Abstract
Innate immunity represents the first line of defense in animals. We report a genome-wide in vivo Drosophila RNA interference screen to uncover genes involved in susceptibility or resistance to intestinal infection with the bacterium Serratia marcescens. We employed first whole-organism gene suppression followed by tissue-specific silencing in gut epithelium or hemocytes to identify several hundred genes involved in intestinal anti-bacterial immunity. Among the pathways identified, we showed that the JAK-STAT signaling pathway controls host defense in the gut by regulating stem cell proliferation and thus epithelial cell homeostasis. Thus, we revealed multiple genes involved in anti-bacterial defense and the regulation of innate immunity.
REPORT
Drosophila melanogaster provides a powerful model that allows the dissection of the innate immune response at the organism level. Drosophila innate immunity is comprised of a humoral and a cellular immune response. The majority of our knowledge on Drosophila immunity is based on injection of non-pathogenic bacteria (1-3); however, this bypasses the initial steps of naturally occurring infections – namely the physical barriers and the local, mucosal immune response. Intestinal immunity is currently the focus of intense research (4). In contrast to the human digestive tract, Drosophila lacks mammalian-like adaptive immunity and so relies entirely upon an innate immune system for protection against invading pathogens.
The intestinal infection model using pathogenic Serratia marcescens allows for the detailed analysis of local intestinal immunity and phagocytosis (5). S. marcescens is a gram-negative, opportunistic pathogen that can infect a range of hosts including Drosophila, Caenorhabditis elegans, and mammals (6, 7). Using ubiquitous RNA interference (RNAi)-mediated suppression, we performed an inducible genome-wide in vivo screen in Drosophila for novel innate immune regulators after S. marcescens infection (Fig. 1A, fig. S1A and B, Suppl. Text). To confirm our experimental approach we assayed various members of the Immune deficiency (IMD) and Toll pathways, the two major fly immune signaling cascades (Fig. 1B) (1, 2, 3). RNAi lines targeting several IMD members resulted in significantly reduced survival upon infection with S. marcescens, whereas suppression of Toll pathway components had a less dramatic effect, supporting previous reports that the immune response to S. marcescens is IMD-dependent and Toll-independent (Fig. 1B) (5). Notably, not all members of the IMD pathway such as imd, rel and ird5 were picked up using our screening criteria, most likely due to inefficient RNAi silencing (Fig. 1B) (8).
Fig. 1.
Analysis of genome-wide in vivo RNAi screen. (A) Total data of all RNAi lines screened for survival after S. marcescens infections. Data were analyzed as the time, in days, when 50% of the total number of flies had died. All data were normalized to the daily LT50 mean of an experimental cohort. In all experiments the cohort ranged from 80-200 lines. Hits were defined by susceptible (red dashed line) and resistant (blue dashed line) cut-offs, i.e. 1.5 standard deviations (SD) below the mean and 2 SD above the mean, respectively, based on the pilot screen and controls. (B). Effect of RNAi knock-down of IMD and Toll pathway components on their survival against S. marcescens infection. SCOREs are shown for each line as described in Methods. The dashed lines indicate the cut-offs used for resistance (+2 SD) and susceptibility (−1.5. SD) candidates. (C) Percentage distribution of gene ontology (GO) annotated genes to biological processes for susceptible candidates.
We assayed 13053 RNAi lines (8) representing 10689 different genes (78% of the genome) against intestinal infection with S. marcescens (fig. S2A and Table S1 and S2). 8.6% (885 genes) were defined as hits, of which the majority (89.3%; 790 genes) were susceptible candidates (fig. S2A and Table S3). On the basis of gene ontology (GO) annotations, susceptible candidates were classified according to their predicted biological processes. Genes involved in signaling, intracellular protein transport, and transcriptional regulation were overly represented among the entire data set (Fig. 1C). We also found marked enrichment for genes that regulate phagocytosis, defense responses, vesicle trafficking, and proteolysis. Several candidate RNAi lines represented genes that have been previously implicated in mounting an effective immune response (9-18) (Table S3).
Our approach also allowed us to identify negative regulators of Drosophila host defense (Fig. 1A). We identified 95 genes (10.7% of the total hits) that confer resistance to S. marcescens infections when silenced (fig. S2A and B, Table S4), none of which had previously been characterized as negative regulators of innate immunity. Thus, our genome wide screen revealed previously known genes associated with Drosophila immunity and more than 800 additional candidate genes implicated in innate immunity; 40% of which had unknown function.
We retested some of our susceptible and resistant RNAi hits in the gut epithelium and the macrophage-like hemocytes, the two major cell types associated with our infection model, using cell-type specific driver lines, NP1-GAL4 and HML-GAL4 respectively (5, 19). We prioritized genes of interest by selecting the primary hits that have mammalian (mouse and/or human) orthologues. Of the 358 susceptible hits tested with the HML-GAL4 driver, RNAi against 98 genes (27%) resulted in significantly reduced survival as compared to RNAi controls indicating that these genes function in hemocytes to combat intestinal S. marcescens infections (Fig. 2A, fig. S3A and Table S5). Using the NP1-GAL4 driver (fig. S4), of the 337 genes tested, RNAi against 129 genes (38%) resulted in significantly reduced survival, suggesting that these genes play an important role in host intestinal defense (Fig. 2B, fig. S3B and Table S6). Of the resistance hits, 37 HML-GAL4 RNAi candidates (79%) and 28 NP1-GAL4 RNAi candidates (61%) exhibited markedly enhanced survival (Fig. 2C and D, fig. S3 and Tables S7-S9). 54 candidate genes functioned in both hemocytes and gut (fig. S3). Multiple susceptibility as well as resistance genes were tested 3-15 independent times, using ≥2 RNAi transformants to exclude position effects and second independent RNAi hairpins to confirm the target gene when available (Fig. 2A-D, fig. S3 and Tables S5-S8). To exclude a potential developmental phenotype, we have tested most candidate lines by feeding flies on a sugar diet in the absence of bacteria (Table S9). Thus, we have identified multiple regulators in hemocytes and/or gut epithelium that confer susceptibility or resistance to S. marcescens infections.
Fig. 2.
Mapping and validation of conserved hits in the gut and hemocytes. (A and B) Survival graphs showing susceptible hits tested 3-15 times with several transformants and hairpins in hemocytes (A) and gut epithelium (B). The kenny mutant line (key Mut) is shown as a positive control.Values are mean ± SEM, n ≥ 3 experiments with 20 flies in each. *, p < 0.05 (Welch t-test). (C and D) Survival graphs showing resistant hits tested 3-15 times with several transformants and hairpins in hemocytes (C) and gut epithelium (D). The kenny mutant line (key Mut) is shown as a positive control. Bars represent mean ± SEM, n ≥ 3 experiments with 20 flies in each. *,P < 0.05 (Welch t-test). (E) Statistically enriched biological processes superimposed upon a sketch depicting a gut epithelial cell with their corresponding p-value in the gut associated with S. marcescens infection are shown. Green indicates processes to which susceptible candidates are exclusively attributed. Red indicates processes to which resistant candidates are exclusively attributed. Blue indicates processes to which both susceptible and resistant candidates can attributed. See also Table S10 for annotation of genes involved in each process. All processes shown display P < 0.05 (Fischer Test).
Using gene ontology enrichment analysis, we classified our tissue-specific candidates into statistically significant biological processes. In the intestinal tract, intracellular processes such as endocytosis and exocytosis, proteolysis, vesicle-mediated transport, and stress response all appeared significantly enriched (Fig. 2E, fig. S5-7 and Table S10). We also observed a marked enhancement of genes associated with immune system development, growth, stem cell division and cell death, suggesting an important role of these processes in the gut during S. marcescens infection. In hemocytes, ontology enrichment analysis revealed a strong enrichment in several processes linked to phagocytosis including endocytosis, response to external stimuli and vesicle trafficking (Figs. S8-10 and Table S11). In both cell types, deregulation of the stress response as well as amine/nitrogen metabolism resulted in enhanced resistance to S. marcescens challenge (Fig. 2E and fig. S8).
We next performed Kegg pathway analysis to identify enriched gene sets that might be involved in S. marcescens infections. Kegg profiling on the susceptible genome-wide candidates (Table S12) showed the importance of the IMD pathway in our infection model and also pointed to a possible role of Notch and transforming growth factor-β signaling, pathways, which have previously been difficult to study in an infection setting due to a lack of adult viable mutants (22, 23). Moreover, our analysis revealed prominent involvement of the Janus kinase-Signal transducer and activator and transcription (JAK-STAT) pathway during S. marcescens infection. In Drosophila the JAK-STAT pathway plays an important role in haematopoiesis, stress responses, stem cell proliferation and anti-viral immunity but a role in the defense against natural bacterial pathogens is unknown (24-27). We therefore sought to validate our analysis and focused on how JAK-STAT signaling regulates the host response during S. marcescens infection.
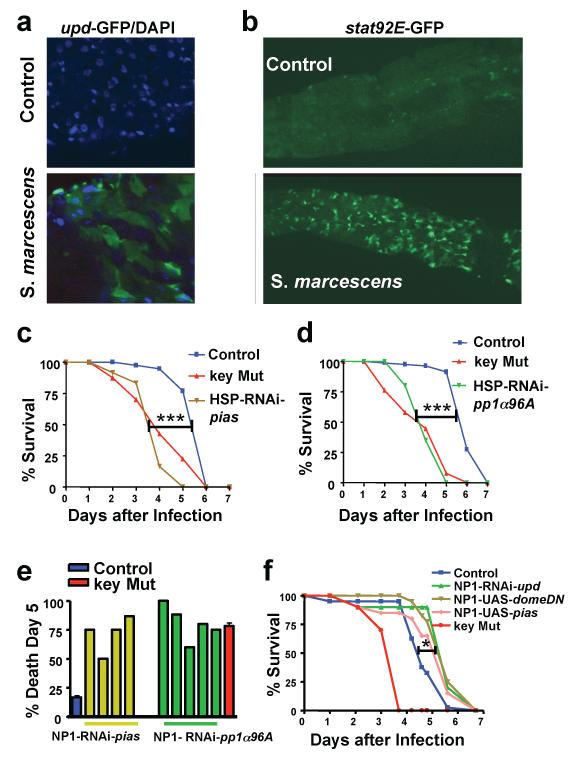
To investigate whether the JAK-STAT pathway is activated during S. marcescens infection, we used transgenic reporter lines (25, 28, 29) in which GFP is expressed under the control of unpaired (upd) and upd-3, which encode two ligands for Domeless (the receptor of the JAK-STAT pathway). We observed upd-GFP and upd3-GFP expression in the gut of S. marcescens infected flies (Fig. 3A and figs. S11 and S12). Moreover, we demonstrated intestinal activation of the JAK-STAT pathway by using a stat92E-binding-site-GFP reporter line (Fig. 3B) (28, 29). Upon ligation of UPD or UPD3 to Domeless, Stat92E translocates to the nucleus and activates reporter GFP gene expression (26). To confirm the relevance of JAK-STAT activation for S. marcescens infections, we performed global (Fig. 3, C and D) and gut-specific (Fig. 3E) RNAi-mediated silencing of PIAS (also called Su(var)-10) and PP1α96A, two negative regulators of JAK/STAT signaling (30, 31). In both RNAi lines, we observed significantly earlier death compared to control flies (Fig. 3, C-E). The role of PP1α96A in intestinal immunity was also validated using a sensitized background (fig. S12). In contrast, partial pathway inhibition via gut specific over-expression of PIAS (NP1-UAS-pias), dominant-negative domeless (NP1-UAS-domeDN), or RNAi-mediated silencing of the domeless ligand, UPD (NP1-RNAi-upd) significantly increased the survival of Serratia-challenged flies (Fig. 3F). Thus, the JAK-STAT pathway activation in the gut negatively regulates survival in response to an intestinal S. marcescens infection.
Fig. 3.
The JAK/STAT pathway controls S. marcescens susceptibility in the gut. (A) GFP (green) and DAPI (blue) expression in the gut of transgenic upd-GFP flies on day 4 after infection with S. marcescens at 25°C compared to control, non-pathogenic conditions. Also shown is nuclear DAPI (blue) staining. (B) GFP (green) expression in the gut of transgenic stat92E-GFP flies under S. marcescens-infected and control conditions on day 4 at 25°C. (C) Survival curves of S. marcescens-infected RNAi lines against the negative JAK-STAT pathway regulator PIAS driven by the ubiquitously-expressed HSP-GAL4 driver compared to control and key mutant flies. (D) Survival curves of S. marcescens-infected RNAi lines targeting the negative JAK-STAT regulator PP1α96A driven by the ubiquitously-expressed HSP-GAL4 driver compared to control and key mutant flies. (E) Survival graph representing individual tests of RNAi-mediated silencing of PIAS and PP1α96A specifically in the gut (NP1 driver) after S. marcescens challenge at 29°C, compared to control and key mutant flies. (F) Survival curves of NP1-RNAi-upd, NP1-UAS-pias, and NP1-UAS-domeDN lines at 29°C compared to control and key mutant flies following S. marcescens feeding. * P ≤ 0.05; *** P ≤ 0.0001 (Logrank test). upd, unpaired; DN, dominant-negative.
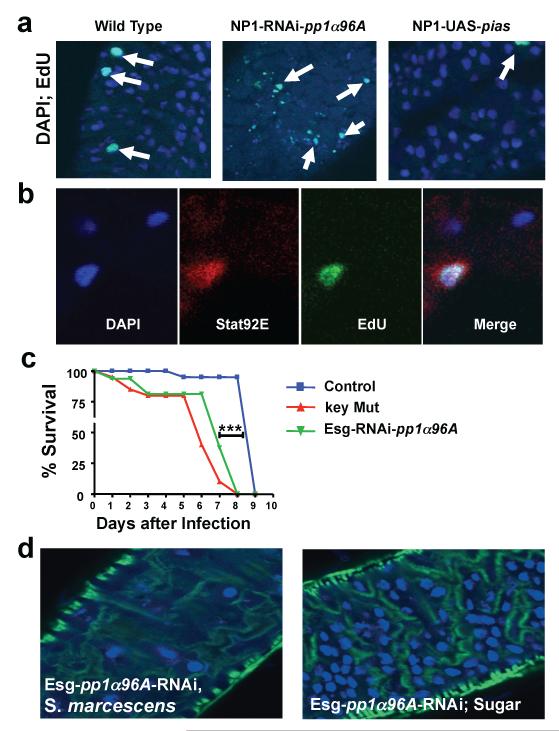
To elucidate a possible mechanism in which JAK-STAT is involved in host defense against S. marcescens, we analyzed the effects of infection on gut epithelium. Infected flies exhibited massive death of intestinal epithelial cells (fig. S14A) and compensatory proliferation (fig. S14B and C). Enhanced JAK-STAT signaling, through the use of NP1-RNAi- pp1α96A flies, resulted in a marked reduction in the number of large, polyploid nuclei, which signify differentiated enterocytes (32), after five days of infection (Fig. 4A). Epithelial morphology (fig. S15A) as well as survival on normal food (fig. S15B) were comparable between control, NP1-RNAi-pp1α96A, NP1-UAS-pias, and NP1-UAS-domeDN fly lines. We next assessed whether JAK-STAT signaling affected cellular proliferation of the epithelium. We found that DNA synthesis in epithelial cells was reduced when JAK-STAT signaling was impaired and significantly increased by silencing pp1α96A in the gut, both in the presence and absence of infection (Fig. 4A and fig. S16). Thus, JAK-STAT signaling enhances epithelial cell death and positively regulates compensatory proliferation of intestinal cells, also after S. marcescens infection.
Fig. 4.
Impaired epithelial integrity and control of intestinal stem cell homeostasis upon S. marcescens challenge. (A) Analysis of gut epithelium integrity using DAPI (blue) and intestinal proliferation using EdU (green) staining; EdU was injected into flies just 3 hours prior to dissections. Samples were assayed on day 5 after S. marcescens infection at 25°C. (B) Representative confocal image showing JAK-STAT pathway activation in EdU-positive nuclei in an intestinal stem cell using the stat92E-GFP reporter line. Data are from day 5 following S. marcescens challenge. In a total of 3 experiments and 39 gut dissections, we detected 13 cells with small nuclei (DAPI – blue) that were positive both for 10×STAT-GFP (red) and positive for EdU (green) staining in the region anterior to the copper cells while no such cells were observed in 42 non-infected control guts (p<0,003 Student T-test). EdU was injected 3h prior to dissection. (C) Survival curves of S. marcescens-infected Drosophila in which PP1α96A is specifically silenced in intestinal stem cells of adult flies using escargot (Esg)-GAL4;tubulinGal80ts at 25°C. Control and key mutant lines are shown for comparison. *** P < 0.0001 (Logrank Test). (D) Integrity of gut epithelium in Esg-pp1a96A-RNAi lines kept under non-pathogenic conditions or 5 days after S. marcescens infections at 25°C. Nuclei were visualized with DAPI (blue) and actin visualised with phalloidin (green).
We next examined whether the JAK-STAT pathway was affecting intestinal cell homeostasis specifically through the resident stem cell compartment. Basal intestinal stem cells (ISCs) can be distinguished from apical enterocytes on the basis of a characteristic smaller nuclear morphology (32, 33). Using the stat92E-GFP reporter line to image JAK-STAT activation, the JAK-STAT pathway was selectively induced in the ISCs and not in mature enterocytes (fig. S17). Moreover, upon infection of stat92E-GFP flies with S. marcescens, we observed GFP expression also in small, EdU-positive cells suggesting that JAK-STAT signaling regulates ISC proliferation during S. marcescens infection (Fig. 4B). To definitively demonstrate that this pathway acts in gut stem cells and that this compartment controls susceptibility to S. marcescens infections, we silenced pp1α96A in adult ISCs using an escargot-GAL4 driver line. Escargot is a specific marker of ISCs (32). ISC-specific suppression of PP1α96A resulted in early lethality in response to S. marcescens infection whereas flies remained viable under non-pathogenic conditions (Fig. 4C and fig. S18). Furthermore, the guts of escargot-GAL4-pp1α96A-RNAi flies showed a phenotype similar to that obtained using the gut-specific NP1 driver, namely severely depleted mature enterocytes (Fig. 4, A and D). Thus, our data demonstrate that JAK-STAT signaling is required for ISC homeostasis and implicates ISCs as a critical component of host defense to mucosal S. marcescens infections.
Our global experimental approach allows a comprehensive dissection of the biological processes that may regulate host defense to a bacterial infection at the organism level. Besides revealing previously known immune pathways, we uncovered more than 800 additional genes, many of which were of unknown function. Furthermore, our data demonstrate that host defense may involve many processes that are not limited to classical innate immune response pathways, as exemplified here by the role of the JAK-STAT pathway in the regulation of epithelial homeostasis in response to infection. In addition, we validate and map conserved candidates to intestinal cells and hemocytes thus allowing us to define a blueprint of processes involved in host defense against S. marcescens infection. As all genes analyzed here have been conserved during evolution, it is likely that some of the processes that are important in flies are also relevant to mammalian host defense (20, 21).
Supplementary Material
Acknowledgments
34. We thank all members of our laboratories and the VDRC for helpful discussions and technical support. We are grateful to M. Novatchakova and M. Lafarge for expert technical help. We thank the Drosophila Resource Center of the National Institute of Genetics of Japan for midgut Gal4 driver stocks and B. Mathey-Prévot for the HML-GAL4 line. We thank J. Mutterer for help with confocal microscopy. This work is supported financially by the CNRS, a NIH Program grant PO1 AI44220, and a DROSELEGANS grant from the Agence Nationale de la Recherche (MIME). The DF laboratory is a “Equipe FRM”, awarded by the Fondation pour la Recherche Médicale. JMP is supported by IMBA, EuroThymaide, an FWF-SFB grant, an advanced ERC grant, and the Austrian Ministry of Science. RMS is supported by the WWTF and the DFG (Ha-1628/8-1). The screen was supported by Boehringer Ingelheim.
References and Notes
- 1.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. Cell. 1996 Sep 20;86:973. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 2.Ferrandon D, Imler JL, Hetru C, Hoffmann JA. Nat Rev Immunol. 2007 Nov;7:862. doi: 10.1038/nri2194. [DOI] [PubMed] [Google Scholar]
- 3.Lemaitre B, Hoffmann J. Annu Rev Immunol. 2007;25:697. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 4.Artis D. Nat Rev Immunol. 2008 Jun;8:411. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- 5.Nehme NT, et al. PLoS Pathog. 2007 Nov;3:e173. doi: 10.1371/journal.ppat.0030173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flyg C, Kenne K, Boman HG. J Gen Microbiol. 1980 Sep;120:173. doi: 10.1099/00221287-120-1-173. [DOI] [PubMed] [Google Scholar]
- 7.Kurz CL, et al. Embo J. 2003 Apr 1;22:1451. doi: 10.1093/emboj/cdg159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dietzl G, et al. Nature. 2007 Jul 12;448:151. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 9.Cheng LW, et al. Proc Natl Acad Sci U S A. 2005 Sep 20;102:13646. doi: 10.1073/pnas.0506461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stroschein-Stevenson SL, Foley E, O’Farrell PH, Johnson AD. PLoS Biol. 2006 Jan;4:e4. doi: 10.1371/journal.pbio.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stuart LM, et al. Nature. 2007 Jan 4;445:95. doi: 10.1038/nature05380. [DOI] [PubMed] [Google Scholar]
- 12.Boutros M, et al. Science. 2004 Feb 6;303:832. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- 13.Lloyd TE, et al. Neuron. 2000 Apr;26:45. doi: 10.1016/s0896-6273(00)81136-8. [DOI] [PubMed] [Google Scholar]
- 14.Boutros M, Agaisse H, Perrimon N. Dev Cell. 2002 Nov;3:711. doi: 10.1016/s1534-5807(02)00325-8. [DOI] [PubMed] [Google Scholar]
- 15.Lu Y, Wu LP, Anderson KV. Genes Dev. 2001 Jan 1;15:104. doi: 10.1101/gad.856901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park JM, et al. Genes Dev. 2004 Mar 1;18:584. doi: 10.1101/gad.1168104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vodovar N, et al. Proc Natl Acad Sci U S A. 2005 Aug 9;102:11414. doi: 10.1073/pnas.0502240102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wojcik C, DeMartino GN. J Biol Chem. 2002 Feb 22;277:6188. doi: 10.1074/jbc.M109996200. [DOI] [PubMed] [Google Scholar]
- 19.Kocks C, et al. Cell. 2005 Oct 21;123:335. [Google Scholar]
- 20.Cadwell K, et al. Nature. 2008 Nov 13;456:259. [Google Scholar]
- 21.Saitoh T, et al. Nature. 2008 Nov 13;456:264. [Google Scholar]
- 22.Lai EC. Development. 2004 Mar;131:965. doi: 10.1242/dev.01074. 2004. [DOI] [PubMed] [Google Scholar]
- 23.Raftery LA, Sutherland DJ. Dev Biol. 1999 June 15;210:251. doi: 10.1006/dbio.1999.9282. [DOI] [PubMed] [Google Scholar]
- 24.Agaisse H, Perrimon N. Immunol Rev. 2004 Apr;198:72. doi: 10.1111/j.0105-2896.2004.0133.x. [DOI] [PubMed] [Google Scholar]
- 25.Agaisse H, Petersen UM, Boutros M, Mathey-Prevot B, Perrimon N. Dev Cell. 2003 Sep;5:441. doi: 10.1016/s1534-5807(03)00244-2. [DOI] [PubMed] [Google Scholar]
- 26.Buchon N, Broderick NA, Poidevin M, Pradervand S, Lemaitre B. Cell Host Microbe. 2009 Feb 19;5:200. doi: 10.1016/j.chom.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Singh SR, Liu W, Hou XS. Cell Stem Cell. 2007;1:191–203. doi: 10.1016/j.stem.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bach EA, et al. Gene Expr Patterns. 2007 Jan;7:323. doi: 10.1016/j.modgep.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 29.Tsai YC, Sun YH. Genesis. 2004 Jun;39:141. doi: 10.1002/gene.20035. [DOI] [PubMed] [Google Scholar]
- 30.Betz A, Lampen N, Martinek S, Young MW, Darnell JE., Jr. Proc Natl Acad Sci U S A. 2001 Aug 14;98:9563. doi: 10.1073/pnas.171302098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller P, Kuttenkeuler D, Gesellchen V, Zeidler MP, Boutros M. Nature. 2005 Aug 11;436:871. doi: 10.1038/nature03869. [DOI] [PubMed] [Google Scholar]
- 32.Micchelli CA, Perrimon N. Nature. 2006 Jan 26;439:475. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- 33.Ohlstein B, Spradling A. Nature. 2006 Jan 26;439:470. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.