Abstract
Cellular metabolism is a series of tightly linked oxidations and reductions that must be balanced. Recycling of intracellular electron carriers during fermentation often requires substrate conversion to undesired products, while respiration demands constant addition of electron acceptors. The use of electrode-based electron acceptors to balance biotransformations may overcome these constraints. To test this hypothesis, the metal-reducing bacterium Shewanella oneidensis was engineered to stoichiometrically convert glycerol into ethanol, a biotransformation that will not occur unless two electrons are removed via an external reaction, such as electrode reduction. Multiple modules were combined into a single plasmid to alter S. oneidensis metabolism: a glycerol module, consisting of glpF, glpK, glpD, and tpiA from Escherichia coli, and an ethanol module containing pdc and adh from Zymomonas mobilis. A further increase in product yields was accomplished through knockout of pta, encoding phosphate acetyltransferase, shifting flux toward ethanol and away from acetate production. In this first-generation demonstration, conversion of glycerol to ethanol required the presence of an electrode to balance the reaction, and electrode-linked rates were on par with volumetric conversion rates observed in engineered E. coli. Linking microbial biocatalysis to current production can eliminate redox constraints by shifting other unbalanced reactions to yield pure products and serve as a new platform for next-generation bioproduction strategies.
IMPORTANCE
All reactions catalyzed by whole cells or enzymes must achieve redox balance. In rare cases, conversion can be achieved via perfectly balanced fermentations, allowing all electron equivalents to be recovered in a single product. In most biotransformations, organisms must produce a mixture of acids, gasses, and/or alcohols, and no amount of enzyme or strain engineering can overcome this fundamental requirement. Stoichiometric conversion of glycerol, a waste product from biodiesel transesterification, into ethanol and CO2 with no side products represents such an impossible fermentation, due to the more reduced state of glycerol than of ethanol and CO2. The unbalanced conversion of glycerol to ethanol has been viewed as having only two solutions: fermenting glycerol to ethanol and potentially useful coproducts or “burning off” excess electrons via careful introduction of oxygen. Here, we use the glycerol-to-ethanol example to demonstrate a third strategy, using bacteria directly interfaced to electrodes.
Introduction
The need to efficiently convert feedstocks into products or fuels, combined with the development of tools for metabolic engineering, has produced many novel strains able to catalyze useful fermentations. However, the need to achieve redox balance limits the range of possible bioconversions, and the synthesis of mixtures of end products increases bioseparation costs. One possible solution to this issue is to harness the ability of bacteria able to transfer electrons to electrodes. Some microorganisms have previously been shown to utilize exogenous redox mediators (e.g., thionine, neutral red, or ferricyanide) to balance redox reactions (1–4). The recent discovery that certain bacteria can natively transfer electrons directly to electrodes (5, 6), a process called extracellular electron transfer, has enabled a strategy for balancing bioconversions. Extracellular electron transfer allows bacteria to utilize an electrode as an external sink for electrons, eliminating the necessity for the production of undesired side products, thereby facilitating stoichiometric conversion of substrate to product while simultaneously generating electrical current.
To demonstrate the principle of such an electrobiocatalyst, we sought to achieve production of ethanol (and CO2) from glycerol without the need to excrete redox-balancing end products, such as formate or 1,2 propanediol, using the facultative anaerobe Shewanella oneidensis. S. oneidensis possesses the ability to respire electrons to insoluble substrates, such as electrodes (7, 8). With a well-studied, diverse metabolism and a sequenced genome (9) amenable to genetic manipulations, S. oneidensis is well suited to demonstrate a concept applicable to a variety of environmentally diverse electrode-respiring organisms (5, 6).
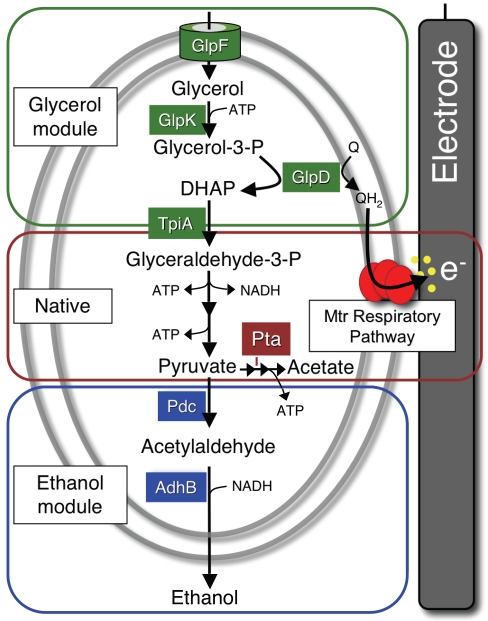
Unlike common industrial hosts, such as Escherichia coli, S. oneidensis is naturally equipped for electron transfer to electrode surfaces. Importantly, it localizes a well-characterized protein complex (MtrCAB) to span the periplasm and outer membrane, in addition to an inner membrane tetraheme protein (CymA) capable of transferring electrons from the respiratory quinone pool to MtrCAB (7, 10, 12). The MtrCAB complex can directly reduce insoluble substrates (7, 12), though its activity is dramatically increased in the presence of low concentrations of flavins (12, 13). Flavins have recently been shown to be secreted by Shewanella (14, 15) and are themselves reduced by the Mtr pathway (16). These electron transfer capabilities are the foundation for the demonstration of a process to enable an otherwise-unbalanced fermentation utilizing bacteria interfaced with an electrode. Two genetic modules were engineered into S. oneidensis to allow for glycerol utilization and ethanol production in a manner that would feed directly into this electrode respiration machinery (Fig. 1).
FIG 1 .
Metabolic modules added to S. oneidensis to enable electrode-dependent conversion of glycerol to ethanol. The glycerol utilization module from E. coli (section outlined in green [top]) and the ethanol production module from Z. mobilis (blue section [bottom]) were combined with native metabolic pathways (red section [middle]) for stoichiometric non-redox-balanced conversion of glycerol to ethanol. Metabolic substrates, intermediates, and products are denoted in black. The genes from E. coli and Z. mobilis engineered into S. oneidensis are represented by the proteins that they encode: GlpF, glycerol transporter; GlpK, glycerol kinase; GlpD, glycerol-3-phosphate dehydrogenase; Pdc, pyruvate decarboxylase; and AdhB, alcohol dehydrogenase. Phosphotransacetylase (Pta) is shown from native metabolism. Electrons not redox balanced within the cell are subsequently transferred through the Mtr pathway to the electrode. Yellow dots represent flavins secreted naturally from cells to accelerate extracellular transfer of electrons (e−) to the electrode.
Here, we report the electrode-dependent conversion of glycerol to ethanol and CO2 in an engineered strain of S. oneidensis, and while our work focuses on solving this specific unbalanced redox reaction, in principle, our strategy can be broadly applied to any reaction where the substrate is more reduced than the desired product(s).
RESULTS
Engineering of S. oneidensis for glycerol uptake and utilization.
No Shewanella isolates tested to date have been shown to utilize glycerol as a sole carbon and energy source (17), and growth experiments confirmed this inability in S. oneidensis strain MR-1. To create a respiratory pathway linked to quinone reduction, unlike fermentative pathways developed for E. coli (18), three genes were predicted to be required: glpF, glpK, and glpD, which encode a glycerol facilitator, a glycerol kinase, and a membrane-bound quinone-linked glycerol-3-phosphate dehydrogenase, respectively (19). When these genes were introduced under the control of a lac promoter, glycerol kinase and glycerol dehydrogenase activities were detected in whole-cell lysates, but no growth or utilization of glycerol was observed by whole cells under any condition tested (data not shown). Only after introduction of tpiA, which encodes a triosephosphate isomerase responsible for the isomerization of dihydroxyacetone phosphate (DHAP) and 3-phosphoglyceraldehyde, were glycerol consumption and cell growth observed. A requirement for increased TpiA was consistent with the known competitive inhibition of GlpD by DHAP (20) and the use of Entner-Doudoroff glycolysis by S. oneidensis (21), which requires only low TpiA activity for gluconeogenic flux during utilization of its preferred substrate (lactate) as a carbon source.
Introduction of these four constitutively expressed genes from E. coli (Fig. 1, glycerol module) enabled S. oneidensis to utilize glycerol as a sole carbon and energy source. It is interesting to note that glpD encodes a flavoenzyme characterized as the aerobic dehydrogenase of E. coli (22), while another enzyme complex (encoded by glpABC) is used for the anaerobic utilization of glycerol by E. coli. S. oneidensis was able to utilize glycerol both aerobically and anaerobically with the GlpD dehydrogenase, suggesting that it is compatible with the menaquinone-dependent transfer of electrons via CymA to electrodes in S. oneidensis (11). Moreover, addition of antibiotics was not necessary to maintain the plasmid in cultures growing with glycerol due to the selective pressure presented by the carbon source.
Engineering of S. oneidensis for ethanol production.
Ethanol production has yet to be observed in S. oneidensis, despite the presence of numerous putative, annotated alcohol dehydrogenases in its genome. Attempts to increase pyruvate levels in S. oneidensis via deletion of acetate-producing pathways, as well as expression of typical acetyl coenzyme A (acetyl-CoA)-dependent alcohol dehydrogenases from E. coli, proved unsuccessful. Even in organisms with functional alcohol dehydrogenases, such as E. coli, it is well established that expression of two genes from Zymomonas mobilis (pdc, encoding pyruvate decarboxylase, and adh, encoding alcohol dehydrogenase) can significantly increase ethanol production rates (23, 24). The effectiveness of this route has been attributed to the high affinity of Pdc for pyruvate (25, 26). When pdc and adh were cloned and expressed constitutively in S. oneidensis (Fig. 1, ethanol module), pyruvate decarboxylase and alcohol dehydrogenase activities were observed in crude extracts, and ethanol production from lactate was observed in cell suspensions (data not shown).
Combination of glycerol utilization and ethanol production modules for conversion of glycerol into ethanol.
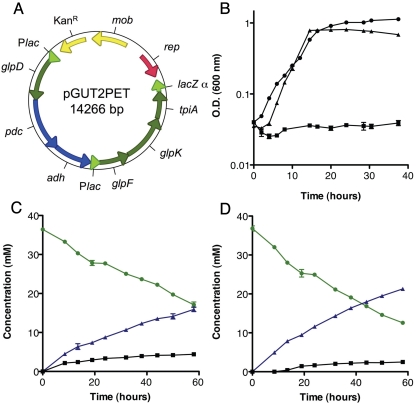
Glycerol utilization and ethanol production modules were incorporated into a single plasmid, encoding a total of six genes in two operons, each driven by a lac promoter. However, because Shewanella obtains more energy via substrate-level phosphorylation (by directing flux to acetate excretion) than via ethanol production (Fig. 1) (27), recombination events between lac promoter regions eliminating pdc and adh were common in initial constructs. However, by altering the gene order on the plasmid by cloning pdc and adh directly downstream of glpD, the selective pressure to maintain glycerol metabolism also maintained ethanol production even in Shewanella strains with native recombination mechanisms, resulting in the construct pGUT2PET (Fig. 2A). A series of 10 transfers of aerobic growth with glycerol yielded an adapted isolate of S. oneidensis with pGUT2PET that grew at rates comparable to those of wild-type E. coli provided with glycerol (Fig. 2B).
FIG 2 .
Glycerol utilization and ethanol production in engineered S. oneidensis strains. (A) Plasmid map of pGUT2PET. (B) Aerobic growth of S. oneidensis with pGUT2PET (solid triangles), S. oneidensis with an empty vector (solid squares), and E. coli K-12 (solid circles) on glycerol. Anaerobic resting cell fumarate batch reactions with the wild-type strain with pGUT2PET (C) and the ∆pta mutant with pGUT2PET (D). (C and D) Glycerol (green line and solid circles), ethanol (blue line and solid triangles), and acetate (black line and solid squares) concentrations were determined by analyzing culture supernatants. Error bars represent standard deviations of results from at least three independent experiments.
The pathway introduced by pGUT2PET was designed to enable conversion of glycerol to ethanol only when a mechanism was available for respiratory removal of two electrons. To demonstrate anaerobic glycerol conversion to ethanol and examine its requirement for an external electron sink, batch cultures with or without the soluble electron acceptor fumarate were used to evaluate metabolic flux. When resting cells were incubated in sealed anaerobic tubes in the absence of an electron acceptor, no fermentative conversion of glycerol to any end product was detected. However, when fumarate was available, cells consumed 19.3 ± 0.6 mM glycerol and produced 15.9 ± 1.0 mM ethanol and 4.4 ± 0.2 mM acetate as sole end products (Fig. 2C; Table 1). Recovery of electrons via fumarate reduction was stoichiometric, demonstrating that expression of these six genes was sufficient to enable conversion of glycerol to ethanol, with 82% of the carbon flux directed to ethanol and 23% to acetate.
TABLE 1 .
Total change in substrates and products from batch culture and bioreactor experiments with strains containing pGUT2PET
| Electron acceptor (expt) | Substrate/product | Change (mM) in production ina: | |
|---|---|---|---|
| WT (MR-1) | Δpta strain | ||
| Fumarate (batch) | Glycerol | −19.3 ± 0.6 | −24.3 ± 1.0 |
| Acetate | +4.4 ± 0.2 (23) | +2.5 ± 0.1 (10) | |
| Ethanol | +15.9 ± 1.0 (82) | +21.3 ± 0.5 (88) | |
| Electrode (bioreactor) | Glycerol | −36.1 ± 1.4 | −32.8 ± 1.5 |
| Acetate | +9.1 ± 1.1 (25) | +4.9 ± 1.3 (15) | |
| Ethanol | +26.9 ± 1.6 (75) | +27.8 ± 0.5 (85) | |
WT, wild type. Percentages of carbon flux going to acetate or ethanol are shown in parentheses.
In E. coli, a variety of pathways (including the succinate, formate, lactate, 1,2-propanediol, and acetate production pathways) must be eliminated to increase ethanol yields. In contrast, Shewanella excreted only acetate as a contaminating product (Fig. 2C), consistent with its anaerobic ATP generation strategy (27). Therefore, to enhance flux to ethanol, an in-frame deletion of the pta gene (encoding phosphate acetyltransferase), which is required for conversion of acetyl-CoA to acetyl-phosphate, was constructed in S. oneidensis (28–30). As predicted, transformation of the pGUT2PET plasmid into the ∆pta background resulted in a 43% decrease in acetate production and a 33% increase in ethanol production compared to levels in the wild type (Fig. 2D; Table 1). These results indicated that standard approaches to metabolic engineering could be utilized with Shewanella.
Balancing engineered metabolic pathways with electrodes.
Our ultimate goal was to utilize electrode-linked redox balancing to eliminate the production of any side products. As S. oneidensis/pGUT2PET strains were unable to convert glycerol to ethanol in the absence of electron acceptors, electrode-dependent conversion of glycerol to ethanol could be tested in a bioreactor containing an electrode poised at an oxidizing potential (Fig. 3A). In these strictly anaerobic reactors, a carbon fiber electrode was poised with a potentiostat to act as a constant electron acceptor at +0.4 V versus the standard hydrogen electrode, allowing electrons to be collected and returned to the reactor via a Pt counter electrode.
FIG 3 .
Bioelectrochemical conversion of glycerol to ethanol. (A) Three-electrode bioreactor setup for electrode-dependent conversion of glycerol to ethanol (EtOH). Glycerol consumption and ethanol production in the wild-type strain with pGUT2PET (B) and the ∆pta strain with pGUT2PET (C) on a graphite electrode. Glycerol (green line and solid circles) and acetate (black line and solid squares) concentrations were determined by HPLC. Error bars represent standard deviations of results from at least three independent experiments. Ethanol concentrations (blue line and solid triangles) were predicted based on average reaction stoichiometries for glycerol consumption and coulombic yields from three independent experiments. Abiotic or nonpoised electrode control maintained a constant glycerol concentration (open squares).
In electrode bioreactors, no glycerol consumption was observed by the wild-type or ∆pta strain containing pGUT2PET when the electrode was disconnected. Fermentation was not stimulated by vigorous flushing with oxygen-free nitrogen or argon, which will enable the fermentation of glycerol by E. coli (31), and no glycerol was consumed in sterile reactors when electrodes were held at an oxidizing potential (Fig. 3B and C).
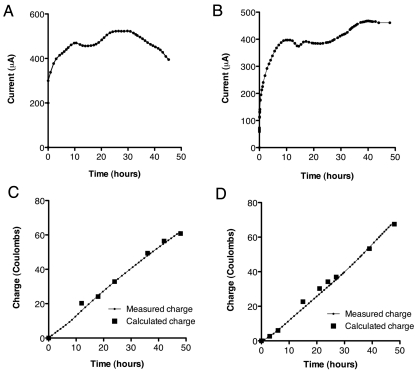
As predicted, inoculation of S. oneidensis/pGUT2PET (Fig. 4A) or the ∆pta strain containing pGUT2PET (Fig. 4B) into chambers containing electrodes poised at an oxidizing potential resulted in immediate anodic current flux and consumption of glycerol (Fig. 3B and C). Conversion of glycerol under these conditions was absolutely dependent upon the oxidizing electrode, demonstrating that this bioconversion was enabled by the flux of electrons to the electrode. Moreover, when additional glycerol was added or when the medium was removed and replaced with fresh glycerol-containing medium, current production and glycerol consumption immediately resumed, showing that cells remained attached to the electrode and that continuous operation was possible (data not shown).
FIG 4 .
Electron balance (coulombic efficiency) for conversion of glycerol to ethanol. (A, B) Representative chronoamperometry of current produced from the conversion of glycerol to ethanol in three-electrode bioreactors (n = 3) inoculated with the wild-type strain with pGUT2PET (A) and the ∆pta strain with pGUT2PET (B). At time zero, ~1.0 OD of cells was added to the reactor. (C, D) Determining the coulombic efficiency of engineered pathways. Representative data of real-time, continuously measured charges and total calculated charges from the stoichiometric conversion of glycerol to ethanol for the wild-type strain with pGUT2PET (C) and the ∆pta strain with pGUT2PET (D). The measured charge was determined from current data in panel A or B and continuously measured during the experiment. The calculated charge is based upon the stoichiometry of the reaction mechanism and was determined when the samples used to obtain the data in panels A and B were extracted for HPLC analysis. See the text for a detailed description.
As observed in incubations with fumarate, only ethanol and a small amount of acetate accumulated during glycerol conversion; no succinate, malate, lactate, formate, or other end products were detected for the wild type (Fig. 3B) or the ∆pta strain (Fig. 3C) containing pGUT2PET. Mutants lacking pta exhibited a 46% decrease in acetate production compared to wild-type cultures when respiring to electrodes (Fig. 3C; Table 1). Continuous sparging with gas did not enhance conversion, nor was it essential for conversion of glycerol to ethanol in electrochemical reactors (data not shown). To improve analytical measurements of coulombic yield by removing a potential electron donor (H2), a low rate (1 ml/min) of gas sparging was employed in subsequent experiments. As this flushing also stripped some ethanol from the medium, volatile products were captured in a dry ice trap to confirm that ethanol was the only end product of glycerol bioconversion, as had been observed in our other experiments. In a subsequent series of experiments with the ∆pta strain, we used water-jacketed reflux and/or dry ice traps on top of flushed reactors and observed 78% ± 5.6% ethanol and 20% ± 9.4% acetate production from glycerol (total carbon balance, 98% ± 3.6%; n = 3).
To further confirm that growth in electrochemical reactors led to stoichiometric glycerol-to-ethanol conversion and to eliminate the possibility that glycerol was being completely oxidized to CO2 or other unknown compounds, a comparison of predicted and actual electron recoveries of the engineered glycerol-to-ethanol pathway was performed. Electrochemical measurements record every electron equivalent excreted, allowing current output to be integrated over time (Fig. 4A and B) and compared with changes in glycerol and acetate levels over the same period. Electron amounts recorded agreed with the pathway in Fig. 1, where glycerol conversion to ethanol releases 2 electrons and acetate production releases 6 electrons. Comparisons between predicted and measured charge transfer values in every experiment deviated no more than 10% (Fig. 4C), and the ∆pta strain with pGUT2PET behaved similarly (Fig. 4D). As only endpoint measurements were possible via dry ice trapping of gas-phase metabolites, ethanol production values at intermediate time points shown in Fig. 3B and C are based on levels calculated from nonvolatile metabolite measurements and coulombic recovery. Taken together, the carbon and electron recovery data showed that the outcomes of the engineered glycerol-to-ethanol pathway were identical when fumarate or an electrode was used as the electron acceptor.
DISCUSSION
The field of biocatalysis relies heavily on certain yeasts (Saccharomyces spp., Pichia spp., and Candida spp.) and bacteria (E. coli and Zymomonas spp.) to convert feedstocks into fuels (32). E. coli has been touted as an efficient and easily modifiable biocatalyst, with ethanol titers reaching 40 g/liter (33) and with volumetric productivities of strains engineered to ferment glycerol to ethanol, with H2 as a coproduct, as high as 4.7 mmol/liter/h (31). With most microbe-electrode systems, the fact that the key reaction occurs at a surface makes electrode surface area a crucial factor. These experiments with our first engineered strains of Shewanella in nonoptimized reactors contained only 3 cm2 electrode/ml, yet they already approached volumetric production rates of 1 mmol ethanol/liter/h. Recent work has shown that high-surface-area electrodes, such as treated carbon brush electrodes, can easily achieve surface/volume ratios on the order of 30 to 70 cm2/ml (34) and dramatically increase rates of current collection from Shewanella.
Electrode-linked microbial catalysis has many potential benefits; it can act as an electrochemical “lever” to drive an unfavorable reaction, allow generation of electricity via operation of a microbial fuel cell, or serve as reducing equivalents for additional product synthesis (35). In the case of glycerol, the shift from aerobic oxidation (36) to anaerobic fermentation, with formate or H2 as an end product (18, 37), represents a 3.5-fold decrease in the ∆G available to the cell. This low energy yield explains why anaerobic strategies are greatly enhanced by stripping of inhibitory by-products, such as H2. In contrast to what occurs during fermentation, use of an oxidizing electrode can accelerate metabolism, as it directly increases the thermodynamic driving force and avoids oxidative losses (and costs) associated with oxygen (36). In a microbial fuel cell-like reactor, passing these captured electrons to oxygen offers the possibility for additional energy recovery of −150 to −80 kJ/mol glycerol, depending on the set potential of the anode.
The challenges facing large-scale bioelectrochemical production of electricity, fuels, and chemicals have been summarized in recent reviews (38, 39). In general, the low cost of electricity and the low volumetric rates of electricity generated by natural bacterial communities present significant barriers to adoption of microbial fuel cells. However, when synthesis of higher-value fuels or chemicals is the goal, the economics become more favorable (39). In addition, electrons flowing from oxidative reactions can be boosted by a small (0.25- to 0.5-V) potential, which significantly increases overall volumetric rates of electron flow and can be used to power H2- or CH4-evolving microbial electrolysis chambers (5, 40) at energetic yields superior to those of water-splitting reactors. More recent work suggests that valuable bio-oxidations could be coupled in this way to electrodes used as electron donors for a wider variety of processes (41–44), such as was recently demonstrated by acetogenesis by biofilms of Sporomusa ovata on graphite electrodes (45).
It has previously been proposed that electrodes may alter fermentative pathways, but these strategies required high concentrations of toxic redox mediators to extract electrons from fermentative cells unable to transfer electrons to their outer surfaces for electrode respiration (1, 2, 46). S. oneidensis proved a tractable organism for linking native electron transfer ability to synthetic biology. Our work here shows that the landscape of metabolic engineering and synthetic biology strategies for biofuel and bioproduct synthesis (47) can be expanded through the use of engineered electrode-interfaced bacteria.
MATERIALS AND METHODS
Bacterial strains, culturing, growth, and reagents.
S. oneidensis strain MR-1 was previously isolated from Lake Oneida in New York State (48). All strains described in this study can be found in Table 2. Overnight cultures were inoculated from single colonies freshly streaked from a frozen stock into Luria-Bertani (LB) medium (supplemented with 50 µg/ml kanamycin [Km] when required for plasmid maintenance) and incubated for 16 h. Shewanella basal medium (SBM) containing 5 ml/liter of vitamins and trace minerals was used where specified below as described previously (49) and supplemented with 0.05% Casamino Acids. Anaerobic cultures were placed in Balch anaerobic tubes sealed with butyl rubber stoppers and flushed with nitrogen for 15 minutes (50). All cultures were maintained at 30°C and shaken continuously at 200 rpm. All molecular biology enzymes were obtained from New England Biolabs (Ipswich, MA), TOPO TA cloning vectors were from Invitrogen (Carlsbad, CA), and PCR cleanup, gel extraction, and plasmid preparation kits were from Qiagen (Valencia, CA). All other chemicals were obtained from Sigma (St. Louis, MO).
TABLE 2 .
Bacterial strains, vectors, and primers used in this study
| Strain, vector, or gene and primera | Relevant characteristic(s) | Reference(s) or source |
|---|---|---|
| Strains | ||
| S. oneidensis | ||
| MR-1 | Isolated from Lake Oneida, NY | 17, 48 |
| JG612 | ∆pta deletion derivative of MR-1 | 27 |
| E. coli | ||
| K-12 | Laboratory stock | |
| UQ950 | E. coli DH5α for cloning | 57 |
| WM3064 | DAP auxotroph donor strain for conjugationb | 57 |
| Vectors | ||
| pBBR1MCS-2 | 5.0-kb broad-host-range vector for cloning; Kmr | 51 |
| pPET | pBBR1MCS-2 containing pdc and adh (cloned from Zymomonas mobilis, pLOI297) | This study |
| pGUT2 | pBBR1MCS-2 containing glpD, glpF, glpK, and tpiA (cloned from E. coli K-12) | This study |
| pGUT2PET | pBBR1MCS-2 containing glpD, glpF, glpK, and tpiA (cloned from E. coli K-12) and pdc and adh (cloned from Z. mobilis, pLOI297) | This study |
| Primers for cloning | ||
| glpD | ||
| J1 KpnI | GGGGTACCACGAAAGTGAATGAGGGCAGCA | This study |
| J2 XhoI | CCGCTCGAGCAGGCCAGATTGAAATCTGA | |
| glpFK | ||
| J3 XbaI | GCTCTAGAAGCATGCCTACAAGCATCGTG | This study |
| J4 NotI | ATAAGAATCGGGCCGCTGCGGCATAAACGCTTCATTCG | |
| tpiA | ||
| J5 SacI | NNGAGCTCCGCTTATAAGCGTGGAGA | This study |
| J6 SacI | NNGAGCTCGAAAGTAAGTGCCGGATATG | |
| glpABC | ||
| J7 HindIII | CCCAAGCTTGCGCGAAATCAAACAATTCA | This study |
| J8 EcoRI | CGGAATTCATACATTGGGCACGGAATCG | |
| pUCmod Fwd | ||
| J9 XhoI | NNCTCGAGCCCGACTGGAAAGCGC | This study |
| pUCmod Rev | ||
| J10 SacI | NNNGAGCTCACATGCGGTGTGAAATACCG | This study |
| pBBR1MCS-2 Rev | ||
| J11 XhoI | NNNCTCGAGCTCTAGAACTAGTGGATCCC | This study |
Fwd, forward; Rev, reverse.
DAP, diaminopimelic acid.
Plasmid construction.
Oligonucleotides used are listed in Table 2. To clone glpD, genomic DNA of E. coli K-12 was used as a PCR template with primers J1 and J2. PCR products were cloned into pBBR1MCS-2 (51), creating JF3. To clone glpF and glpK, genomic DNA of E. coli K-12 was used as a PCR template with primers J3 and J4 to clone the native glpFK operon. PCR products were cloned into modified pUC19, previously described (52), creating JF4. JF4 was used as a PCR template with primers J9 and J10, designed to incorporate the previously cloned genes in addition to the lac promoter preceding glpF. PCR products were cloned into JF3, creating JF5. To clone tpiA, genomic DNA of K-12 was used as a PCR template with primers J5 and J6. PCR products were cloned into JF5, creating pGUT2. Plasmid pLOI297 (53) obtained from the ATCC (ATCC 68239), which contains pdc and adhB cloned from Z. mobilis, was digested with BamHI and EcoRI. The fragment containing these genes was cloned into pBBR1MCS-2, creating pPET. Plasmid pPET was used as template for a PCR with primer J11 and the standard M13 reverse primer. PCR products were A-tailed and then cloned into the TOPO TA vector, creating JF7. JF7 was digested with XhoI (at a site introduced by the J11 primer), yielding a 3.3-kb band containing a lac promoter, pdc, and adhB, and then cloned into pGUT2, creating pGUT2PET (Fig. 2A). In every case, vector inserts were sequenced to verify accuracy and orientation.
Growth on glycerol.
Strains were grown overnight aerobically in LB medium supplemented with Km (when appropriate), washed twice with SBM, and resuspended in SBM. The cells were then inoculated to an optical density at 600 nm (OD600) of ~0.05 into SBM medium containing 50 mM glycerol.
Resting cell assays.
Strains were grown overnight aerobically in LB medium supplemented with Km, washed twice with SBM, and resuspended in SBM. For measuring the conversion of lactate to ethanol, cells were then inoculated to an OD600 of ~0.8 into a culture containing 50 mM lactate and 50 mM fumarate and made anaerobic. For measuring the conversion of glycerol to ethanol, washed cells were inoculated to an OD600 of ~0.8 into an anaerobic culture tube containing 40 mM glycerol and 60 mM fumarate. Periodically, 0.2-ml aliquots were removed and centrifuged, and supernatants were immediately frozen at −80°C for high-performance liquid chromatography (HPLC) analysis.
HPLC analysis.
Metabolites were quantified by HPLC (all components were from Shimadzu Scientific) equipped with a UV–visible-light (Vis) detector and refractive index detector. The system consisted of an SCL-10A system controller, LC-10AT liquid chromatograph, SIL-10AF autoinjector, RID-10A refractive index detector, SPD-10A UV-Vis detector, and CTO-10A column oven. Separation of compounds was performed as described previously (37) with an Aminex HPX-87H guard column and an HPX-87H cation-exchange column (Bio-Rad [Hercules, CA]). The mobile phase consisted of 0.005 N H2SO4, set at a flow rate of 0.4 ml/min. The column was maintained at 42°C, and the injection volume was 50 µl.
Bioreactor analysis.
Bioreactors were constructed as previously described, with modifications (54). The counter electrode, housed in a glass capillary tube with dialysis tubing at one end, facilitated ion movement but inhibited gas transfer to avoid any utilization of stray H2 produced at the counter electrode and allow precise accounting of electron recovery. Isolation of the counter electrode was not necessary for routine glycerol conversion to ethanol. Strains were grown overnight in SBM supplemented with 50 mM glycerol and resuspended in 1 ml of SBM containing 50 mM glycerol and 4 µM riboflavin. The cell suspension was added to 11 ml of the same anaerobic medium in the bioreactor, which was continuously flushed at the counter electrode with nitrogen gas. The electrodes were maintained at an oxidizing potential (+0.44 V versus the standard hydrogen electrode [SHE]) using a 16-channel VMP1 potentiostat (Bio-Logic SA [Knoxville, TN]). Current production was monitored over time; 0.2-ml samples were taken periodically for HPLC analysis. To obtain carbon balance measurements, bioreactor exhaust ports were fitted with a dry ice trap or a stainless steel exhaust condenser to capture or reflux volatized ethanol. Dry ice traps were maintained at −72°C with a mixture of dry ice and ethanol. Condensers were connected to a circulating water bath (Thermo Scientific [Waltham, MA]) set at −2°C and filled with antifreeze.
Enzyme assays.
The activity assay for glycerol-3-phosphate dehydrogenase was performed as previously described (55). Fifty-milliliter cultures of cells to be tested were grown in LB medium supplemented with 50 µg/ml Km overnight, shaken, and incubated (37°C for E. coli and 30°C for S. oneidensis). The cultures were centrifuged at 6,000 × g for 15 minutes, and cells were resuspended in 1 ml of 0.1 M sodium phosphate buffered to pH 7.5 (PBS). Cells were then sonicated for 1 min on ice. Sonicated samples were centrifuged at 15,000 × g for 10 min. Glycerol kinase (56), alcohol dehydrogenase (23) and pyruvate decarboxylase (24) activities in cell lysates were determined as previously described, and cell lysates were prepared as described above.
ACKNOWLEDGMENTS
This work was supported by the College of Biological Sciences, by the BioTechnology Institute, a Cargill Higher Education Initiative award to D.R.B. and J.A.G., and by a Discovery grant, DG-0015-08, awarded to D.R.B. and J.A.G. from the Initiative for Renewable Energy and the Environment (Institute on the Environment) at the University of Minnesota.
Footnotes
Citation Flynn, J. M., D. E. Ross, K. A. Hunt, D. R. Bond, and J. A. Gralnick. 2010. Enabling unbalanced fermentations by using engineered electrode-interfaced bacteria. mBio 1(5):e00190-10. doi:10.1128/mBio.00190-10.
REFERENCES
- 1. Emde R., Swain A., Schink B. 1989. Anaerobic oxidation of glycerol by Escherichia coli in an amperometric poised-potential culture system. Appl. Microbiol. Biotechnol. 32:170–175 [Google Scholar]
- 2. Park D. H., Zeikus J. G. 1999. Utilization of electrically reduced neutral red by Actinobacillus succinogenes: physiological function of neutral red in membrane-driven fumarate reduction and energy conservation. J. Bacteriol. 181:2403–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sakai S., Yagishita T. 2007. Microbial production of hydrogen and ethanol from glycerol-containing wastes discharged from a biodiesel fuel production plant in a bioelectrochemical reactor with thionine. Biotechnol. Bioeng. 98:340–348 [DOI] [PubMed] [Google Scholar]
- 4. Steinbusch K. J., Hamelers H. V., Schaap J. D., Kampman C., Buisman C. J. 2010. Bioelectrochemical ethanol production through mediated acetate reduction by mixed cultures. Environ. Sci. Technol. 44:513–517 [DOI] [PubMed] [Google Scholar]
- 5. Logan B. E. 2009. Exoelectrogenic bacteria that power microbial fuel cells. Nat. Rev. Microbiol. 7:375–381 [DOI] [PubMed] [Google Scholar]
- 6. Lovley D. R. 2008. The microbe electric: conversion of organic matter to electricity. Curr. Opin. Biotechnol. 19:564–571 [DOI] [PubMed] [Google Scholar]
- 7. Hartshorne R. S., Reardon C. L., Ross D., Nuester J., Clarke T. A., Gates A. J., Mills P. C., Fredrickson J. K., Zachara J. M., Shi L., Beliaev A. S., Marshall M. J., Tien M., Brantley S., Butt J. N., Richardson D. J. 2009. Characterization of an electron conduit between bacteria and the extracellular environment. Proc. Natl. Acad. Sci. U. S. A. 106:22169–22174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hau H. H., Gralnick J. A. 2007. Ecology and biotechnology of the genus Shewanella. Annu. Rev. Microbiol. 61:237–258 [DOI] [PubMed] [Google Scholar]
- 9. Heidelberg J. F., Paulsen I. T., Nelson K. E., Gaidos E. J., Nelson W. C., Read T. D., Eisen J. A., Seshadri R., Ward N., Methe B., Clayton R. A., Meyer T., Tsapin A., Scott J., Beanan M., Brinkac L., Daugherty S., DeBoy R. T., Dodson R. J., Durkin A. S., Haft D. H., Kolonay J. F., Madupu R., Peterson J. D., Umayam L. A., White O., Wolf A. M., Vamathevan J., Weidman J., Impraim M., Lee K., Berry K., Lee C., Mueller J., Khouri H., Gill J., Utterback T. R., McDonald L. A., Feldblyum T. V., Smith H. O., Venter J. C., Nealson K. H., Fraser C. M. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20:1118–1123 [DOI] [PubMed] [Google Scholar]
- 10. Ross D. E., Ruebush S. S., Brantley S. L., Hartshorne R. S., Clarke T. A., Richardson D. J., Tien M. 2007. Characterization of protein-protein interactions involved in iron reduction by Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 73:5797–5808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shi L., Squier T. C., Zachara J. M., Fredrickson J. K. 2007. Respiration of metal (hydr)oxides by Shewanella and Geobacter: a key role for multihaem c-type cytochromes. Mol. Microbiol. 65:12–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ross D. E., Brantley S. L., Tien M. 2009. Kinetic characterization of OmcA and MtrC, terminal reductases involved in respiratory electron transfer for dissimilatory iron reduction in Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 75:5218–5226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baron D., LaBelle E., Coursolle D., Gralnick J. A., Bond D. R. 2009. Electrochemical measurement of electron transfer kinetics by Shewanella oneidensis MR-1. J. Biol. Chem. 284:28865–28873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marsili E., Baron D. B., Shikhare I. D., Coursolle D., Gralnick J. A., Bond D. R. 2008. Shewanella secretes flavins that mediate extracellular electron transfer. Proc. Natl. Acad. Sci. U. S. A. 105:3968–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. von Canstein H., Ogawa J., Shimizu S., Lloyd J. R. 2008. Secretion of flavins by Shewanella species and their role in extracellular electron transfer. Appl. Environ. Microbiol. 74:615–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Coursolle D., Baron D. B., Bond D. R., Gralnick J. A. 2010. The Mtr respiratory pathway is essential for reducing flavins and electrodes in Shewanella oneidensis. J. Bacteriol. 192:467–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Venkateswaran K., Moser D. P., Dollhopf M. E., Lies D. P., Saffarini D. A., MacGregor B. J., Ringelberg D. B., White D. C., Nishijima M., Sano H., Burghardt J., Stackebrandt E., Nealson K. H. 1999. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int. J. Syst. Bacteriol. 49:705–724 [DOI] [PubMed] [Google Scholar]
- 18. Gonzalez R., Murarka A., Dharmadi Y., Yazdani S. S. 2008. A new model for the anaerobic fermentation of glycerol in enteric bacteria: trunk and auxiliary pathways in Escherichia coli. Metab. Eng. 10:234–245 [DOI] [PubMed] [Google Scholar]
- 19. da Silva G. P., Mack M., Contiero J. 2009. Glycerol: a promising and abundant carbon source for industrial microbiology. Biotechnol. Adv. 27:30–39 [DOI] [PubMed] [Google Scholar]
- 20. Schryvers A., Lohmeier E., Weiner J. H. 1978. Chemical and functional properties of native and reconstituted forms of membrane bound, aerobic glycerol-3-phosphate dehydrogenase of Escherichia coli. J. Biol. Chem. 253:783–788 [PubMed] [Google Scholar]
- 21. Scott J. H., Nealson K. H. 1994. A biochemical study of the intermediary carbon metabolism of Shewanella putrefaciens. J. Bacteriol. 176:3408–3411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Austin D., Larson T. J. 1991. Nucleotide-sequence of the glpD gene encoding aerobic sn-glycerol 3-phosphate dehydrogenase of Escherichia coli K-12. J. Bacteriol. 173:101–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Conway T., Sewell G. W., Osman Y. A., Ingram L. O. 1987. Cloning and sequencing of the alcohol dehydrogenase-II gene from Zymomonas mobilis. J. Bacteriol. 169:2591–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ingram L. O., Conway T., Clark D. P., Sewell G. W., Preston J. F. 1987. Genetic engineering of ethanol production in Escherichia coli. Appl. Environ. Microbiol. 53:2420–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brau B., Sahm H. 1986. Cloning and expression of the structural gene for pyruvate decarboxylase of Zymomonas mobilis in Escherichia coli. Arch. Microbiol. 144:296–301 [Google Scholar]
- 26. Bringer-Meyer S., Schimz K. L., Sahm H. 1986. Pyruvate decarboxylase from Zymomonas mobilis. Isolation and partial characterization. Arch. Microbiol. 146:105–110 [Google Scholar]
- 27. Hunt K. A., Flynn J. M., Naranjo B., Shikhare I. D., Gralnick J. A. 2010. Substrate-level phosphorylation is the primary source of energy conservation during anaerobic respiration of Shewanella oneidensis strain MR-1. J. Bacteriol. 192:3345–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Castaño-Cerezo S., Pastor J. M., Renilla S., Bernal V., Iborra J. L., Cánovas M. 2009. An insight into the role of phosphotransacetlyase (pta) and the acetate/acetyl-CoA node in Escherichia coli. Microb. Cell Fact. 8:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Contiero J., Beatty C., Kumari S., DeSanti C. L., Strohl W. R., Wolfe A. 2000. Effects of mutations in acetate metabolism on high-cell-density growth of Escherichia coli. J. Ind. Microbiol. Biotechnol. 24:421–430 [Google Scholar]
- 30. Kakuda H., Hosono K., Shiroishi K., Ichihara S. 1994. Identification and characterization of the ackA (acetate kinase A)-pta (phosphotransacetylase) operon and complementation analysis of acetate utilization by an ackA-pta deletion mutant of Escherichia coli. J. Biochem. 116:916–922 [DOI] [PubMed] [Google Scholar]
- 31. Yazdani S. S., Gonzalez R. 2008. Engineering Escherichia coli for the efficient conversion of glycerol to ethanol and co-products. Metab. Eng. 10:340–351 [DOI] [PubMed] [Google Scholar]
- 32. Dellomonaco C., Fava F., Gonzalez R. 2010. The path to next generation biofuels: successes and challenges in the era of synthetic biology. Microb. Cell Fact. 9:3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yomano L. P., York S. W., Zhou S., Shanmugam K. T., Ingram L. O. 2008. Re-engineering Escherichia coli for ethanol production. Biotechnol. Lett. 30:2097–2103 [DOI] [PubMed] [Google Scholar]
- 34. Watson V. J., Logan B. E. 2010. Power production in MFCs inoculated with Shewanella oneidensis MR-1 or mixed cultures. Biotechnol. Bioeng. 105:489–498 [DOI] [PubMed] [Google Scholar]
- 35. Rabaey K., Rozendal R. A. 2010. Microbial electrosynthesis—revisiting the electrical route for microbial production. Nat. Rev. Microbiol. 8:706–716 [DOI] [PubMed] [Google Scholar]
- 36. Trinh C. T., Srienc F. 2009. Metabolic engineering of Escherichia coli for efficient conversion of glycerol to ethanol. Appl. Environ. Microbiol. 75:6696–6705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dharmadi Y., Murarka A., Gonzalez R. 2006. Anaerobic fermentation of glycerol by Escherichia coli: a new platform for metabolic engineering. Biotechnol. Bioeng. 94:821–829 [DOI] [PubMed] [Google Scholar]
- 38. Logan B. E. 2010. Scaling up microbial fuel cells and other bioelectrochemical systems. Appl. Microbiol. Biotechnol. 85:1665–1671 [DOI] [PubMed] [Google Scholar]
- 39. Foley J. M., Rozendal R. A., Hertle C. K., Lant P. A., Rabaey K. 2010. Life cycle assessment of high-rate anaerobic treatment, microbial fuel cells, and microbial electrolysis cells. Environ. Sci. Technol. 44:3629–3637 [DOI] [PubMed] [Google Scholar]
- 40. Liu H., Grot S., Logan B. E. 2005. Electrochemically assisted microbial production of hydrogen from acetate. Environ. Sci. Technol. 39:4317–4320 [DOI] [PubMed] [Google Scholar]
- 41. Gregory K. B., Bond D. R., Lovley D. R. 2004. Graphite electrodes as electron donors for anaerobic respiration. Environ. Microbiol. 6:596–604 [DOI] [PubMed] [Google Scholar]
- 42. Gregory K. B., Lovley D. R. 2005. Remediation and recovery of uranium from contaminated subsurface environments with electrodes. Environ. Sci. Technol. 39:8943–8947 [DOI] [PubMed] [Google Scholar]
- 43. Strycharz S. M., Woodard T. L., Johnson J. P., Nevin K. P., Sanford R. A., Loffler F. E., Lovley D. R. 2008. Graphite electrode as a sole electron donor for reductive dechlorination of tetrachlorethene by Geobacter lovleyi. Appl. Environ. Microbiol. 74:5943–5947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wrighton K. C., Virdis B., Clauwaert P., Read S. T., Daly R. A., Boon N., Piceno Y., Andersen G. L., Coates J. D., Rabaey K. Bacterial community structure corresponds to performance during cathodic nitrate reduction. ISME J., in press. [DOI] [PubMed] [Google Scholar]
- 45. Nevin K. P., Woodard T. L., Franks A. E., Summers Z. M., Lovley D. R. 2010. Microbial electrosynthesis: feeding microbes electricity to convert carbon dioxide and water to multicarbon extracellular organic compounds. mBio 1(2):e00103–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang X., Shanmugam K. T., Ingram L. O. 2010. Fermentation of glycerol to succinate by metabolically engineered strains of Escherichia coli. Appl. Environ. Microbiol. 76:2397–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Clomburg J. M., Gonzalez R. 2010. Biofuel production in Escherichia coli: the role of metabolic engineering and synthetic biology. Appl. Microbiol. Biotechnol. 86:419–434 [DOI] [PubMed] [Google Scholar]
- 48. Myers C. R., Nealson K. H. 1988. Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science 240:1319–1321 [DOI] [PubMed] [Google Scholar]
- 49. Hau H. H., Gilbert A., Coursolle D., Gralnick J. A. 2008. Mechanism and consequences of anaerobic respiration of cobalt by Shewanella oneidensis strain MR-1. Appl. Environ. Microbiol. 74:6880–6886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Balch W. E., Fox G. E., Magrum L. J., Woese C. R., Wolfe R. S. 1979. Methanogens: re-evaluation of a unique biological group. Microbiol. Rev. 43:260–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kovach M. E., Elzer P. H., Hill D. S., Robertson G. T., Farris M. A., Roop R. M., Peterson K. M. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176 [DOI] [PubMed] [Google Scholar]
- 52. Schmidt-Dannert C., Umeno D., Arnold F. H. 2000. Molecular breeding of carotenoid biosynthetic pathways. Nat. Biotechnol. 18:750–753 [DOI] [PubMed] [Google Scholar]
- 53. Alterthum F., Ingram L. O. 1989. Efficient ethanol production from glucose, lactose, and xylose by recombinant Escherichia coli. Appl. Environ. Microbiol. 55:1943–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Marsili E., Rollefson J. B., Baron D. B., Hozalski R. M., Bond D. R. 2008. Microbial biofilm voltammetry: direct electrochemical characterization of catalytic electrode-attached biofilms. Appl. Environ. Microbiol. 74:7329–7337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kistler W. S., Lin E. C. 1971. Anaerobic l-alpha-glycerophosphate dehydrogenase of Escherichia coli: its genetic locus and its physiological role. J. Bacteriol. 108:1224–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hayashi S. I., Lin E. C. 1967. Purification and properties of glycerol kinase from Escherichia coli. J. Biol. Chem. 242:1030–1035 [PubMed] [Google Scholar]
- 57. Saltikov C. W., Newman D. K. 2003. Genetic identification of a respiratory arsenate reductase. Proc. Natl. Acad. Sci. U. S. A. 100:10983–10988 [DOI] [PMC free article] [PubMed] [Google Scholar]