values, E[error%] = 0, and the SE of error% given the predicted value equals ∼2.2%. Then the expected absolute error equals E[|error%|] ≈ 1.8%, and |error%| > 4.4% indicates significant bias (P < 0.05; two-sided, not accounting for multiple testing).
≈ 0.022 (Stuart and Ord 1994). Therefore, when predictions equal the true values, E[error%] = 0, and the SE of error% given the predicted value equals ∼2.2%. Then the expected absolute error equals E[|error%|] ≈ 1.8%, and |error%| > 4.4% indicates significant bias (P < 0.05; two-sided, not accounting for multiple testing).
Table 7.
Precision of predicted SEs for groups composed of two families
| Error% |
||||||
|---|---|---|---|---|---|---|
| Scheme | 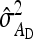 |
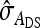 |
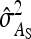 |
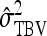 |
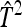 |
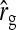 |
| Basic | 3 | 2 | −2 | −3 | 0 | 2 |
| Alt. 1 | 1 | −5 | −2 | −1 | 5 | 147 |
| Alt. 2 | 4 | −1 | 1 | −3 | −1 | 140 |
| Alt. 3 | 1 | 2 | −1 | −1 | 5 | 101 |
| Alt. 4 | 5 | 7 | 1 | −3 | −2 | 95 |
| Alt. 5 | 2 | −2 | −2 | −1 | 5 | 121 |
| Alt. 6 | 3 | 1 | −2 | −3 | 1 | 1 |
| Alt. 7 | 4 | 3 | 1 | −5 | −3 | 117 |
| Alt. 8 | 3 | 2 | −1 | −3 | 1 | 2 |
| Alt. 9 | −1 | 2 | 4 | 0 | 4 | −1 |
| Alt. 10 | 0 | 1 | 6 | 3 | 6 | 1 |
| Alt. 11 | 2 | 3 | −1 | −2 | −1 | −1 |
| Alt. 12 | 4 | 1 | −2 | −3 | 3 | 3 |
| Alt. 13 | 2 | 2 | 1 | −2 | 1 | 3 |
| Alt. 14 | 5 | 1 | −3 | −4 | 1 | 1 |
| Alt. 15 | 3 | 7 | −1 | 0 | 4 | 4 |
| Alt. 16 | 2 | 2 | 3 | 2 | 5 | 2 |
| Alt. 17 | 0 | 1 | 4 | 7 | 10 | −3 |
| Alt. 18 | 0 | 6 | −1 | 3 | 6 | 7 |
| Alt. 19 | 8 | −1 | 1 | −4 | −5 | −44 |
| Alt. 20 | 8 | 6 | 4 | 2 | 1 | −36 |
See Table 5 for description of schemes. Error% = 100% × (predicted − empirical)/empirical. Empirical SEs were based on stochastic simulations with 1000 replicates. With 1000 replicates, the relative SE of the empirical SE equals 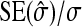 ≈
≈ 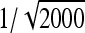 ≈ 0.022 (Stuart and Ord 1994). Therefore, when predictions equals the true values, E[error%] = 0, and the SE of error% given the predicted value equals ∼2.2%. Then the expected absolute error equals
≈ 0.022 (Stuart and Ord 1994). Therefore, when predictions equals the true values, E[error%] = 0, and the SE of error% given the predicted value equals ∼2.2%. Then the expected absolute error equals  ≈ 1.8%, and |error%| > 4.4% indicates significant bias (P < 0.05; two-sided, not accounting for multiple testing).
≈ 1.8%, and |error%| > 4.4% indicates significant bias (P < 0.05; two-sided, not accounting for multiple testing).
OPTIMUM DESIGNS
Optimum family sizes for 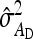 ,
, 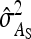 ,
, 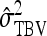 , and
, and 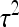 :
:
Robertson (1959) showed that the optimum family size for estimating an intraclass correlation, 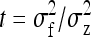 , equals
, equals
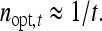 |
(14) |
Since (classical) heritability follows directly from the intraclass correlation, 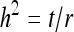 ,
, 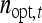 also represents the optimum family size for estimating heritability. With respect to optimum family size, IGEs introduce two complications. First, with IGEs there are multiple genetic parameters, each of which may be of interest, and optimum family size may differ among parameters. Second, the parameters
also represents the optimum family size for estimating heritability. With respect to optimum family size, IGEs introduce two complications. First, with IGEs there are multiple genetic parameters, each of which may be of interest, and optimum family size may differ among parameters. Second, the parameters 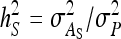 and
and 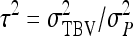 are not true intraclass correlations, because the denominators of those ratios differ from the
are not true intraclass correlations, because the denominators of those ratios differ from the 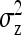 used in the ANOVA for those parameters (Tables 3 and 4). As a consequence, Robertson's
used in the ANOVA for those parameters (Tables 3 and 4). As a consequence, Robertson's 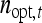 cannot be applied directly to
cannot be applied directly to 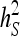 and
and 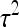 .
.
Approximate expressions for optimum family sizes for 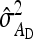 ,
, 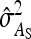 , and
, and  can be obtained as follows. The
can be obtained as follows. The  originates mainly from uncertainty in
originates mainly from uncertainty in 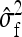 rather than
rather than 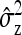 . Thus the optimum family size for estimating
. Thus the optimum family size for estimating 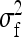 is close to the optimum family size for estimating t. Hence, from Equation 14, the approximate optimum number of records per family for estimating a genetic variance equals
is close to the optimum family size for estimating t. Hence, from Equation 14, the approximate optimum number of records per family for estimating a genetic variance equals 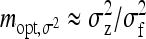 . Thus, for schemes with groups composed at random, where n = m (Table 3), approximate optimum family size equals
. Thus, for schemes with groups composed at random, where n = m (Table 3), approximate optimum family size equals
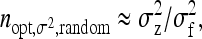 |
(15) |
and for schemes with groups composed of two families, where n = 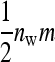 (Table 4), approximate optimum family size equals
(Table 4), approximate optimum family size equals
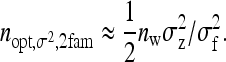 |
(16) |
These results can be applied for the parameter of interest by using the appropriate 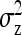 and
and 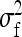 from Table 3 or 4. Finally, under the assumptions given above, optimum family size for
from Table 3 or 4. Finally, under the assumptions given above, optimum family size for 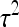 is approximately equal to that for
is approximately equal to that for 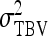 ,
,
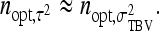 |
(17) |
To evaluate precision of Equations 15 and 16, optimum family sizes were obtained numerically using Equation 7 by varying n from 1 through T, and storing the family size yielding the lowest predicted SE encountered in this range. Results showed that Equations 15 and 16 are very accurate. For all schemes in Table 5, the relative difference between the numerically obtained minimum SE and the result from Equations 15 and 16 was smaller than 1% (results not shown).
An impression of optimum family sizes can be obtained by using estimates available in the literature. Using estimates of Bergsma et al. (2008) for growth rate in fattening pigs with  , optimum half-sib family sizes for groups composed at random are
, optimum half-sib family sizes for groups composed at random are 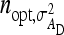 = 19 for DGE, and
= 19 for DGE, and 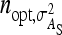 = 200 for IGE. For groups composed of two families, those values are
= 200 for IGE. For groups composed of two families, those values are 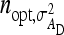 = 29 and
= 29 and 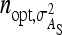 = 69. Using estimates of Ellen et al. (2008; line WB) for survival time in cannibalistic laying hens with
= 69. Using estimates of Ellen et al. (2008; line WB) for survival time in cannibalistic laying hens with  , optimum half-sib family sizes are
, optimum half-sib family sizes are 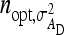 = 42 and
= 42 and 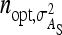 = 113 for groups composed at random, and
= 113 for groups composed at random, and 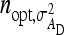 = 52 and
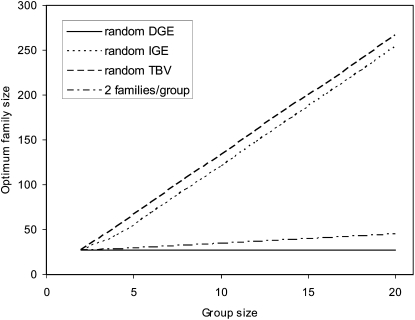
= 52 and 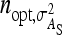 = 82 for groups composed of two families. These values suggest that estimation of IGEs using groups composed at random may require rather large families. Moreover, the difference between optimum family size for DGEs vs. IGEs is much larger with groups composed at random than with groups composed of two families. This pattern is typical, as illustrated in Figure 1. Hence, when interest is in both DGEs and IGEs, the use of groups composed of two families is attractive, because optimum family sizes are more similar for both parameters.
= 82 for groups composed of two families. These values suggest that estimation of IGEs using groups composed at random may require rather large families. Moreover, the difference between optimum family size for DGEs vs. IGEs is much larger with groups composed at random than with groups composed of two families. This pattern is typical, as illustrated in Figure 1. Hence, when interest is in both DGEs and IGEs, the use of groups composed of two families is attractive, because optimum family sizes are more similar for both parameters.
Figure 1.—
Optimum half-sib family size for schemes with groups either composed at random or consisting of two families. A value of d = 0.5 was used for the entire figure, and therefore, the phenotypic variance is the same for all group sizes. (See Bijma 2010, the accompanying article in this issue, and section “Dilution of IGEs”.) With 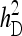 =
= 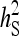 = 0.3,
= 0.3, 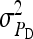 =
= 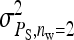 = 1.
= 1. 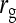 =
= 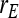 = 0, r = 0.25. For schemes with two families per group, optimum family size was identical for
= 0, r = 0.25. For schemes with two families per group, optimum family size was identical for 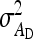 ,
, 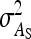 , and
, and 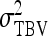 .
.
Optimum group size
The above equations shown that the SEs of estimated genetic parameters depend on group size. Relative performance of experimental designs may, therefore, depend on group size as well, and statistical power can be improved by optimizing group size.
Dilution of IGEs:
The effect of group size on the SE of estimated genetic parameters is complicated by a potential effect of group size on the magnitude of IGEs (Arango et al. 2005; Hadfield and Wilson 2007; Bijma 2010). An individual's IGE on a single recipient may become smaller in larger groups, because its total IGE is divided over more group mates. The degree of dilution, meaning the decrease of indirect effects in larger groups, will depend on the trait of interest. With competition for a finite amount of feed per group, for example, an individual consuming 1 kg has an average indirect effect of 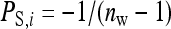 on each of its group mates. Hence, the indirect effect is inversely proportional to the number of group mates, indicating full dilution. The other extreme of no dilution may be illustrated by a highly contagious infectious disease, where an individual may infect all its group mates, irrespective of group size. Here the indirect effect each group mate receives is independent of group size, indicating no dilution.
on each of its group mates. Hence, the indirect effect is inversely proportional to the number of group mates, indicating full dilution. The other extreme of no dilution may be illustrated by a highly contagious infectious disease, where an individual may infect all its group mates, irrespective of group size. Here the indirect effect each group mate receives is independent of group size, indicating no dilution.
Following Bijma (2010), dilution of indirect effects was modeled as
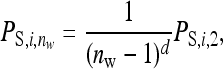 |
(18) |
where  is the indirect effect of individual i in a group of nw members,
is the indirect effect of individual i in a group of nw members, 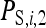 is the indirect effect of i when it has a single group member (i.e., when nw = 2), and d is the degree of dilution. With no dilution, d = 0, indirect effects do not depend on group size,
is the indirect effect of i when it has a single group member (i.e., when nw = 2), and d is the degree of dilution. With no dilution, d = 0, indirect effects do not depend on group size, 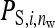 =
= 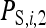 , as with the infectious disease. With full dilution, d = 1, indirect effects are inversely proportional to the number of group members,
, as with the infectious disease. With full dilution, d = 1, indirect effects are inversely proportional to the number of group members, 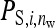 =
= 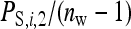 , as with the finite amount of feed. The magnitude of d can be estimated empirically (Arango et al. 2005; Bijma 2010; Canario et al. 2010). Dilution affects the relationship of phenotypic variance with group size; for d < 0.5 phenotypic variance increases with group size, whereas for d > 0.5 phenotypic variance decreases with group size (Bijma 2010). It is assumed here that IGEs and nongenetic indirect effects are diluted in the same manner as phenotypic indirect effects (i.e., Equation 18 applies also to
, as with the finite amount of feed. The magnitude of d can be estimated empirically (Arango et al. 2005; Bijma 2010; Canario et al. 2010). Dilution affects the relationship of phenotypic variance with group size; for d < 0.5 phenotypic variance increases with group size, whereas for d > 0.5 phenotypic variance decreases with group size (Bijma 2010). It is assumed here that IGEs and nongenetic indirect effects are diluted in the same manner as phenotypic indirect effects (i.e., Equation 18 applies also to 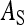 and
and 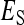 ).
).
The following results illustrate the effect of group size on the SEs of estimated variance components, which will depend on the degree of dilution. Results were very similar for fixed and optimum family sizes and are presented for optimum family sizes only, first for groups composed at random, and subsequently for groups composed of two families.
Groups composed at random:
Depending on the situation, the size of an experiment may be limited either by the total number of individuals or by the number of groups. Results are presented first for a fixed number of individuals and subsequently for a fixed number of groups.
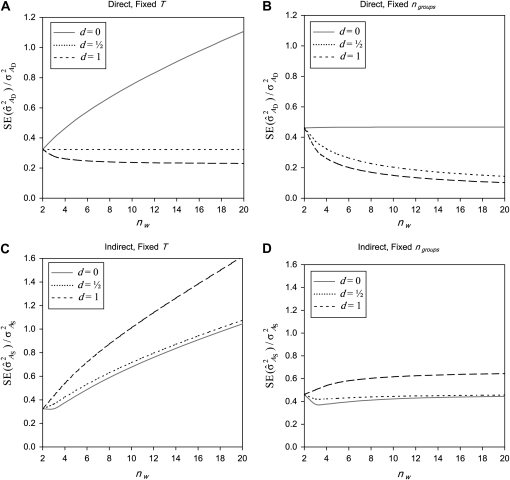
Fixed number of individuals (T):
With fixed T, the number of groups decreases as group size increases, 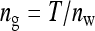 . The effect of group size on SEs differs between DGEs and IGEs. For DGEs, SEs increased with group size when d < 0.5 and decreased with group size when d > 0.5 (Figure 2A). This relationship originates from the impact of dilution on phenotypic variance, which increases with group size for d < 0.5.
. The effect of group size on SEs differs between DGEs and IGEs. For DGEs, SEs increased with group size when d < 0.5 and decreased with group size when d > 0.5 (Figure 2A). This relationship originates from the impact of dilution on phenotypic variance, which increases with group size for d < 0.5.
Figure 2.—
Relationship between SEs and group size, for groups composed at random. (A) Direct genetic variance, T = 2000. (B) Direct genetic variance, ngroups = 500. (C) Indirect genetic variance, T = 2000. (D) Indirect genetic variance, ngroups = 500. (A–D)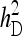 =
= 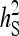 = 0.3,
= 0.3, 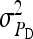 =
= 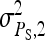 = 1.
= 1. 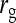 =
= 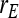 = 0, r = 0.25 (half-sibs). Family size is optimum (Equation 15). Because
= 0, r = 0.25 (half-sibs). Family size is optimum (Equation 15). Because 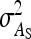 varies with group size when d ≠ 0, results focus on the relative SE,
varies with group size when d ≠ 0, results focus on the relative SE, 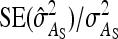 .
.
For IGEs, SEs increased with group size, particularly with strong dilution (Figure 2C). When d = 0, SEs were identical for groups of 2 and 3 individuals. This relationship originates from two mechanisms. First, group size affects the variance of the record of interest, 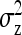 (Table 3), higher
(Table 3), higher 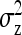 yielding higher SEs. With d = 0,
yielding higher SEs. With d = 0, 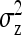 was minimal for groups of ∼3 individuals, explaining lowest SEs for those group sizes. Second, with dilution,
was minimal for groups of ∼3 individuals, explaining lowest SEs for those group sizes. Second, with dilution, 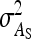 decreases with group size, which decreases the intraclass correlation (Equation 25b) and therefore increases SEs. Hence, with strong dilution, smaller groups are considerably better. Results for total genetic variance were similar to those for indirect genetic variance, smaller groups yielding lower SEs (results not shown).
decreases with group size, which decreases the intraclass correlation (Equation 25b) and therefore increases SEs. Hence, with strong dilution, smaller groups are considerably better. Results for total genetic variance were similar to those for indirect genetic variance, smaller groups yielding lower SEs (results not shown).
Fixed number of groups (ng):
With a fixed number of groups, the total number of individuals increases as group size increases, T = 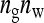 . The effect of group size on SEs differs between DGEs and IGEs. For direct effects, relative SEs decreased with group size, except for d = 0, in which case SEs were nearly independent of group size (Figure 2B). This relationship originates from the increase of T with group size, which is counterbalanced by an increase of
. The effect of group size on SEs differs between DGEs and IGEs. For direct effects, relative SEs decreased with group size, except for d = 0, in which case SEs were nearly independent of group size (Figure 2B). This relationship originates from the increase of T with group size, which is counterbalanced by an increase of 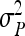 when d = 0. Hence, for d > 0,
when d = 0. Hence, for d > 0, 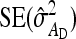 can be decreased substantially by using more individuals.
can be decreased substantially by using more individuals.
For IGEs, SEs were lowest for groups of two or three individuals (Figure 2D), and increased slightly with larger group sizes. For d = 0, groups of three individuals were clearly better than groups of two individuals. This relationship originates from an increase in phenotypic variance and dilution of IGEs, which counterbalances the benefit of greater T. Results for total genetic variance were similar to those for indirect genetic variance.
In conclusion, to estimate the variance due to IGEs while using groups composed at random, it is optimal to have small groups of two or three individuals. This result applies irrespective of whether the number of individuals or the number of groups is the limiting factor in the experimental design.
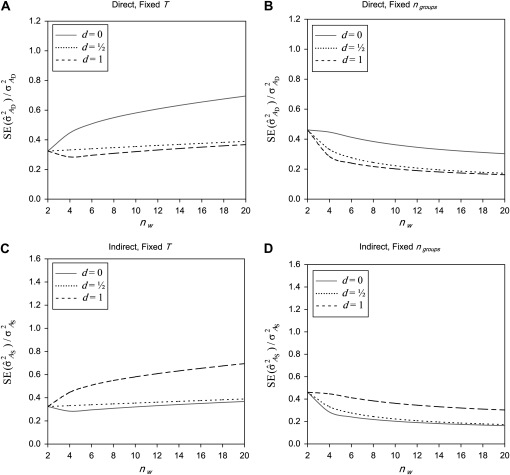
Groups composed of two families:
Results are presented first for a fixed number of individuals and subsequently for a fixed number of groups.
Fixed number of individuals (T):
For direct effects, SEs mostly increased with group size, particularly for d = 0 (Figure 3A). This result originates from an increase in 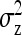 with group size, which reduces the intraclass correlation and thus increases the SE of
with group size, which reduces the intraclass correlation and thus increases the SE of 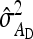 in larger groups. Also for IGEs, SEs increased with group size, particularly for d = 1 (Figure 3C). This relationship originates from two mechanisms. First, with dilution (d > 0),
in larger groups. Also for IGEs, SEs increased with group size, particularly for d = 1 (Figure 3C). This relationship originates from two mechanisms. First, with dilution (d > 0), 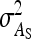 decreases with group size, which increases the relative SE. Hence, stronger dilution favors smaller groups. Second, when groups are composed of two families, an increase in group size tends to increase the intraclass correlation, particularly when
decreases with group size, which increases the relative SE. Hence, stronger dilution favors smaller groups. Second, when groups are composed of two families, an increase in group size tends to increase the intraclass correlation, particularly when 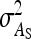 is small, which favors larger groups. Compared to groups composed at random, the second mechanism limits the increase of SEs with group size (Figure 3C vs. Figure 2C). Results for total genetic variance were similar to those for indirect genetic variance (not shown).
is small, which favors larger groups. Compared to groups composed at random, the second mechanism limits the increase of SEs with group size (Figure 3C vs. Figure 2C). Results for total genetic variance were similar to those for indirect genetic variance (not shown).
Figure 3.—
Relationship between SEs and group size, for groups composed of two families. (A) Direct genetic variance, T = 2000. (B) Direct genetic variance, ngroups = 500. (C) Indirect genetic variance, T = 2000. (D) Indirect genetic variance, ngroups = 500. (A–D) 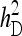 =
= 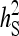 = 0.3,
= 0.3, 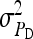 =
= 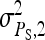 = 1.
= 1. 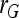 =
= 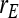 = 0, r = 0.25 (half-sibs). Family size is optimum (Equation 16). Because
= 0, r = 0.25 (half-sibs). Family size is optimum (Equation 16). Because 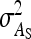 varies with group size when d ≠ 0, results focus on the relative SE,
varies with group size when d ≠ 0, results focus on the relative SE, 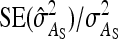 .
.
Fixed number of groups (ng):
For direct effects, relative SEs decreased with group size, particularly for d = 1 (Figure 3B). This relationship originates mainly from the increase of T with group size. Also for IGEs, relative SEs decreased with group size (Figure 3D). Benefits of increasing group size were largest for small d and small 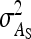 . For example, with d = 0 and
. For example, with d = 0 and 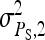 = 0.2, the
= 0.2, the 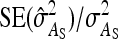 decreased from 0.51 for nw = 2 to 0.10 for nw = 20 (results not shown in Figure 3). Hence, when IGEs are expected to be small and the number of groups is the limiting factor in the experimental design, then the use of large groups, each composed of two families, is a powerful scheme.
decreased from 0.51 for nw = 2 to 0.10 for nw = 20 (results not shown in Figure 3). Hence, when IGEs are expected to be small and the number of groups is the limiting factor in the experimental design, then the use of large groups, each composed of two families, is a powerful scheme.
Figures 2 and 3 show that the impact of dilution is opposite for DGEs vs. IGEs. For direct effects, SEs are lowest with full dilution, whereas for IGEs, SEs are lowest with no dilution. Moreover, Figure 2, A and B, vs. Figure 2, C and D, shows that, for random groups and d > 0.5, there is a conflict between optimum group sizes for direct genetic variance, which are large, and optimum group sizes for indirect genetic variance, which are small. This conflict is largely absent when groups are composed of two families, particularly when the number of groups is limiting (Figure 3, A and B, vs. Figure 3, C and D).
In conclusion, when groups are composed of two families, optimum group size to estimate the variance due to IGEs depends on the factor limiting the size of the experiment; small groups tend to be optimal when the number of individuals is limiting, whereas large groups tend to be optimal when the number of groups is limiting.
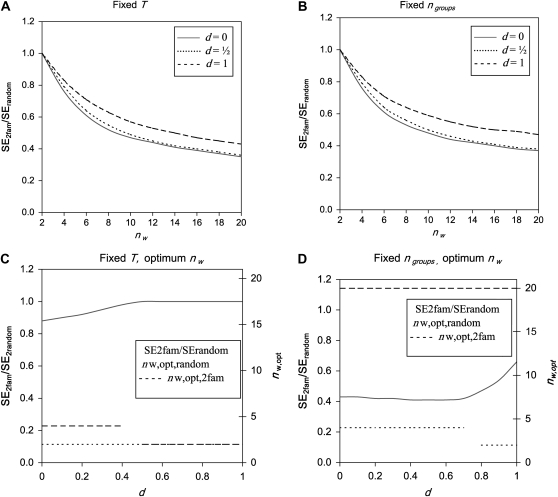
Group composition:
To compare both experimental designs, predicted SEs were compared between schemes with groups composed at random and schemes with groups composed of two families, for all schemes listed in Table 3 (results not shown). Differences between both designs were small for DGEs. For IGEs, schemes with groups composed of two families were superior, particularly when groups were large (Table 5, Alt. 10). Predicted SEs were identical for both designs when groups consisted of two individuals.
Figure 4, A and B, shows that schemes with groups composed of two families become increasingly superior when group size increases, superiority being largest for d = 0. Figure 4, C and D, shows the relative performance of both schemes when compared at optimum group sizes, as a function of d. When the total number of individuals is limiting, superiority of two-family schemes is limited and only present for d < 0.5 (Figure 4C). When the number of groups is limiting, superiority of two-family schemes is substantial (Figure 4D). In that case, optimum group size for two-family schemes is large (nw was limited to a maximum of 20 in Figure 4), whereas optimum group size for random schemes is small, ranging from two to four. In conclusion, if number of groups is the limiting factor in the experiment and it is possible to use groups of more than two members, then there is substantial benefit of putting two families in each group rather than using groups composed at random.
Figure 4.—
Ratio of 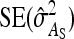 for schemes with two families per group over that for schemes with random groups. (A) For fixed T = 2000 B. For fixed
for schemes with two families per group over that for schemes with random groups. (A) For fixed T = 2000 B. For fixed 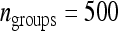 . (C) For fixed T = 2000 and optimum group size. (D) For fixed
. (C) For fixed T = 2000 and optimum group size. (D) For fixed 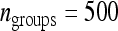 and optimum group size. (A–D)
and optimum group size. (A–D) 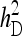 =
= 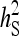 = 0.3,
= 0.3, 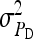 =
= 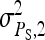 = 1.
= 1. 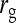 =
= 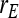 = 0, r = 0.25 (half-sibs). Family size is optimum (Equations 15 and 16).
= 0, r = 0.25 (half-sibs). Family size is optimum (Equations 15 and 16).
DISCUSSION
IGEs attract increasing attention, as illustrated by the increasing number of publications (e.g., Bleakley and Brodie 2009; Wilson et al. 2009; Chenoweth et al. 2010; Hsu et al. 2010; Mcglothlin et al. 2010; Muir et al. 2010). This development is warranted, given the large impact of IGEs on both the direction and magnitude of response to genetic selection and the increasing empirical evidence of their existence (see Introduction). Thus IGEs exist and cannot be ignored. Accurate knowledge of the genetic parameters of IGEs and of their relationship to DGEs is required to quantify the consequences for response to selection, in natural, agricultural, and laboratory populations.
This work has provided the tools to quantify the required size of experiments aiming to identify IGEs and to optimize experimental designs. The expressions for SEs of estimated genetic parameters are accurate for the variance of DGEs and IGEs, for their covariance, and for the total genetic variance. The expression for the SE of the estimated genetic correlation between DGEs and IGEs requires further improvement. An expression for optimum family size was provided as well, and optimum group size was investigated numerically. For IGEs, schemes with groups composed of two families were clearly better than schemes with groups composed at random. The R-package SE.IGE was developed, which predicts SEs and identifies optimum family and group sizes for each parameter.
Results suggest that, when optimum designs are implemented, presence of IGEs can be demonstrated successfully when the number of individuals is at least ∼1000–2000, or the number of groups is at least ∼250–500 (Figures 2 and 3; obviously, this will depend on the true parameters, which are unknown a priori). For example, when using the estimates of Bergsma et al. (2008), the P-value for the variance of IGEs is just below 5% when T = 2000 in a half-sib design with two families per group and groups of eight members. Using the estimates of Ellen et al. (2008; line WB), T = 2000, and in groups of four members, the corresponding P-value is below 5% for a half-sib design and below 1% for a full-sib design. Those experimental sizes are within the feasible range for many species in agriculture and aquaculture and for the smaller laboratory species. This work has considered a simple population structure, where relatedness is the same for all families and members of different families are unrelated. Extension to more complex family structures, such as full-sib families within half-sib families, would be useful.
This work has compared two experimental designs, showing that schemes with groups composed of two families yield lower SEs for the variance of IGEs than schemes with groups composed at random. The mechanism is as follows. With 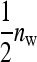 family members in a group, the phenotypes of the group mates belonging to the other family contain
family members in a group, the phenotypes of the group mates belonging to the other family contain 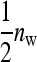 IGEs from the focal family. With a random group composition, in contrast, the phenotypes of the group mates contain only a single IGE from the focal family. Hence, the two-family scheme has the effect of increasing the family component in the phenotype, which increases the intraclass correlation. This increase in intraclass correlation can be observed in the expression for
IGEs from the focal family. With a random group composition, in contrast, the phenotypes of the group mates contain only a single IGE from the focal family. Hence, the two-family scheme has the effect of increasing the family component in the phenotype, which increases the intraclass correlation. This increase in intraclass correlation can be observed in the expression for 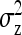 , which has a factor
, which has a factor 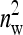 in the denominator (Table 4). Since larger effects can be estimated more precisely, this reduces the
in the denominator (Table 4). Since larger effects can be estimated more precisely, this reduces the 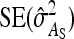 . This effect is counteracted by a difference in the effective number of records per family, which equals m = n for random schemes but
. This effect is counteracted by a difference in the effective number of records per family, which equals m = n for random schemes but 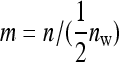 for two-family schemes (Tables 3 and 4). Hence, moving from random schemes to two-family schemes increases the relative size of the effect by a factor of
for two-family schemes (Tables 3 and 4). Hence, moving from random schemes to two-family schemes increases the relative size of the effect by a factor of 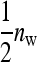 , but reduces the effective number of records by the same factor. Investigation of Equation 7, however, reveals a positive net effect.
, but reduces the effective number of records by the same factor. Investigation of Equation 7, however, reveals a positive net effect.
Although a formal proof of optimality of the two-family design is difficult, the above suggests that this design is probably optimal or near optimal. Consider the full range of schemes with 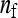 families per group, each family contributing
families per group, each family contributing 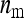 members, so that
members, so that 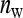 =
= 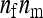 . The extremes of this range are given by taking all members from different families,
. The extremes of this range are given by taking all members from different families,  =
= 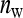 and
and 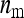 = 1, which is near identical to the random scheme, and by the two-family scheme,
= 1, which is near identical to the random scheme, and by the two-family scheme, 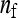 = 2 and
= 2 and 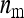 =
= 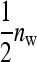 . (With a single family per group,
. (With a single family per group, 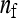 = 1 and
= 1 and 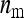 =
= 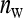 , genetic parameters of DGEs and IGEs are statistically nonidentifiable.) Within this range, the size of the family effect in the phenotypes of the group mates of that family equals
, genetic parameters of DGEs and IGEs are statistically nonidentifiable.) Within this range, the size of the family effect in the phenotypes of the group mates of that family equals 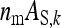 , and the number of effective records per family equals
, and the number of effective records per family equals 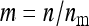 . Investigation of Equation 7 shows that increasing the size of the effect while reducing the effective number of records by the same factor reduces the SE of
. Investigation of Equation 7 shows that increasing the size of the effect while reducing the effective number of records by the same factor reduces the SE of 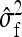 (unless m is very small). Hence, this suggests that using the highest possible number of family members per group is optimal, which is the two-family scheme.
(unless m is very small). Hence, this suggests that using the highest possible number of family members per group is optimal, which is the two-family scheme.
In any power calculation, predicted SEs depend on the true values of the parameters to be estimated. Hence, application of prediction equations presented here requires prior assumptions on those true values. Such prior assumptions may be obtained from knowledge of traits underlying the indirect effects, e.g., from the intensity of behavioral interactions among individuals and the heritability of such behaviors or from existing estimates of genetic parameters taken from the literature. The expressions for SEs presented here are formulated in terms of the true parameters for total direct and indirect effect, 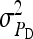 ,
, 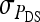 , and
, and 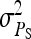 , and the corresponding genetic parameters,
, and the corresponding genetic parameters, 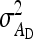 ,
, 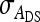 , and
, and 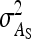 . However, while the genetic parameters are statistically identifiable, the parameters for total direct and indirect effect are not. The reason is that the nongenetic parameters
. However, while the genetic parameters are statistically identifiable, the parameters for total direct and indirect effect are not. The reason is that the nongenetic parameters 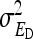 ,
, 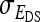 , and
, and 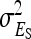 are nonidentifiable, because there exist multiple combinations of those parameters yielding the same nongenetic variances between and within groups. The between-group variance equals
are nonidentifiable, because there exist multiple combinations of those parameters yielding the same nongenetic variances between and within groups. The between-group variance equals 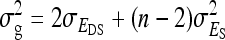 , whereas the within-group variance equals
, whereas the within-group variance equals 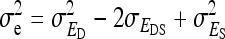 . Hence, the literature will include estimates of
. Hence, the literature will include estimates of 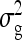 and
and 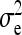 , rather than
, rather than 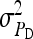 ,
, 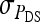 , and
, and 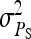 (e.g., Appendix C). This issue is resolved by reformulating the expressions in Tables 3 and 4 in terms of
(e.g., Appendix C). This issue is resolved by reformulating the expressions in Tables 3 and 4 in terms of 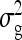 and
and 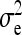 , using
, using
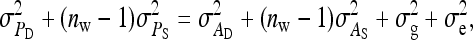 |
(19) |
and
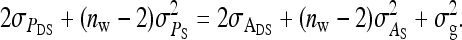 |
(20) |
Hence, expression presented here can be implemented using prior values taken from the literature, on the condition that authors present not only the estimated genetic parameters, but also the estimated between-group and residual variance.
Acknowledgments
I thank Seteng Alemu and Jørgen Ødegård for helpful discussion on this topic, and Albart Coster for assistance with R. The R-package SE.IGE can be downloaded from the repository of R-packages, CRAN http://cran.r-project.org/package=SE.IGE, following the usual method to install R packages. This research was financially supported by the Dutch science council (Nederlandse organisatie voor Wetenschappelijk Onderzoek) and was coordinated by the Netherlands Technology Foundation (Stichting Technische Wetenschappen).
Note added in proof: See P. Bijma (pp. 1029–1031) in this issue, for a related work.
APPENDIX A
This appendix shows the derivations of Equations 7, 9, 12, and 13.
Equation 7:
With one-way ANOVA, expected mean squares are (Stuart et al. 1999)
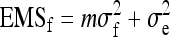 |
(A1a) |
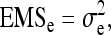 |
(A1b) |
so that the estimated family variance equals
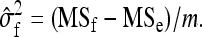 |
(A2) |
The variance of 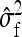 follows from the variances of the mean squares, using the general result that (Stuart et al. 1999)
follows from the variances of the mean squares, using the general result that (Stuart et al. 1999)
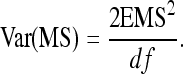 |
(A3) |
Combining Equations A1–A3 yields
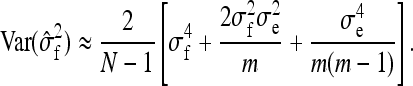 |
(A4) |
The last term in this expression uses (T−1)/T ≈ 1. Finally, using 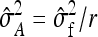 , r denoting additive genetic relatedness among family members, yields Equation 7. The r equals ½ for full-sib families and ¼ for half-sib families (Falconer and Mackay 1996).
, r denoting additive genetic relatedness among family members, yields Equation 7. The r equals ½ for full-sib families and ¼ for half-sib families (Falconer and Mackay 1996).
Equation 9:
The derivation of  is based on an approximation of the variance of an estimated covariance. Consider df paired observations on two random variables, say x and y, following a bivariate normal distribution with zero mean,
is based on an approximation of the variance of an estimated covariance. Consider df paired observations on two random variables, say x and y, following a bivariate normal distribution with zero mean, 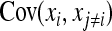 =
= 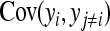 =
= 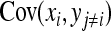 = 0, and
= 0, and 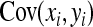 =
= 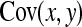 =
= 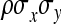 , where
, where 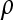 denotes the correlation between x and y within a pair. (With respect to a covariance, the assumption of zero mean is equivalent to assuming the mean known. Hence, this derivation ignores that the mean has to be estimated from the sample.) Interest is in
denotes the correlation between x and y within a pair. (With respect to a covariance, the assumption of zero mean is equivalent to assuming the mean known. Hence, this derivation ignores that the mean has to be estimated from the sample.) Interest is in 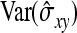 . With
. With
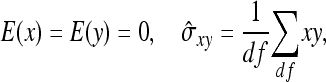 |
so that  =
=  . Next, we use
. Next, we use  =
= 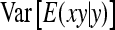 +
+ 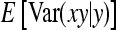 (Stuart and Ord 1994). Under normality,
(Stuart and Ord 1994). Under normality, 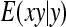 =
= 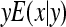 =
= 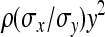 . From the moments of the normal distribution (Stuart and Ord 1994),
. From the moments of the normal distribution (Stuart and Ord 1994), 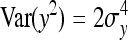 , so that
, so that 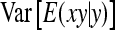 =
= 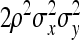 . Moreover, under normality,
. Moreover, under normality, 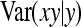 =
= 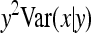 =
= 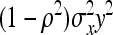 , so that
, so that 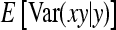 =
= 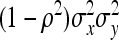 . Combining both terms and substituting
. Combining both terms and substituting 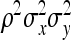 =
= 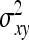 yields
yields
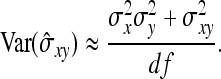 |
(A5) |
Application of this result to the direct-social genetic covariance requires knowledge of 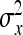 ,
, 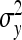 , and
, and 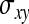 . Because DGE and IGEs are not directly observed, one cannot simply substitute those parameters by the true genetic parameters. Rather, the
. Because DGE and IGEs are not directly observed, one cannot simply substitute those parameters by the true genetic parameters. Rather, the 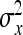 and
and 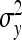 represent the “effective variance” of the “record” providing information on DGE and IGE, respectively. Those effective variances can be backcalculated from the SEs of the estimated variances of x and y. Using the general result that
represent the “effective variance” of the “record” providing information on DGE and IGE, respectively. Those effective variances can be backcalculated from the SEs of the estimated variances of x and y. Using the general result that 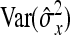 =
= 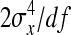 , the effective variances are
, the effective variances are 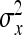 =
= 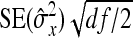 , and
, and 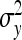 =
= 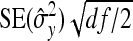 . Moreover, the
. Moreover, the 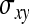 in Equation A5 represents the covariance between the records providing information on the DGE and IGE, respectively. Because covariances are not affected by taking averages, the true value of
in Equation A5 represents the covariance between the records providing information on the DGE and IGE, respectively. Because covariances are not affected by taking averages, the true value of 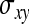 can be used. Substituting the above results into Equation A5 yields
can be used. Substituting the above results into Equation A5 yields
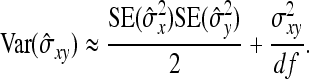 |
(A6) |
Applying this result to DGEs and IGEs, substituting df = N−1, and taking the square root yields Equation 9.
Equations 12:
The estimated genetic correlation equals
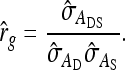 |
This derivation ignores covariances among 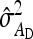 ,
, 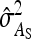 , and
, and 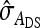 . Then
. Then 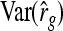 can be predicted using a first-order Taylor-series approximation of a ratio, say x/y, giving
can be predicted using a first-order Taylor-series approximation of a ratio, say x/y, giving 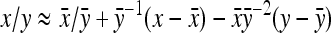 . Assuming
. Assuming 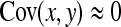 ,
,
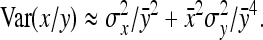 |
(A7) |
Substituting 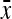 =
= 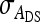 ,
,  =
= 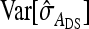 ,
,  =
= 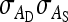 , and
, and 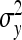 =
= 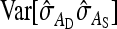 and taking the square root yield Equation 12.
and taking the square root yield Equation 12.
Equation 13:
Application of Equation 12 requires 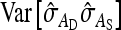 , the variance of a product. Consider the product
, the variance of a product. Consider the product 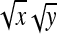 , where x corresponds to
, where x corresponds to 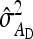 , and y corresponds to
, and y corresponds to 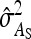 . Using
. Using 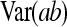 =
= 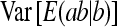 +
+ 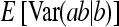 , and assuming
, and assuming 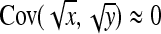 and
and 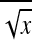 ≈
≈ 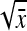 , yields
, yields 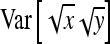 ≈
≈ 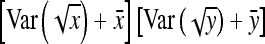 −
− 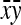 . Next, a first-order Taylor-series approximation for the square root of a variable yields
. Next, a first-order Taylor-series approximation for the square root of a variable yields 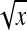 ≈
≈ 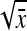 +
+ 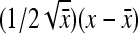 , so that
, so that 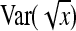 ≈
≈ 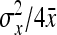 . Analogously
. Analogously 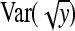 ≈
≈ 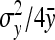 . Substituting those results yields
. Substituting those results yields
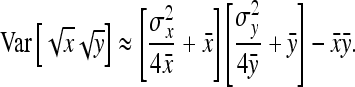 |
(A8) |
Substituting 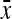 =
= 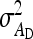 ,
, 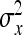 =
= 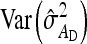 ,
, 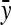 =
= 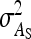 , and
, and 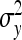 =
= 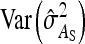 yields Equation 13.
yields Equation 13.
APPENDIX B
This appendix shows the derivation of results presented in Tables 3 and 4.
Table 3, 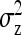 , and
, and 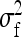 for groups composed at random:
for groups composed at random:
When groups are composed at random with respect to family, group mates are genetically independent. The effective number of records in this design equals the number of individuals per family (see derivations below), so that
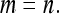 |
(B1) |
From Equation 1, phenotypic variance equals
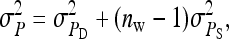 |
(B2) |
and the covariance between phenotypes of group mates equals
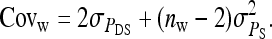 |
(B3) |
Direct genetic variance:
The DGE of a family is expressed in the phenotypes of the family members themselves. Hence, in terms of Equation 6, the record of interest for DGEs is the phenotypic value of the lth member of family k,
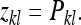 |
(B4) |
Thus the full- and between-family variances are
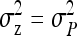 |
(B5a) |
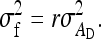 |
(B5b) |
Indirect genetic variance:
The IGE of an individual is expressed in the phenotypes of its group mates. Hence, the group mates of the members of a family provide information on the IGE of that family. Thus the record of interest for IGEs is the mean phenotypic value of the nw − 1 group mates of the lth member of family k,
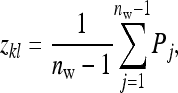 |
(B6) |
j denoting the group mates of l. From Equation B6, the full variance equals
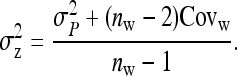 |
(B7a) |
The expected value of 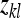 , given family k, equals the IGE of family k,
, given family k, equals the IGE of family k, 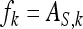 . Thus the between-family variance equals
. Thus the between-family variance equals
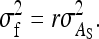 |
(B7b) |
Total heritable variance:
An individual's total heritable effect equals TBVi = 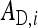 +
+ 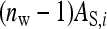 , of which the
, of which the 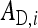 is expressed in its own phenotype, and the
is expressed in its own phenotype, and the 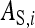 in the phenotypes of each of its nw − 1 group mates. Hence, the summed phenotypes of the focal individual and its nw − 1 group mates expresses the TBV of the focal individual. Thus the record of interest for TBVs is the sum of phenotypic values of all nw individuals belonging to the group of the lth member of family k,
in the phenotypes of each of its nw − 1 group mates. Hence, the summed phenotypes of the focal individual and its nw − 1 group mates expresses the TBV of the focal individual. Thus the record of interest for TBVs is the sum of phenotypic values of all nw individuals belonging to the group of the lth member of family k,
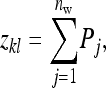 |
(B8) |
j denoting group members of l, including l itself. From Equation B8, the full variance equals
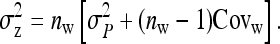 |
(B9a) |
The expected value of zkl, given family k, equals the TBV of family k, 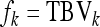 . Thus the between-family variance equals
. Thus the between-family variance equals
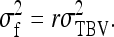 |
(B9b) |
Table 4, 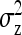 , and
, and 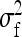 for groups composed of two families:
for groups composed of two families:
In this design, each group consists of members of two families, each contributing 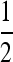 nw individuals (Table 2). The effective number of records in this design equals the number of groups to which a family contributes (see derivations below), so that
nw individuals (Table 2). The effective number of records in this design equals the number of groups to which a family contributes (see derivations below), so that
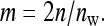 |
(B10) |
Phenotypic variance depends on relatedness. Relatedness within group equals r between members of the same family and zero between members of different families. From
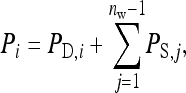 |
phenotypic variance equals
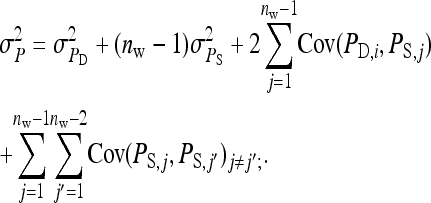 |
Only covariances between family member are nonzero. Since i has 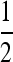 nw − 1 family members in the same group,
nw − 1 family members in the same group,
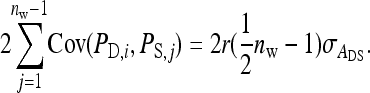 |
In
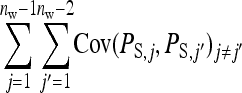 |
there are 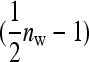 members of the family of i together contributing
members of the family of i together contributing 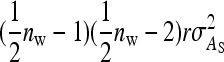 , and
, and 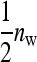 members of the other family together contributing
members of the other family together contributing  , yielding a total of
, yielding a total of 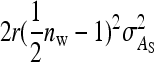 . Collecting terms yields
. Collecting terms yields
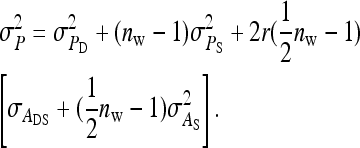 |
(B11) |
The covariance between phenotypes of group mates belonging to the same family follows from
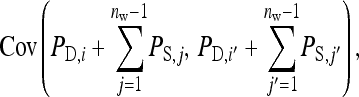 |
where i and i′ belong to the same family, and j and j′ denote the group mates of i and i′, respectively. This covariance can be split into a term due to unrelated individuals, which is the same as for groups composed at random and equals 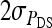 +
+ 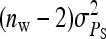 , and a remaining term due to relatedness. Carefully collecting terms shows that components due to relatedness are: (1)
, and a remaining term due to relatedness. Carefully collecting terms shows that components due to relatedness are: (1) 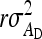 from
from 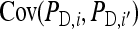 , (2)
, (2) 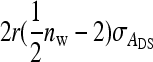 from the covariance between DGEs and IGEs among members of the same family, (3)
from the covariance between DGEs and IGEs among members of the same family, (3) 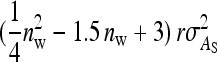 from IGEs of the focal family, and (4)
from IGEs of the focal family, and (4) 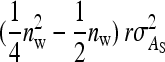 from IGEs of the other family. Collecting terms yields
from IGEs of the other family. Collecting terms yields
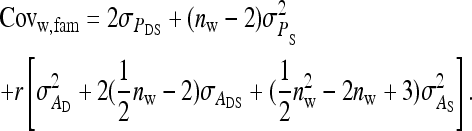 |
(B12) |
The covariance between phenotypes of group mates belonging to different families can be derived analogously. Components due to unrelated individuals contribute 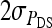 +
+  . Components due to relatedness are
. Components due to relatedness are 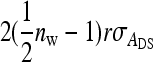 from the covariances between DGEs and IGEs of family members and
from the covariances between DGEs and IGEs of family members and 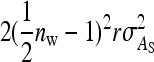 from the covariances between IGEs of family members. Collecting terms yields
from the covariances between IGEs of family members. Collecting terms yields
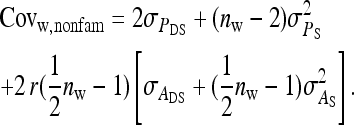 |
(B13) |
Moreover, the variance of the mean phenotype of the 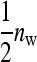 group mates belonging to the same family equals
group mates belonging to the same family equals
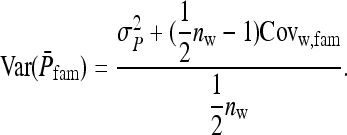 |
(B14) |
Direct genetic variance:
With two families per group, the mean phenotype of the 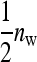 group mates of a family depends not only on the DGE of that family, but also on its IGE. Given family k, the expected mean of the
group mates of a family depends not only on the DGE of that family, but also on its IGE. Given family k, the expected mean of the 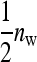 members of family k in group l equals
members of family k in group l equals 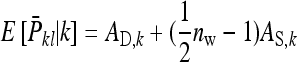 . The
. The 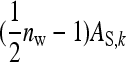 arises because each family member has
arises because each family member has 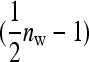 group mates belonging to its own family. Moreover, given family k, the expected mean phenotype of the other family in the group, say k′, equals
group mates belonging to its own family. Moreover, given family k, the expected mean phenotype of the other family in the group, say k′, equals 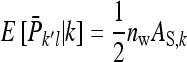 . A linear combination of both expected means yields an estimate of the DGE of family k,
. A linear combination of both expected means yields an estimate of the DGE of family k, 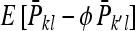 =
= 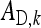 , where
, where
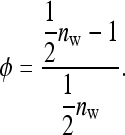 |
(B15) |
Thus the record of interest for DGEs is
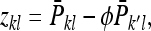 |
(B16) |
where 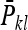 is the mean phenotypic value of the
is the mean phenotypic value of the 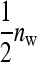 member of family k in the lth group to which family k contributes, and
member of family k in the lth group to which family k contributes, and 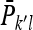 is the mean phenotypic value of the
is the mean phenotypic value of the 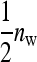 members of the other family in that group. Hence, l indexes the 1 through
members of the other family in that group. Hence, l indexes the 1 through 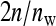 groups to which a family contributes, so that the number of records equals
groups to which a family contributes, so that the number of records equals 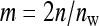 . From Equation B16, the full variance equals
. From Equation B16, the full variance equals
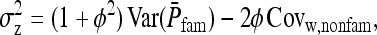 |
(B17a) |
and the between-family variance equals
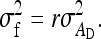 |
(B17b) |
Indirect genetic variance:
As shown above, given family k, the expected mean phenotype of the other family in group l equals 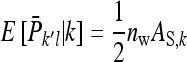 , so that
, so that 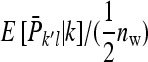 =
=  . Thus the record of interest for IGEs is
. Thus the record of interest for IGEs is
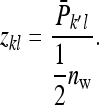 |
(B18) |
The full variance, therefore, equals
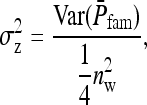 |
(B19a) |
and the between-family variance equals
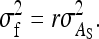 |
(B19b) |
Total heritable variance:
In any group, each of the 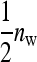 members of family k receives a contribution
members of family k receives a contribution 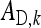 +
+ 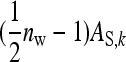 to its phenotype. Moreover, each of the
to its phenotype. Moreover, each of the 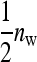 members of the other family, k′, receives a contribution
members of the other family, k′, receives a contribution 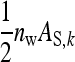 . Hence, given family k, the expected value of the sum of phenotypes of all nw members of group l equals
. Hence, given family k, the expected value of the sum of phenotypes of all nw members of group l equals 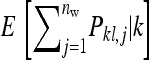 =
= 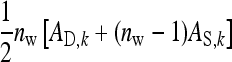 =
= 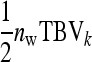 , where j denotes members of the lth group to which family k contributes. Thus the record of interest for the TBVs is
, where j denotes members of the lth group to which family k contributes. Thus the record of interest for the TBVs is
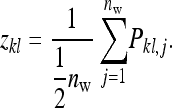 |
(B20) |
The full variance, therefore, equals
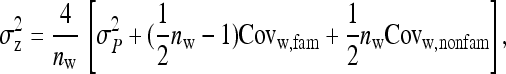 |
(B21a) |
and the between-family variance equals
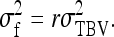 |
(B21b) |
APPENDIX C
Stochastic simulation and data analysis:
Simulated population structures are described in the main text. DGEs and IGEs were sampled from a bivariate normal distribution,
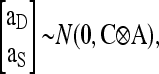 |
where aD and aS are vectors of DGEs and IGEs for all individuals, respectively,
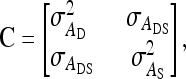 |
and A is the so-called numerator relationship matrix, containing coefficients of additive genetic relatedness between all individuals (Emik and Terril 1949; Henderson 1976). Elements Aij of A equaled r when i and j were from the same family, and zero otherwise.
Variance components were estimated from simulated data using restricted maximum likelihood (ReML) as implemented in ASReml (Gilmour et al. 2006), with the model (Arango et al. 2005; Muir 2005; Bijma et al. 2007b)
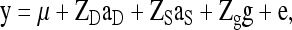 |
(C1) |
where y is a vector of phenotypes of all individuals, μ the overall mean, ZD an incidence matrix linking phenotypes of individuals to their own DGE, ZS an incidence matrix linking phenotypes of individuals to the IGEs of their group mates, and Zg an incidence matrix linking phenotypes of individuals to their group. The g is a vector of random group effects with 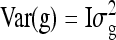 , and e is a vector of residuals with
, and e is a vector of residuals with 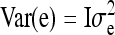 , I denoting an identity matrix. Results showed that variance components estimated using Equation C1 were unbiased. Fitting model C1 yields estimates of the genetic parameters of interest,
, I denoting an identity matrix. Results showed that variance components estimated using Equation C1 were unbiased. Fitting model C1 yields estimates of the genetic parameters of interest, 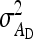 ,
, 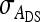 , and
, and 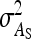 , and of the nongenetic parameters
, and of the nongenetic parameters 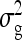 and
and 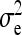 . Variance components for nongenetic direct and indirect effects,
. Variance components for nongenetic direct and indirect effects, 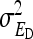 ,
, 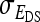 , and
, and 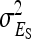 are not statistically identifiable, and therefore not fitted (see discussion).
are not statistically identifiable, and therefore not fitted (see discussion).
The g accounts for the covariance between group mates arising from nonheritable indirect effects, 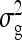 =
= 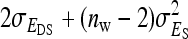 (Bergsma et al. 2008). If
(Bergsma et al. 2008). If 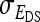 is negative and sufficiently large, the
is negative and sufficiently large, the 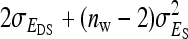 can be a negative value, so that the nongenetic covariance between group mates is negative, and fitting a random group effect is inappropriate. In those cases, therefore, the random group effect was omitted from the model, and residuals were allowed to be correlated within group, using
can be a negative value, so that the nongenetic covariance between group mates is negative, and fitting a random group effect is inappropriate. In those cases, therefore, the random group effect was omitted from the model, and residuals were allowed to be correlated within group, using 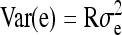 , where Rii = 1, Rij = ρ when i and j are in the same group, and Rij = 0 when i and j are in different groups. The ρ represents the correlation between residuals of group mates, which is estimated from the data (Bijma et al. 2007b). Both models are equivalent when ρ > 0 (Bergsma et al. 2008). When ρ > 0, we fitted random group effects rather than correlated residuals, because it was computationally less demanding and showed better convergence.
, where Rii = 1, Rij = ρ when i and j are in the same group, and Rij = 0 when i and j are in different groups. The ρ represents the correlation between residuals of group mates, which is estimated from the data (Bijma et al. 2007b). Both models are equivalent when ρ > 0 (Bergsma et al. 2008). When ρ > 0, we fitted random group effects rather than correlated residuals, because it was computationally less demanding and showed better convergence.
References
- Agrawal, A. F., E. D. Brodie, III and M. J. Wade, 2001. On indirect genetic effects in structured populations. Am. Nat. 158 308–323. [DOI] [PubMed] [Google Scholar]
- Arango, J., I. Misztal, S. Tsuruta, M. Cubertson and W. Herring, 2005. Estimation of variance components including competitive effects of Large White growing gilts. J. Anim. Sci. 83 1241–1246. [DOI] [PubMed] [Google Scholar]
- Bergsma, R., E. Kanis, E. F. Knol and P. Bijma, 2008. The contribution of social effects to heritable variation in finishing traits of domestic pigs (Sus scrofa). Genetics 178 1559–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijma, P., 2010. Multilevel selection 4: modeling the relationship of indirect genetic effects and group size. Genetics 186 1029–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijma, P., and M. J. Wade, 2008. The joint effects of kin, multilevel selection and indirect genetic effects on response to genetic selection. J. Evol. Biol. 21 1175–1188. [DOI] [PubMed] [Google Scholar]
- Bijma, P., W. M. Muir and J. A. M. van Arendonk, 2007. a Multilevel selection 1: quantitative genetics of inheritance and response to selection. Genetics 175 277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijma, P., W. M. Muir, E. D. Ellen, J. B. Wolf and J. A. M. van Arendonk, 2007. b Multilevel selection 2: estimating the genetic parameters determining inheritance and response to selection. Genetics 175 289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleakley, B. H., and E. D. Brodie, III, 2009. Indirect genetic effects influence antipredator behavior in guppies: estimates of the coefficient of interaction psi and the inheritance of reciprocity. Evolution 63 1796–1806. [DOI] [PubMed] [Google Scholar]
- Canario, L., N. Lundeheim and P. Bijma, 2010. Pig growth is affected by social genetic effects and social litter effects that depend on group size. Proceedings of the 9th World Congress on Genetics Applied to Livestock Production. German Society of Animal Science, Leipzig, Germany, p. 87.
- Cantet, R. J. C., and E. P. Cappa, 2008. On identifiability of (co)variance components in animal models with competition effects. J. Anim. Breed. Genet. 125 371–381. [DOI] [PubMed] [Google Scholar]
- Cappa, E. P., and R. J. C. Cantet, 2008. Direct and competition additive effects in tree breeding: Bayesian estimation from an individual mixed tree model. Silvae Genet. 57 45–57. [Google Scholar]
- Chen, C. Y., S. D. Kachman, R. K. Johnson, S. Newman and L. D. Van Vleck, 2008. Estimation of genetic parameters for average daily gain using models with competition effects. J. Anim. Sci. 86 2525–2530. [DOI] [PubMed] [Google Scholar]
- Chenoweth, S. F., H. D. Rundle and M. W. Blows, 2010. Experimental evidence for the evolution of indirect genetic effects: changes in the interaction effect coefficient, Ψ, due to sexual selection. Evolution 64 1849–1856. [DOI] [PubMed] [Google Scholar]
- Cheverud, J. M., 1984. Evolution by kin selection: a quantitative genetic model illustrated by maternal performance in mice. Evolution 38 766–777. [DOI] [PubMed] [Google Scholar]
- Cheverud, J. M., and A. J. Moore, 1994. Quantitative genetics and the role of the environment provided by relatives in the evolution of behavior, pp. 60–100 in Quantitative Genetic Studies of Behavioral Evolution, edited by C. R. B. Boake. University of Chicago Press, Chicago.
- Clutton-Brock, T. H., 2002. Breeding together: kin selection, reciprocity and mutualism in cooperative animal societies. Nature 373 209–216. [Google Scholar]
- Darwin, C., 1859. On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life, Ed.1. John Murray, London. [PMC free article] [PubMed]
- Dickerson, G. E., 1947. Composition of hog carcasses as influenced by heritable differences in rate and economy of gain. Iowa Agric. Exp. Stn. Res. Bull. 354 492–524. [Google Scholar]
- Ellen, E. D., J. Visscher, J. A. M. van Arendonk and P. Bijma, 2008. Survival of laying hens: genetic parameters for direct and associative effects in three purebred layer lines. Poultry Sci. 87 233–239. [DOI] [PubMed] [Google Scholar]
- Emik, L. O., and C. E. Terril, 1949. Systematic procedures for calculating inbreeding coefficients. J. Hered. 40 51–55. [DOI] [PubMed] [Google Scholar]
- Falconer, D. S., 1965. Maternal effects and selection response, pp. 763–774 in Genetics Today, Proceedings of the XIth International Congress on Genetics, Vol. 3, edited by S. J. Geerts. Pergamon, Oxford.
- Falconer, D. S., and T. F. C. Mackay, 1996. Introduction to Quantitative Genetics. Pearson Education, Harlow, UK.
- Frank, S. A., 1998. Foundations of Social Evolution. Princeton University Press, Princeton, NJ.
- Gilmour, A. R., B. J. Gogel, B. R. Cullis and R. Thompson, 2006. ASReml User Guide, Release 2.0, VSN International, Hemel Hempstead, UK.
- Griffing, B., 1967. Selection in reference to biological groups. I. Individual and group selection applied to populations of unordered groups. Aust. J. Biol. Sci. 20 127–142. [PubMed] [Google Scholar]
- Griffing, B., 1976. Selection in reference to biological groups. VI. Analysis of full sib groups. Genetics 82 723–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffing, B., 1981. a A theory of natural-selection incorporating interaction among individuals. 1. The modeling process. J. Theor. Biol. 89 635–658. [DOI] [PubMed] [Google Scholar]
- Griffing, B., 1981. b A theory of natural-selection incorporating interaction among individuals. 2. Use of related groups. J. Theor. Biol. 89 659–677. [DOI] [PubMed] [Google Scholar]
- Hadfield, J. D., and A. J. Wilson, 2007. Multilevel selection 3: modeling the effects of interacting individuals as a function of group size. Genetics 177 667–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, W. D., 1964. The genetical evolution of social behavior. J. Theor. Biol. 7 1–52. [DOI] [PubMed] [Google Scholar]
- Henderson, C. R., 1976. A simple method for the inverse of a numerator relationship matrix used in prediction of breeding values. Biometrics 32 69–83. [Google Scholar]
- Hsu, W. L., R. K. Johnson and L. D. VanVleck, 2010. Effect of pen mates on growth, backfat depth, and longissimus muscle area of swine. J. Anim. Sci. 88 895–902. [DOI] [PubMed] [Google Scholar]
- Keller, L., 1999. Levels of Selection in Evolution. Princeton University Press, Princeton, NJ.
- Kirkpatrick, M., and R. Lande, 1989. The evolution of maternal characters. Evolution 43 485–503. [DOI] [PubMed] [Google Scholar]
- Kruuk, L. E. B., 2004. Estimating genetic parameters in natural populations using the ‘animal model.’ Philos. Trans. R. Soc. Lond. B Biol. Sci. 359 873–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., and B. Walsh, 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA.
- McGlothlin, J. W., and E. D. Brodie, III, 2009. How to measure indirect genetic effects: the congruence of trait-based and variance-partitioning approaches. Evolution 63 1785–1795. [DOI] [PubMed] [Google Scholar]
- McGlothlin, J. W., A. J. Moore, J. B. Wolf and E. D. Brodie, III, 2010. Interacting phenotypes and the evolutionary process. III. Social evolution. Evolution 64 2558–2574. [DOI] [PubMed] [Google Scholar]
- Moore, A. J., E. D. Brodie, III and J. B. Wolf, 1997. Interacting phenotypes and the evolutionary process: I. Direct and indirect genetic effects of social interactions. Evolution 51 1352–1362. [DOI] [PubMed] [Google Scholar]
- Mousseau, T. A., and C. W. Fox, eds., 1998. Maternal effects as adaptations. Oxford University Press, New York.
- Muir, W. M., 1996. Group selection for adaptation to multiple-hen cages: selection program and direct responses. Poult. Sci. 75 447–458. [DOI] [PubMed] [Google Scholar]
- Muir, W. M., 2005. Incorporation of competitive effects in forest tree or animal breeding programs. Genetics 170 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir, W. M., and A. Schinckel, 2002. Incorporation of competitive effects in breeding programs to improve productivity and animal well being. Proceedings of the 7th World Congress on Genetics Applied to Livestock Production, Montpellier, France, communication no. 14–07.
- Muir, W. M., P. Bijma, C. Y. Chen, I. Misztal and M. J. Wade, 2010. Advances in selection theory including experimental demonstrations: the theory of selection with social competition. Proceedings of the 9th World Congress on Genetics Applied to Livestock Production. German Society of Animal Science, Leipzig, Germany, p. 141.
- Mutic, J. J., and J. B. Wolf, 2007. Indirect genetic effects from ecological interactions in Arabidopsis thaliana. Mol. Ecol. 16 2371–2381. [DOI] [PubMed] [Google Scholar]
- Patterson, H. D., and R. Thompson, 1971. Recovery of interblock information when block sizes are unequal. Biometrika 58 545–554. [Google Scholar]
- Petfield, D., S. F. Chenoweth, H. D. Rundle and M. W. Blows, 2005. Genetic variance in female condition predicts indirect genetic variance in male sexual display traits. Proc. Natl. Acad. Sci. USA 102 6045–6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, A., 1959. Experimental design in the evaluation of genetic parameters. Biometrics 15 219–226. [Google Scholar]
- Stuart, A., and J. K. Ord, 1994. Kendall's Advanced Theory of Statistics, Vol. 1, Distribution Theory. Hodder Education, London.
- Stuart, A., J. K. Ord and S. Arnold, 1999. Kendall's Advanced Theory of Statistics, Vol. 2A, Classical Inference and the Linear Model, Ed.6. Arnold, London.
- Van Vleck, L. D., and J. P. Cassady, 2004. Modification of MTDFREML to estimate variance due to genetic competition effects. J. Anim. Sci. 82(Suppl. 2) 38 (Abstr.). [Google Scholar]
- Van Vleck, L. D., and J. P. Cassady, 2005. Unexpected estimates of variance components with a true model containing genetic competition effects. J. Anim. Sci. 83 68–74. [PubMed] [Google Scholar]
- Willham, R. L., 1963. The covariance between relatives for characters composed of components contributed by related individuals. Biometrics 19 18–27. [Google Scholar]
- Wilson, A. J., U. Gelin, M-C. Perron and D. Réale, 2009. Indirect genetic effects and the evolution of aggression in a vertebrate system. Proc. R. Soc. B 276 533–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, E. O., 1975. Sociobiology: The New Synthesis. Belknap Press of Harvard University Press, Cambridge, MA.
- Wolf, J. B., E. D. Brodie, III, A. J. Cheverud, A. J. Moore and M. J. Wade, 1998. Evolutionary consequences of indirect genetic effects. Trends Ecol. Evol. 13 64–69. [DOI] [PubMed] [Google Scholar]