Abstract
Bacteria are social organisms that display distinct behaviors/phenotypes when present in groups. These behaviors include the abilities to construct antibiotic-resistant sessile biofilm communities and to communicate with small signaling molecules (quorum sensing [QS]). Our understanding of biofilms and QS arises primarily from in vitro studies of bacterial communities containing large numbers of cells, often greater than 108 bacteria; however, in nature, bacteria often reside in dense clusters (aggregates) consisting of significantly fewer cells. Indeed, bacterial clusters containing 101 to 105 cells are important for transmission of many bacterial pathogens. Here, we describe a versatile strategy for conducting mechanistic studies to interrogate the molecular processes controlling antibiotic resistance and QS-mediated virulence factor production in high-density bacterial clusters. This strategy involves enclosing a single bacterium within three-dimensional picoliter-scale microcavities (referred to as bacterial “lobster traps”) defined by walls that are permeable to nutrients, waste products, and other bioactive small molecules. Within these traps, bacteria divide normally into extremely dense (1012 cells/ml) clonal populations with final population sizes similar to that observed in naturally occurring bacterial clusters. Using these traps, we provide strong evidence that within low-cell-number/high-density bacterial clusters, QS is modulated not only by bacterial density but also by population size and flow rate of the surrounding medium. We also demonstrate that antibiotic resistance develops as cell density increases, with as few as ~150 confined bacteria exhibiting an antibiotic-resistant phenotype similar to biofilm bacteria. Together, these findings provide key insights into clinically relevant phenotypes in low-cell-number/high-density bacterial populations.
IMPORTANCE
Prokaryotes are social organisms capable of coordinated group behaviors, including the abilities to construct antibiotic-resistant biofilms and to communicate with small signaling molecules (quorum sensing [QS]). While there has been significant effort devoted to understanding biofilm formation and QS, few studies have examined these processes in high-density/low-cell-number populations. Such studies have clinical significance, as many infections are initiated by small bacterial populations (<105) that are organized into dense clusters. Here, we describe a technology for studying such bacterial populations in picoliter-sized porous cavities (referred to as bacterial “lobster traps”) capable of capturing a single bacterium and tracking growth and behavior in real time. We provide evidence that small changes in the size of the bacterial cluster as well as flow rate of the surrounding medium modulate QS in Pseudomonas aeruginosa. We also demonstrate that as few as ~150 confined bacteria are needed to exhibit an antibiotic-resistant phenotype similar to biofilm bacteria.
INTRODUCTION
Bacteria engage in numerous social behaviors, including formation of antibiotic-resistant sessile biofilm communities and coordination of group activities via quorum sensing (QS), a process in which gene transcription is controlled within a population by small signaling molecules. Discovery of these social behaviors has led to a renaissance in bacteriology, as “sociomicrobiology” has become one of the most studied themes in microbiology over the last 20 years (1). Just as studies of multicellular organisms depend on well-defined, in vitro models containing small numbers of cells, advancing the understanding of bacterial social behaviors requires observation and manipulation of small, structured bacterial populations. This is especially true because bacterial group behaviors frequently are localized to aggregates of only a few thousand cells (2–4). Moreover, clusters containing 101 to 105 cells are important clinically for seeding many infections, with a single aggregate often containing an infective dose of a bacterial pathogen (5–12). Indeed, it has been proposed that these clusters are the primary means of transmission of many pathogens, including Pseudomonas aeruginosa, Staphylococcus aureus, and Vibrio cholerae (5, 6, 8, 11–18).
Attempts to study small numbers of bacteria within ultralow-volume containers (19–21) often produce conditions not conducive to cell growth, a basic hallmark of physiologic relevance. Although some techniques for trapping bacteria have allowed cells to double at rates similar to those of batch cultures (22–25), these approaches have not offered capabilities for organizing cells in three dimensions, commonly do not provide a means to precisely control mass transport through the cell population, and in some cases rely on periodic exchange of cells between the enclosed population and its surrounding medium.
In this report, we describe a versatile strategy for capturing individual bacteria within three-dimensional (3D), picoliter-scale microcavities defined by permeable, photo-cross-linked-protein walls. Unlike materials used in conventional microfabrication, the protein-based barriers used here support efficient transfer of nutrients, waste products, and other bioactive small molecules, enabling bacteria to grow into small clonal populations of tunable size and density that can be phenotypically evaluated in real time. The power of this approach to address diverse problems in sociomicrobiology is demonstrated by examining both QS and antibiotic resistance in the opportunistic pathogen P. aeruginosa. Here, we provide strong empirical evidence that QS depends not only on population density but also on population size and the convective rate of solution surrounding a microcolony. We also demonstrate that, surprisingly, clusters containing as few as 150 confined bacterial cells can develop antibiotic resistance for reasons other than mass transport rates through the microcolonies.
RESULTS AND DISCUSSION
Fabrication of dynamic picoliter-sized bacterial “lobster traps.”
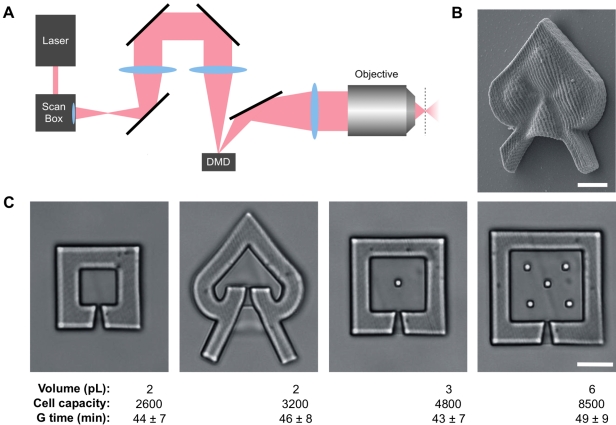
To study the social behavior of small bacterial clusters, we developed a means to sequester desired numbers of cells in porous traps of desired size and shape. In our approach, traps for capturing single bacteria were fabricated from the protein bovine serum albumin (BSA) using a dynamic mask-based multiphoton lithography (MPL) technique (Fig. 1A) capable of creating arbitrary 3D architectures. In protein MPL, laser-initiated covalent cross-linking of amino acid side chains is limited to a 3D voxel (<1 µm3) that is raster scanned in a series of stacked horizontal planes to produce 3D microstructures (Fig. 1B) (26–30). By placing an electronic, reflective photomask in a plane conjugate to the fabrication plane, the scanning laser beam can be modulated to produce cross sections for virtually any desired structure. Notably, it is possible to produce photo-cross-linked BSA structures that have a mechanical stiffness similar to that of some conventional soft lithographic materials (e.g., polydimethylsiloxane [PDMS]) (31).
FIG 1 .
Construction of bacterial lobster traps. (A) Schematic showing the optical setup of the dynamic mask-based multiphoton lithography technique. An electronic mask is placed in a plane conjugate to the fabrication plane (i.e., having a one-to-one spatial correspondence), with modulation of laser intensity to create a protein microstructure representing the negative of the original mask. After each horizontal scan, the focal volume is stepped further into the fabrication solution to produce a 3D structure in a layer-by-layer fashion. The broken line represents the fabrication plane. DMD, digital micromirror device. (B) Scanning electron microscopy (SEM) image of a trap constructed via cross-linking of bovine serum albumin (BSA). Bar, 5 µm. (C) Differential interference contrast (DIC) micrographs of traps used in this study with basic figures of merit. The objects within the square traps (right two panels) are pillars that support roofs, which are difficult to observe in these images because of their high degree of transparency. Bar, 10 µm. Volume, cell capacity, and generation time (G time, means ± standard deviations), were calculated as described in Materials and Methods.
Two microcavity geometries (square and heart-shaped) were constructed; these microcavities had internal volumes ranging from ~2 to 6 picoliters (Fig. 1C), with funneled entry pores to promote bacterial entry while limiting the exit rate. Fabrication of the bacterial traps [see Movie S1 in the supplemental material] was performed on the cover glass of a single-pass flow cell, allowing bacteria within traps to be studied under various flow velocities. P. aeruginosa was introduced into a channel, and flow was halted to allow motile cells to swim into the lumens of the traps. After brief loading, most traps contained zero or one bacterium, meaning that after growth, filled microcavities generally contained clonal populations. Care was taken to limit the number of bacteria attached outside the traps (see Materials and Methods), ensuring that high densities of cells existed only within the microcavities.
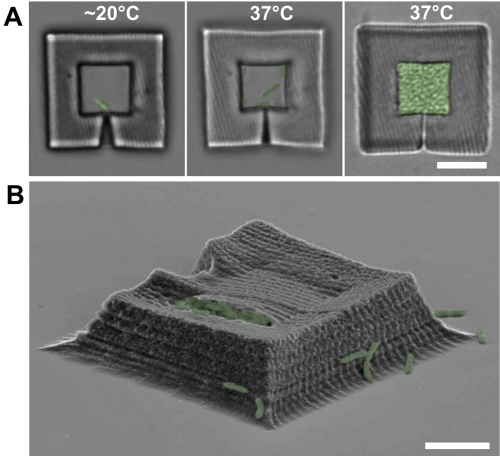
In earlier studies, we described the use of protein-based microcavities of similar geometry to form small aggregates of Escherichia coli (28). However, it was not possible to eliminate escape of cells after their entry and growth within cavities or to modulate solution flow rates around the cavity, limitations that prevented the application of this approach to controlled studies on QS and antibiotic resistance. In a critical extension to this technology, we discovered that BSA matrixes formed using MPL undergo irreversible expansion when heated (i.e., they do not contract if cooled after the temperature is raised), an attribute that enables narrow entry pores to be closed on command. In the current studies, a microcavity could be fabricated, loaded with a bacterium under ambient conditions (18 to 22°C), and then subjected to heated (37°C) medium to pinch off entry pores as the trap walls expanded. In this manner, cells could be sequestered in microcavities of definable size and shape (Fig. 2A). This controlled, dynamic manipulation of porous corrals for single cells could not have been achieved using virtually any other method.
FIG 2 .
Capturing a bacterium in a trap and monitoring growth. (A) DIC micrographs of bacterial traps chronicling the capture and growth of an individual P. aeruginosa bacterium. A cell swims into the lumen of a trap (left), the entry pore is constricted by increasing the temperature to 37°C (middle), and the bacteria divide normally to fill the trap with a picoliter-sized colony of P. aeruginosa (right). Bar, 10 µm. (B) SEM image of a trap filled with P. aeruginosa. The tear in the roof occurred during SEM preparation. The bacteria in all images are shown in green (false color). Bar, 5 µm.
Bacterial growth and transport properties within protein traps.
The use of photo-cross-linked BSA was critical to supporting the requisite exchange of nutrients and waste products that could support growth at normal (i.e., batch) rates. Once cells were captured within traps (see Movie S2 in the supplemental material), flow was initiated, and bacterial growth was assessed via microscopy and direct cell counting. P. aeruginosa divided and filled traps in 6 to 10 h (Fig. 2A and B). The mean generation time of P. aeruginosa within traps (46 ± 7 min) was indistinguishable from that observed in laboratory flasks (40 min) (32) and in the rat peritoneum (50 min) (32) and similar to that in the human lung (~100 min) (33). Moreover, P. aeruginosa generation time was not significantly affected by the size or geometry of the trap (Fig. 1C) or by the flow rate through the channel, which suggests that the enclosure walls were porous to nutrients and waste products.
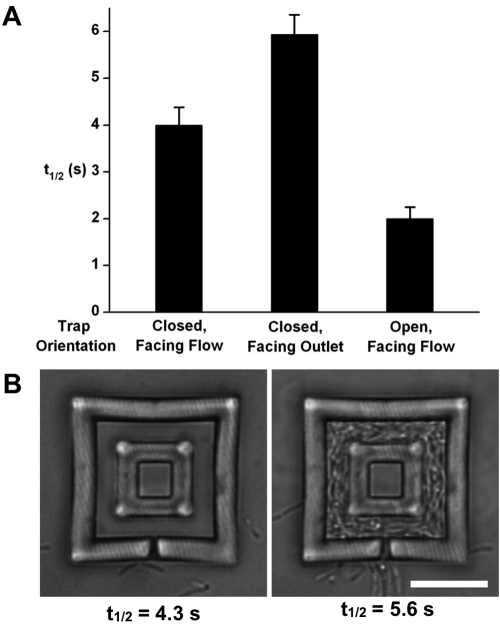
To directly assess whether bioactive small molecules cross the porous trap walls, a fluorescent derivative of the antibiotic gentamicin (fluorescein-gentamicin conjugate [fluorescein-gentamicin]) was perfused through the flow channel. Fluorescein-gentamicin readily diffused into the cavities of the traps, reaching equilibrium within seconds (Fig. 3A), a result that verifies that traps are porous to biologically relevant small molecules. To ensure that dense populations of bacteria within traps do not dramatically alter mass transfer rates, we also measured diffusion of fluorescein-gentamicin through a trap packed with P. aeruginosa into an unoccupied, inner microcavity (Fig. 3B). Here, the inner cavity was bordered on all sides (other than its glass floor) by a dense shell of bacteria, a nested microscopic geometry that could not have been fabricated using conventional lithographic methods. Fluorescein-gentamicin entered the inner cavity rapidly, reaching half the equilibrium concentration (t1/2) in 5.6 s, a period slightly longer than that required to reach the inner cavity in the absence of cells (4.3 s). These results demonstrate that, although clustered cells do not pose an impenetrable barrier to the diffusion of small molecules, they impede molecular transport to a small degree.
FIG 3 .
Traps are permeable to small molecules. (A) Fluorescein-gentamicin conjugate (fluorescein-gentamicin) rapidly diffuses into unfilled traps. The orientation describes the direction that the sealed entry pore faces. t1/2 is the time required for the concentration within a trap to reach half of the equilibrium concentration. Values are means plus standard deviations (error bars) (n ≥ 8). (B) Fluorescein-gentamicin diffuses through an unoccupied trap into a sealed inner microcavity of a two-layered structure (left). Filling the trap with a dense aggregate of cells slows diffusion by ~20% (right). Standard deviations are 0.3 s (n = 5) and 0.6 s (n = 12) for the left and right structures, respectively. Sealed entry pores of these structures face flow. Bar, 10 µm.
Population size and external flow rate modulate QS in P. aeruginosa cell clusters.
Despite the biological importance of QS in hundreds of bacterial species, there is considerable controversy regarding the environmental factors monitored during QS. The most common QS model (“basic QS”) asserts that population density is the sole determinant of extracellular-signal concentrations and, therefore, is exclusively responsible for coordinating group activities. However, two alternative models, diffusion sensing and efficiency sensing, propose that the mass transfer properties surrounding a cell impact group activities (34, 35), a concept implicit in some QS models (36, 37). The diffusion-sensing model suggests that QS relies on mass transfer alone, essentially independent of cell density (35). Recent evolutionary evidence, however, suggests that cell density is at least one component affecting QS behaviors (38, 39). The efficiency-sensing model proposes a melding of basic QS and diffusion-sensing hypotheses (34), positing that cells produce signaling molecules not only to monitor cell density but also to spatially assess population size and mass transfer rates through their environment.
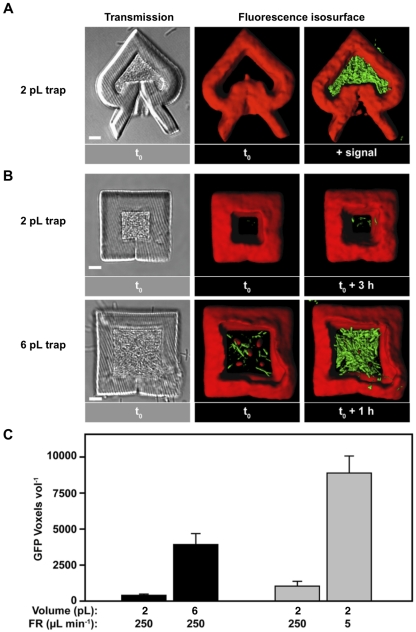
To differentiate between these models, microcavities were loaded with a P. aeruginosa strain containing the gene encoding green fluorescent protein (GFP) under the control of a QS-responsive promoter. This strain displays green fluorescence when the signal 3-oxododecanoyl homoserine lactone (3OC12-HSL) reaches a critical concentration, thus allowing GFP to be used as a marker to assess whether a P. aeruginosa population is communicating via QS. When this bacterium was grown in a heart-shaped, 2-picoliter trap positioned within a high-flow-rate channel (250 µl min−1), essentially no green fluorescence was observed via confocal microscopy, even when the traps were filled to near capacity. Importantly, however, introduction of 3OC12-HSL into the flow cell yielded high GFP levels within an hour (Fig. 4A), indicating that although bacterially generated signal concentrations do not reach inducing levels under the high-flow conditions initially tested, bacteria in 2-pl traps are metabolically active and capable of responding to QS signals.
FIG 4 .
P. aeruginosa quorum sensing (QS) gene expression is dependent on population size and flow rate. (A) P. aeruginosa carrying an rsaL::gfp fusion is captured inside a heart-shaped, 2-picoliter trap and exposed to an external flow rate of 250 µl min−1. After filling traps to near capacity (left), P. aeruginosa exhibits no detectable green fluorescent protein (GFP) expression (middle). High GFP expression is observed in less than 1 h after exogenous addition of the rsaL-inducing signal 3-oxododecanoyl homoserine lactone (3OC12-HSL) (+ signal) (right). Bar, 5 µm. (B) P. aeruginosa carrying an rsaL::gfp fusion is captured inside 2-pl and 6-pl traps and exposed to a flow rate of 250 µl min−1. When filled to near capacity (left), little expression is observed in 2-pl traps, while significant GFP expression is observed in 6-pl traps. Even after an additional incubation period of up to several hours (right), the 2-pl trap yields lower expression than the 6-pl trap. Bars, 5 µm. (C) Quantification of GFP expression inside traps of different sizes at a constant flow rate (black bars) and inside 2-pl traps at different flow rates (gray bars). The number of GFP-positive voxels inside the trap per unit volume is shown on the y axis. The flow rate (FR) is shown below the bars. The values are means plus standard errors of the means (error bars) (n ≥ 3).
Although cells within heart-shaped, 2-pl traps did not display substantial QS at flow rates of 250 µl min−1, we hypothesized that increasing the population size would increase steady-state concentrations of 3OC12-HSL signal within a colony to levels necessary to induce QS. To test this hypothesis, QS was assessed in P. aeruginosa populations containing different numbers of bacteria while holding cell density and flow rate constant. Increasing the trap size from 2 to 6 pl—and, thus, the nominal population size from 2,600 to 8,500 cells—resulted in significant GFP expression in large traps, while GFP was again nearly undetectable in smaller traps (Fig. 4B and C). These data demonstrate that population size is a critical parameter influencing QS, a finding consistent with reaction diffusion theory and the efficiency-sensing model of QS (40, 41).
Our unique abilities to manipulate small clonal populations of bacteria in contact with a dynamic microenvironment allowed us to evaluate the role in QS of a second, fundamental efficiency-sensing parameter, external flow rate. Were flow rate a critical factor influencing QS, one would predict that QS within trap populations should be influenced by the flow rate when cell density and population size are held constant. Specifically, slower flow velocities should reduce mass transfer rates of QS signals from populations within traps into the extraluminal volume, leading to enhanced QS-dependent gene expression. Supporting this prediction, reduction of the flow rate from 250 to 5 µl min−1 resulted in an ~6-fold increase in QS-dependent GFP expression for populations within 2-pl traps (Fig. 4C). Collectively, these data provide empirical evidence that three fundamental efficiency-sensing parameters (population size, population density, and mass transfer) impact QS gene expression. From a general perspective, it may seem intuitive that population size and external flow rate both have the capacity to modulate quorum sensing; however, methodologies have not previously existed for systematically and independently examining these parameters in small, defined bacterial populations.
Antibiotic resistance in small P. aeruginosa populations.
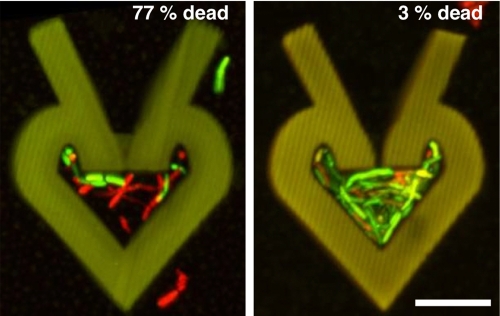
A second, critical problem in sociomicrobiology was addressed using this technology, the onset of biofilm-like antibiotic resistance in small aggregate populations. The porous nature of traps allowed us to study how low-number bacterial populations respond to antibiotic exposure, an attribute of clinical importance. To examine whether cell density impacts antibiotic resistance in low-cell-number populations, P. aeruginosa was grown to high (~225 cells pl−1) and low (~20 cells pl−1) cell densities and treated for 2 h with the gentamicin antibiotic at the MIC (see Materials and Methods). At these densities, traps were not filled to capacity, and bacteria were actively growing. The cells within high-density populations containing as few as ~150 cells displayed decreased susceptibility to gentamicin (3% dead) compared to those growing at low density (77% dead) (Fig. 5), indicating the potential for extremely small bacterial communities to develop antibiotic resistance profiles similar to those of biofilm bacteria. Because the diffusion of fluorescein-gentamicin through dense P. aeruginosa populations is rapid (see above [Fig. 3B]), this decreased susceptibility to antibiotic is caused by phenotypic changes that take place at higher cell densities rather than by limits on mass transfer. This unexpected finding raises important questions regarding the potential onset of biofilm-like properties as small bacterial clusters seed new sites of infections.
FIG 5 .
High-density populations display increased antibiotic resistance. 3D confocal reconstructions of traps dosed for 2 h with gentamicin at the MIC show that only 3% (±2% [standard deviation]) of cells growing at a higher density (right) are dead compared to 77% (±11% [standard deviation]) for those growing at a lower density (left). Dead and live cells appear red and green, respectively, and n ≥ 16 for each dosing condition. Bar, 10 µm.
Lobster traps provide new capabilities for probing bacterial group behaviors.
These studies describe a new strategy for probing bacterial social behaviors. Idiosyncratic environments are developed for bacteria in which growth conditions can be dynamically controlled and individual environmental perturbations can be introduced down to the single-cell level. As a consequence, phenotypic and genotypic responses to intrinsic and extrinsic stimuli can be monitored for small bacterial ensembles at high densities. Most models of bacterial group behavior and evolution ultimately rely on understanding the interactions between bacteria in small populations under physiologically relevant conditions, and this unique approach provides the means to test these models in detail. The traps and additional structures for directing and modulating growth will have clear utility for examining other challenging problems, such as polymicrobial interactions. Importantly, such studies are not limited to motile bacteria, as nonmotile cells can be introduced into traps using flow-based methodology and optical trapping (see Fig. S1 in the supplemental material). Finally, by understanding the organization of bacterial communities and their underlying interactions and behaviors from the moment of microcolony formation, it should prove possible to develop new interventions for pathogenic microbes, such as the development of surfaces that have intrinsic antimicrobial capabilities.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
P. aeruginosa PAO1 carrying the rsaL-gfp reporter plasmid pGJB5 was used in all QS studies. P. aeruginosa PAO1 constitutively expressing gfp from pMRP9-1 was used in all antibiotic resistance experiments (42). Planktonic cultures were grown aerobically at 37°C in tryptic soy broth (TSB) and incubated with antibiotics for plasmid selection (gentamicin [100 µg ml−1] for QS; carbenicillin [300 µg ml−1] for antibiotic resistance) or maintenance (gentamicin [10 µg ml−1] for QS). One-third-strength TSB was used for all flow cell experiments.
Microstructure fabrication.
Photo-cross-linked-protein traps were fabricated using a dynamic mask-based multiphoton lithography technique described in detail elsewhere (27, 29). Briefly, the output of a mode-locked titanium-sapphire (Ti-S) laser (Tsunami; Spectra-Physics) operating at 740 nm was raster scanned using a confocal scan box (MRC600; Bio-Rad) and aligned on a digital micromirror device (DMD) (Texas Instruments) (0.55-in. supervideo graphics array [SVGA]), which served as an electronic reflectance mask. The beam was adjusted to overfill the back aperture of an oil immersion objective (Zeiss 100× Fluar, 1.3 numerical aperture [NA]) situated on a Zeiss Axiovert inverted microscope system. The power of the laser measured at the back aperture of the objective ranged from 50 to 60 mW. Structures were fabricated using 400 mg ml−1 BSA (Equitech-Bio) and 8.5 mM rose bengal (Sigma) as a photosensitizer in 20 mM HEPES–0.1 M NaCl buffer (pH 7.4) on no. 1 coverglass (VWR) within a single-pass flow cell system (42). Vertical steps of 0.5 µm were made using a motorized focus driver (H122 focus motor; Prior Scientific) between sequential scanned horizontal planes. The walls and roofs of all structures were nominally 4.5 µm and 1.5 µm thick, respectively, although the actual thickness of roofs was up to 2-fold greater due to the oblong shape of the fabrication voxel.
Two-layered microcavities were designed to have a closed inner cavity within a larger shell cavity that could be filled with bacteria around the sides and top of the inner chamber (Fig. 3B). Here, a nominal spacing of 4.5 µm was created between the walls of the inner and outer microcavities and between the roof of the inner microcavity and the ceiling of the outer microcavity. Support pillars—2-µm-thick square posts—were fabricated between each of the four corners of the inner microcavity roof and the outer microcavity ceiling.
Flow cell system, inoculation, and growth conditions.
A single-pass continuous culture system was prepared as previously described with some modifications (42). An inoculation inlet was made 25 mm from the flow cell inlet using a T-connector and a Luer-Lok connector and cap. Luer-Lok connectors were also added upstream of the bubble trap to allow the system to be separated into two parts and remain sterile. For introduction of P. aeruginosa into the flow cell channels, exponential-phase bacteria were diluted to an optical density at 600 nm (OD600) of 0.01 in TSB and vortexed for 2 min, and 2 ml of the bacterial solution was added to a petri dish for 2 min. This process minimized the number of highly adherent bacteria inoculated into the flow cell channel and greatly reduced the number of bacteria attached to the coverslip outside the traps. The bacterial solution (1.5 ml), backfilled into a Luer-Lok syringe, was used to inoculate a flow cell channel containing ~40 to 60 traps.
Flow cells containing P. aeruginosa were incubated under static conditions at the ambient temperature (~18 to 22°C) for ~15 min to allow entry of P. aeruginosa into the traps via swimming through an opening ~1 µm in diameter. Then the flow rate was increased to ~6 ml min−1 for 1 min to remove bacteria that may have attached to the channel walls. The temperature within the flow cell channel was raised by filling the channel with one-third-strength TSB at 37°C using a peristaltic pump (Watson-Marlow). The medium was heated before it entered the flow cell channel by one of two methods. (i) Silicon tubing was laid on Briskheat flexible electric heating tape (Barnstead/Thermolyne) powered by a variable transformer (Variac) and calibrated to maintain a temperature of 37°C in the fabrication region of the flow cell. (ii) The temperature was raised to 37°C by placing the flow cell system inside a microscope incubator (In Vivo Scientific). After 5 min of heated flow, the flow cell was manually agitated for 30 s to reduce cell attachment to the cover glass, and flow was continued. The number of cells in each structure was monitored over time during the initial 5 h of growth to calculate the generation time.
Electron microscopy.
Samples were prepared for scanning electron microscopy (SEM) by immersion (15 min per solution) in the following series of solutions: 5% glutaraldehyde (Ted Pella, Redding, CA), HEPES-NaCl buffer (pH 7.4), HEPES-NaCl buffer (pH 7.4), deionized water, 50% ethanol (EtOH), 100% EtOH, 50% EtOH–50% methanol (MeOH), 100% MeOH, and 100% MeOH. After air drying overnight, the samples were sputter coated with Pd/Pt to a nominal thickness of 12 nm and imaged using a Supra 40VP electron microscope (Zeiss).
Diffusion measurements.
The fluorescein-gentamicin conjugate (fluorescein-gentamicin) was prepared by reacting a 5-fold molar excess of gentamicin sulfate with 6-(fluorescein-5-carboxamido) hexanoic acid, succinimidyl ester (AnaSpec) in a 0.2 M sodium bicarbonate buffer (pH 9.0) for 2 h. The fluorescein-gentamicin conjugate was confirmed by mass spectrometry.
Fluorescein-gentamicin was added to the flow stream via the inoculation inlet T-connector, and medium and the conjugate were delivered at a flow rate of 500 µl min−1. Diffusion of fluorescein-gentamicin into cavities was measured via a series of two-photon point measurements using a titanium-sapphire (Ti-S) beam operated at 780 nm. The Ti-S beam was split into two probe beams of equal power (10 mW each) at the back aperture of the objective. One probe was positioned in front of the structure and one inside an empty cavity, and the two beams were used to simultaneously monitor fluorescence at the two positions. Two-photon-excited fluorescence was collected using the 100× objective, passed though a dichroic mirror and a BG-39 filter (Chroma, Rockingham, VT) and detected using a 12-bit 1,392- by 1,040-element charge-coupled-device (CCD) camera (CoolSnap HQ; Photometrics). Fluorescence time series were acquired using Metamorph software (Universal Imaging, Sunnyvale, CA), and signal was analyzed using ImageJ. A time course of normalized fluorescence for each position was created by subtracting the background level and then calculating the ratio of the intensity at a given time point to the maximum intensity reached at the position.
Antibiotic susceptibility experiments.
The cells were grown within traps at 37°C and 250 µl min−1 flow to high density (5.5 h growth) and low density (3.5 h). The cells in traps were then dosed with the MIC of gentamicin (1.6 µg ml−1) diluted into the growth medium under flow conditions for 2 h. Gentamicin susceptibility was determined by staining with a Live/Dead BacLight bacterial viability staining kit (Molecular Probes). The stain solution of SYTO 9 (green fluorescence indicates live cells) and propidium iodide (red emission indicates dead cells) was prepared in a 5-ml solution of 50% TSB and 50% deionized water. The stain solution was injected into a flow cell containing bacterial traps and incubated for 30 min with the flow arrested. Live and dead cells were first visualized using wide-field fluorescence in “green” and “red” channels followed by confocal microscopy. For high-density experiments, the number of red cells was counted from the wide-field fluorescence images for each trap (and verified in a subset of experiments using confocal fluorescence images) and then divided by the total number of cells (calculated to be in a structure at the time of the antibiotic dose based on the generation time) to obtain the dead percentage. For low-density experiments, a 3D confocal reconstruction of each structure was used to count the number of red and green cells and the ratio of the two was used as the dead percentage. The traps in high-density studies contained 220 ± 100 cells pl−1 (290 ± 140 cells trap−1 [mean ± standard deviation], calculated based on the generation time for individual structures at the beginning of the 2-h dose), and the traps in low-density experiments contained 21 ± 13 cells pl−1 (27 ± 17 cells trap−1, directly counted after viability stain).
Microscopy and analysis of QS reporter fluorescence in traps.
For QS experiments, scanning confocal laser microscopy (see below) images were acquired under low-flow (5 µl min−1) and high-flow (250 µl min−1) conditions at the ambient temperature 6 to 12 h postinoculation. For low-flow conditions, the flow rate was maintained at 250 µl min−1 for 5 h postinoculation at which time it was reduced to 5 µl min−1. In all studies, the traps were not filled to capacity when the flow rate was reduced. Approximately 6 hours postinoculation, the flow cell was removed from the heat source, and the flow cell system was transported to the confocal microscope. During the 10 to 15 min required to transport specimens, the flow cell system was not flowing.
Differential interference contrast (DIC) images of the traps were obtained using an Axiovert microscope equipped with a 100×, 1.3-NA Fluar objective (Carl Zeiss, Germany) and a 12-bit, 1,392- by 1,040-element CCD camera (CoolSnap HQ; Photometrics). Fluorescence and transmission confocal images were acquired as a z-series using an SP2 acousto-optical beam splitter (AOBS) confocal microscope equipped with a 63×, 1.4-NA objective and argon ion and orange HeNe lasers (Leica Microsystems, Germany). Green-channel images were acquired using 488-nm excitation and emission centered at 515 nm (35-nm slit width), while red-channel images were collected using 594-nm excitation and emission centered at 625 nm (44-nm slit width). The height of each trap and the total voxels of GFP-positive cells were determined using Imaris 5.7.0 software (Bitplane AG, Switzerland). For the data in Fig. 4C, the traps were analyzed when the roof was distended 4 to 6 µm to ensure that the traps were filled with bacteria. For the values shown in Fig. 1C and the data in Fig. 4B and C, nominal volumes were calculated from confocal scans of filled traps. Approximate cell capacities were determined by dividing this trap volume by the nominal volume of P. aeruginosa (assumed to be a cylinder with the dimensions 0.75 µm by 1.5 µm). The generation time (given as mean ± standard error of the mean; n ≥ 9 for each trap) was calculated by directly counting cells within the traps for approximately six generations.
Within Imaris, the IsoSurface mode of the Surpass module was used to generate isosurfaces from red-channel stacks. The threshold for the red-channel isosurfaces was determined manually. Because traps emit in both the red and green channels, red isosurfaces were used to eliminate trap fluorescence from the green-channel stacks, generating a modified green channel containing GFP-positive cells (and not the trap). The total green voxels from GFP-positive cells within each trap were determined by generating an isosurface image of the modified green channel. Background green fluorescence within the trap cavity was determined for traps filled to near capacity with non-GFP-positive bacteria. Background in the green channel was very low in the trap cavity (~2 digitization units on an 8-bit scale), allowing values greater than 4 digitization units to be confidently considered GFP positive. All images shown in Fig. 4B were included in the data shown in Fig. 4C. The 2-pl trap was selected because it illustrates that some of the smaller boxes can develop visible GFP expression at time points substantially after time zero (t0). GFP expression in the displayed 6-pl trap was in the upper range of those observed.
ACKNOWLEDGMENTS
We gratefully acknowledge the financial support of the National Institutes of Health (grant 5R01AI075068 to M.W. and grant 5R03AI081216 to J.B.S.); the Welch Foundation (grant F-1331 to J.B.S.); and the Norman Hackerman Advanced Research Program (grant 003658-0273-2007 to J.B.S.). M.W. is a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Disease. J.B.S. and M.W. are Fellows of the Institute for Cell and Molecular Biology.
We thank Eric Ritschdorff for assistance with diffusion measurements, members of the Whiteley lab for critical reading of the manuscript, Matthew Ramsey for help with microscopy, and Phil Stewart for discussions regarding reaction diffusion theory.
Footnotes
Citation Connell, J. L., A. K. Wessel, M. R. Parsek, A. D. Ellington, M. Whiteley, et al. 2010. Probing prokaryotic social behaviors with bacterial “lobster traps”. mBio 1(4):e00202-10. doi:10.1128/mBio.00202-10.
SUPPLEMENTAL MATERIAL
REFERENCES
- 1. Parsek M. R., Greenberg E. P. 2005. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 13:27–33 [DOI] [PubMed] [Google Scholar]
- 2. De Kievit T. R., Gillis R., Marx S., Brown C., Iglewski B. H. 2001. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl. Environ. Microbiol. 67:1865–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Velicer G. J., Vos M. 2009. Sociobiology of the myxobacteria. Annu. Rev. Microbiol. 63:599–623 [DOI] [PubMed] [Google Scholar]
- 4. Yarwood J. M., Bartels D. J., Volper E. M., Greenberg E. P. 2004. Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 186:1838–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Faruque S. M., Biswas K., Udden S. M., Ahmad Q. S., Sack D. A., Nair G. B., Mekalanos J. J. 2006. Transmissibility of cholera: in vivo-formed biofilms and their relationship to infectivity and persistence in the environment. Proc. Natl. Acad. Sci. U. S. A. 103:6350–6355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hall-Stoodley L., Stoodley P. 2005. Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol. 13:7–10 [DOI] [PubMed] [Google Scholar]
- 7. Heydorn A., Nielsen A. T., Hentzer M., Sternberg C., Givskov M., Ersboll B. K., Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407 [DOI] [PubMed] [Google Scholar]
- 8. Kamruzzaman M., Udden S. M., Cameron D. E., Calderwood S. B., Nair G. B., Mekalanos J. J., Faruque S. M. 2010. Quorum-regulated biofilms enhance the development of conditionally viable, environmental Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 107:1588–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kolenbrander P. E. 1995. Coaggregations among oral bacteria. Methods Enzymol. 253:385–397 [DOI] [PubMed] [Google Scholar]
- 10. Schaber J. A., Triffo W. J., Suh S. J., Oliver J. W., Hastert M. C., Griswold J. A., Auer M., Hamood A. N., Rumbaugh K. P. 2007. Pseudomonas aeruginosa forms biofilms in acute infection independent of cell-to-cell signaling. Infect. Immun. 75:3715–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stoodley P., Wilson S., Hall-Stoodley L., Boyle J. D., Lappin-Scott H. M., Costerton J. W. 2001. Growth and detachment of cell clusters from mature mixed-species biofilms. Appl. Environ. Microbiol. 67:5608–5613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yildiz F. H. 2007. Processes controlling the transmission of bacterial pathogens in the environment. Res. Microbiol. 158:195–202 [DOI] [PubMed] [Google Scholar]
- 13. Donnenberg M. S., Whittam T. S. 2001. Pathogenesis and evolution of virulence in enteropathogenic and enterohemorrhagic Escherichia coli. J. Clin. Invest. 107:539–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Frick I. M., Morgelin M., Bjorck L. 2000. Virulent aggregates of Streptococcus pyogenes are generated by homophilic protein-protein interactions. Mol. Microbiol. 37:1232–1247 [DOI] [PubMed] [Google Scholar]
- 15. Nataro J. P., Jansen A. 2005. Aggregation and dispersal on mucosal surfaces, p. 253 – 263 . In Nataro J. P., Cohen P. S., Mobley H. L. T., Weiser J. H. (ed.), Colonization of mucosal surfaces. ASM Press, Washington, DC. [Google Scholar]
- 16. Ochiai K., Kikuchi K., Fukushima K., Kurita-Ochiai T. 1999. Co-aggregation as a virulent factor of Streptococcus sanguis isolated from infective endocarditis. J. Oral Sci. 41:117–122 [DOI] [PubMed] [Google Scholar]
- 17. Ochiai K., Kurita-Ochiai T., Kamino Y., Ikeda T. 1993. Effect of co-aggregation on the pathogenicity of oral bacteria. J. Med. Microbiol. 39:183–190 [DOI] [PubMed] [Google Scholar]
- 18. Reid G., Bruce A. W., Llano M., McGroarty J. A., Blake M. 1990. Bacterial aggregation in sepsis. Curr. Microbiol. 20:185–190 [Google Scholar]
- 19. Baca H. K., Ashley C., Carnes E., Lopez D., Flemming J., Dunphy D., Singh S., Chen Z., Liu N., Fan H., López G. P., Brozik S. M., Werner-Washburne M., Brinker C. J. 2006. Cell-directed assembly of lipid-silica nanostructures providing extended cell viability. Science 313:337–341 [DOI] [PubMed] [Google Scholar]
- 20. Boedicker J. Q., Vincent M. E., Ismagilov R. F. 2009. Microfluidic confinement of single cells of bacteria in small volumes initiates high-density behavior of quorum sensing and growth and reveals its variability. Angew. Chem. Int. Ed. Engl. 48:5908–5911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carnes E. C., Lopez D. M., Donegan N. P., Cheung A., Gresham H., Timmins G. S., Brinker C. J. 2010. Confinement-induced quorum sensing of individual Staphylococcus aureus bacteria. Nat. Chem. Biol. 6:41–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Balaban N. Q., Merrin J., Chait R., Kowalik L., Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625 [DOI] [PubMed] [Google Scholar]
- 23. Balagadde F. K., You L., Hansen C. L., Arnold F. H., Quake S. R. 2005. Long-term monitoring of bacteria undergoing programmed population control in a microchemostat. Science 309:137–140 [DOI] [PubMed] [Google Scholar]
- 24. Cho H., Jonsson H., Campbell K., Melke P., Williams J. W., Jedynak B., Stevens A. M., Groisman A., Levchenko A. 2007. Self-organization in high-density bacterial colonies: efficient crowd control. PLoS Biol. 5:e302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Groisman A., Lobo C., Cho H., Campbell J. K., Dufour Y. S., Stevens A. M., Levchenko A. 2005. A microfluidic chemostat for experiments with bacterial and yeast cells. Nat. Methods 2:685–689 [DOI] [PubMed] [Google Scholar]
- 26. Basu S., Wolgemuth C. W., Campagnola P. J. 2004. Measurement of normal and anomalous diffusion of dyes within protein structures fabricated via multiphoton excited cross-linking. Biomacromolecules 5:2347–2357 [DOI] [PubMed] [Google Scholar]
- 27. Kaehr B., Shear J. B. 2007. Mask-directed multiphoton lithography. J. Am. Chem. Soc. 129:1904–1905 [DOI] [PubMed] [Google Scholar]
- 28. Kaehr B., Shear J. B. 2008. Multiphoton fabrication of chemically responsive protein hydrogels for microactuation. Proc. Natl. Acad. Sci. U. S. A. 105:8850–8854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nielson R., Kaehr B., Shear J. B. 2009. Microreplication and design of biological architectures using dynamic-mask multiphoton lithography. Small 5:120–125 [DOI] [PubMed] [Google Scholar]
- 30. Pitts J. D., Campagnola P. J., Epling G. A., Goodman S. L. 2000. Submicron multiphoton free-form fabrication of proteins and polymers: studies of reaction efficiencies and applications in sustained release. Macromolecules 33:1514–1523 [Google Scholar]
- 31. Khripin C. Y., Brinker C. J., Kaehr B. 2010. Mechanically tunable multiphoton fabrication protein hydrogels investigated using atomic force microscopy. Soft Matter 6:2842–2848 [Google Scholar]
- 32. Mashburn L. M., Jett A. M., Akins D. R., Whiteley M. 2005. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J. Bacteriol. 187:554–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang L., Haagensen J. A., Jelsbak L., Johansen H. K., Sternberg C., Hoiby N., Molin S. 2008. In situ growth rates and biofilm development of Pseudomonas aeruginosa populations in chronic lung infections. J. Bacteriol. 190:2767–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hense B. A., Kuttler C., Muller J., Rothballer M., Hartmann A., Kreft J. U. 2007. Does efficiency sensing unify diffusion and quorum sensing? Nat. Rev. Microbiol. 5:230–239 [DOI] [PubMed] [Google Scholar]
- 35. Redfield R. J. 2002. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 10:365–370 [DOI] [PubMed] [Google Scholar]
- 36. Brown S. P., Johnstone R. A. 2001. Cooperation in the dark: signalling and collective action in quorum-sensing bacteria. Proc. Biol. Sci. 268:961–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Horswill A. R., Stoodley P., Stewart P. S., Parsek M. R. 2007. The effect of the chemical, biological, and physical environment on quorum sensing in structured microbial communities. Anal. Bioanal. Chem. 387:371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Diggle S. P., Griffin A. S., Campbell G. S., West S. A. 2007. Cooperation and conflict in quorum-sensing bacterial populations. Nature 450:411–414 [DOI] [PubMed] [Google Scholar]
- 39. Rumbaugh K. P., Diggle S. P., Watters C. M., Ross-Gillespie A., Griffin A. S., West S. A. 2009. Quorum sensing and the social evolution of bacterial virulence. Curr. Biol. 19:341–345 [DOI] [PubMed] [Google Scholar]
- 40. Stewart P. S. 2003. Diffusion in biofilms. J. Bacteriol. 185:1485–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stewart P. S., Robertson C. R. 1989. Microbial growth in fixed volume: studies with entrapped Escherichia coli. Appl. Microbiol. Biotechnol. 30:34–40 [Google Scholar]
- 42. Davies D. G., Parsek M. R., Pearson J. P., Iglewski B. H., Costerton J. W., Greenberg E. P. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295–298 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.