Abstract
Clustered, regularly interspaced short palindromic repeats (CRISPR) provide bacteria and archaea with sequence-specific, acquired defense against plasmids and phage. Because mobile elements constitute up to 25% of the genome of multidrug-resistant (MDR) enterococci, it was of interest to examine the codistribution of CRISPR and acquired antibiotic resistance in enterococcal lineages. A database was built from 16 Enterococcus faecalis draft genome sequences to identify commonalities and polymorphisms in the location and content of CRISPR loci. With this data set, we were able to detect identities between CRISPR spacers and sequences from mobile elements, including pheromone-responsive plasmids and phage, suggesting that CRISPR regulates the flux of these elements through the E. faecalis species. Based on conserved locations of CRISPR and CRISPR-cas loci and the discovery of a new CRISPR locus with associated functional genes, CRISPR3-cas, we screened additional E. faecalis strains for CRISPR content, including isolates predating the use of antibiotics. We found a highly significant inverse correlation between the presence of a CRISPR-cas locus and acquired antibiotic resistance in E. faecalis, and examination of an additional eight E. faecium genomes yielded similar results for that species. A mechanism for CRISPR-cas loss in E. faecalis was identified. The inverse relationship between CRISPR-cas and antibiotic resistance suggests that antibiotic use inadvertently selects for enterococcal strains with compromised genome defense.
IMPORTANCE
For many bacteria, including the opportunistically pathogenic enterococci, antibiotic resistance is mediated by acquisition of new DNA and is frequently encoded on mobile DNA elements such as plasmids and transposons. Certain enterococcal lineages have recently emerged that are characterized by abundant mobile DNA, including numerous viruses (phage), and plasmids and transposons encoding multiple antibiotic resistances. These lineages cause hospital infection outbreaks around the world. The striking influx of mobile DNA into these lineages is in contrast to what would be expected if a self (genome)-defense system was present. Clustered, regularly interspaced short palindromic repeat (CRISPR) defense is a recently discovered mechanism of prokaryotic self-defense that provides a type of acquired immunity. Here, we find that antibiotic resistance and possession of complete CRISPR loci are inversely related and that members of recently emerged high-risk enterococcal lineages lack complete CRISPR loci. Our results suggest that antibiotic therapy inadvertently selects for enterococci with compromised genome defense.
INTRODUCTION
Enterococcus faecalis and Enterococcus faecium rank among the leading causes of antibiotic-resistant hospital-acquired bacterial infections (1–3). Resistance to last-line drugs, such as vancomycin (4), is common, and enterococci are now disseminating this resistance to methicillin-resistant Staphylococcus aureus (MRSA) (5–7). The first vancomycin-resistant E. faecalis isolate in the United States, V583 (8), is a member of clonal complex 2 (CC2) (9), a cluster of hospital-adapted lineages that emerged as early as 1981 (10). It was the first Enterococcus isolate for which a genome sequence was determined (11). Mobile elements constitute one-quarter of its genome and include three independently replicating plasmids, three chromosomally integrated plasmid remnants, seven prophage, and a pathogenicity island (PAI) (11, 12). The E. faecium hospital-adapted lineage CC17 (13), which emerged as early as 1982 (14), is similarly characterized by an abundance of exogenously acquired genes, including insertion sequences, phage, and antibiotic resistance genes (15).
Enterococci are natural inhabitants of the digestive tracts of humans and other mammals (16). Genome analysis of the natively antibiotic-sensitive human oral isolate of E. faecalis, OG1RF, revealed that this strain lacks most of the mobile elements and externally acquired DNA found in the hospital-adapted strain V583 (17). Instead, two clustered, regularly interspaced short palindromic repeat (CRISPR) loci were identified (17). CRISPR is a prokaryotic sequence-specific defense system that provides a type of acquired immunity (18, 19). Mechanistic details are emerging, but in general, a small segment of an invading mobile element is incorporated into the CRISPR array between roughly palindromic repeats (20, 21). This mobile element segment is then transcribed and processed at the palindromes to generate a small RNA termed crRNA (22). crRNA targets CRISPR-associated nucleases, encoded by the CRISPR-associated genes (cas genes), to incoming mobile elements from which the spacers derive (20, 22, 23). CRISPR defense appears to be widespread in prokaryotes, with ~90% of archaeal and ~45% of bacterial genomes possessing convincing CRISPR loci (24). Of the two CRISPR loci discovered in the E. faecalis OG1RF genome, one possesses the associated cas nuclease genes (CRISPR1-cas) and one is an orphan locus lacking cas genes (CRISPR2) (17). The hospital-adapted V583 strain possesses only the orphan locus CRISPR2, lacking the functional cas genes required for CRISPR defense (25).
Because of the potential for limiting entry of mobile elements, it was of interest to determine whether a correlation exists between the presence of CRISPR loci and the emergence of antibiotic-resistant enterococcal lineages. By examining enterococcal draft genomes to develop consensus information on the occurrence of CRISPR loci, and expanding the study to a historical collection of isolates that extends coverage through antibiotic and preantibiotic eras, we found a strong correlation between the absence of CRISPR-cas loci and the emergence of MDR enterococcal strains. We hypothesize that widespread antibiotic use has selected for enterococcal strains able to readily acquire novel traits—those with compromised genome defense—ultimately leading to the emergence of enterococcal lineages replete with antibiotic resistances and other mobile traits.
RESULTS
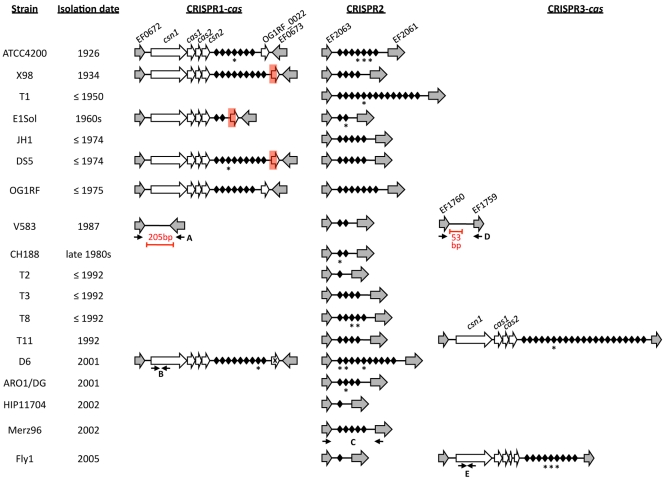
CRISPR distribution in 16 E. faecalis draft genomes.
We recently collaborated in an effort to sequence the genomes of 28 enterococci, including 16 E. faecalis strains representing deep phylogenetic nodes in the E. faecalis multilocus sequence typing (MLST) dendrogram (26). CRISPRfinder (27) was used to identify putative CRISPR loci in the 16 draft E. faecalis genomes. The orphan CRISPR2 locus, consisting only of palindromes and spacers, and not cas genes, was identified in all 16 E. faecalis genomes and was invariably located between homologues of E. faecalis V583 open reading frames (ORFs) EF2063 and EF2061 (Fig. 1). CRISPR2 repeat palindromes were highly conserved, in every case being nearly or perfectly identical to the E. faecalis OG1RF CRISPR1/2 repeat sequence (see Table S1 in the supplemental material). In contrast, the occurrence of the CRISPR1-cas locus varied (Fig. 1). Of the 16 genome sequences analyzed, 5 (ATCC 4200, X98, E1Sol, DS5, and D6) possessed CRISPR1-cas loci, always located between homologues of the V583 ORFs EF0672 and EF0673 (Fig. 1).
FIG 1 .
CRISPR loci in E. faecalis draft genomes. Strains are organized by date of isolation, from oldest to most recent. Homologues of E. faecalis V583 genes are shown as grey arrows and with V583 ORF assignments. CRISPR locus-specific genes are shown as white arrows, and CRISPR spacers are represented by black diamonds. CRISPR spacers with identity to mobile elements are starred. Note that CRISPR repeats are not shown. Red rectangles denote deletions in the X98, E1Sol, and DS5 CRISPR1-cas loci. An “X” denotes a frameshift mutation in the D6 CRISPR1-cas region corresponding to OG1RF_0022. The E. faecalis V583 CRISPR2 locus and the OG1RF CRISPR1-cas and CRISPR2 loci were previously reported (17, 25). Red bars and text denote novel DNA sequences present in V583. Primer sets used in CRISPR profiling are labeled with uppercase letters: A, CRISPR1-cas flanking primers; B, CRISPR1-cas csn1 screening primers; C, CRISPR2 screening primers; D, CRISPR3-cas flanking primers; E, CRISPR3-cas csn1 screening primers. The figure is not drawn to scale.
The putative Cas proteins encoded by these loci are highly conserved, as are the palindromic repeat sequences (see Tables S1 and S2 in the supplemental material). As for E. faecalis OG1RF CRISPR1-cas, these loci are of the Nmeni subtype, a CRISPR-cas organizational structure found only in vertebrate-associated commensals and pathogens (28). OG1RF CRISPR1-cas includes an ORF 3′ to the CRISPR1 repeat-spacer array, OG1RF_0022, that has no homologue (17) and is not common to Nmeni subtype loci (28). This ORF varies among strains with CRISPR1-cas, with DS5, E1Sol, and X98 possessing a 435-bp deletion extending 255 bp into it (Fig. 1). A frameshift occurs in the homologue of OG1RF_0022 in strain D6 (Fig. 1). Given the variability in this region, this ORF may be dispensable for CRISPR function.
A novel CRISPR locus, CRISPR3-cas, was identified in the genomes of strains Fly1 and T11, occurring between homologues of the E. faecalis V583 ORFs EF1760 and EF1759 (Fig. 1). CRISPR3-cas also exhibits an organization consistent with Nmeni subtype CRISPR loci (28) (see Table S3 in the supplemental material). However, CRISPR3-cas is distinct from CRISPR1-cas and CRISPR2 in palindrome repeat sequence (see Fig. S1 in the supplemental material) and is distinct from CRISPR1-cas in cas gene content. Nmeni subtype loci possess the universal CRISPR marker gene cas1, the core CRISPR gene cas2, and the Nmeni subtype-specific gene csn1, while possession of the Nmeni subtype-specific gene csn2 varies (28). CRISPR1-cas possesses csn2, the fourth and final ORF in the inferred cas operon (17) (Fig. 1). Curiously, the fourth ORF in the inferred E. faecalis T11 CRISPR3-cas operon (Fig. 1) does not encode a protein with significant identity to Csn2 (TIGR01866) (28) or to any other conserved protein domains (Table S3). In Fly1, the region corresponding to this unknown ORF is interrupted by a frameshift mutation that results in two truncated ORFs (Fig. 1).
CRISPR spacer identities with mobile elements.
No identity was reported between E. faecalis OG1RF CRISPR spacer sequences and known mobile element sequences, or any other sequences deposited in GenBank (17), and thus no evidence supporting a role for CRISPR in E. faecalis self-defense exists. Having a larger data set of 140 CRISPR spacers from 16 additional genomes, we queried the NCBI nonredundant nucleotide database to identify elements with possible identity to E. faecalis CRISPR spacer sequences. Because spacer sequences are small (30 bp), only hits with >90% identity (27 of 30 nucleotides) were considered significant. By this criterion, identities to 38 of 140 E. faecalis CRISPR spacers were found. Of the 38 hits, 19 were identical to CRISPR2 spacers of the previously sequenced E. faecalis OG1RF and V583 genomes and 19 were derived from mobile elements (Table 1). These elements include the pheromone-responsive type plasmids pAD1/pTEF1 and pCF10/pTEF2, enterococcal phage and prophage, and plasmids integrated within the E. faecalis V583 genome (Table 1). Identities to certain mobile elements were more common than others: five strains (ATCC4200, D6, T8, Fly1, and T11) possess spacers with identity to pheromone-responsive plasmids, and five strains (ATCC 4200, D6, T8, Fly1, and ARO1/DG) possess spacer sequences with identities to different regions of E. faecalis V583 prophage 6.
TABLE 1 .
E. faecalis CRISPR spacer identities to mobile genetic elements
| Strain | CRISPR and spacer no.a | Sequence identityb | Representative Blastn hit | Area of identity in Blastn hit | Identical Blastn hits |
|---|---|---|---|---|---|
| ATCC 4200 | 1-4 | 29/30 | Enterococcus phage φEF24C | EFP_gp114 hypothetical protein | |
| 2-4 | 29/30 | V583 prophage 6c | EF2813 tail tape measure protein | ||
| 2-5 | 30/30 | V583 prophage 6 | EF2836_EF2387 intergenic region | ||
| 2-6 | 29/30 | V583 pTEF2 | EF_B0043 ssb-6 | E. faecalis pCF10 | |
| 29/30 | E. faecalis pMG2200 | ORF54 hypothetical protein BAH02364.1 | E. faecalis pBEE99; pYI14 | ||
| DS5 | 1-3 | 30/30 | Enterococcus phage φFL2B | gp34-gp35 intergenic region | Enterococcus phage φFL2A |
| 29/30 | Enterococcus phage φFL3B | gp37-gp38 intergenic region | Enterococcus phage φFL3A; φEf11 | ||
| 28/30 | Enterococcus phage φFL1C | gp39-gp40 intergenic region | Enterococcus phage φFL1B; φFL1A | ||
| D6 | 1-8 | 29/30 | V583 VR1c-integrated plasmid | EF0133-EF0134 intergenic region | |
| 29/30 | V583 VR11-integrated plasmid | EF2539-EF2540 intergenic region | |||
| D6 | 2-1 | 29/30 | V583 prophage 3 | EF1486 endolysin | |
| D6 | 2-2 | 29/30 | V583 prophage 6 | EF2834 hypothetical protein | |
| D6 | 2-5 | 30/30 | V583 pTEF2 | EF_B0047 membrane protein, putative | E. faecalis pCF10 |
| T1 | 2-5 | 27/30 | Enterococcus phage φEF24C | EFP_gp116 hypothetical protein | |
| E1Sol | 2-2 | 30/30 | V583 VR11-integrated plasmid | EF2535 nucleotidyltransferase domain protein | |
| CH188 | 2-1 | 28/30 | V583 prophage 1 | EF0334 portal protein | Enterococcus phage φFL4A |
| T8d | 2-3 | 30/30 | V583 prophage 6 | EF2836-EF2387 intergenic region | |
| T8e | 2-4 | 29/30 | V583 pTEF2 | EF_B0043 ssb-6 | E. faecalis pCF10 |
| 29/30 | E. faecalis pMG2200 | ORF54 hypothetical protein BAH02364.1 | E. faecalis pBEE99; pYI14 | ||
| ARO1/DG | 2-2 | 30/30 | V583 prophage 6 | EF2823 terminase, large subunit, putative | |
| Fly1 | 3-4 | 30/30 | V583 prophage 6 | EF2838 DNA replication protein | |
| Fly1 | 3-5 | 29/30 | V583 pTEF1 | EF_A0022-EF_A0024 intergenic region | E. faecalis pAM373; pAD1 |
| Fly1 | 3-6 | 29/30 | V583 prophage 6 | EF2825 conserved hypothetical protein | |
| T11 | 3-6 | 30/30 | V583 pTEF1 | EF_A0083 rep-1 | E. faecalis pAD1 |
The CRISPR locus (1, 2, or 3) followed by the spacer number is shown. CRISPR spacers are numbered in consecutive order from left to right as shown in Fig. 1.
Sequence identity is shown as the number of base pairs with sequence identity in GenBank/total number of base pairs in spacer.
V583 variable regions (VR) are from a report by McBride et al. (10).
This spacer is identical to ATCC 4200 CRISPR2 spacer 5.
This spacer is identical to ATCC 4200 CRISPR2 spacer 6.
E. faecalis CRISPR-cas and acquired antibiotic resistance.
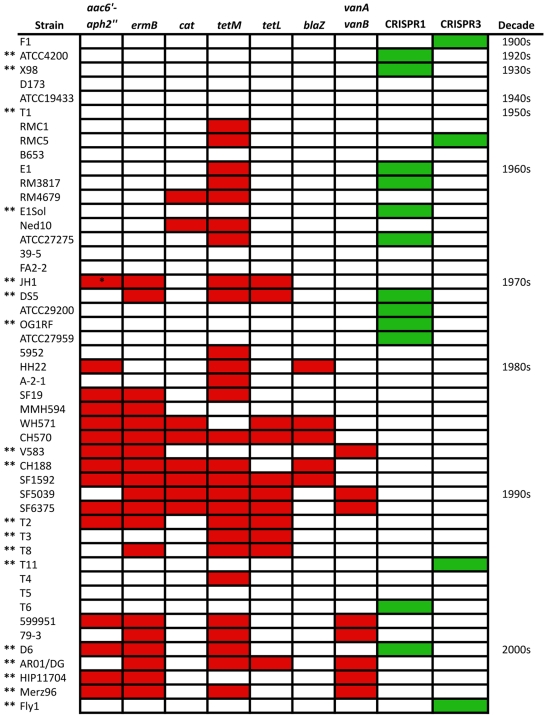
Examining the entire genomes of 16 E. faecalis strains allowed us to determine that, when present, CRISPR loci always occurred at conserved chromosomal positions: the CRISPR1-cas locus occurred between EF0672 and EF0673 homologues, CRISPR2 between EF2063 and EF2061 homologues, and CRISPR3-cas between EF1760 and EF1759 homologues. Based on this information, we extended the analysis of E. faecalis CRISPR by examining 29 additional isolates, including strains with isolation dates prior to the mid-1970s (Fig. 2; for additional isolation date and strain source information, see Data Set S1 in the supplemental material). E. faecalis strains were first screened for internal regions of CRISPR1 csn1 and CRISPR3 csn1 by PCR. Strains negative by PCR were rescreened with primers annealing outside the conserved CRISPR1-cas or CRISPR3-cas locus positions, and products from these reactions were sequenced to confirm CRISPR absence. Using this approach, positive PCR results indicating the presence or absence of CRISPR were obtained for all strains tested. CRISPR2 was detected by amplification of the conserved locus in all 29 E. faecalis strains and confirmed by DNA sequencing. CRISPR content for E. faecalis V583, OG1RF, and HH22, as previously reported (17, 25), was included in the analysis.
FIG 2 .
CRISPR-cas and acquired antibiotic resistance in a historical collection of E. faecalis strains. E. faecalis strains are listed by date of isolation, from oldest to most recent. Acquired antibiotic resistance is shown in red, and CRISPR-cas presence is shown in green. Antibiotic resistance (tetracycline [tetL and tetM], erythromycin [ermB], gentamicin [aac6′-aph2′′], chloramphenicol [cat], ampicillin [blaZ], and vancomycin [vanA and vanB]) was previously profiled (red squares) (10). A single asterisk indicates that gentamicin resistance is conferred by a 3′-5′′-aminoglycoside phosphotransferase in this strain; double asterisks denote E. faecalis strains for which draft or complete genome sequences are available.
All 48 E. faecalis strains possess an orphan CRISPR2 locus (see Table S1 in the supplemental material). In contrast, CRISPR-cas distribution varies among antibiotic-sensitive and antibiotic-resistant strains (Fig. 2). One-third of the E. faecalis strains in our collection (16/48) possess a complete CRISPR-cas locus (CRISPR1-cas or CRISPR3-cas), and all but two of these either lack acquired antibiotic resistance genes or possess tetM, a gene commonly disseminated by the self-mobilizable conjugative transposon Tn916 (29, 30). Strikingly, of the MDR E. faecalis strains in this collection (defined as resistant to two or more antibiotics), all except two lack CRISPR-cas loci (n = 22), including all vancomycin-resistant strains (n = 8) (Fig. 2).
We used a combination of statistical analyses to evaluate the hypothesis that E. faecalis strains with CRISPR-cas possess significantly fewer acquired antibiotic resistance genes than those lacking CRISPR-cas. We first tallied acquired antibiotic resistance genes for each strain and performed a one-tailed Wilcoxon rank sum test to address the null hypothesis that there is no difference in the distributions of acquired antibiotic resistance genes between strains that possess CRISPR-cas and strains that lack CRISPR-cas. Because antibiotic resistance genes are often coacquired on elements conferring resistance to multiple antibiotics, we performed the Wilcoxon rank sum test using progressively stricter models for coacquisition, in which potentially coacquired genes were counted as resistance to one antibiotic, instead of two (see Data Set S1 in the supplemental material). We found that, irrespective of the model used, the null hypothesis could be rejected (P < 0.001), indicating that the distributions of acquired antibiotic resistance significantly differ between CRISPR-cas-positive and -negative E. faecalis strains. Probability values did not change for a model in which tetM and ermB, which can be codisseminated by Tn916 and related elements (30), are coacquired (P values of 0.0003 if acquired jointly and 0.0004 if acquired separately) (Data Set S1). If aac6′-aph2′′ and blaZ are also assumed to be coacquired, as was suggested by the patterns of occurrence of antibiotic resistance in 106 E. faecalis strains (10), the P value increases nominally to 0.0005 (Data Set S1). Finally, if ermB and vancomycin resistance are assumed to be coacquired (10), in addition to tetM/ermB and aac6′-aph2′′/blaZ coacquisition, the P value increases to 0.0008 (Data Set S1), remaining highly significant. We additionally used a one-tailed Fisher exact test with a 2 × 2 contingency table comparing cooccurrences of CRISPR-cas and antibiotic resistance to address the null hypothesis that CRISPR-cas presence is unrelated to antibiotic resistance. The P value of 0.007 is highly significant (P < 0.01), which led us to reject the null hypothesis (Data Set S1). The results of these analyses demonstrate that a statistically significant relationship between the presence or absence of CRISPR-cas and acquired antibiotic resistance exists.
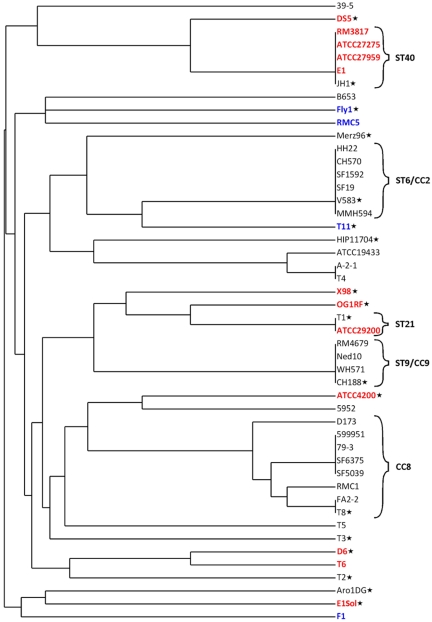
To examine the phylogenetic distribution of CRISPR-cas loci in E. faecalis, we generated an MLST dendrogram of the strains used for CRISPR analysis (10) (Fig. 3). CRISPR-cas loci are discontinuously distributed across this MLST dendrogram, and CRISPR-cas distribution varies even between strains of a single sequence type (e.g., ST21 and ST40 lineages) (Fig. 3). In other lineages, including ST6 (a CC2 lineage; 6 strains), ST9 (a CC9 lineage; 4 strains), and CC8 (8 strains) lineages, CRISPR-cas loci are uniformly absent. All three lineages are associated with MDR (9, 10); vancomycin resistance and β-lactamase production first emerged in the United States in ST6 strains of CC2 (10), and the CC2 and CC9 lineages are highly associated with nosocomial infections and hospital endemicity (9). Loss of CRISPR-cas in founders of these lineages may have precipitated their success in acquiring traits facilitating hospital adaptation.
FIG 3 .
CRISPR-cas distribution across the E. faecalis MLST dendrogram. An MLST-based dendrogram of the 48 strains utilized in CRISPR profiling was generated using the E. faecalis MLST database (see Materials and Methods). Strains indicated in red possess CRISPR1-cas, and those in blue possess CRISPR3-cas. Stars denote strains for which draft or complete genome sequences are available. ST, sequence type; CC, clonal complex.
CRISPR-cas in E. faecium.
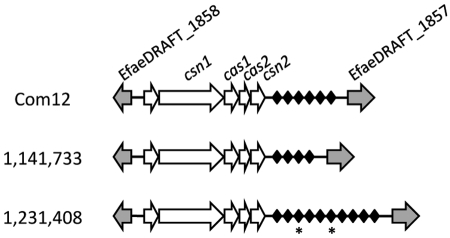
No CRISPR loci were identified in seven recently reported E. faecium draft genomes (31). We therefore examined the CRISPR locus content in eight additional E. faecium genomes (26). The E. faecium strains originated from human clinical samples (strains represented by numbers in Fig. 4 and 5) and from the feces of healthy human volunteers (Com12 and Com15). All were isolated in the early to mid-2000s. We identified an E. faecium CRISPR-cas locus (EfmCRISPR1-cas) in three genomes (Fig. 4). This locus is found between homologues of EfaeDRAFT_1858 and EfaeDRAFT_1857 from the previously sequenced clinical isolate E. faecium DO draft genome (GenBank accession no. ACIY00000000), which itself lacks CRISPR (25). EfmCRISPR1-cas possesses a palindromic repeat sequence that is divergent from those of the E. faecalis CRISPR-cas loci (see Fig. S1 in the supplemental material). The four predicted Cas proteins encoded by EfmCRISPR1-cas are consistent with the Nmeni subtype (Table S4). However, EfmCRISPR-cas differs from the E. faecalis Nmeni subtype by the presence of an ORF of unknown function 5′ to the csn1 gene which is conserved and unique to the three strains possessing EfmCRISPR-cas (Fig. 4). A Blastn comparison of the 20 EfmCRISPR1-cas spacers to sequences in the NCBI nonredundant nucleotide database reveals identity between two spacers and previously identified clostridial and lactococcal phages (Table 2).
FIG 4 .
EfmCRISPR1-cas loci in E. faecium draft genomes. Homologues of E. faecium DO genes are shown as grey arrows and with DO ORF assignments. EfmCRISPR1-cas locus-specific genes are shown as white arrows, and CRISPR spacers are represented by black diamonds. CRISPR spacers with identity to mobile elements are starred. Note that CRISPR repeats are not shown. The figure is not drawn to scale.
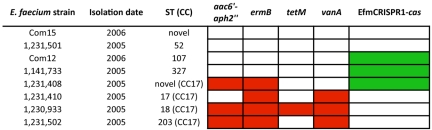
FIG 5 .
MLST, EfmCRISPR1-cas, and acquired antibiotic resistance in E. faecium draft genomes. Acquired antibiotic resistance is shown in red, and CRISPR-cas presence is shown in green. Antibiotic resistance (tetracycline [tetM], erythromycin [ermB], gentamicin [aac6′-aph2′′], and vancomycin [vanA]), EfmCRISPR1-cas, and ST profiles were generated by genomic analysis.
TABLE 2 .
E. faecium EfmCRISPR1-cas spacer identities to mobile genetic elements
| Strain | Spacera | Sequence identityb | Representative BLASTN hit | Area of homology |
|---|---|---|---|---|
| 1,231,408 | 3 | 28/30 | Clostridium novyi NT genome | NT01CX_2197, putative phage antirepressor |
| 1,231,408 | 6 | 28/30 | Lactococcus lactis subsp. lactis KF147 genome | LLKF_1066, phage protein, HNH endonuclease |
CRISPR spacers are numbered in consecutive order from left to right as shown in Fig. 4.
Sequence identity is shown as the number of base pairs with sequence identity in GenBank/total number of base pairs in spacer.
MLST phylogeny and acquired antibiotic resistance profiles were determined for the eight E. faecium strains. Four strains belong to the hospital-adapted CC17, being either ST17 or double-locus variants of ST17 (Fig. 5; see Table S5 in the supplemental material). As expected for CC17 strains (1), all are MDR. Three of the CC17 strains possess vanA, encoding vancomycin resistance, while the fourth CC17 strain constitutes a novel sequence type variant and lacks vanA. The remaining four strains belong to other sequence types and lack acquired antibiotic resistance genes (Fig. 5). Although the E. faecium analysis was smaller in sample size than that for E. faecalis, the distribution of EfmCRISPR1-cas in these genomes supports the conclusion that MDR enterococcal strains generally lack CRISPR-cas; in this case, three of four MDR strains lack EfmCRISPR1-cas, and these three strains are vancomycin-resistant members of the hospital-adapted CC17 lineage.
Mechanism for CRISPR-cas variability.
It was previously proposed that deletion of a Streptococcus thermophilus Nmeni subtype CRISPR-cas may occur through recombination (32). Nmeni subtype CRISPR-cas loci possess a conserved but imperfect CRISPR repeat upstream of the cas operon, meaning that a Nmeni CRISPR-cas locus is flanked on both the 5′ and 3′ ends by repetitive sequences (28, 32). It is possible that recombination could occur at these sites, resulting in deletion (32). We analyzed the E. faecalis genome sequences corresponding to the locations where deletion of CRISPR1-cas or CRISPR3-cas would have occurred. Interestingly, E. faecalis strains lacking CRISPR-cas possess small, common sequences at these sites. E. faecalis V583 possesses 205 bp of unique sequence when aligned with the E. faecalis D6 CRISPR1-cas and flanking region and 53 bp of unique sequence compared to the Fly1 CRISPR3-cas region (Fig. 1). These 205-bp and 53-bp sequences are highly conserved in all E. faecalis strains lacking CRISPR1-cas or CRISPR3-cas (see Fig. S2 and S3 in the supplemental material). This would not be predicted for CRISPR-cas loss by homologous recombination and excision.
We recently reported the low-frequency mobilization and conjugal transfer of large regions of chromosomal DNA from E. faecalis V583 donors to E. faecalis OG1RF recipients, resulting in transconjugant strains with hybrid V583-OG1RF genomes (33). This transfer was found to be dependent on either of two pheromone-responsive plasmids resident in V583, pTEF1 or pTEF2, and occurred as the result of plasmid integration and chromosome mobilization. Chromosome-to-chromosome movement of the PAI, a vancomycin resistance transposon, capsule genes, and other V583 genes to OG1RF recipients was observed. It is important to note that OG1RF does not possess CRISPR spacers with identity to pTEF1, pTEF2, or other V583 sequences (17), and in these experiments, CRISPR1-cas did not block incoming DNA from V583. Because the OG1RF CRISPR1-cas locus occurs ~41.5 kb 3′ to the site of PAI incorporation, it was of interest to determine whether the CRISPR1-cas locus in PAI-containing transconjugants had been displaced by incoming DNA from V583 donors. We used five strains (TC1, TC3, TC4, TC5, and TC12) representing the five classes of PAI transconjugants (A to E) identified in our previous study (33). These transconjugants possess the V583 PAI and various amounts of flanking DNA, representing a total acquisition of 285 to 857 kb of V583 donor chromosome (33). The five PAI-containing transconjugants were screened for the occurrence of the OG1RF CRISPR1-cas by PCR. Strikingly, all five transconjugants lacked CRISPR1-cas and instead possessed at this locus the conserved 205-bp sequence found in the V583 donor (see Fig. S2 in the supplemental material). Thus, in a single conjugative transfer event, a novel hybrid strain that simultaneously becomes antibiotic resistant, acquires the PAI, and is rendered deficient for CRISPR-cas can be generated.
DISCUSSION
We examined 48 E. faecalis strains from a historical collection and 8 recent E. faecium isolates to determine the relationship between CRISPR and the emergence of multidrug resistance in enterococci. We found that CRISPR1-cas and CRISPR3-cas loci are variable among E. faecalis strains, but, interestingly, an orphan CRISPR2 locus occurred in all E. faecalis strains tested. Selective pressure for maintenance of CRISPR2 in a CRISPR-cas-deficient E. faecalis background is unclear, as is the origin of CRISPR2 (i.e., whether this locus is functionally and evolutionarily linked to CRISPR1-cas, or once independently possessed cas genes that have been lost). CRISPR1-cas and CRISPR2 repeat sequences are identical (17), and because Cas proteins interact with CRISPR repeat sequences following transcription (22), it is likely that CRISPR1-cas and CRISPR2 are functionally linked. Nevertheless, genome sequence analysis finds that CRISPR-cas-deficient E. faecalis strains lack any detectable csn1 or csn1-like gene, which is essential for protection mediated by a Nmeni subtype CRISPR-cas locus (20). The lack of required functional genes in strains with the CRISPR2 orphan locus, together with the abundance of antibiotic resistance genes in these strains, indicates that CRISPR2 alone does not confer immunity in E. faecalis hosts. Ultimately, a functional analysis of the CRISPR1-cas and CRISPR2 loci will be required to confirm this hypothesis.
By analyzing a large data set, we were able to detect identities between CRISPR spacers and sequences on known mobile elements, including pheromone-responsive plasmids and phage. Enterococci have a highly coevolved relationship with the narrow-host-range pheromone-responsive plasmids, which encode machinery to utilize extracellular signals produced by enterococcal cells to induce conjugation functions and to manipulate signal production in enterococcal cells in which they reside (34). The results of this study and our recent work (33) support a role for pheromone-responsive plasmids as important drivers of enterococcal genome plasticity, capable of promoting their own transfer, mobilizing chromosomally encoded antibiotic resistance and virulence determinants, and now also causing displacement of CRISPR-cas.
That E. faecalis CRISPR loci contain spacers with identity to enterococcal mobile elements, and the distribution of these spacers, suggests that certain elements, such as pheromone-responsive plasmids, are frequently encountered by E. faecalis and/or have a propensity to be incorporated into CRISPR loci as spacers. It is interesting that no CRISPR spacers have yet been identified with sequence identity to conjugative transposons such as Tn916, which are also vectors of antibiotic resistance in the enterococci (29). CRISPR elements have been shown experimentally to confer immunity to plasmids and phages (20–23), although many mechanistic details remain unknown. To our knowledge, there is no experimental evidence that CRISPR defense confers protection from conjugative transposons. The observation that the tetM gene, commonly disseminated by Tn916 and related conjugative transposons (29, 30), is present in E. faecalis strains possessing CRISPR-cas (Fig. 2) suggests that conjugative transposons may evade this defense. Spacers targeting the Inc18 plasmid family, plasmids that are significant for their role in the dissemination of vancomycin resistance genes from enterococci to MRSA (6, 7), are also absent. Tn916 and the Inc18 plasmid family are broad host range in nature (29, 30), and the notable lack of spacers with identity to these elements may reflect the relative rarity of interspecies transfer or the relative inefficiency of transfer of elements lacking mechanisms for effective pair formation, such as the pheromone-induced aggregation mechanism.
Of the 48 E. faecalis and 8 E. faecium strains examined in this study, 7 E. faecalis and 2 E. faecium strains lacked CRISPR-cas and also lacked antibiotic resistance. Draft genome sequences are not available for six of these nine strains, so it remains a formal possibility that novel CRISPR-cas loci may be found in those strains. Alternatively, it may be that the absence of CRISPR-cas (particularly in strains isolated before, or early in, the age of antibiotics, such as E. faecalis D173 [isolated in 1939], ATCC 19433 [isolated before 1942], and T1 [isolated before 1950] [10]) facilitated acquisition of a mobile element carrying traits other than antibiotic resistance (for example, hemolysin/bacteriocin production and resistance or new metabolic properties), which provided a selective advantage.
Mobile elements often provide an accessory pool of genes that enhance survival in select environments. It is possible that elements such as the pheromone-responsive plasmids and putative enterococcal self-defense mechanisms such as CRISPR-cas act, respectively, to diversify and to stabilize the enterococcal genome, and that the dynamic between these opposing forces is important for the ultimate success of these microbes. Based on the data presented here, we speculate that modern antibiotic therapy has shifted this dynamic toward the facile acquisition of foreign elements conferring antibiotic resistance, among other things, decreasing genome stability/increasing plasticity, and enabling the colonization of new habitats, including the antibiotic-laden hospital environment.
MATERIALS AND METHODS
Genomes, bacterial strains, and media.
Draft genome data for 16 E. faecalis and 8 E. faecium strains were generated in collaboration with the Broad Institute (26) and accessed at the Enterococcus database website (http://www.broadinstitute.org/annotation/genome/enterococcus_faecalis/MultiHome.html). E. faecalis strains used for CRISPR profiling are shown in Fig. 2 and in Data Set S1 in the supplemental material and have previously been described with respect to isolation dates, antibiotic resistance profiles, and MLST data (10). E. faecalis OG1RF transconjugants TC1, TC3, TC4, TC5, and TC12, which received the PAI and other chromosomal markers from E. faecalis V583 in conjugative transfers, have been previously described (33). E. faecalis was routinely cultured at 37°C on brain heart infusion (BHI) agar.
CRISPR locus identification in draft genome sequences.
CRISPR loci were identified in 16 E. faecalis and 8 E. faecium draft genomes (26) using CRISPRfinder (http://crispr.u-psud.fr/Server/CRISPRfinder.php) (27). Sequence data from the Broad Institute Enterococcus database were downloaded, and CRISPR repeat-spacer arrays were manually annotated in MacVector to confirm the presence of CRISPR. In E. faecalis strains Fly1, HIP11704, and T2, the CRISPR2 locus was detected by annotation of the intergenic region between homologues of the E. faecalis V583 ORFs EF2063 and EF2061 and not by CRISPRfinder. Conserved domains in putative CRISPR3 and EfmCRISPR1 Cas proteins were identified by Blastp of the NCBI nonredundant protein database. CRISPR spacer sequences were compared to GenBank sequences using Blastn of the NCBI nonredundant nucleotide database with default parameters for short input sequences. Sequence alignments were generated with ClustalW in MacVector.
CRISPR locus screening by PCR.
Locations of screening primers used in this study relative to CRISPR locus positions are shown in Fig. 1. Twenty-nine E. faecalis strains were screened for CRISPR loci by colony PCR. Colonies were suspended in 25 µl Tris-EDTA (TE) buffer, boiled at 98 to 100°C for 5 min, and pelleted by centrifugation, and 2 µl supernatant was used as the template per standard PCR with Taq polymerase (New England Biolabs). Primers amplifying a 676-bp region of the 16S-B rRNA gene (For-5′-CAT GCA AGT CGA ACG CTT CT-3′, Rev-5′-CCA TAT ATC TAC GCA TTT CAC-3′) were used in positive-control reactions for each colony lysate. To screen for CRISPR1-cas loci, primers amplifying a 783-bp internal region of the E. faecalis D6 CRISPR1-cas csn1 homologue EFLG_01963 (For-5′-CAG AAG ACT ATC AGT TGG TG-3′, Rev-5′-CCT TCT AAA TCT TCT TCA TAG-3′) were used. To screen for CRISPR3-cas loci, primers amplifying a 258-bp internal region of the E. faecalis Fly1 CRISPR3-cas csn1 homologue EFKG_00787 (For-5′-GCT GAA TCT GTG AAG TTA CTC-3′, Rev-5′-CTG TTT TGT TCA CCG TTG GAT-3′) were used. To identify CRISPR2 loci, primers nested in EF2063 and EF2061 homologues flanking the E. faecalis Merz96 CRISPR2 locus (For-5′-CTG GCT CGC TGT TAC AGC T-3′, Rev-5′-GCC AAT GTT ACA ATA TCA AAC A-3′) were used. CRISPR2 products were submitted for DNA sequencing at the Massachusetts General Hospital DNA Sequencing Core Facility, and CRISPR2 loci were manually annotated in MacVector for all 29 E. faecalis strains.
Because divergence of csn1 genes may have led to false-negative PCR results, E. faecalis strains with negative PCR results were further screened with primers flanking the conserved locations of the CRISPR1-cas and CRISPR3-cas loci, between homologues of EF0672-EF0673 and EF1760-EF1759, respectively. Colony PCR was performed as described above with primer pairs flanking the E. faecalis CRISPR1-cas region (For-5′-GCG ATG TTA GCT GAT ACA AC-3′ and Rev-5′-CGA ATA TGC CTG TGG TGA AA-3′; expected product size of 315 bp for strains lacking CRISPR1-cas) and flanking the E. faecalis CRISPR3-cas region (For-5′-GAT CAC TAG GTT CAG TTA TTT C-3′ and Rev-5′-CAT CGA TTC ATT ATT CCT CCA A-3′; expected product size of 224 bp for strains lacking CRISPR3-cas). E. faecalis strains for which CRISPR-cas loci were not detected in genomic analyses (Fig. 1) were similarly screened. E. faecalis OG1RF, Fly1, and V583 were included as positive and negative controls where appropriate. All PCR products were sequenced to confirm the absence of CRISPR loci.
E. faecalis OG1RF transconjugants were screened for the presence of CRISPR1-cas (33) by PCR using a primer nested in EF0672 (For-5′-GCG ATG TTA GCT GAT ACA AC-3′) and a primer nested in CRISPR1-cas csn1 (Rev-5′-CTT CAC CAA CTG ATA GTC TTC-3′). E. faecalis OG1RF was included as a positive control. Strains were additionally screened with CRISPR1-cas flanking primers as described above, and products were sequenced to confirm the absence of CRISPR1-cas.
Analysis of E. faecalis CRISPR distribution.
To test whether the acquired antibiotic resistance gene contents of CRISPR-cas-positive and -negative strains significantly differed, data were analyzed by the nonparametric Wilcoxon rank sum test and the Fisher exact test. MLST data for E. faecalis strains have been previously reported (10), and an MLST-based phylogenetic tree of strains used in this study was generated using the E. faecalis MLST database (http://efaecalis.mlst.net/).
Analysis of E. faecium genomes.
MLST of E. faecium strains was performed by concatenating sequences of the adk, atpA, ddl, gdh, gyd, purK, and ptsS alleles for each strain and comparing them to known sequences in the E. faecium MLST database (35) (http://efaecium.mlst.net/). Single- and double-locus variants of ST17 were assigned to the CC17 lineage. Antibiotic resistance genes were identified by Blastp queries of the Broad Institute Enterococcus group database with reference enterococcal resistance proteins from the NCBI protein database. Accession numbers for the proteins used in the Blastp queries are as follows: for TetM (Enterococcus faecium), accession no. ADA62733; TetL (Enterococcus faecium), AAL92527; VanA (Enterococcus faecium), ACC93633; VanB (Enterococcus faecium), AAQ12894; ErmB (Enterococcus faecium), AAF86219; Cat (Enterococcus faecium), AAF64429; BlaZ (Enterococcus faecalis HH22), AAA24777; and Aac6′-Aph2′′, P0A0C2. Genes encoding VanB, Cat, and BlaZ were not detected in draft E. faecium genomes in this database.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants R01AI072360 and P01AI083214 to M.S.G. and T32EY007145 to K.L.P.
Biostatistical consulting support was obtained from Douglas Hayden of the Harvard Catalyst | The Clinical and Translational Science Center, and with helpful input from Michael Caparon, Washington University.
The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of Harvard Catalyst, Harvard University, or its affiliated academic health care centers.
We gratefully thank Ian Jorgeson, Janet Manson, and members of the Broad Institute and the Gilmore laboratory for helpful discussions.
Footnotes
Citation Palmer, K. L., and M. S. Gilmore. 2010. Multidrug-resistant enterococci lack CRISPR-cas. mBio 1(4):e00227-10. doi:10.1128/mBio.00227-10.
SUPPLEMENTAL MATERIAL
REFERENCES
- 1. Top J., Willems R., Bonten M. 2008. Emergence of CC17 Enterococcus faecium: from commensal to hospital-adapted pathogen. FEMS Immunol. Med. Microbiol. 52:297–308 [DOI] [PubMed] [Google Scholar]
- 2. Werner G., Coque T. M., Hammerum A. M., Hope R., Hryniewicz W., Johnson A., Klare I., Kristinsson K. G., Leclercq R., Lester C. H., Lillie M., Novais C., Olsson-Liljequist B., Peixe L. V., Sadowy E., Simonsen G. S., Top J., Vuopio-Varkila J., Willems R. J., Witte W., Woodford N. 2008. Emergence and spread of vancomycin resistance among enterococci in Europe. Euro. Surveill. 13(47):p ii=19046 [PubMed] [Google Scholar]
- 3. Hidron A. I., Edwards J. R., Patel J., Horan T. C., Sievert D. M., Pollock D. A., Fridkin S. K. 2008. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect. Control Hosp. Epidemiol. 29:996–1011 [DOI] [PubMed] [Google Scholar]
- 4. Kak V., Chow J. W. 2002. Acquired antibiotic resistances in enterococci, p. 355–383 In Gilmore M. S., The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC [Google Scholar]
- 5. Weigel L. M., Clewell D. B., Gill S. R., Clark N. C., McDougal L. K., Flannagan S. E., Kolonay J. F., Shetty J., Killgore G. E., Tenover F. C. 2003. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302:1569–1571 [DOI] [PubMed] [Google Scholar]
- 6. Flannagan S. E., Chow J. W., Donabedian S. M., Brown W. J., Perri M. B., Zervos M. J., Ozawa Y., Clewell D. B. 2003. Plasmid content of a vancomycin-resistant Enterococcus faecalis isolate from a patient also colonized by Staphylococcus aureus with a VanA phenotype. Antimicrob. Agents Chemother. 47:3954–3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhu W., Clark N. C., McDougal L. K., Hageman J., McDonald L. C., Patel J. B. 2008. Vancomycin-resistant Staphylococcus aureus isolates associated with Inc18-like vanA plasmids in Michigan. Antimicrob. Agents Chemother. 52:452–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sahm D. F., Kissinger J., Gilmore M. S., Murray P. R., Mulder R., Solliday J., Clarke B. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ruiz-Garbajosa P., Bonten M. J., Robinson D. A., Top J., Nallapareddy S. R., Torres C., Coque T. M., Canton R., Baquero F., Murray B. E., del Campo R., Willems R. J. 2006. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J. Clin. Microbiol. 44:2220–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McBride S. M., Fischetti V. A., Leblanc D. J., Moellering R. C., Jr., Gilmore M. S. 2007. Genetic diversity among Enterococcus faecalis. PLoS One 2:e582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Paulsen I. T., Banerjei L., Myers G. S., Nelson K. E., Seshadri R., Read T. D., Fouts D. E., Eisen J. A., Gill S. R., Heidelberg J. F., Tettelin H., Dodson R. J., Umayam L., Brinkac L., Beanan M., Daugherty S., DeBoy R. T., Durkin S., Kolonay J., Madupu R., Nelson W., Vamathevan J., Tran B., Upton J., Hansen T., Shetty J., Khouri H., Utterback T., Radune D., Ketchum K. A., Dougherty B. A., Fraser C. M. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071–2074 [DOI] [PubMed] [Google Scholar]
- 12. Shankar N., Baghdayan A. S., Gilmore M. S. 2002. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature 417:746–750 [DOI] [PubMed] [Google Scholar]
- 13. Willems R. J., Top J., van Santen M., Robinson D. A., Coque T. M., Baquero F., Grundmann H., Bonten M. J. 2005. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg. Infect. Dis. 11:821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Galloway-Pena J. R., Nallapareddy S. R., Arias C. A., Eliopoulos G. M., Murray B. E. 2009. Analysis of clonality and antibiotic resistance among early clinical isolates of Enterococcus faecium in the United States. J. Infect. Dis. 200:1566–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leavis H. L., Willems R. J., van Wamel W. J., Schuren F. H., Caspers M. P., Bonten M. J. 2007. Insertion sequence-driven diversification creates a globally dispersed emerging multiresistant subspecies of E. faecium. PLoS Pathog. 3:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tannock G. W., Cook G. 2002. Enterococci as members of the intestinal microflora of humans, p. 101–132 In Gilmore M. S., The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC [Google Scholar]
- 17. Bourgogne A., Garsin D. A., Qin X., Singh K. V., Sillanpaa J., Yerrapragada S., Ding Y., Dugan-Rocha S., Buhay C., Shen H., Chen G., Williams G., Muzny D., Maadani A., Fox K. A., Gioia J., Chen L., Shang Y., Arias C. A., Nallapareddy S. R., Zhao M., Prakash V. P., Chowdhury S., Jiang H., Gibbs R. A., Murray B. E., Highlander S. K., Weinstock G. M. 2008. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 9:R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marraffini L. A., Sontheimer E. J. 2010. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 11:181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Horvath P., Barrangou R. 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327:167–170 [DOI] [PubMed] [Google Scholar]
- 20. Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D. A., Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712 [DOI] [PubMed] [Google Scholar]
- 21. Deveau H., Barrangou R., Garneau J. E., Labonte J., Fremaux C., Boyaval P., Romero D. A., Horvath P., Moineau S. 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 190:1390–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brouns S. J., Jore M. M., Lundgren M., Westra E. R., Slijkhuis R. J., Snijders A. P., Dickman M. J., Makarova K. S., Koonin E. V., van der Oost J. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marraffini L. A., Sontheimer E. J. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grissa I., Vergnaud G., Pourcel C. 2007. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Horvath P., Coute-Monvoisin A. C., Romero D. A., Boyaval P., Fremaux C., Barrangou R. 2009. Comparative analysis of CRISPR loci in lactic acid bacteria genomes. Int. J. Food Microbiol. 131:62–70 [DOI] [PubMed] [Google Scholar]
- 26. Palmer K. L., Carniol K., Manson J. M., Heiman D., Shea T., Young S., Zeng Q., Gevers D., Feldgarden M., Birren B., Gilmore M. S. 2010. High-quality draft genome sequences of 28 Enterococcus sp. isolates. J. Bacteriol. 192:2469–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Grissa I., Vergnaud G., Pourcel C. 2007. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35:W52–W57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haft D. H., Selengut J., Mongodin E. F., Nelson K. E. 2005. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 1:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weaver K. E., Rice L. B., Churchward G. 2002. Plasmids and transposons, p. 219–263 In Gilmore M. S., The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC [Google Scholar]
- 30. Roberts A. P., Mullany P. 2009. A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol. 17:251–258 [DOI] [PubMed] [Google Scholar]
- 31. van Schaik W., Top J., Riley D. R., Boekhorst J., Vrijenhoek J. E., Schapendonk C. M., Hendrickx A. P., Nijman I. J., Bonten M. J., Tettelin H., Willems R. J. 2010. Pyrosequencing-based comparative genome analysis of the nosocomial pathogen Enterococcus faecium and identification of a large transferable pathogenicity island. BMC Genomics 11:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Horvath P., Romero D. A., Coute-Monvoisin A. C., Richards M., Deveau H., Moineau S., Boyaval P., Fremaux C., Barrangou R. 2008. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 190:1401–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Manson J. M., Hancock L. E., Gilmore M. S. 2010. Mechanism of chromosomal transfer of Enterococcus faecalis pathogenicity island, capsule, antimicrobial resistance, and other traits. Proc. Natl. Acad. Sci. U. S. A. 107:12269–12274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Clewell D. B., Dunny G. 2002. Conjugation and genetic exchange in enterococci, p. 265–300 In Gilmore M. S., The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC [Google Scholar]
- 35. Homan W. L., Tribe D., Poznanski S., Li M., Hogg G., Spalburg E., Van Embden J. D., Willems R. J. 2002. Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 40:1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.